Background: Staphylococcal nuclease domain-containing 1 (SND1) is overexpressed in human hepatocellular carcinoma (HCC).
Results: SND1 augments tumor angiogenesis by activating NF-κB, resulting in the induction of miR-221, which subsequently induces angiogenin and CXCL16.
Conclusion: A novel pathway activated by SND1 is identified as contributing to tumor angiogenesis.
Significance: SND1 promotes hepatocarcinogenesis by multiple ways indicating that small molecule inhibitors of SND1 might be effective therapies for HCC.
Keywords: Angiogenesis, Cancer Biology, Chemokines, Liver Cancer, NF-κB, CXCL16, SND1, Angiogenin, miR-221
Abstract
Staphylococcal nuclease domain-containing 1 (SND1) is a multifunctional protein that is overexpressed in multiple cancers, including hepatocellular carcinoma (HCC). Stable overexpression of SND1 in Hep3B cells expressing a low level of SND1 augments, whereas stable knockdown of SND1 in QGY-7703 cells expressing a high level of SND1 inhibits establishment of xenografts in nude mice, indicating that SND1 promotes an aggressive tumorigenic phenotype. In this study we analyzed the role of SND1 in regulating tumor angiogenesis, a hallmark of cancer. Conditioned medium from Hep3B-SND1 cells stably overexpressing SND1 augmented, whereas that from QGY-SND1si cells stably overexpressing SND1 siRNA significantly inhibited angiogenesis, as analyzed by a chicken chorioallantoic membrane assay and a human umbilical vein endothelial cell differentiation assay. We unraveled a linear pathway in which SND1-induced activation of NF-κB resulted in induction of miR-221 and subsequent induction of angiogenic factors Angiogenin and CXCL16. Inhibition of either of these components resulted in significant inhibition of SND1-induced angiogenesis, thus highlighting the importance of this molecular cascade in regulating SND1 function. Because SND1 regulates NF-κB and miR-221, two important determinants of HCC controlling the aggressive phenotype, SND1 inhibition might be an effective strategy to counteract this fatal malady.
Introduction
Staphylococcal nuclease domain-containing 1 (SND1), a multifunctional nuclease containing four staphylococcal nuclease domains and a tudor domain, regulates a variety of cellular functions (1). As a coactivator it facilitates transcriptional activity of STAT5, STAT6, and c-Myb (2–4). SND1 interacts with spliceosomal small nuclear ribonucleoproteins via its tudor domain and accelerates the kinetics of spliceosome assembly and splicing activity, thereby facilitating pre-mRNA splicing (5). SND1 is a component of the RNA-induced silencing complex (RISC)4 and, consequently, regulates RNAi-mediated gene silencing (6). It is also involved in stabilizing specific mRNA, leading to increased translation (7). SND1 is cleaved by caspases during drug-induced apoptosis (8). A non-cleavable SND1 mutant increased cell viability, and knocking down SND1 promoted drug-induced apoptosis (8). It was demonstrated that enzymatic activity of SND1 is required for its anti-apoptotic activity.
Although SND1 is known to regulate cell viability, its role in the carcinogenic process is not well understood. Antisense inhibition of SND1 in B lymphoblasts results in cell death (9). Proteomic profiling identified high SND1 expression in metastatic breast cancer cells and also in tumor samples of metastatic breast cancer patients (10). SND1 is overexpressed in human colon cancers, and overexpression of SND1 in rat intestinal epithelial cells resulted in loss of contact inhibition and promoted cell proliferation via activation of the Wnt signaling pathway (11). SND1 overexpression has been detected in prostate cancer, and siRNA inhibition of SND1 inhibited viability of prostate cancer cells (12). Our recent studies document that SND1 interacts with the oncogene astrocyte-elevated gene 1 (AEG-1) in RISC and that SND1 is overexpressed in human hepatocellular carcinoma (HCC) (13). The overexpression of both AEG-1 and SND1 in human HCC cells confers increased RISC activity that facilitates degradation of tumor suppressor mRNAs by oncogenic miRNAs such as miR-221, thereby promoting the hepatocarcinogenic process (13). Stable overexpression of SND1 in Hep3B cells expressing a low level of SND1 augments, whereas stable knockdown of SND1 in QGY-7703 cells expressing a high level of SND1 inhibits establishment of xenografts in nude mice, indicating that SND1 promotes an aggressive tumorigenic phenotype (13). Breast cancer cells overexpress SND1, which interacts with AEG-1, and the importance of SND1 in breast cancer metastasis has been delineated recently (14).
Tumor angiogenesis plays a critical role in the development and progression of HCC (15). HCC is one of the most vascular solid cancers, associated with a high propensity for vascular invasion, and its growth relies on the formation of new blood vessels by diverse angiogenic factors (16). The chemokine CXCL16 is overexpressed in a diverse array of cancers and promotes invasion, angiogenesis, and metastasis (17). Soluble CXCL16 binds with its cognate receptor CXCR6 and activates multiple signal transduction pathways, including Akt/mammalian target of rapamycin (mTOR) and NF-κB (17). Angiogenin, originally discovered in a human colon cancer cell line, is a liver-derived polypeptide that shows strong angiogenic activity in vivo and regulates angiogenesis under both physiological and pathological conditions (18). Angiogenin is highly expressed in tumor tissue, and elevated serum Angiogenin concentration is observed in patients with various malignancies, suggesting its involvement in neovascularization (19–21). In human HCC, overexpression of angiogenin correlates with increased vascularity (22).
