Abstract
Transition metals are essential components of important biomolecules, and their homeostasis is central to many life processes. Transmembrane transporters are key elements controlling the distribution of metals in various compartments. However, due to their chemical properties, transition elements require transporters with different structural-functional characteristics from those of alkali and alkali earth ions. Emerging structural information and functional studies have revealed distinctive features of metal transport. Among these are the relevance of multifaceted events involving metal transfer among participating proteins, the importance of coordination geometry at transmembrane transport sites, and the presence of the largely irreversible steps associated with vectorial transport. Here, we discuss how these characteristics shape novel transition metal ion transport models.
Keywords: ATPases, Copper, Metals, Protein/Metal Ion Interaction, Transport Metals, Zinc
Introduction
Micronutrient transition metals (manganese, iron, cobalt, nickel, copper, zinc, molybdenum, and tungsten) serve catalytic and structural functions as prosthetic groups in metalloproteins. In these roles, they are required for a number of diverse physiological processes, ranging from gene transcription to respiration (1). However, despite their essential roles and ubiquitous presence, metals can cause deleterious effects by catalyzing the production of free radicals or by simply impairing metalloenzyme functions by substituting for the optimal metal cofactors. Consequently, organisms strive to maintain a tightly controlled homeostasis of these elements through the coordinated action of transmembrane transporters; chaperone, sequestering, and storage molecules; and metal-responsive transcriptional regulators (2–4). These components distribute the ions to appropriate targets and maintain adequate metal quotas, keeping the cellular compartments essentially free of unsequestered metals (3, 5, 6).
This minireview focuses on the structural and functional aspects of transmembrane transporters that participate in the homeostasis of transition metals. The current understanding of ion transmembrane transport is rooted in 6 decades of research characterizing alkali (H+, Na+, and K+) and alkali earth (Mg2+ and Ca2+) channels, carriers, and pumps. These ions are free (hydrated) and abundant in biological systems. Therefore, their transport mechanisms are shaped by electrochemical gradients and governed by their reversible interaction with transmembrane transport sites constituted by polar amino acid side chains (7). Consideration of the physicochemical differences between alkali/alkali earth and transition metal ions quickly reveals that the existing models describing ion transmembrane translocation cannot explain the mechanism of transport of transition metals. In this context, emerging paradigms for the transport of uncomplexed metal ions are discussed here. Transporters of metal complexes (siderophore-metal, heme, etc.) will not be considered because their selectivity and mechanism might not be determined by the bound metal, but rather by the coordinating molecules (8–10).
General Characteristics of Transition Metal Transport
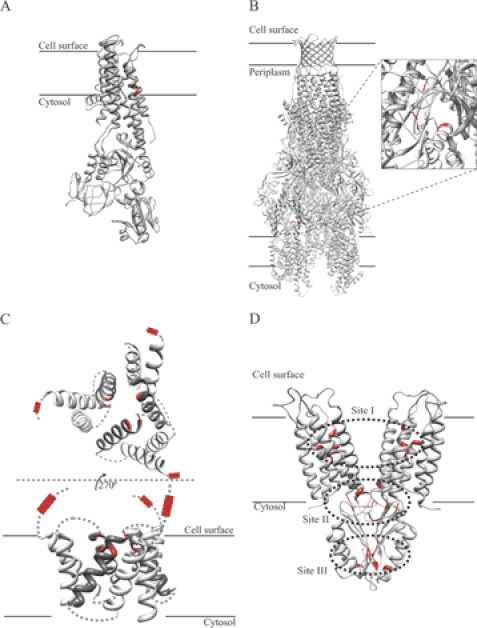
A number of families of carriers and pumps responsible for metal influx and efflux from various subcellular compartments have been identified: ATP-binding cassette-type ATPases, PIB-type ATPases, ZIP (Zrt/Irt-like protein), Ctr (copper uptake), Nramp (natural resistance-associated macrophage protein), RND (resistance-nodulation-cell division), and cation diffusion facilitator (CDF)2 transporters, among others (6, 11–13). These are polytopic membrane proteins that show diverse structural arrangements of transmembrane segments (TM), combined in some cases with regulatory and catalytic hydrophilic domains. High-resolution structures of model members of some of these families have been reported (Fig. 1) (14, 15). Although the presence of an ion path across the membrane is a logical feature of these transporters, additional distinctive characteristics are surfacing, e.g. the presence of putative docking regions where chaperone proteins and/or chelating molecules might deliver the metal substrate to the transporter or, alternatively, receive the metal subsequently after its translocation (see below) (14, 16–19). However, although this docking will contribute to the in vivo substrate selectivity of transporters, the metal coordination during transport is the defining feature that determines the functional capabilities of these proteins (20, 21).
FIGURE 1.
Structures and metal-binding sites of metal transporters. A, structure of the Cu+-ATPase CopA (Protein Data Bank code 3RFU), with amino acids forming the two transmembrane Cu+-binding sites indicated in red. B, CusABC model assembled with Swiss-PDBViewer using the CusAB (code 3NE5) and CusC (code 3PIK) structures. The magnified section indicates the Cu+-binding sites (red) in a CusA monomer. C, side and apical views of modeled Ctr transporters (provided by Dr. Vincent Unger, Northwestern University). Extracytoplasmic and transmembrane Met-rich Cu+-binding sites are indicated in red. Darker helices correspond to those involved in transmembrane metal binding. D, structure of the CDF transporter YiiP (code 2QFI). Red amino acids indicate Zn2+-binding sites. Dotted areas indicate the three metal-binding sites in each YiiP monomer.
