Background: NAD+-dependent deacetylase SIRT3 is essential for the prevention of age-related hearing loss during caloric restriction.
Results: Oxidative stress resistance by SIRT3 was mediated through IDH2. SIRT3 deacetylates IDH2 at lysine 413 and stimulates activity by as much as 44-fold.
Conclusion: Increased SIRT3 expression protects cells from oxidative stress through IDH2 activation.
Significance: Our results provide the mechanism by which SIRT3 regulates IDH2.
Keywords: Histone Deacetylase, Metabolic Regulation, Mitochondrial Metabolism, Redox Regulation, Sirtuins, Deacetylation, Isocitrate Dehydrogenase
Abstract
Mitochondria play a central role in oxidative energy metabolism and age-related diseases such as cancer. Accumulation of spurious oxidative damage can cause cellular dysfunction. Antioxidant pathways that rely on NADPH are needed for the reduction of glutathione and maintenance of proper redox status. The mitochondrial matrix protein isocitrate dehydrogenase 2 (IDH2) is a major source of NADPH. Previously, we demonstrated that the NAD+-dependent deacetylase SIRT3 was essential for the prevention of age-related hearing loss in mice fed a calorically restricted diet. Here we provide direct biochemical and biological evidence establishing an exquisite regulatory relationship between IDH2 and SIRT3 under acute and chronic caloric restriction. The regulated site of acetylation was mapped to Lys-413, an evolutionarily invariant residue. Site-specific, genetic incorporation of Nϵ-acetyllysine into position 413 of IDH2 revealed that acetylated IDH2 displays a dramatic 44-fold loss in activity. Deacetylation by SIRT3 fully restored maximum IDH2 activity. The ability of SIRT3 to protect cells from oxidative stress was dependent on IDH2, and the deacetylated mimic, IDH2K413R variant was able to protect Sirt3−/− mouse embryonic fibroblasts from oxidative stress through increased reduced glutathione levels. Together these results uncover a previously unknown mechanism by which SIRT3 regulates IDH2 under dietary restriction. Recent findings demonstrate that IDH2 activities are a major factor in cancer, and as such, these results implicate SIRT3 as a potential regulator of IDH2-dependent functions in cancer cell metabolism.
Introduction
Mitochondria play a central role in energy metabolism and are a source of reactive oxygen species (ROS)2 generated through normal respiration activity (1, 2). Cellular damage caused by aberrant ROS is considered a major factor in many age-related diseases. Isocitrate dehydrogenase 2 (IDH2) is a critical component of the mitochondrial antioxidant pathway through its ability to generate NADPH from oxidative decarboxylation of isocitrate to α-ketoglutarate (3, 4). NADPH is necessary for the regeneration of reduced glutathione (GSH), the major antioxidant responsible for preventing ROS damage. In addition to a critical role in maintaining a proper redox state, IDH2 and its cytoplasmic counterpart IDH1 recently have received great attention because of their involvement in reductive glutamine metabolism in cancer cells and because mutated forms of IDH1 and IDH2 produce an oncogenic metabolite 2-hydroxyglutarate, which alters the epigenome (5–10).
Protein acetylation is now recognized as a major post-translational modification for regulating protein function. Proteomic analysis indicated that IDH2 is one of many proteins in the mitochondria displaying acetylation at lysine residues (11–14), suggesting that IDH2 is controlled by acetylation. Recent evidence supports the role of the NAD+-dependent protein deacetylase SIRT3 in regulating the function of several mitochondrial proteins (2, 11, 15–24). SIRT3 regulates key metabolic pathways in response to nutrient deprivation and caloric restriction, including fatty acid oxidation, oxidative phosphorylation, and the urea cycle (2, 15–20, 25). In fasted or calorie-restricted mice, SIRT3 expression is induced severalfold (2, 17, 26). Caloric restriction (CR) extends the lifespan and delays the onset of age-associated phenotypes in diverse species through a reduction of oxidative damage in multiple tissues (27–30). SIRT3 is essential for the prevention of age-related hearing loss under CR as mice lacking Sirt3−/− failed to manifest the benefits of CR, displaying similar hearing loss to mice on a control diet (2). Although Sirt3−/− mice showed no response to CR, Sirt3−/− mice on a CR diet exhibited higher levels of NADPH, of reduced glutathione, and of IDH2 activity in mitochondria. These observations led us to postulate that IDH2 may be a direct in vivo target of SIRT3 (2). Correlative evidence supported the idea, but the significance and mechanism of regulation remained unknown. Here we provide detailed biochemical and biological evidence that elucidate the exquisite regulatory relationship between SIRT3 and IDH2. Using in vivo and in vitro studies, the site of regulatory (de)acetylation was mapped to Lys-413 of IDH2. In response to calorie and glucose restriction, we demonstrate that SIRT3 deacetylates IDH2 on Lys-413, stimulating catalysis by 25-fold. We show that protection against oxidant stress afforded by SIRT3 depends on IDH2, which contributes ≥25% of the total NADPH in the mitochondria. Moreover, the deacetylated mimic IDH2K413R is able to protect Sirt3−/− MEFs from oxidant stress through increased mitochondrial GSH:GSSG. This study uncovers a previously unknown mechanism by which SIRT3 regulates IDH2 under acute and chronic caloric restriction. In addition to regulation of mitochondrial redox status, these results implicate SIRT3 as a general regulator of IDH2 functions, particularly in cancer cell metabolism.