In this study, we document that SND1 promotes angiogenesis by augmenting the production of angiogenin and CXCL16. SND1 activates NF-κB, resulting in increased expression of miR-221, a known onco-miRNA for human HCC (23). We demonstrate that SND1-induced miR-221 is involved in regulating increased expression of angiogenin and CXCL16. Thus, we unravel a previously unknown angiogenic pathway dependent on SND1 that might be critical in regulating progression and metastasis of HCC.
MATERIALS AND METHODS
Cell Lines, Culture Conditions, and Adenovirus
The generation of the Hep3B-Con, Hep3B-SND1–17, QGY-Consi, and QGY-SND1si-12 clones was described (13). Hep3B cells were obtained from the ATCC and were cultured as recommended. The human HCC cell line QGY-7703 was developed at Fudan University, Shanghai, obtained from Dr. Zhao-zhong Su, and cultured as described (13). Human vascular endothelial cells (HUVEC) were obtained from Lonza and were cultured according to the provided protocol. Ad.vec and Ad.IκBαmt32 construction and transduction were described (24).
Plasmids, siRNAs, Transfection, and Luciferase Assay
The construction of miR-221 expression plasmid was described (25). The anti-miR-221 expression plasmid was obtained from GeneCopoeia. 3κB-Luc, containing three tandem NF-κB binding sites upstream of the luciferase gene, was described (24). Control siRNA and siRNAs for angiogenin and CXCL16 were obtained from Santa Cruz Biotechnology. All constructs were transfected into HCC cells using Lipofectamine 2000 according to the manufacturer's protocol. For the luciferase assay, cells were treated with TNF-α (10 ng/ml) for 12 h, and luciferase activity was measured as described (24).
Immunohistochemistry
Immunohistochemistry in formalin-fixed paraffin-embedded tumor sections was performed as described (13). The primary antibodies were anti-SND1 (rabbit polyclonal, 1:100, Prestige antibodies® powered by Atlas antibodies from Sigma) and anti-CD31 (1:200, mouse monoclonal antibodies, Dako). The signals were developed by avidin-biotin-peroxidase complexes with a diaminobenzidine (DAB) substrate solution (Vector Laboratories). Images were analyzed using an Olympus microscope.
CAM Assay
Cells were seeded on the CAM surface of 9-day-old chick embryos according to established protocols (26). One week after inoculation, the neovasculature was examined and photographed. Angiogenesis was quantified by counting the blood vessel branch points under a stereo microscope. The angiogenic index was calculated by subtracting the number of branch points from the branching in the control group.
Capillary-like Tube Formation Assay
Tube formation by HUVECs was performed using a Cultrex basement membrane extract (R&D Systems) as described (27). HUVECs were seeded in 96-wells plates at 5 × 104 cells/well, treated with the conditioned medium collected from SND1-overexpressing and SND1 knockdown HCC cells, and incubated at 37 °C overnight. Images were captured using an inverted microscope (Nikon), and the degree of network formation was quantified using an image analyzer (National Institutes of Health Image).
Human Angiogenesis Array
The expression levels of 55 angiogenesis-associated proteins were analyzed using conditioned media from cells cultured in serum-free condition with the human angiogenesis antibody array kit (R&D Systems, Proteome ProfilerTM) according to the manufacturer's instructions.
ELISA
The expression of CXCL16 and Angiogenin was analyzed in supernatants of cells cultured in serum-free condition using ELISA kits (R&D Systems, QuantakineR) according to the manufacturer's instructions.
Preparation of Nuclear Fractions and Western Blot Analysis
Nuclear fractions were prepared as described (24). Western blot analysis was performed using primary antibodies, anti-p65 (rabbit polyclonal antibody, 1:200, Santa Cruz Biotechnologies), anti-nucleolin (mouse monoclonal antibody, 1:200, Santa Cruz Biotechnology), anti-phospho-IκBα (rabbit polyclonal antibody, 1:1000, Cell Signaling Technology), anti-IκBα (rabbit polyclonal antibody, 1:1000, Cell Signaling Technology), anti-phospho-IKKα (rabbit polyclonal antibody, 1:1000, Cell Signaling Technology), and anti-IKKα (rabbit polyclonal antibody, 1:1000, Cell Signaling Technology) as described (28).
Total RNA Extraction and Real-time PCR Assay
Total RNA was extracted using the miRNAeasy mini kit (Qiagen). Real-time PCR was performed using an ABI 7900 fast real-time PCR system and Taqman miRNA expression assays according to the manufacturer's protocol (Applied Biosystems).
Nude Mice Xenograft Studies
QGY-7703 cells were infected with Ad.vec or Ad.IκBαmt32 ex vivo at a multiplicity of infection of 1000 viral particle (vp)/cell, and after 18 h, subcutaneous xenografts were established in the flanks of athymic nude mice using 1 × 106 cells (13). Tumor volume was measured twice weekly with a caliper and calculated using the formula π/6 × larger diameter × (smaller diameter)2. The animals were sacrificed after 4 weeks. The experiments were performed twice with six mice per group in each experiment.
Statistical Analysis
Data were represented as the mean ± S.E. and analyzed for statistical significance using one-way analysis of variance followed by a Newman-Keuls test as a post-hoc test. A p value of < 0.05 was considered significant.