Biologically relevant transition metals (manganese, iron, cobalt, copper, and zinc) are located in the d-block of the periodic table (with most in period 4, groups 7–12), i.e. their electronic d-shell is incomplete (except for zinc). These elements are considered soft Lewis acids (22). They present high binding stability constants in aqueous media (fm−1 range) when coordinated by soft Lewis base ligands such as thiolate (sulfur) and imidazolium (nitrogen). These differ from alkali/alkali earth metals that behave as hard Lewis acids. These rather prefer coordination by smaller hard Lewis bases such as carboxylate (oxygen), forming ionic adducts that have lower binding stability constants (1). Consequently, it is expected that transition metal-binding sites involved in the transient association during translocation across the membrane would be mostly constituted by fitting intermediate (N) or soft (S) bases (1). Furthermore, the outer shell electronic configurations would favor particular coordination geometries for the various metal substrates. Well known metal coordination architectures are largely based on the characterization of organometallic complexes and of metal sites within soluble metalloproteins, where the static prosthetic group remains bound for the life of the protein. In contrast, although binding the metal with very high affinities, transmembrane transport sites only transiently interact with the substrate during transport. Moreover, these sites must present the flexibility to allow the vectorial ion release (i.e. across the permeability barrier) upon minimal coordination shifts (17, 20, 23). Consequently, novel ligands and metal coordination architectures might be expected at the transmembrane transport/binding sites (transmembrane metal-binding sites (TM-MBS)) of transition metal transporters. An example of these features is the coordination of Cu+ by PIB-ATPases or Ctr proteins. Although this metal has a tetragonal (e.g. superoxide dismutase) or tetrahedral (e.g. plastocyanin) coordination in metalloproteins and a linear coordination in regulatory cytosolic metal-binding domains (e.g. N-terminal metal-binding domain (N-MBD) Cu+-ATPases), it has a trigonal planar coordination during transport (17, 24, 25). Moreover, the unexpected coordination by one or more oxygen-containing side chains is observed in these sites. Consequently, we hypothesize that a specific metal coordination in the TM-MBSs is required for transport and that evolution has selected some coordination states over others. An additional outcome of a selectivity based on acid-base Lewis chemistry and coordination geometry is the capability of these transporters to bind and, in some cases, translocate non-physiological ligands at a greater extent than alkali/alkali earth transporters. Cu+ transporters can translocate Ag+ and probably Au+; Pb2+ and Cd2+ function as substrates of Zn2+-ATPases, and these can also bind non-transported metals (Cu2+, Co2+, and Ni2+) with similar affinity (26–30).
The tight metal binding to the transport sites has an immediate consequence, which is the observed slow transmembrane transport rates. Alkali/alkali earth ion transport rates range from 109-107 ions/s for the classical Na+ or K+ channels (31) to ∼200 ions/s for ion pumps (32). In contrast, estimated turnover rates for Cu+-ATPases, Zn2+-transporting CDFs, or the Cu+ carrier CTR1 are <10 ions/s (26, 28, 33).3 Available functional determinations suggest that these low transport rates are associated with the slow release of metal by the transporter. However, it might be argued that, in some cases, the reported metal transport rates are underestimated because metal transport assays are frequently performed in the absence of a post-transport “receiving” molecule, i.e. an accepting metal-sequestering molecule preventing release of free metal. Although this might be the case, it appears unlikely that the presence of accepting molecules in the reaction would substantially increase the transport rates (>10-fold) to levels comparable with those of alkali/alkali earth transporters.
Toward explaining the slow transport, it can be speculated that the minimal cellular requirements for transition metals have not represented a significant selective pressure for the evolution of faster metal transporters. Alternatively, alkali/alkali earth metals participate in dynamic events requiring significant mass/charge redistribution (signal transduction and osmotic and electrical balance). This has probably driven the selection of fast transporting molecules. Moreover, the necessary absence of free metals in cellular compartments constrains the overall transport rate to the availability of chaperone/sequestering molecules. These characteristics of metal homeostasis explain the absence of transition metal “channels,” where the ions travel at rates close to that of diffusion.
These common features of metal transport (substrate transfer through protein/protein interaction, specificity of substrate, and relatively slow transport rates) are best illustrated by discussing recent developments in the structural analysis of some of these transporters. Although, in most instances, the structures have been obtained from bacterial transporters, it is expected that their characteristic features are also shared with eukaryotic transporters, albeit with minor changes. Similarly, most of the information considered in this minireview refers to Cu+ transporters because most of the structural/mechanistic work has been done in Cu+-transporting systems.
Metal Transport by PIB-ATPases
PIB-ATPases are polytopic membrane proteins (Fig. 1A). Sharing a common core structure and catalytic mechanism, they belong to the superfamily of P-ATPases (21). They are present in all life kingdoms, and most sequenced genomes contain several members of the PIB family with different substrate specificities or distinct functional roles (18, 21, 34). In eukaryotic cells, PIB-ATPases are present in almost all organelles (vacuole, trans-Golgi network, chloroplast, and plasma membrane, among others) (35–37), where they assist in metal detoxification and metalloprotein synthesis (37, 38), as also occur in bacteria (34). Early studies based on bioinformatic, genetic, and biochemical analyses suggested the following distinct metal specificities: Cu+ (PIB-1), Zn2+ (PIB-2), Cu2+ (PIB-3), and Co2+ (PIB-4) (18, 21, 39). These are determined by highly conserved amino acids present in three TMs constituting the TM-MBSs. The involved TMs flank the catalytic cytosolic loop where ATP binding and hydrolysis occur (21, 39), providing a structural link for cytosolic metal export coupled to ATP hydrolysis and enzyme phosphorylation as described for well characterized members of the P-ATPase superfamily (21, 40, 41).