EXPERIMENTAL PROCEDURES
In Vivo Site-specific Incorporation of Acetyllysine into Proteins
To generate a homogeneously acetylated IDH2 construct for analysis, we used a three-plasmid system described by Neumann et al. (31, 32). This system allows for the site-specific incorporation of Nϵ-acetyllysine by way of a Methanosarcina barkeri acetyl-lysyl-tRNA synthetase/tRNACUA pair that recognizes an amber codon. To avoid the isolation of truncated forms of IDH2Ac, wild-type IDH2 was cloned into pTEV-9 (pET-21b backboned with tobacco etch virus cleavage site), producing a C-terminal His6-tagged construct. By incorporating an amber codon at Lys-413 (AAG to TAG by site-directed mutagenesis), we produced pTEV-IDH2 that encodes for a homogeneous pool of IDH2Ac. Both proteins were produced and purified as described below.
The three plasmids (TEV-9 with expressed gene, pCDF pylT-1, and pAcKRS) were transformed into electrocompetent Escherichia coli BL21 DE3 cells (laboratory collection). Overnight cultures were subcultured 1:100 into 2 liters of 2× YT containing spectinomycin (50 μg/ml) + kanamycin (50 μg/ml) + ampicillin (150 μg/ml). Cultures were grown at 37 °C with shaking to an A600 of 0.6, induced with 0.4 mm isopropyl-1-thio-β-d-galactopyranoside in addition to 2 mm Nϵ-acetyllysine (Sigma-Aldrich) and 20 mm nicotinamide to inhibit the activity of E. coli deacetylases. Cells were grown overnight at 22 °C, harvested by centrifugation, and resuspended in 20 ml of binding buffer (20 mm nicotinamide, 20 mm sodium phosphate at pH 7.5, containing 500 mm NaCl and 20 mm imidazole) containing lysozyme (1 mg/ml), DNase I (25 μg/ml), and PMSF (0.5 mm). Cells were lysed by sonication, and clarified cell lysate was obtained after centrifugation (15 min, 10,000 rpm) and filtration. Samples were loaded onto a 5-ml HisTrap Ni+ column attached to an ÄKTA FPLC system (GE Healthcare) and purified. The rabbit polyclonal antibody used to detect Lys-413 acetylation was generated with a synthetic peptide corresponding to the human IDH2 sequence SGAMT(ac)KDLAGC (GeneTel Laboratories LLC, Madison, WI).
Measurement of NADPH and GSH:GSSG
NADPH levels were determined by the method of Zerez et al. (33). Briefly, 200 μl of the mitochondrial lysate was mixed with 180 μl of a nicotinamide solution (10 mm nicotinamide, 20 mm NaHCO3, 100 mm Na2CO3) and underwent three freeze-thaw cycles to extract NADP+ and NADPH. To destroy NADP+ in the sample, 90 μl of the lysate was incubated in a heating block for 30 min at 60 °C. Twenty-five microliters of each unheated and heated sample was mixed with 225 μl of a reaction mixture (100 mm Tris, 5 mm EDTA, 0.5 μm thiazolyl blue tetrazolium bromide, 2 μm phenazine ethosulfate, 1.3 units of glucose-6-phosphate dehydrogenase, pH 8.0) and incubated in a water bath for 5 min at 37 °C. The reaction mixture was transferred to each well of a 96-well plate, and 1 μl of 1 mm glucose 6-phosphate was added to initiate the reaction. The absorbance was read at 570 nm every 10 s for 3 min in a microplate reader (Synergy H4, BioTek). All samples were run in duplicate. The reaction rates were calculated, and NADPH levels were determined as the ratio of NADPH (heated sample) to the total of NADP+ and NADPH (unheated sample). The GSH:GSSG ratio was determined using the GSH:GSSG-Glo assay kit (Promega).
Generation of Stable Cell Pools
HEK293 or immortal Sirt3+/+ or Sirt3−/− MEF cells were initially cultured in DMEM supplemented with 10% FBS prior to their use in establishing stable transfections. SIRT3 MEFS were kindly provided by Leonard Guarente (34). To establish stable Sirt3-expressing cells, HEK293 cells were transfected with pCDNA3-Sirt3-FLAG by calcium phosphate transfection. A vector control cell line was established by the same method using pCDNA3. To establish stable IDH2-expressing cells, pCDNA3.1-IDH2-FLAG, K413R, and K413Q plasmids were transfected into Sirt3+/+ or Sirt3−/− MEF by TurboFect in vitro transfection reagent (Fermentas). After transfection, cells were selected in the medium containing G418 (1.5 mg/ml) for 10 days. The antibiotic-resistant clones were selected, expanded, and further cultured in medium supplemented with adequate amounts of antibiotics. To generate stable IDH2 knockdown cell pools in HEK293 cells stably expressing SIRT3 protein, 30 pmol of siRNA for IDH2 mRNA (Sigma) was transfected into HEK293 by TurboFect reagent. After 36 h of transfection, mitochondria were isolated (2) from 9 × 106 HEK293 cells. The mitochondrial fraction was used to measure NADPH concentration.