RESULTS
SND Induces Angiogenesis
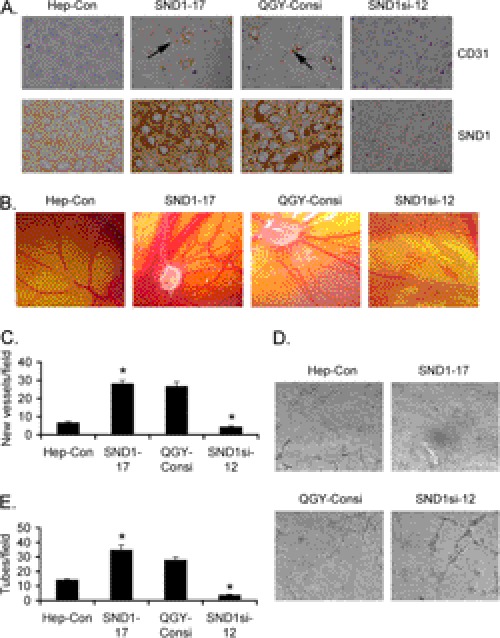
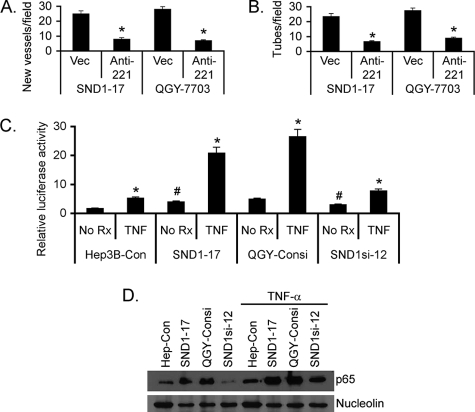
We have documented recently that Hep3B and QGY-7703 cells express low and high levels of SND1, respectively (13). Stable overexpression of SND1 in Hep3B cells significantly augments, whereas stable knockdown of SND1 in QGY-7703 cells significantly inhibits formation of subcutaneous xenografts in athymic nude mice (13). These findings were observed in multiple clones of SND1-overexpressing Hep3B cells as well as SND1 knockdown QGY-7703 cells, and these clones have been characterized in detail (13). Because angiogenesis is an integral component of aggressive tumors, we analyzed the role of SND1 in facilitating tumor angiogenesis. Immunohistochemical analysis revealed significantly high levels of CD31, a marker for microvessels denoting increased angiogenesis, in tumors from the Hep3B-SND1–17 clone compared with that from the control Hep3B clone (Hep3B-Con) (Fig. 1A). As a corollary, a significant decrease in CD31 expression was observed in tumors from the QGY-SND1si-12 clone compared with that from the QGY-Consi clone, expressing control scrambled siRNA (Fig. 1A). The proangiogenic property of SND1 was characterized further using a CAM assay by implanting SND1-overexpressing and SND1 knockdown clones in 9-day-old chick embryos. After 1 week, neovascularization was examined, photographed, and quantified. Significantly marked outgrowth of new blood vessels from the tumor core region was observed in the Hep3B-SND1–17 and QGY-Consi clones when compared with the Hep3B-Con and QGY-SND1si-12 clones, respectively (Fig. 1, B and C). These findings were extended further by endothelial cell tube formation assays. HUVECs, cultured on basement membrane extract, rapidly align, extend processes into the matrix, and finally form capillary-like structures surrounding a central lumen. Conditioned medium (CM) from the Hep3B-SND1–17 and QGY-Consi clones markedly enhanced tube formation when compared with the CM from the Hep3B-Con and QGY-SND1si-12 clones, respectively (Fig. 1, D and E). These findings confirm that SND1 is a potent inducer of angiogenesis.
FIGURE 1.
SND1 induces angiogenesis. A, tumor sections from subcutaneous xenografts established in nude mice by Hep3B-Con, Hep3B-SND1–17, QGY-Consi, and QGY-SND1si-12 cells were stained for CD31 (upper panel) and SND1 (lower panel). B, the indicated cells were implanted in CAM, and neovascularization was photographed. C, graphical representation of new blood vessel formation in CAM when the indicated cells were implanted. The experiments were performed twice using eight eggs per group. Data represent mean ± S.E. *, p < 0.05. D, HUVECs were treated with conditioned media from the indicated cells, and tube formation was photographed. E, graphical representation of tube formation by HUVECs treated with conditioned media from the indicated cells. The experiments were performed twice using triplicates per group. Data represent mean ± S.E. *, p < 0.05.
SND1 Induces Angiogenesis by Up-regulating Angiogenin and CXCL16
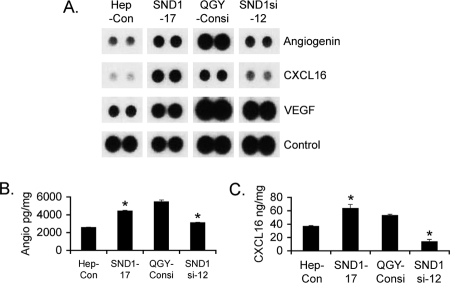
To identify the angiogenic factors induced by SND1, we screened a human angiogenesis array using CM from the Hep3B-Con, Hep3B-SND1–17, QGY-Consi, and QGY-SND1si-12 clones. Markedly increased levels of angiogenin and CXCL16 and moderate increases in VEGF were observed in the Hep3B-SND1–17 and QGY-Consi clones when compared with the Hep3B-Con and QGY-SND1si-12 clones, respectively (Fig. 2A and supplemental Fig. S1). In addition, insulin-like growth factor binding protein-3 (IGFBP3) and PAI-1 (SERPINE1) also showed induction by SND1, albeit at a lower level. Because the most robust changes were observed for angiogenin and CXCL16, we further interrogated the roles of these molecules in SND1-induced angiogenesis. The induction of angiogenin and CXCL16 by SND1 was confirmed by ELISA using CM from the cells. Indeed, both angiogenin and CXCL16 levels were significantly higher in the Hep3B-SND1–17 and QGY-Consi clones when compared with the Hep3B-Con and QGY-SND1si-12 clones, respectively (Fig. 2, B and C).