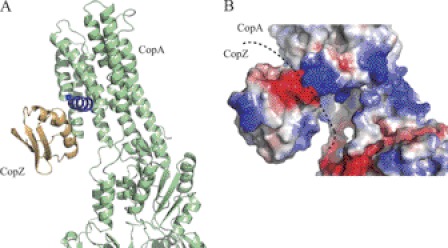
The metal selectivity of PIB-ATPases is the outcome of hierarchical multifaceted events. Ultimately, these yield the binding of the correct substrate to the TM-MBS (Fig. 2). Functional and structural studies of the Archaeoglobus fulgidus Cu+-ATPase CopA indicated that, in a first step, substrate specificity is determined in vivo by the interaction between a soluble metal-loaded chaperone and the metal-accepting ATPase (17). In vitro assays using purified proteins showed that the Cu+·chaperone complex (Cu+·A. fulgidus CopZ (AfCopZ)) was responsible for metal transfer to the ATPase TM-MBSs. In A. fulgidus, the chaperone cannot be substituted in this role by the homologous N-MBDs usually present in Cu+-ATPases, although it seems to be possible in yeast (42). The interaction between the Cu+-delivering protein and the ATPase is likely determined by a specific geometry in both interfaces that assists the positioning of the metal in the proximity of the ATPase metal-accepting sites. The atomic resolution structure of Legionella pneumophila CopA supports this model (14). In this structure, the first and second TMs form a platform on which the chaperone would likely dock to transfer the metal (Fig. 3A). This interaction is determined by electrostatic forces, in which the negatively charged face of the chaperone would interact with the positively charged docking “platform” (Fig. 3B). This would orient the chaperone-bound Cu+ toward three conserved amino acids (Met, Asp, and Glu) located at the cytoplasmic entrance of the metal transmembrane path. The electropositive exposed surface of the platform also precludes a postulated interaction of the cytoplasmic N-MBDs present in these ATPases, delivering Cu+ to TM-MBSs. It has been shown that N-MBDs have the same electrostatic charge as the ATPase platform and an opposite charge to the chaperone (43). In fact, the electrostatic complementation of N-MBDs and Cu+ chaperones contributes to their interactions and subsequent Cu+ exchange.
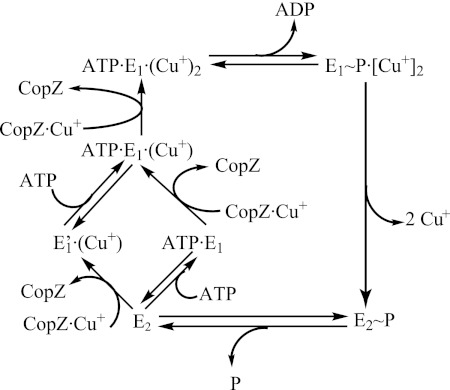
FIGURE 2.
Catalytic cycle of a Cu+-ATPase. Cu+ binding to two TM-MBSs is required for catalytic phosphorylation by ATP (E1∼P·Cu2+). Note the irreversibility of Cu+ transfer from the chaperone (CopZ) to the transport site and that full occupancy is reached only in the presence of ATP. Metal is released after a conformational change (to E2∼P) leading TM-MBSs to open to the vesicular/extracellular medium. E2 → E1 transition is accelerated by ATP (or ADP) acting in a modulatory mode (low affinity). See Ref. 46 for more details.
FIGURE 3.
Cu+ chaperone/Cu+-ATPase interaction. A, docking was modeled using ClusPro (83). A. fulgidus CopA (green) was modeled after L. pneumophila CopA (Protein Data Bank code 3RFU), whereas the model of the C-terminal domain of A. fulgidus CopZ (ochre) was built using Enterococcus hirae CopZ (code 1CPZ) as a template. The CopA platform for interaction with CopZ is shown in blue. B, surface charges in the predicted docking of CopZ with CopA. Positive and negative charge densities are indicated in blue and red, respectively.
Testing this model, we calculated the polar binding energies involved in the docking of Cu+·AfCopZ·AfCopA (complex 1) and Cu+·AfN-MBD·AfCopA (complex 2) in the platform region of the ATPase. This approach estimates the stability of the complexes in a salt (0.15 m) aqueous solution (see legend of Fig. 3 for details) (44). The energy values are obtained by solving the Poisson-Boltzmann equation through semi-numerical/semi-analytical methods. The polar binding energy of complex 1 was −11.11 kcal/mol, whereas that of complex 2 was +26.12 kcal/mol. This suggests a lack of stability of a hypothetical intramolecular complex 2 and supports previous data indicating a regulatory role for N-MBDs rather than one delivering Cu+ to TM-MBSs (17, 45, 46). More importantly, this analysis points out the likely favorable interaction of the Cu+ chaperone and the ATPase. Interestingly, the interaction requires the metal-bound chaperone, as indicated by the unsuccessful docking of the apo-chaperone with the ATPase. This is in agreement with biochemical data showing that AfCopZ does not compete with Cu+·AfCopZ in inhibiting the Cu+ transfer to the ATPase (17). This results in a unidirectional metal movement that yields a stoichiometric Cu+ transfer to TM-MBSs (Cu+·CopZ + CopA → CopZ + Cu+·CopA) (Fig. 2) (17, 46). However, the chaperone metal sites can potentially bind other metals (47), which in turn might enable docking with the ATPase and subsequent metal delivery. A second layer of specificity provided by the metal coordination at the TM-MBSs then becomes relevant.