Cytotoxicity Assay
The three stable HEK293 cell pools (vector, SIRT3, and SIRT3 with knockdown IDH2 lines) or Sirt3+/+ or Sirt3−/− MEF cell pools of stably expressing IDH2K413Q IDH2K413R were first grown on a 96-well plate at a density of 1 × 104 cells/well before oxidant treatment and subsequent assessment of cell viability, using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (35). After overnight culture, 25 μm menadione or 1 mm hydrogen peroxide was applied to the cells in serum-free DMEM, and cells were incubated for an additional 48 h at 37 °C. After 48 h of oxidant treatment, culture medium was aspirated before 200 μl of MTT (1 mg/ml) was added and further incubated for 4 h at 37 °C. The MTT solution was discarded by aspirating, and the resulting formazan product converted by the viable cells was dissolved in 150 μl of dimethyl sulfoxide. The absorbance was read in an ELISA plate reader at 595 nm (Synergy H4, BioTek). Cell viability is expressed as a percentage of the absorbance measured in the untreated control cells.
RESULTS
Mapping Functional Acetylation Sites in IDH2
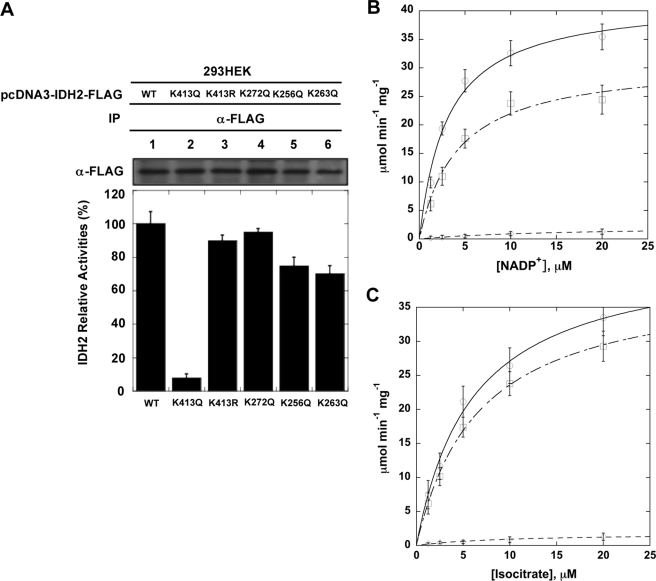
Recent proteomic studies reported that 13 different lysine residues in IDH2 were modified by acetylation (12–14); however, it was unknown which sites played a critical role in regulating IDH2 activity. Because IDH2 is an evolutionarily conserved protein, we speculated that important regulatory sites targeted by acetylation might be conserved. Sequence alignments from diverse species revealed that lysines 256, 263, and 413 are invariant, allowing us to focus our inquiry on those residues (supplemental Fig. S1). We replaced lysine 256, 263, and 413 with either glutamine or arginine (Fig. 1A). As a control, we also mutated Lys-272, which is not conserved but was reported as acetylated. The Lys to Arg mutation retains a positive charge and is often utilized as a deacetylated mimic, whereas Lys to Gln abolishes the positive charge and can act as a surrogate of acetylation (18, 23). The IDH2 variants were overexpressed in HEK293 cells and immunoprecipitated, and IDH2 activity assays were performed. The assay continuously monitors the formation of NADPH as an increase in absorbance at 340 nm (36). When compared with WT IDH2, substitutions at Lys-256, Lys-263, and Lys-272 displayed no significant change in activity. Strikingly, activity of the K413Q mutant decreased by 20-fold, whereas the corresponding K413R mutant displayed minimal change (Fig. 1 and supplemental Table S1). Sequence alignment of five human IDH enzymes revealed that Lys-413 is conserved among NADP+-dependent IDH1 and IDH2, but not in NAD+-dependent IDH3. These observations suggest that a positive charge at position Lys-413 is critical for NADP+ binding and/or IDH2 catalytic activity.
FIGURE 1.
A, maintenance of unacetylated Lys-413 is necessary for IDH2 activity. Wild-type IDH2 and K413R, K413Q, K272Q, K256Q, and K263Q mutants were expressed in HEK293 cells. Proteins were purified by immunoprecipitation (IP), IDH2 levels were normalized for protein, and activity assays were performed. Wild-type IDH2 activity was set as 100%. Bars and error bars represent mean and S.D. of triplicate assays. B and C, kinetic comparison of wild-type IDH2 and variants. Lys-413 was mutated to glutamine or arginine and bacterially expressed, recombinant proteins were purified, and steady-state kinetic analyses were performed. Comparison of IDH2 (open circles) and variants K413R (open squares) and K413Q (open diamonds) shows that the Lys-413 is important in catalysis.