FIGURE 2.
SND1 induces Angiogenin and CXCL16. A, a human angiogenesis array was screened using CM from the indicated cells. Representative expression levels of angiogenin, CXCL16, and VEGF are shown. The experiment was performed twice. Angiogenin (B) and CXCL16 (C) levels in the conditioned media of the indicated cells are as determined by ELISA. The cytokine levels were adjusted to per mg of total cellular protein. The experiments were performed twice using triplicates per group. Data represent mean ± S.E. *, p < 0.05.
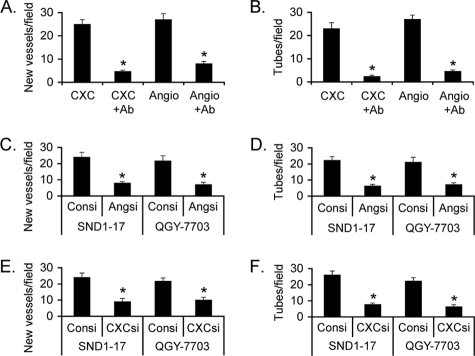
We confirmed the role of CXCL16 and angiogenin in the process of angiogenesis. CAM was treated with either CXCL16 or angiogenin to induce angiogenesis, and then the effects of these factors were blocked by corresponding neutralizing antibody. Inhibition of CXCL16 or angiogenin by its respective antibody profoundly inhibited neovascularization in CAM (Fig. 3A). Similarly, treatment of HUVECs with CXCL16 or angiogenin significantly augmented tube formation that was markedly inhibited by the corresponding neutralizing antibody (Fig. 3B), confirming that both CXCL16 and Angiogenin play critical roles in regulating tumor angiogenesis. An isotype control antibody did not affect HUVEC tube formation, and angiogenin antibody did not affect CXCL16-induced tube formation and vice versa (supplemental Fig. S2). Next, we analyzed the involvement of angiogenin and CXCL16 in mediating SND1-induced angiogenesis. Hep3B-SND1–17 and QGY-7703 cells were transfected with either control siRNA or with angiogenin siRNA, and the CM was evaluated in CAM and HUVEC differentiation assays. Compared with the control, scrambled siRNA, angiogenin siRNA markedly inhibited neovascularization in CAM and HUVEC tube formation by the CM (Fig. 3, C and D, respectively). Similar findings were also obtained using CXCL16 siRNA (Fig. 3, E and F), confirming the key roles of angiogenin and CXCL16 in SND1-induced angiogenesis. A combination of angiogenin and CXCL16 siRNA further inhibited tube formation when compared with either siRNA alone (supplemental Fig. S3).
FIGURE 3.
Angiogenin and CXCL16 are required for SND1-induced angiogenesis. A, CAM was treated either with CXCL16 or angiogenin with or without corresponding neutralizing antibody, and neovascularization was quantified. The experiments were performed twice with eight eggs per group. B, HUVECs were treated either with CXCL16 or angiogenin with or without corresponding neutralizing antibody, and tube formation was quantified. The experiments were performed twice using triplicates per group. Hep3B-SND1–17 and QGY-7703 cells were transfected either with control siRNA or angiogenin siRNA, and the CM was subjected to CAM assay (C) and tube formation assay (D). Hep3B-SND1–17 and QGY-7703 cells were transfected either with control siRNA or CXCL16 siRNA, and the CM was subjected to CAM assay (E) and tube formation assay (F). The experiments were performed twice using triplicates per group. Data represent mean ± S.E. *, p < 0.05.
SND1-induced Angiogenesis Involves miR-221
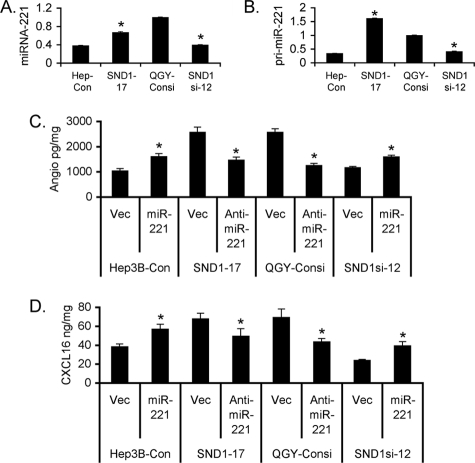
In our previous studies we documented that as a component of RISC, SND1 facilitates miRNA-mediated gene regulation, especially the expression of genes that are regulated by miR-221 (13). Because both SND1 and miR-221 are overexpressed in HCC and because they complement each other (13, 23), we determined whether miR-221 plays any role in mediating SND1-induced angiogenesis. Compared with the Hep3B-Con clone there was a significant increase in the mature miR-221 level in the Hep3B-SND1–17 clone (Fig. 4A). Similarly, the miR-221 level in QGY-SND1si-12 was significantly less than that in the QGY-Consi clone (Fig. 4A). As a corollary, the pri-miR-221 level was higher in the Hep3B-SND1–17 and QGY-Consi clones when compared with that in the Hep3B-Con and QGY-SND1si-12 clones, respectively, demonstrating that SND1 induces miR-221 at the transcriptional level (Fig. 4B). Because angiogenin and CXCL16 are major mediators of SND1-induced angiogenesis, we checked the role of miR-221 in regulating angiogenin and CXCL16 expression. Overexpression of miR-221 in the Hep3B-Con and QGY-SND1si-12 clones significantly increased, whereas knockdown of miR-221 by anti-miR-221 in the Hep3B-SND1–17 and QGY-Consi clones significantly decreased both angiogenin and CXCL16 levels (Fig. 4C & 4D) indicating that miR-221 regulates the expression of these angiogenic factors. Indeed, knockdown of miR-221 by anti-miR-221 in the Hep3B-SND1–17 and QGY-7703 cells significantly inhibited neovascularization in CAM and HUVEC differentiation by the CM from the treated cells (Fig. 5, A and B).