Biochemical studies have shown the trigonal coordination of Cu+ at the TM-MBSs (48, 49). As mentioned above, this unique geometry is distinctly associated with transport sites. When the activation of Cu+-ATPases by various ions in the absence of chaperones is tested in vitro, the ATPases apparently accept only Cu+ and similar ions (Ag+ and Au+) but not others such as Zn2+, Cu2+, Co2+, and Ni2+ (26). Thus, independent of the chaperone, the ATPase TM-MBS selects the transported metals. Studies of the Escherichia coli Zn2+-ATPase ZntA might better support these ideas. The ZntA TM-MBS binds transported substrates (Zn2+, Cd2+, and Pb2+) as well as non-transported divalent heavy metals (Cu2+, Co2+, and Ni2+) with similar affinity (20, 30). Although Cu2+, Co2+, and Ni2+ tightly bind the enzyme, these metals cannot induce the required enzyme conformation that enables the catalytic hydrolysis of ATP. This suggests that the geometry of coordination and metal–ligand bond distances play an important role in the activation of PIB-ATPases. As a corollary, metal coordination geometry, rather than binding affinity, is the determinant of transport specificity. Similar phenomena are observed in metal regulatory proteins (2, 50).
Metal release from the protein occurs upon the major conformational change. Here, PIB-ATPases also highlight a common feature of metal transporters: a slower transport rate compared with closely related alkali/alkali earth-transporting ATPases. This is especially evident in those Cu+-ATPases of the FixI/CopA2 subgroup (18, 34). These ATPases present the slowest transport rates, most probably to couple metal transport with the export of metal-accepting apoproteins. In multicellular organisms, a similar mechanism seems to be in place (16).
RND Metal Transporters
The members of the RND superfamily are tripartite transporters widespread in Gram-negative bacteria (6). This superfamily contains seven subfamilies with different substrate specificities. These include antimicrobial agents, organic solvents, and heavy metals, among other molecules. In all cases, the substrate appears to be transported from the periplasm to the extracellular space. The systems span the periplasmic space with a cytoplasmic membrane protein (RND), an outer membrane porin, and a periplasmic membrane fusion protein (MFP) bridging the inner and outer membrane components. A H+ antiport is used to satisfy the energetic requirements of the substrate efflux (51).
The best characterized RND heavy metal transporter is the E. coli Cu+/Ag+ efflux CusCFBA system (52–55). The corresponding operon encodes the three characteristic proteins of these systems: RND (CusA), the outer membrane porin (CusC), and the MFP (CusB). These proteins are arranged in a multimeric form with trigonal symmetry: a CusA trimer contacts a CusB hexamer, which interacts with a CusC trimer (Fig. 1B) (55, 56). In the most likely model, metal transport is initiated by the binding of periplasmic Cu+ to the N-terminal domain of CusB (57), where it is again coordinated in planar trigonal geometry by three Met residues (55). The sequence of this site corresponds to the transported metal substrate. For instance, in the MFP of Zn2+-transporting ZneABC, the metal is coordinated by two His residues and one Glu residue (58). In this case, an additional ligand should be involved to achieve the tetradentate Zn2+ coordination common to Zn2+ transporters and metalloproteins (23, 59).4 The fate of the metal after binding CusB has not been established. It has been proposed that metal binding to CusB causes a conformational change that might position the metal closer to the metal-binding site of CusA in the plasma membrane (58, 60). This site (constituted by three Met residues) is in a periplasmic cleft formed by the loops between TM1 and TM2 and between TM7 and TM8 (Fig. 1B) (54). Following this model, the subsequent step involving the transfer from CusA to other components of the system has not been established.
Because free Cu+ is toxic, the metal has to be provided by a periplasmic chaperone to CusB. For this purpose, the CusCFBA operon also encodes a periplasmic metal-binding protein, CusF. In a role analogous to cytoplasmic CopZ, CusF appears to function as a periplasmic Cu+ chaperone that delivers the metal to CusABC (57). Cu+ coordination in E. coli CusF is achieved by a trigonal coordination by two Met residues and one His residue (52). The presence of Met in the CusF binding site solves the likely oxidation of periplasmic –SH groups if Cys were part of this site. In vitro, CusF transfers metal directly to metal-binding sites in the N-terminal domain of CusB (61).
Evidently, a central element of the transport mechanism of RND systems is the transfer of metals between different protein components. For instance, Cu+ transfer from CusF to CusB is considered part of the metal transport pathway. However, CusF exchanges Cu+ with CusB with Keq ∼ 1 (60). Thus, Cu+ transfer is far from unidirectional. If this is part of the transport pathway, then a largely irreversible step should occur later in the translocation process. Furthermore, metal occupancy of CusB rather than the level of Cu+·CusF would control the transport rate. Alternatively, it could be postulated that Cu+ binding to CusB might have a regulatory effect and that CusF might be able to transfer metal directly to CusC, as cytoplasmic chaperones deliver Cu+ to the TM-MBSs of Cu+-ATPases. Supporting the latter model, in some cases, CusF and CusB are fused in a single protein, as seems to be the case for SilB, the MFP subunit of SilABC, a RND system in Cupriavidus metallidurans CH4 (62). In this protein, the MFP has an extended C-terminal metallochaperone domain closely related to CusF. Some organisms even appear to lack CusB (Legionella longbeachae, several Pseudomonas and Shigella species, Xanthomonas campestris, etc.) while maintaining the other elements of the system.5 This suggests that CusB is not an essential component of the system.
Although RND metal efflux systems seem to be primarily responsible for detoxification of periplasmic metals (57), it has been suggested that the system would also transport cytosolic metal across the plasma membrane (63). In this model, Cu+ would follow a Met “shuttle” in CusA. Experiments testing the transport into reconstituted liposomes show that CusA transports Ag+ in favor of a large gradient (0.5 mm Ag+ in the cytoplasmic side) in a pH-dependent fashion. However, despite the large gradient, transport is quickly inhibited (20 s). The mechanism of this transport, how Cu+ is transferred to CusC, and the role of a cytoplasmic Cu+ efflux system in addition to the ubiquitous Cu+-ATPase are not clear.