To investigate the mechanism by which Lys-413 acetylation might reduce IDH2 activity, we recombinantly expressed and purified human wild-type IDH2 and the K413R and K413Q mutants from E. coli. Steady-state kinetic analysis of IDH2 and variants (Fig. 1, B and C) indicated that substitution of Lys-413 with arginine did not appreciably alter the Km values for isocitrate and NADP+; however, the K413Q variant exhibited a dramatic 20-fold decrease in Vmax when compared with WT, whereas the K413R variant was unaffected in Vmax (supplemental Table S1). Also, the K413Q variant displayed an ∼5-fold increase in the Km value for NADP+ but no significant change in the Km value for isocitrate. These results support an inhibitory effect of Lys-413 acetylation on IDH2 activity.
Genetic Incorporation of Acetyllysine at Amino Acid Position 413
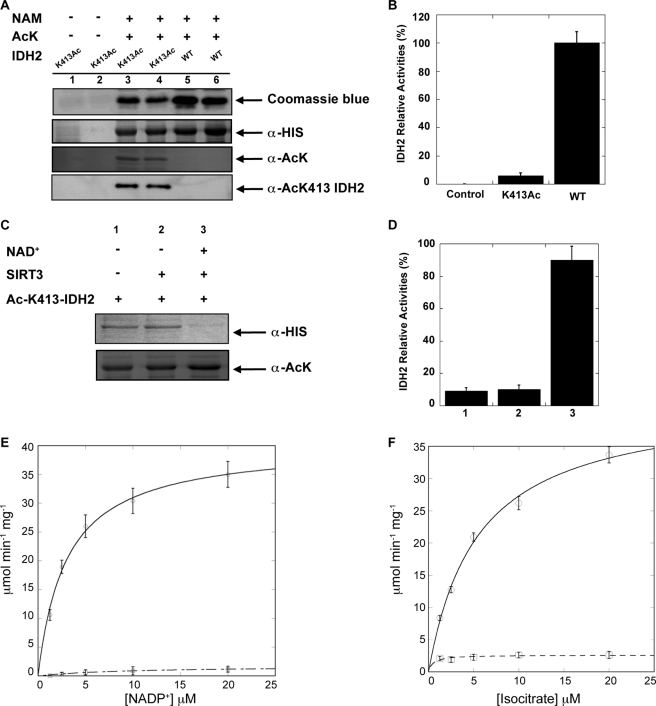
Although arginine/glutamine substitutions can provide general insight into possible functional effects of protein acetylation, there is sufficient evidence that such strategies do not reveal the true consequences of acetylation (37). To directly determine the effect of acetylation, we utilized site-specific, genetic incorporation of Nϵ-acetyllysine at position 413 of IDH2. The incorporation of Nϵ-acetyl-lysine was genetically encoded through an amber stop codon (TAG) at amino acid position 413. The mutant version of IDH2 was coexpressed in E. coli with an orthogonal Nϵ-acetyllysine-tRNA synthetase/tRNACUA pair in the presence of Nϵ-acetyllysine. Only with Nϵ-acetyllysine in the growth medium was the full-length protein formed, and incorporated acetyllysine was verified by immunoblotting with an anti-acetyllysine antibody (Fig. 2A).
FIGURE 2.
A, in vivo site-specific incorporation of acetylated Lys-413 in IDH2. IDH2 and IDH2K413Ac were recombinantly expressed and purified and analyzed by 12% SDS-PAGE or detected in total lysates by Western blot with an anti-His6 antibody or anti-pan acetyllysine antibody. NAM, nicotinamide; AcK, acetyllysine. B, relative IDH2 activity assays using purified IDH2 and IDH2K413Ac. C and D, SIRT3 specifically deacetylates IDH2K413Ac and rescues activity. C, recombinant IDH2K413Ac from A was incubated with purified recombinant SIRT3 with or without NAD+ at 37 °C for 1 h. Acetylation status was assessed by Western blotting with anti-acetyllysine antibody. An anti-His Western blot shows that equivalent IDH2 protein levels were used. D, IDH2 activity assays were performed using the corresponding IDH2K413Ac or unacetylated IDH2 from C. E and F, steady-state kinetic analysis of wild-type IDH2 and IDH2K413Ac. Recombinant proteins were purified by nickel affinity chromatography, and kinetic assays were performed as described under “Experimental Procedures.” Comparison of wild-type IDH2 (open circles) and IDH2K413Ac (open squares) reveals that acetylation greatly affects Vmax and Vmax/Km. Data are means ± S.E.
Acetylation of Lys-413 Decreases Catalysis and SIRT3 Reactivates IDH2 upon Deacetylation
IDH2K413Ac was purified, and catalytic activity was assessed, yielding an enzyme with decreased activity by greater than 10-fold when compared with wild-type enzyme (Fig. 2B). Next, we determined whether SIRT3 could deacetylate Lys-413 and recover IDH2 activity. Using an in vitro deacetylation assay, we demonstrated that only when SIRT3 and the co-substrate NAD+ are present does SIRT3 remove the acetyl group from IDH2K413Ac (Fig. 2C). Corresponding IDH2 activity assays revealed that deacetylated IDH2 recovers full catalytic activity when compared with WT (Fig. 2D).