FIGURE 4.
SND1-induced miRNA-221 regulates angiogenin and CXCL16 expression. A and B, analysis of miRNA-221 (A) and pri-miR-221 (B) expression in the indicated cells. C, Hep3B-Con and QGY-SND1si-12 cells were transfected with either control empty vector or miR-221 expression constructs, whereas Hep3B-SND1–17 and QGY-Consi cells were transfected with either empty vector or anti-miR-221 expression construct, and the angiogenin level was measured in the culture supernatant after 48 h by ELISA. D, the treatment was performed as in C, and CXCL16 level was measured in the culture supernatant after 48 h by ELISA. The experiments were performed twice using triplicates per group. Data represent mean ± S.E. *, p < 0.05.
FIGURE 5.
SND1 activates NF-κB to induce miR-221 expression. Hep3B-SND1–17 and QGY-7703 cells were transfected with either empty vector or anti-miR-221 expression construct, and the CM was subjected to CAM assay (A) and tube formation assay (B). C, the indicated cells were transfected with either the empty pGL3-basic vector or 3κB-Luc along with the Renilla luciferase construct and treated with TNF-α (10 ng/ml) for 12 h. Firefly luciferase activity was normalized by Renilla luciferase activity and further by activity of the pGL3-basic vector. Data represent mean ± S.E. *, p < 0.05. D, nuclear fractions from the indicated cells, treated or not treated with TNF-α for 12 h, were subjected to Western blot analysis for the p65 subunit of NF-κB and nucleolin as a loading control.
SND1 Activates NF-κB to Induce miR-221
Because the miR-221 level has been shown to be regulated by NF-κB (29), which is also a major proangiogenic factor, we checked the involvement of NF-κB in SND1-mediated miR-221 induction, leading to angiogenesis. To check NF-κB activity, Hep3B-Con, Hep3B-SND1–17, QGY-Consi, and QGY-SND1si-12 cells were transfected with a reporter vector, 3κB-Luc, containing three tandem NF-κB binding sites upstream of the luciferase gene along with a Renilla luciferase expression construct, treated with TNF-α, a known activator of NF-κB, and then luciferase activity was determined (28). Firefly luciferase activity was corrected by Renilla luciferase activity. The basal NF-κB activity was significantly higher in Hep3B-SND1–17 and QGY-Consi cells compared with that in Hep3B-Con and QGY-SND1si-12 cells, respectively (Fig. 5C). However, upon treatment with TNF-α, NF-κB activity was markedly increased in Hep3B-SND1–17 and QGY-Consi cells compared with that in Hep3B-Con and QGY-SND1si-12 cells (Fig. 5C), indicating that SND1 significantly potentiates NF-κB activity. In nuclear fractions of Hep3B-SND1–17 and QGY-Consi cells, the level of the p65 subunit of NF-κB was significantly higher compared with that in Hep3B-Con and QGY-SND1si-12 cells, respectively, and this phenomenon was further accentuated upon treatment with TNF-α (Fig. 5D). Increased phosphorylation of IKKα and IκBα was observed in the Hep3B-SND1–17 clone compared with the Hep3B-Con clone, and a corresponding decrease was observed in the QGY-SND1si-12 clone when compared with the QGY-Consi clone, indicating that SND1 augments a canonical pathway of NF-κB activation (supplemental Fig. S4).
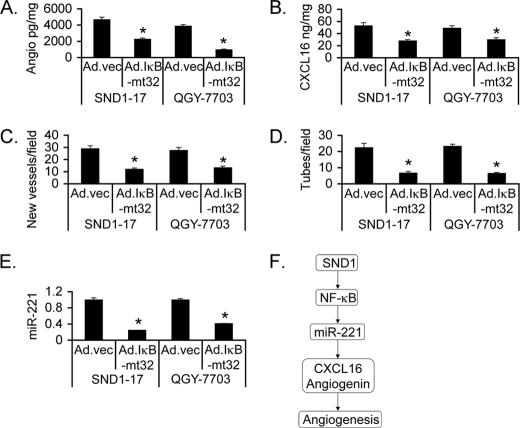
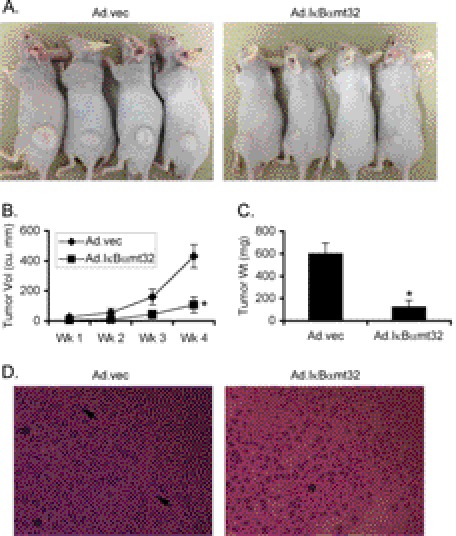
The role of NF-κB in SND1-induced angiogenesis was analyzed using an adenovirus expressing the mt32IκBα superrepressor (IκBαmt32), Ad.IκBαmt32, which inhibits IκBα degradation and subsequent NF-κB nuclear translocation (30). Inhibition of NF-κB significantly abrogated angiogenin and CXCL16 generation by Hep3B-SND1–17 and QGY-7703 cells (Fig. 6, A and B). CM of Hep3B-SND1–17 and QGY-7703 cells infected with Ad.IκBαmt32 induced significantly less neovascularization in CAM and HUVEC tube formation compared with CM of cells infected with control empty adenovirus (Ad.vec) (Fig. 6, C and D). Compared with Ad.vec, Ad.IκBαmt32 significantly inhibited miR-221 levels in Hep3B-SND1–17 and QGY-7703 cells, demonstrating the transcriptional regulation of miR-221 by NF-κB (Fig. 6E). The role of NF-κB in regulating tumor growth was confirmed by in vivo assays. QGY-7703 cells were treated with either Ad.vec or Ad.IκBαmt32 and were subcutaneously implanted into athymic nude mice 18 h later before induction of any in vitro cytopathic effect. In a 4-week assay, tumor growth was markedly inhibited upon treatment with Ad.IκBαmt32 versus Ad.vec (Fig. 7, A--C). Histological analysis of the tumors show mitotic cells and radiating blood vessels in the Ad.vec-treated group, whereas Ad.IκBαmt32-treated tumors were significantly hypocellular with areas of necrosis (Fig. 7D).