Ctr Family of Eukaryotic Cu+ Transporters
The Ctr family transporters are found exclusively in eukaryotes, where they enable the flux of Cu+ into the cytoplasm, either facilitating its incorporation from the extracellular space or mobilizing the vacuolar stores (64). Their importance is highlighted, for instance, by the embryonic lethal phenotype resulting from the Ctr1 gene knock-out in mice (65). Ctr proteins are homotrimers. Monomers are 140–400-amino acid proteins with three TMs (TM1–3) and frequently present an extracellular N-terminal Met-rich motif (MXXM/MXM) (12). TM2 contains a conserved MXXXM motif that faces a path at the center of the trimer (Fig. 1C) (25). The final structure appears as a channel or “pore,” with a conical side narrower at the extracellular/luminal side of the protein. Consistent with this channel-like structure, it has been proposed that Cu+ uptake through Ctr transporters is driven by a passive, membrane potential-dependent mechanism (12). However, this model still needs to be supported by strong experimental evidence and has to take into account the role of metal-accepting chaperones.
The functional Ctr transporter complex can stably bind two Cu+ ions/trimer (25). One of these sites is within the Ctr pore and is constituted by a Met residue from each monomer, thus providing a trigonal planar coordination as observed in Cu+ sites of Cu+-ATPases and RND transporters. The second site has not been identified, but likely candidates are the N-terminal Met-rich region or the HCH motif at the C terminus (25). The functional roles of the N-terminal region, amino acids in the transmembrane region, and C-terminal HCH motifs have not been well defined. Structural data suggest that Met in TM2 should play an important role in the Cu+ transport across the membrane, providing a mechanism of selectivity through an appropriate geometry of coordination (25). However, mutation of transmembrane Met located in TM2 does not abolish metal flux, although it decreases the rate of transport (66). On the other hand, in some cases, the N-terminal Met-rich region seems to be essential for metal transport (66), where it could play a role in binding extracellular Cu+. Based on relatively limited experimental evidence, two models have been proposed for the transport mechanism: a “channel-like” model in which Cu+ would interact weakly with ligands facing the inner face of the pore (66) and a model in which Cu+ would be translocated, passing through several binding sites composed by “essential” residues accommodated by hierarchical affinities in the Cu+ pathway (25, 67). Several findings such as differential trypsin digestion in the presence of Cu+ (68), the C-terminal interaction likely coupled to Cu+ transport activity (67), and molecular dynamic simulations (69) point to a Ctr metal ion transport mechanism involving structural conformational changes.
As cells strive to prevent the presence of free Cu+, part of a Ctr transport mechanism is the metal delivery to specific Cu+ chaperones that would carry the ion to appropriate targets. In vitro studies have shown that the C-terminal domain of Saccharomyces cerevisiae Ctr1 would interact with the corresponding Cu+ chaperone Atox1 (70). This is a relevant finding to elucidate how the secretory pathway may acquire Cu+. However, it does not address whether Ctr1 is directly involved in the metallation of other cytosolic Cu+ chaperones such as CCS or if it supplies Cu+ to cellular labile metal-sequestering pools such as glutathione.
CDFs
CDF transporters are ubiquitous membrane proteins responsible for the cytosolic efflux of divalent cations coupled to the influx of H+ or Na+ (71). In eukaryotes, they are localized in the plasma membrane and in organelles (vacuole, endoplasmic reticulum, Golgi, etc.) (72–74), where they participate in metal detoxification, metalloprotein assembly, and packaging of secretory vesicles (75–77). The functional forms of the transporters are homodimers. The topology of the subunit, six TMs and a cytoplasmic hydrophilic C-terminal domain, is well conserved among all family members (Fig. 1D). In addition, all eukaryotic and some bacterial CDFs present a His-rich cytosolic region between TM4 and TM5 (78, 79).
A significant understanding of the mechanism of CDFs emerged from the biochemical characterization and structural studies of the E. coli Zn2+ transporter YiiP (15, 23, 79). YiiP is a homodimer of two 33-kDa subunits in 2-fold symmetry (Fig. 1D) (15, 79). It presents several high-affinity Zn2+/Cd2+-binding sites with seemingly different coordination geometry (28, 80). These are located in the transmembrane region (site I), the membrane-cytosol interface not fully conserved in all CDFs (site II), and the C-terminal domain (site III) (15, 80).
Site I binds the transported metal (15, 23). This site defines the selectivity of YiiP for Zn2+/Cd2+ over Fe2+, Mn2+, Ni2+, and Co2+. The site is composed of two Asp residues in TM2, one His residue, and one Asp residue in TM5, binding the metal with tetrahedral coordination. Mutation of these residues prevents metal transport (23, 81, 82). Kinetic evidence suggests that once Zn2+ is bound to site I, it is quickly extruded, ensuring a unidirectional, largely irreversible transport mechanism (28). This analysis also highlights the relatively low transport rate of metal transporters (2.6 s−1 for Zn2+ transport by E. coli ZitB) (28).
Although it has been speculated that the conserved C-terminal domain (site III) might act as a metallochaperone (15), there is no experimental evidence for this hypothesis. Site III appears to be involved in YiiP dimerization and consequent activation (80). Zn2+ is bound tetrahedrally by amino acids from each monomer (Fig. 1D). Metal binding in this site contributes to the stabilization of the interaction between the C-terminal domains of each monomer. This appears to be a regulatory mechanism by which the functional transporter is assembled when excess substrate is present.