To provide molecular insight into the regulatory mechanism of IDH2 reversible acetylation, a steady-state kinetic analysis was performed with WT and IDH2K413Ac. Substrate saturation curves were determined with NADP+ (Fig. 2E) and isocitrate (Fig. 2F) as the varied substrate with either IDH2 or IDH2K413Ac. When compared with IDH2, this kinetic analysis revealed that IDH2K413Ac displays a 15-fold (average from supplemental Table S1) decrease in Vmax and a 44-fold decrease in the apparent second order rate constant Vmax/Km with NADP+ as substrate. (Fig. 2, E and F). Interestingly, the Km values for isocitrate were similar between the acetylated and unacetylated enzyme (supplemental Table S1). These results demonstrate for the first time the molecular and functional consequence of Lys-413 acetylation on IDH2 activity.
SIRT3-dependent Deacetylation of IDH2 Suppresses Cellular ROS Stress
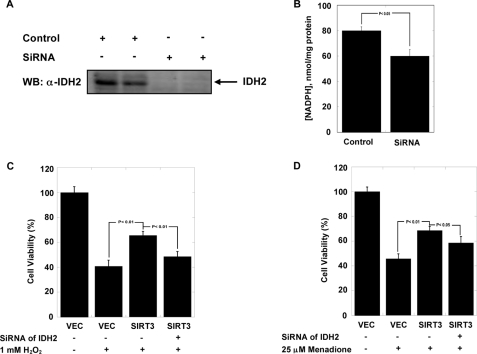
Previous studies implicate IDH2-produced NADPH as a protective mechanism against cellular oxidative stress (3). We demonstrated that SIRT3 regulates the ability of cells to resist ROS damage from exogenous and endogenous ROS (2). We proposed that SIRT3 deacetylates IDH2 and stimulates an increase in NADPH, which is utilized by the mitochondrion antioxidant system to combat ROS. To determine how much IDH2 contributes to the steady-state levels of NADPH, we used siRNA to knock down endogenous IDH2 in HEK293 cells (Fig. 3A) and measured the corresponding change in NADPH levels within mitochondria. The results (Fig. 3B) indicate that IDH2 contributes ≥25% of the total NADPH produced in the mitochondria, which is consistent with previous studies showing IDH2 as one of major mitochondrial sources of NADPH (38).
FIGURE 3.
A and B, NADPH concentrations in mitochondria decrease when endogenous IDH2 is knocked down in HEK293 cells. A, Western blotting (WB) with anti-IDH2 antibody confirms IDH2 expression. B, measurements with errors are shown for two different stable cell populations from each type of transfection (SiRNA control, SiRNA of Idh2) (n = 3). p values indicate values significantly different from control (p < 0.05). C and D, IDH2 is critical to mediate the SIRT3-dependent protection in HEK293 cells treated with oxidant stressors hydrogen peroxide (H2O2) (C) or menadione (D). The three different stable cell populations (vector (VEC), SIRT3 stable expression, and IDH2 knockdown in stable SIRT3 cells, supplemental Fig. 2A) were transiently exposed to either 1 mm H2O2 or 25 μm menadione (n = 16). Data are means ± S.E.
To test whether the protective effect of SIRT3 against oxidative stress is dependent on IDH2, we used siRNA to knock down endogenous IDH2 in cells stably overexpressing SIRT3. HEK293 cells were treated with H2O2 or menadione, and cell viability was monitored (Fig. 3, C and D). Consistent with our previous observations (2), expression of SIRT3 offers significant protection from ROS insults such as H2O2 and menadione (Fig. 3, C and D). Here we show that when IDH2 levels are knocked down, the protective effects of SIRT3 expression are eliminated (with H2O2, Fig. 3C) or substantially reduced (with menadione, Fig. 3D). These results provide strong biochemical evidence that IDH2 is a major pathway through which SIRT3 provides defense against ROS.
Acetylation of Lys-413 Is Regulated by SIRT3 in Response to Calorie and Glucose Restriction
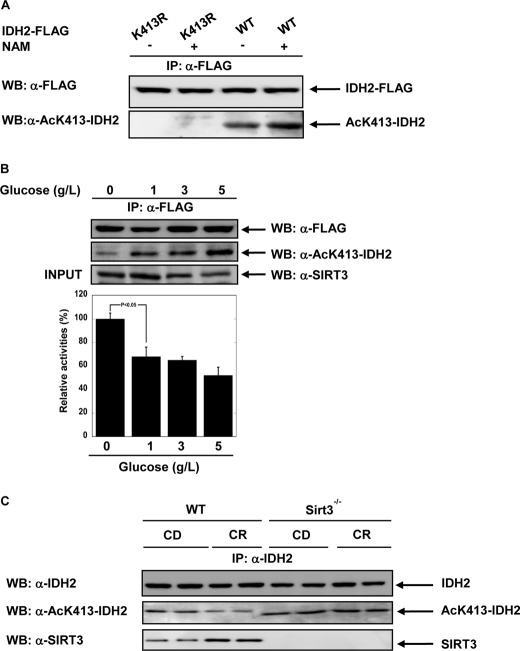
Next, we investigated in vivo whether Lys-413 is a critical acetylation site that modulates the activity of IDH2 in response to glucose or calorie restriction. To determine the in vivo acetylation status of Lys-413, a rabbit polyclonal anti-acetylated Lys-413 IDH2 antibody was generated as outlined under “Experimental Procedures.” Western blot analysis of IDH2 overexpressed in HEK293 cells revealed an immune-reactive band that displayed increased intensity with cells treated with nicotinamide, a general sirtuin inhibitor (Fig. 4A). The anti-acetylated Lys-413 IDH2 antibody, however, did not recognize the IDH2 K413R mutant, indicating that the antibody is specific for IDH2 K413Ac. These data validate the site-specific antibody and confirm that Lys-413 in IDH2 is indeed acetylated when expressed in HEK293 cells.