FIGURE 6.
Inhibition of NF-κB blocks SND1-induced angiogenesis. Hep3B-SND1–17 and QGY-7703 cells were infected with either Ad.vec or Ad.IκBαmt32, and angiogenin (A) and CXCL16 (B) levels were measured in culture supernatants by ELISA. Conditioned media were collected after similar treatment as in A and subjected to CAM assay (C) and tube formation assay (D). Data represent mean ± S.E. *, p < 0.05. E, Hep3B-SND1–17 and QGY-7703 cells were treated as in A, and the miR-221 level was determined by real-time PCR. F, schematic representation of the molecular pathway involved in SND1-induced angiogenesis.
FIGURE 7.
Inhibition of NF-κB profoundly abrogates tumorigenesis in vivo. QGY-7703 cells were treated either with Ad.vec or with Ad.IκBαmt32 and were subcutaneously implanted into athymic nude mice 18 h later. Tumors were allowed to grow for 4 weeks. Six (6) animals per group were used, and the experiment was performed twice. A, tumor-bearing animals at the end of the study. B, measurement of tumor volume. C, measurement of tumor weight at the end of the study. Data represent mean ± S.E. *, p < 0.05. D, histological analysis of the tumor sections. The asterisks in the left panel indicate mitotic cells, whereas the arrows indicate blood vessels. The asterisk in the right panel indicates the area of necrosis.
DISCUSSION
We now unravel a previously uncharacterized pathway involving SND1, NF-κB, miR-221, and angiogenic factors CXCL16 and angiogenin-regulating tumor angiogenesis in HCC (Fig. 6F). All of these components are overexpressed in diverse cancers, and we presently link them together in a defined end point of angiogenesis. The key players identified in this study extensively cross-talk with each other. CXCL16 can be induced by TNF-α treatment via NF-κB activation, and both CXCL16 and angiogenin activate NF-κB, thus establishing a positive feedback regulation (31–33). Both CXCL16 and angiogenin activate Akt/mTOR signaling, which in turn is involved in CXCL16 and angiogenin expression and secretion (34, 35). On the other hand, miR-221 targets phosphatase and tensin homolog (PTEN) and DDIT4, inhibitors of the Akt/mTOR pathway (23). Thus, an expansive protumorigenic signaling network lies downstream of SND1. Compared with normal liver, SND1 expression is markedly elevated in human HCC, and its expression increases with disease progression (13). SND1 might be an important determinant of HCC progression, and targeted inhibition of SND1 could be an effective way to counteract this disease. The observation that SND1 lies upstream of major contributors of HCC, such as NF-κB and miR-221, further strengthens the rationale of SND1 inhibition as a means of treating HCC. 3′, 5′-deoxythymidine bisphosphate is a known inhibitor of SND1 enzymatic activity (6). However, it works at a high micromolar concentration, precluding its use clinically. Thus, there is an important need to develop clinically relevant small-molecule inhibitors of SND1 as potential anti-cancer therapeutics.
Our findings raise several important questions that need to be addressed in follow-up studies. First, what is the underlying mechanism of SND1-induced NF-κB activation? SND1 functions as a coactivator for several transcription factors (2–4). Does SND1 interact with NF-κB and increases its transactivation function? AEG-1, the interacting partner of SND1 (13), interacts with NF-κB and CBP/p300 to augment NF-κB function (28). Upon treatment with TNF-α, AEG-1 translocates into the nucleus where it interacts with NF-κB (9). Although AEG-1/SND1 interaction in the RISC is observed in the cytoplasm (13), it might be possible that upon stimulation, such as by TNF-α, the AEG-1·SND1 complex translocates to the nucleus to interact with NF-κB. However, a coimmunoprecipitation experiment did not detect potential interaction between SND1 and p65 subunit of NF-κB (data not shown), and we observed phosphorylation of IKKα and IκBα upon SND1 overexpression, suggesting activation of a canonical NF-κB signaling pathway by SND1. The mechanism by which SND1 activates IKKα remains to be determined.