Future Directions
In the last few years, the first high-resolution structures of representative members of some of the main metal transporter families have been obtained. Further progress is expected in this direction with the structural characterization of other metal transporter families such as ZIP and Nramp and with further refinements of already determined structures in all their conformational stages. This will help establish the structural and functional determinants that lead to distinct metal transport mechanisms and transport specificity required by the cell to handle fundamental but highly toxic transition metal ions. However, to validate the accuracy of novel models, similar advances in biochemical and biophysical studies will be required. Because of their importance in metalloprotein assembly and, consequently, in overall cell physiology, the determination of the precise interaction mechanism of metal transporters and metal-delivering and metal-accepting chaperones is one of the areas in which significant developments are likely.
Acknowledgments
We thank Dr. Luis Fernández Pacios for polar binding energy calculations and Drs. R. Dempski and M. Emmert for critical reading of the manuscript and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R21AI082484-01 (to J. M. A.). This work was also supported by National Science Foundation Grant MCB-0743901 and National Institute of Food and Agriculture Grant 2010-65108-20606 from the United States Department of Agriculture (to J. M. A.) and by Marie Curie International Reintegration Grant MENOMED and Ramón y Cajal Fellowship RYC-2010-06363 (to M. G.-G.). This is the first article in the Thematic Minireview Series on Metals in Biology 2012.
J. H. Kaplan, personal communication.
D. Raimunda, T. L. Stemmler, and J. M. Argüello, unpublished data.
G. Hernández, J. M. Argüello, and B. Valderrama, unpublished data.
- CDF
- cation diffusion facilitator
- TM
- transmembrane segment
- TM-MBS
- transmembrane metal-binding site
- N-MBD
- N-terminal metal-binding domain
- AfCopZ
- A. fulgidus CopZ
- MFP
- membrane fusion protein.
REFERENCES
- 1. Fraústro da Silva J. J. R., Williams R. J. P. (2001) The Biological Chemistry of the Elements, 2nd Ed., Oxford University Press, New York [Google Scholar]
- 2. Ma Z., Jacobsen F. E., Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robinson N. J., Winge D. R. (2010) Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osman D., Cavet J. S. (2008) Copper homeostasis in bacteria. Adv. Appl. Microbiol. 65, 217–247 [DOI] [PubMed] [Google Scholar]
- 5. Outten C. E., O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 [DOI] [PubMed] [Google Scholar]
- 6. Nies D. H. (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339 [DOI] [PubMed] [Google Scholar]
- 7. Blaustein M. P., Kao J. P. Y., Matteson D. R. (eds) (2011) Cellular Physiology and Neurophysiology, 2nd Ed., Elsevier/Mosby, Philadelphia [Google Scholar]
- 8. Wandersman C., Delepelaire P. (2004) Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58, 611–647 [DOI] [PubMed] [Google Scholar]
- 9. DiDonato R. J., Jr., Roberts L. A., Sanderson T., Eisley R. B., Walker E. L. (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotinamine-metal complexes. Plant J. 39, 403–414 [DOI] [PubMed] [Google Scholar]
- 10. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colangelo E. P., Guerinot M. L. (2006) Put the metal to the petal: metal uptake and transport throughout plants. Curr. Opin. Plant Biol. 9, 322–330 [DOI] [PubMed] [Google Scholar]
- 12. Dumay Q. C., Debut A. J., Mansour N. M., Saier M. H., Jr. (2006) The copper transporter (Ctr) family of Cu+ uptake systems. J. Mol. Microbiol. Biotechnol. 11, 10–19 [DOI] [PubMed] [Google Scholar]
- 13. Nevo Y., Nelson N. (2006) The NRAMP family of metal ion transporters. Biochim. Biophys. Acta 1763, 609–620 [DOI] [PubMed] [Google Scholar]
- 14. Gourdon P., Liu X. Y., Skjørringe T., Morth J. P., Møller L. B., Pedersen B. P., Nissen P. (2011) Crystal structure of a copper-transporting PIB-type ATPase. Nature 475, 59–64 [DOI] [PubMed] [Google Scholar]
- 15. Lu M., Fu D. (2007) Structure of the zinc transporter YiiP. Science 317, 1746–1748 [DOI] [PubMed] [Google Scholar]
- 16. Barry A. N., Otoikhian A., Bhatt S., Shinde U., Tsivkovskii R., Blackburn N. J., Lutsenko S. (2011) The luminal loop Met672–Pro707 of copper-transporting ATPase ATP7A binds metals and facilitates copper release from the intramembrane sites. J. Biol. Chem. 286, 26585–26594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. González-Guerrero M., Argüello J. M. (2008) Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. U.S.A. 105, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raimunda D., González-Guerrero M., Leeber B. W., 3rd, Argüello J. M. (2011) The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals 24, 467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aller S. G., Unger V. M. (2006) Projection structure of the human copper transporter CTR1 at 6-Å resolution reveals a compact trimer with a novel channel-like architecture. Proc. Natl. Acad. Sci. U.S.A. 103, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu J., Dutta S. J., Stemmler A. J., Mitra B. (2006) Metal-binding affinity of the transmembrane site in ZntA: implications for metal selectivity. Biochemistry 45, 763–772 [DOI] [PubMed] [Google Scholar]
- 21. Argüello J. M., Eren E., González-Guerrero M. (2007) The structure and function of heavy metal transport PIB-ATPases. Biometals 20, 233–248 [DOI] [PubMed] [Google Scholar]
- 22. Pearson R. G. (1963) Hard and soft acids and bases. J. Am. Chem. Soc. 85, 3533–3539 [Google Scholar]