FIGURE 4.
A, in cells, IDH2 is acetylated on lysine 413. A specific acetyllysine 413 antibody was generated using the peptide SGAMT(AC)KDLAGC. HEK293 cells were co-transfected by pcDNA3-IDH2-FLAG and pcDNA3-IDH2K413R-FLAG. IDH2 proteins were immunoprecipitated (IP) using FLAG beads. IDH2 Lys-413 acetylation levels were detected using the site-specific antibody. WB, Western blotting; NAM, nicotinamide; AcK, acetyllysine. B, low glucose decreases acetylation level of IDH2 Lys-413 and increases IDH2 activity. IDH2-FLAG was overexpressed in HEK293 cells following treatment with varying glucose levels (5, 3, 1, and 0 g/liter) for 6 h. Cell lysates were resolved by SDS-PAGE and detected by Western blotting with anti-SIRT3 antibody. IDH2-FLAG was immunoprecipitated with anti-FLAG beads, and IDH2 activity was measured and normalized to IDH2 protein levels; quantifications of the amounts of IDH2 were performed by anti-FLAG antibody and anti-acetylated Lys-413 antibody from A. Error bars represent standard error measurement (S.E.) (n = 3), p < 0.05. C, Bottom and middle, Western blot analysis of SIRT3 (bottom) and levels of acetylated lysine 413 (middle) in liver from 5-month-old WT or Sirt3−/− mice fed either control diet (CD) or calorie-restricted diet (CR). Top, endogenous acetylated IDH2 was isolated by immunoprecipitation with anti-IDH2 antibody followed by Western blotting with anti-acetyllysine 413 IDH2 antibody (middle).
Under caloric restriction or fasting, SIRT3 mRNA and protein levels are dramatically increased in mice (2, 39, 40). This led us to inquire whether IDH2 acetylation levels and activity might be directly controlled by glucose availability. We examined the effect of glucose levels on IDH2 acetylation in cultured cells and found that lowering glucose levels led to a decrease in Lys-413 acetylation, as determined by the Lys-413 acetylation-specific antibody (Fig. 4B). Lys-413 acetylation inversely correlated with the measured IDH2 activity (Fig. 4B), in agreement with an inhibitory effect of Lys-413 acetylation on IDH2 activity. Also, consistent with a direct effect by SIRT3, we found that lowered glucose levels increase the protein level of SIRT3 (Fig. 4B). These results suggest an exquisite relationship between nutrient status, SIRT3 levels, and regulated IDH2 activity.
To provide direct evidence that SIRT3 controls IDH2 activity via reversible acetylation of Lys-413, we utilized the site-specific anti-K413Ac antibody and determined acetylation levels of IDH2 from liver mitochondria of WT and Sirt3−/− mice fed control or calorie-restricted diets (Fig. 4C). In CR tissues, the acetylation of Lys-413 is decreased 3-fold when compared with control diet. This change closely matches the 3-fold increase in SIRT3 protein levels under CR (Fig. 4C) (2, 17, 26). In Sirt3−/− mice tissues, hyperacetylation of IDH2 was observed in both control and CR diet conditions, indicating that SIRT3 is required for the CR-induced deacetylation of IDH2 Lys-413.
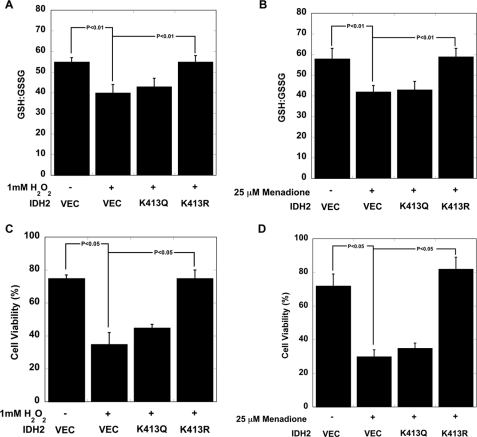
IDH2K413R Rescues Oxidant Damage in Sirt3−/− MEFs
We have shown that substitution of lysine with glutamine mimics an acetylated Lys-413 of IDH2, whereas substitution with an arginine mimics the deacetylated state (Figs. 1, B and C, and 2, E and F). We took advantage of this unique mimicry to investigate, in the absence of SIRT3, whether overexpression of IDH2K413R could protect cells from oxidant stress. For comparison, we separately expressed the IDH2K413Q acetylation mimic. Three stable cell pools expressing IDH2K413R, IDH2K413Q, or a vector control were generated in Sirt3−/− MEFs (34) (Fig. 5A and supplemental Fig. 2B). The levels of GSH:GSSG and cell viability were determined ± H2O2 or ± menadione. In Sirt3−/− MEFs, treatment with H2O2 and menadione caused significant decreases in mitochondrial GSH:GSSG ratios (Fig. 5, A and B). Expression of the IDH2K413R variant in Sirt3−/− MEFs (Fig. 5, B and C) provided maintenance of the GSH:GSSG ratio after H2O2 or menadione treatment. The IDH2K413Q variant was unable to maintain the GSH:GSSG, yielding similar GSH:GSSG values to those of vector control after ROS treatment. The effects on cell viability are more dramatic and consistent with the trends observed for GSH:GSSG changes (Fig. 5, C and D). Expression of the IDH2K413R variant provided complete protection against both oxidant insults (Fig. 5, C and D). In contrast, stable expression of IDH2K413Q offered no significant protection against H2O2 and menadione, consistent with the trends on GSH:GSSG levels. These results support a critical role for IDH2 in maintaining the redox balance in mitochondria and a regulatory function of reversible acetylation at Lys-413.