Second, how does miR-221 induce CXCL16 and angiogenin production? Is there a common negative regulator of both these factors that is a target of miR-221? CXCL16 exists as a transmembrane (TM-CXCL16) form and as a soluble form (sCXCL16), and it is sCXCL16 that promotes angiogenesis and metastasis via interaction with CXCR6 (17). TM-CXCL16 needs to be cleaved by disintegrin-like metalloproteinases ADAM10 and ADAM17 to generate sCXCL16 (17, 31). Tissue inhibitor of metalloproteinase 3 (TIMP3) is an endogenous inhibitor of both ADAM10 and ADAM17 (36). More importantly, TIMP3 is also a target of miR-221 (37). Thus, miR-221 might increase sCXCL16 production by down-regulating TIMP3 and augmenting ADAM10 and ADAM17 activity. Angiogenin is the most up-regulated gene in a transgenic mouse with prostate-specific overexpression of Akt (38). Because miR-221 targets PTEN, leading to the activation of the Akt pathway, this might be a potential mechanism by which miR-221 induces angiogenin expression.
In summary, our previous and current observations from in vivo and in vitro studies established SND1 as a major nodal point in the carcinogenic network. Its overexpression in diverse cancer indications, its ability to regulate various protumorigenic signaling cascades and molecular events, and the presence of a potential druggable enzymatic domain strongly advocates SND1 as a promising target for anti-cancer therapy.
Supplementary Material
This work was supported in part by grants from the Dana Foundation, the James S. McDonnell Foundation, a Massey Cancer Center pilot project grant, and National Cancer Institute Grants R01 CA138540 (to D. S.) and R01 CA134721 (to P. B. F.).

This article contains supplemental Figs. S1–S4.
- RISC
- RNA-induced silencing complex
- HCC
- hepatocellular carcinoma
- HUVEC
- human umbilical vein endothelial cell
- CAM
- chicken chorioallantoic membrane
- CM
- conditioned medium/media.
REFERENCES
- 1. Li C. L., Yang W. Z., Chen Y. P., Yuan H. S. (2008) Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 36, 3579–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leverson J. D., Koskinen P. J., Orrico F. C., Rainio E. M., Jalkanen K. J., Dash A. B., Eisenman R. N., Ness S. A. (1998) Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell 2, 417–425 [DOI] [PubMed] [Google Scholar]
- 3. Paukku K., Yang J., Silvennoinen O. (2003) Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 17, 1805–1814 [DOI] [PubMed] [Google Scholar]
- 4. Yang J., Aittomäki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O. (2002) Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21, 4950–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang J., Välineva T., Hong J., Bu T., Yao Z., Jensen O. N., Frilander M. J., Silvennoinen O. (2007) Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 35, 4485–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caudy A. A., Ketting R. F., Hammond S. M., Denli A. M., Bathoorn A. M., Tops B. B., Silva J. M., Myers M. M., Hannon G. J., Plasterk R. H. (2003) A micrococcal nuclease homologue in RNAi effector complexes. Nature 425, 411–414 [DOI] [PubMed] [Google Scholar]
- 7. Paukku K., Kalkkinen N., Silvennoinen O., Kontula K. K., Lehtonen J. Y. (2008) p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res. 36, 4474–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundström J. F., Vaculova A., Smertenko A. P., Savenkov E. I., Golovko A., Minina E., Tiwari B. S., Rodriguez-Nieto S., Zamyatnin A. A., Jr., Välineva T., Saarikettu J., Frilander M. J., Suarez M. F., Zavialov A., Ståhl U., Hussey P. J., Silvennoinen O., Sundberg E., Zhivotovsky B., Bozhkov P. V. (2009) Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11, 1347–1354 [DOI] [PubMed] [Google Scholar]
- 9. Tong X., Drapkin R., Yalamanchili R., Mosialos G., Kieff E. (1995) The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15, 4735–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ho J., Kong J. W., Choong L. Y., Loh M. C., Toy W., Chong P. K., Wong C. H., Wong C. Y., Shah N., Lim Y. P. (2009) Novel breast cancer metastasis-associated proteins. J. Proteome Res. 8, 583–594 [DOI] [PubMed] [Google Scholar]
- 11. Tsuchiya N., Ochiai M., Nakashima K., Ubagai T., Sugimura T., Nakagama H. (2007) SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 67, 9568–9576 [DOI] [PubMed] [Google Scholar]
- 12. Kuruma H., Kamata Y., Takahashi H., Igarashi K., Kimura T., Miki K., Miki J., Sasaki H., Hayashi N., Egawa S. (2009) Staphylococcal nuclease domain-containing protein 1 as a potential tissue marker for prostate cancer. Am. J. Pathol. 174, 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoo B. K., Santhekadur P. K., Gredler R., Chen D., Emdad L., Bhutia S., Pannell L., Fisher P. B., Sarkar D. (2011) Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 53, 1538–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blanco M. A., Alecković M., Hua Y., Li T., Wei Y., Xu Z., Cristea I. M., Kang Y. (2011) Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J. Biol. Chem. 286, 19982–19992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pang R., Poon R. T. (2006) Angiogenesis and antiangiogenic therapy in hepatocellular carcinoma. Cancer Lett. 242, 151–167 [DOI] [PubMed] [Google Scholar]