- 23. Wei Y., Fu D. (2006) Binding and transport of metal ions at the dimer interface of the Escherichia coli metal transporter YiiP. J. Biol. Chem. 281, 23492–23502 [DOI] [PubMed] [Google Scholar]
- 24. Holm R. H., Kennepohl P., Solomon E. I. (1996) Structural and functional aspects of metal sites in biology. Chem. Rev. 96, 2239–2314 [DOI] [PubMed] [Google Scholar]
- 25. De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., Unger V. M. (2009) Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mandal A. K., Cheung W. D., Argüello J. M. (2002) Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeoglobus fulgidus. J. Biol. Chem. 277, 7201–7208 [DOI] [PubMed] [Google Scholar]
- 27. Hou Z., Mitra B. (2003) The metal specificity and selectivity of ZntA from Escherichia coli using the acylphosphate intermediate. J. Biol. Chem. 278, 28455–28461 [DOI] [PubMed] [Google Scholar]
- 28. Chao Y., Fu D. (2004) Kinetic study of the antiport mechanism of an Escherichia coli zinc transporter, ZitB. J. Biol. Chem. 279, 12043–12050 [DOI] [PubMed] [Google Scholar]
- 29. Espariz M., Checa S. K., Audero M. E., Pontel L. B., Soncini F. C. (2007) Dissecting the Salmonella response to copper. Microbiology 153, 2989–2997 [DOI] [PubMed] [Google Scholar]
- 30. Dutta S. J., Liu J., Hou Z., Mitra B. (2006) Conserved aspartic acid 714 in transmembrane segment 8 of the ZntA subgroup of PIB-type ATPases is a metal-binding residue. Biochemistry 45, 5923–5931 [DOI] [PubMed] [Google Scholar]
- 31. Hille B. (2001) Ion Channels of Excitable Membranes, 3rd Ed., Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 32. Palmgren M. G., Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 33. Sharma R., Rensing C., Rosen B. P., Mitra B. (2000) The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J. Biol. Chem. 275, 3873–3978 [DOI] [PubMed] [Google Scholar]
- 34. González-Guerrero M., Raimunda D., Cheng X., Argüello J. M. (2010) Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 78, 1246–1258 [DOI] [PubMed] [Google Scholar]
- 35. La Fontaine S., Mercer J. F. (2007) Trafficking of the copper-ATPases, ATP7A and ATP7B: role in copper homeostasis. Arch. Biochem. Biophys. 463, 149–167 [DOI] [PubMed] [Google Scholar]
- 36. Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K. K., Pilon M. (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15, 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morel M., Crouzet J., Gravot A., Auroy P., Leonhardt N., Vavasseur A., Richaud P. (2009) AtHMA3, a PIB-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 149, 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan D. S., Stearman R., Dancis A., Dunn T., Beeler T., Klausner R. D. (1995) The Menkes/Wilson disease gene homolog in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. U.S.A. 92, 2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Argüello J. M. (2003) Identification of ion selectivity determinants in heavy metal transport PIB-type ATPases. J. Membr. Biol. 195, 93–108 [DOI] [PubMed] [Google Scholar]
- 40. Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007) The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 41. Kaplan J. H. (2002) Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 [DOI] [PubMed] [Google Scholar]
- 42. Morin I., Gudin S., Mintz E., Cuillel M. (2009) Dissecting the role of the N-terminal metal-binding domains in activating the yeast copper ATPase in vivo. FEBS J. 276, 4483–4495 [DOI] [PubMed] [Google Scholar]
- 43. Arnesano F., Banci L., Bertini I., Ciofi-Baffoni S., Molteni E., Huffman D. L., O'Halloran T. V. (2002) Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res. 12, 255–271 [DOI] [PubMed] [Google Scholar]
- 44. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mandal A. K., Argüello J. M. (2003) Functional roles of metal-binding domains of the Archaeoglobus fulgidus Cu+-ATPase CopA. Biochemistry 42, 11040–11047 [DOI] [PubMed] [Google Scholar]
- 46. González-Guerrero M., Hong D., Argüello J. M. (2009) Chaperone-mediated Cu+ delivery to Cu+ transport ATPases: requirement of nucleotide binding. J. Biol. Chem. 284, 20804–20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wernimont A. K., Huffman D. L., Lamb A. L., O'Halloran T. V., Rosenzweig A. C. (2000) Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat. Struct. Mol. Biol. 7, 766–771 [DOI] [PubMed] [Google Scholar]
- 48. Mandal A. K., Yang Y., Kertesz T. M., Argüello J. M. (2004) Identification of the transmembrane metal-binding site in Cu+-transporting PIB-type ATPases. J. Biol. Chem. 279, 54802–54807 [DOI] [PubMed] [Google Scholar]
- 49. González-Guerrero M., Eren E., Rawat S., Stemmler T. L., Argüello J. M. (2008) Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J. Biol. Chem. 283, 29753–29759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Waldron K. J., Robinson N. J. (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35 [DOI] [PubMed] [Google Scholar]
- 51. Goldberg M., Pribyl T., Juhnke S., Nies D. H. (1999) Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 274, 26065–26070 [DOI] [PubMed] [Google Scholar]
- 52. Loftin I. R., Franke S., Roberts S. A., Weichsel A., Héroux A., Montfort W. R., Rensing C., McEvoy M. M. (2005) A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44, 10533–10540 [DOI] [PubMed] [Google Scholar]