FIGURE 5.
A and B, GSH:GSSG ratios are significantly increased when IDH2K413R was stably overexpressed in Sirt3−/− MEFs. Measurements with errors are shown for the five different stable cell populations from each type of transfection (vector (VEC), IDH2K413Q, and IDH2K413R in Sirt3−/− MEFs) (n = 3), (p < 0.01) in cells treated with oxidants hydrogen peroxide (H2O2) (A) and menadione (B). C and D, IDH2K413R overexpression is sufficient to protect Sirt3−/− MEFs from hydrogen peroxide (H2O2) (C) and menadione (D). Cells were transiently exposed to either 1 mm H2O2 or 25 μm menadione (n = 16) (p < 0.05). Data are means ± S.E.
DISCUSSION
Here we provide the mechanistic basis for SIRT3-dependent regulation of IDH2 under glucose and caloric restriction. We mapped the regulated site of acetylation to Lys-413 using mutational analysis of evolutionarily invariant lysine residues. To provide unequivocal assessment for the effect of acetylation, we utilized site-specific, genetic incorporation of Nϵ-acetyllysine into recombinantly expressed and purified IDH2. When compared with WT enzyme, steady-state kinetic analysis revealed that IDH2K413Ac displays an approximate 15- and 44-fold decrease in Vmax and Vmax/Km (NADP+), respectively, consistent with the kinetics of IDH2K413Q. SIRT3 specifically deacetylated IDH2K413Ac, resulting in full recovery of IDH2 activity. To our knowledge, this is the first use of site-specific genetic incorporation of acetyllysine to perform a detailed mechanistic analysis on the catalytic effects of acetylation. Previous kinetic analysis of NADP+-dependent IDH revealed that catalysis proceeds by a random sequential mechanism where catalysis is more rapid than product release (41). Therefore, the effect of acetylation on Vmax likely reflects altered product release. Similarly, the dramatic drop in Vmax/Kmfor NADP+ might reflect inefficient formation of the catalytically competent conformation. Structural studies of IDH2 from Saccharomyces cerevisiae (57% identity to human IDH2) indicated that the protein undergoes substantial conformational changes during catalysis (42). We hypothesize that acetylation of Lys-413, partially through the ablation of the positively charged binding pocket (supplemental Fig. S3), as well as through potential structural alteration induced by acetylation, prevents the proper binding conformation and positioning of NADP+ for catalysis.
The ability of SIRT3 to modulate IDH2 activity has wide-ranging implications in metabolism, ROS mitigation, and cancer. Consistent with emerging evidence (2, 15–23, 25, 43), SIRT3 promotes oxidative metabolism and mitochondrial reprogramming under lowered energy input (acute and chronic caloric restriction). Stimulation of oxidative metabolic pathways in mitochondria can result in increased production of ROS, which causes damage to cellular components and compromises cell viability (44, 45). The role of IDH2 in the mitochondrial antioxidant pathways suggests a protective role in cancer as it is postulated that decreased cellular damage by ROS slows cancer development (46, 47). We propose that SIRT3 activates the IDH2 pathway as a coordinated program to mitigate increased ROS during periods of increased oxidative metabolism. We demonstrate that SIRT3 can protect cells from mitochondrial oxidative stress and that this effect is almost entirely dependent on IDH2. Cellular knockdown of IDH2 revealed that 25% of mitochondrial NADPH is maintained by IDH2 under nonstressed growth, which is consistent with previous reports that IDH2 is one of two major mitochondrial NADPH producers (48). IDH2 knockdown in cells stably overexpressing SIRT3 led to a complete or nearly complete loss of protection from oxidative stress. Importantly, we show that the Lys-413 deacetylation mimic, IDH2K413R, is able to protect Sirt3−/− MEFs from oxidative stress by increasing GSH:GSSG, providing evidence that IDH2 serves as the critical link between SIRT3 and protection from ROS. Recent studies reported that SIRT3 can also activate superoxide dismutase 2 (SOD2) (21) and Mn-SOD (22, 23). SOD catalyzes the conversion of superoxide to H2O2. Hydrogen peroxide, produced by dismutation of superoxide and other cellular redox chemistries, is the major oxidizer of GSH and protein thiols. Oxidized thiols are repaired by the IDH2-dependent antioxidant pathway involving glutathione reductase. The observation that IDH2 is critical for GSH buffering and cell viability against both H2O2 and menadione supports a critical role for IDH2 regardless of ROS origin. Taken together, SIRT3 is a key factor in a coordinate up-regulation of oxidative metabolism and antioxidant pathways in response to acute and chronic caloric restriction.