- 16. Zhu A. X., Duda D. G., Sahani D. V., Jain R. K. (2011) HCC and angiogenesis. Possible targets and future directions. Nat. Rev. Clin. Oncol. 8, 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng L., Chen N., Li Y., Zheng H., Lei Q. (2010) CXCR6/CXCL16 functions as a regulator in metastasis and progression of cancer. Biochim. Biophys. Acta 1806, 42–49 [DOI] [PubMed] [Google Scholar]
- 18. Tello-Montoliu A., Patel J. V., Lip G. Y. (2006) Angiogenin. A review of the pathophysiology and potential clinical applications. J. Thromb. Haemost. 4, 1864–1874 [DOI] [PubMed] [Google Scholar]
- 19. Hartmann A., Kunz M., Köstlin S., Gillitzer R., Toksoy A., Bröcker E. B., Klein C. E. (1999) Hypoxia-induced up-regulation of angiogenin in human malignant melanoma. Cancer Res. 59, 1578–1583 [PubMed] [Google Scholar]
- 20. Shimoyama S., Yamasaki K., Kawahara M., Kaminishi M. (1999) Increased serum angiogenin concentration in colorectal cancer is correlated with cancer progression. Clin. Cancer Res. 5, 1125–1130 [PubMed] [Google Scholar]
- 21. Shimoyama S., Gansauge F., Gansauge S., Negri G., Oohara T., Beger H. G. (1996) Increased angiogenin expression in pancreatic cancer is related to cancer aggressiveness. Cancer Res. 56, 2703–2706 [PubMed] [Google Scholar]
- 22. Hisai H., Kato J., Kobune M., Murakami T., Miyanishi K., Takahashi M., Yoshizaki N., Takimoto R., Terui T., Niitsu Y. (2003) Increased expression of angiogenin in hepatocellular carcinoma in correlation with tumor vascularity. Clin. Cancer Res. 9, 4852–4859 [PubMed] [Google Scholar]
- 23. Pineau P., Volinia S., McJunkin K., Marchio A., Battiston C., Terris B., Mazzaferro V., Lowe S. W., Croce C. M., Dejean A. (2010) miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 264–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emdad L., Sarkar D., Su Z. Z., Randolph A., Boukerche H., Valerie K., Fisher P. B. (2006) Activation of the nuclear factor κB pathway by astrocyte elevated gene-1. Implications for tumor progression and metastasis. Cancer Res. 66, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 25. Das S. K., Sokhi U. K., Bhutia S. K., Azab B., Su Z. Z., Sarkar D., Fisher P. B. (2010) Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc. Natl. Acad. Sci. U.S.A. 107, 11948–11953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfeifer A., Kessler T., Silletti S., Cheresh D. A., Verma I. M. (2000) Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc. Natl. Acad. Sci. U.S.A. 97, 12227–12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rocnik E. F., Liu P., Sato K., Walsh K., Vaziri C. (2006) The novel SPARC family member SMOC-2 potentiates angiogenic growth factor activity. J. Biol. Chem. 281, 22855–22864 [DOI] [PubMed] [Google Scholar]
- 28. Sarkar D., Park E. S., Emdad L., Lee S-G., Su Z. Z., Fisher P. B. (2008) Molecular basis of NF-κB activation by astrocyte elevated gene-1 (AEG-1). Cancer Res. 1, 1478–1484 [DOI] [PubMed] [Google Scholar]
- 29. Galardi S., Mercatelli N., Farace M. G., Ciafrè S. A. (2011) NF-κB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 39, 3892–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown K., Gerstberger S., Carlson L., Franzoso G., Siebenlist U. (1995) Control of I κ B-α proteolysis by site-specific, signal-induced phosphorylation. Science 267, 1485–1488 [DOI] [PubMed] [Google Scholar]
- 31. Abel S., Hundhausen C., Mentlein R., Schulte A., Berkhout T. A., Broadway N., Hartmann D., Sedlacek R., Dietrich S., Muetze B., Schuster B., Kallen K. J., Saftig P., Rose-John S., Ludwig A. (2004) The transmembrane CXC-chemokine ligand 16 is induced by IFN-γ and TNF-α and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J. Immunol. 172, 6362–6372 [DOI] [PubMed] [Google Scholar]
- 32. Chandrasekar B., Bysani S., Mummidi S. (2004) CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, I κ B kinase, and nuclear factor-κ B and induces cell-cell adhesion and aortic smooth muscle cell proliferation. J. Biol. Chem. 279, 3188–3196 [DOI] [PubMed] [Google Scholar]
- 33. Li S., Yu W., Hu G. F. (2012) Angiogenin inhibits nuclear translocation of apoptosis inducing factor in a Bcl-2-dependent manner. J. Cell. Physiol. 227, 1639–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ibaragi S., Yoshioka N., Kishikawa H., Hu J. K., Sadow P. M., Li M., Hu G. F. (2009) Angiogenin-stimulated rRNA transcription is essential for initiation and survival of AKT-induced prostate intraepithelial neoplasia. Mol. Cancer Res. 7, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J., Lu Y., Wang J., Koch A. E., Zhang J., Taichman R. S. (2008) CXCR6 induces prostate cancer progression by the AKT/mammalian target of rapamycin signaling pathway. Cancer Res. 68, 10367–10376 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., Murphy G. (2000) The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 473, 275–279 [DOI] [PubMed] [Google Scholar]
- 37. Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S. S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P., Gasparini P., Gonelli A., Costinean S., Acunzo M., Condorelli G., Croce C. M. (2009) miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 down-regulation. Cancer Cell 16, 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38. Majumder P. K., Yeh J. J., George D. J., Febbo P. G., Kum J., Xue Q., Bikoff R., Ma H., Kantoff P. W., Golub T. R., Loda M., Sellers W. R. (2003) Prostate intraepithelial neoplasia induced by prostate restricted Akt activation. The MPAKT model. Proc. Natl. Acad. Sci. U.S.A. 100, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.