- 53. Su C. C., Yang F., Long F., Reyon D., Routh M. D., Kuo D. W., Mokhtari A. K., Van Ornam J. D., Rabe K. L., Hoy J. A., Lee Y. J., Rajashankar K. R., Yu E. W. (2009) Crystal structure of the membrane fusion protein CusB from Escherichia coli. J. Mol. Biol. 393, 342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Long F., Su C. C., Zimmermann M. T., Boyken S. E., Rajashankar K. R., Jernigan R. L., Yu E. W. (2010) Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Su C. C., Long F., Zimmermann M. T., Rajashankar K. R., Jernigan R. L., Yu E. W. (2011) Crystal structure of the CusBA heavy metal efflux complex of Escherichia coli. Nature 470, 558–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kulathila R., Kulathila R., Indic M., van den Berg B. (2011) Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS ONE 6, e15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim E. H., Nies D. H., McEvoy M. M., Rensing C. (2011) Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J. Bacteriol. 193, 2381–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. De Angelis F., Lee J. K., O'Connell J. D., 3rd, Miercke L. J., Verschueren K. H., Srinivasan V., Bauvois C., Govaerts C., Robbins R. A., Ruysschaert J. M., Stroud R. M., Vandenbussche G. (2010) Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc. Natl. Acad. Sci. U.S.A. 107, 11038–11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. VanZile M. L., Cosper N. J., Scott R. A., Giedroc D. P. (2000) The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry 39, 11818–11829 [DOI] [PubMed] [Google Scholar]
- 60. Bagai I., Liu W., Rensing C., Blackburn N. J., McEvoy M. M. (2007) Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J. Biol. Chem. 282, 35695–35702 [DOI] [PubMed] [Google Scholar]
- 61. Bagai I., Rensing C., Blackburn N. J., McEvoy M. M. (2008) Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 47, 11408–11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bersch B., Derfoufi K. M., De Angelis F., Auquier V., Ekendé E. N., Mergeay M., Ruysschaert J. M., Vandenbussche G. (2011) Structural and metal binding characterization of the C-terminal metallochaperone domain of membrane fusion protein SilB from Cupriavidus metallidurans CH34. Biochemistry 50, 2194–2204 [DOI] [PubMed] [Google Scholar]
- 63. Su C. C., Long F., Yu E. W. (2011) The Cus efflux system removes toxic ions via a methionine shuttle. Protein Sci. 20, 6–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaplan J. H., Lutsenko S. (2009) Copper transport in mammalian cells: special care for a metal with special needs. J. Biol. Chem. 284, 25461–25465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee J., Prohaska J. R., Thiele D. J. (2001) Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98, 6842–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eisses J. F., Chi Y., Kaplan J. H. (2005) Stable plasma membrane levels of hCTR1 mediate cellular copper uptake. J. Biol. Chem. 280, 9635–9639 [DOI] [PubMed] [Google Scholar]
- 67. Sinani D., Adle D. J., Kim H., Lee J. (2007) Distinct mechanisms for Ctr1-mediated copper and cisplatin transport. J. Biol. Chem. 282, 26775–26785 [DOI] [PubMed] [Google Scholar]
- 68. Eisses J. F., Kaplan J. H. (2002) Molecular characterization of hCTR1, the human copper uptake protein. J. Biol. Chem. 277, 29162–29171 [DOI] [PubMed] [Google Scholar]
- 69. Schushan M., Barkan Y., Haliloglu T., Ben-Tal N. (2010) Cα trace model of the transmembrane domain of human copper transporter 1, motion and functional implications. Proc. Natl. Acad. Sci. U.S.A. 107, 10908–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xiao Z., Loughlin F., George G. N., Howlett G. J., Wedd A. G. (2004) C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: subfemtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 126, 3081–3090 [DOI] [PubMed] [Google Scholar]
- 71. Paulsen I. T., Saier M. H., Jr. (1997) A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156, 99–103 [DOI] [PubMed] [Google Scholar]
- 72. MacDiarmid C. W., Milanick M. A., Eide D. J. (2002) Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 277, 39187–39194 [DOI] [PubMed] [Google Scholar]
- 73. Ellis C. D., Wang F., MacDiarmid C. W., Clark S., Lyons T., Eide D. J. (2004) Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 166, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kirschke C. P., Huang L. (2003) ZnT7, a novel mammalian zinc transporter, accumulates zinc in the Golgi apparatus. J. Biol. Chem. 278, 4096–4102 [DOI] [PubMed] [Google Scholar]
- 75. Haydon M. J., Cobbett C. S. (2007) A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143, 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ellis C. D., MacDiarmid C. W., Eide D. J. (2005) Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J. Biol. Chem. 280, 28811–28818 [DOI] [PubMed] [Google Scholar]
- 77. Lichten L. A., Cousins R. J. (2009) Mammalian zinc transporters: nutritional and physiologic regulation. Annu. Rev. Nutr. 29, 153–176 [DOI] [PubMed] [Google Scholar]
- 78. Blaudez D., Kohler A., Martin F., Sanders D., Chalot M. (2003) Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15, 2911–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wei Y., Li H., Fu D. (2004) Oligomeric state of the Escherichia coli metal transporter YiiP. J. Biol. Chem. 279, 39251–39259 [DOI] [PubMed] [Google Scholar]
- 80. Lu M., Chai J., Fu D. (2009) Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 16, 1063–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wei Y., Fu D. (2005) Selective metal binding to a membrane-embedded aspartate in the Escherichia coli metal transporter YiiP (FieF). J. Biol. Chem. 280, 33716–33724 [DOI] [PubMed] [Google Scholar]
- 82. Anton A., Weltrowski A., Haney C. J., Franke S., Grass G., Rensing C., Nies D. H. (2004) Characteristics of zinc transport by two bacterial cation diffusion facilitators from Ralstonia metallidurans CH34 and Escherichia coli. J. Bacteriol. 186, 7499–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis I. Ch., Vajda S. (2010) Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]