Recent studies describe previously unappreciated functions of IDH that suggest a major role in cancer cell metabolism. First, there are known active-site mutations that result in decreased rates of α-ketoglutarate formation, but increased ability to form an oncogenic product, 2-hydroxyglutarate (5–7). Second, IDH2 is critical for the metabolic reprogramming that allows tumor cells to utilize glutamine for macromolecular biosynthesis required for cell growth (8–10). In the latter case, IDH2 catalyzes the reductive carboxylation of α-ketoglutarate to isocitrate, the reverse direction of the normal IDH2-catalyzed reaction.
Here we have uncovered a previously unknown mechanism by which SIRT3 regulates IDH2 under dietary restriction. In addition to the antioxidant functions described in the current study, these results implicate SIRT3 as a major regulator of IDH2 function and cancer cell metabolism. Several recent studies suggest that SIRT3 acts as a tumor suppressor (34, 49, 50), whereas others report a positive correlation with cancer and SIRT3 expression (51). Given this regulatory connection between these enzymes and their effects in cancer cell metabolism, future investigations will be important to elucidate the role of SIRT3-dependent modulation of IDH2 activity in cancer progression.
Supplementary Material
Acknowledgments
We thank Leonard Guarente for SIRT3 MEFs, Jason Chin for site-specific incorporation vectors, Tom Prolla and James Vann for supplying MEFs, and members of the Denu laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065386 (to J. M. D.). This work was also supported by the National Science Foundation Graduate Research Fellowship Program (to K. E. D.).
This article was selected as a Paper of the Week.

This article contains supplemental Table S1 and Figs. S1–S3.
- ROS
- reactive oxygen species
- CR
- caloric restriction
- IDH2
- isocitrate dehydrogenase 2
- MEF
- mouse embryonic fibroblast
- SOD2
- superoxide dismutase 2
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1. Singh K. K. (2006) Mitochondria damage checkpoint, aging, and cancer. Ann. N.Y. Acad. Sci. 1067, 182–190 [DOI] [PubMed] [Google Scholar]
- 2. Someya S., Yu W., Hallows W. C., Xu J., Vann J. M., Leeuwenburgh C., Tanokura M., Denu J. M., Prolla T. A. (2010) Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jo S. H., Son M. K., Koh H. J., Lee S. M., Song I. H., Kim Y. O., Lee Y. S., Jeong K. S., Kim W. B., Park J. W., Song B. J., Huh T. L. (2001) Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 276, 16168–16176 [DOI] [PubMed] [Google Scholar]
- 4. Lee J. H., Yang E. S., Park J. W. (2003) Inactivation of NADP+-dependent isocitrate dehydrogenase by peroxynitrite: implications for cytotoxicity and alcohol-induced liver injury. J. Biol. Chem. 278, 51360–51371 [DOI] [PubMed] [Google Scholar]
- 5. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Vander Heiden M. G., Su S. M. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., Cross J. R., Fantin V. R., Hedvat C. V., Perl A. E., Rabinowitz J. D., Carroll M., Su S. M., Sharp K. A., Levine R. L., Thompson C. B. (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gross S., Cairns R. A., Minden M. D., Driggers E. M., Bittinger M. A., Jang H. G., Sasaki M., Jin S., Schenkein D. P., Su S. M., Dang L., Fantin V. R., Mak T. W. (2010) Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 207, 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., DeBerardinis R. J. (2012) Reductive carboxylation supports growth in tumor cells with defective mitochondria. Nature 481, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., Stephanopoulos G. (2012) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., Thompson C. B. (2011) Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 13. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 14. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008) A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hallows W. C., Lee S., Denu J. M. (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y., Cimen H., Han M. J., Shi T., Deng J. H., Koc H., Palacios O. M., Montier L., Bai Y., Tong Q., Koc E. C. (2010) NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J. Biol. Chem. 285, 7417–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hallows W. C., Yu W., Smith B. C., Devries M. K., Ellinger J. J., Someya S., Shortreed M. R., Prolla T., Markley J. L., Smith L. M., Zhao S., Guan K. L., Denu J. M. (2011) Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu X., Brown K., Hirschey M. D., Verdin E., Chen D. (2010) Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 12, 662–667 [DOI] [PubMed] [Google Scholar]
- 22. Chen Y., Zhang J., Lin Y., Lei Q., Guan K. L., Zhao S., Xiong Y. (2011) Tumor suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 12, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tao R., Coleman M. C., Pennington J. D., Ozden O., Park S. H., Jiang H., Kim H. S., Flynn C. R., Hill S., Hayes McDonald W., Olivier A. K., Spitz D. R., Gius D. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates Mn-SOD activity in response to stress. Mol. Cell 40, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 25. Guan K. L., Xiong Y. (2011) Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 36, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi T., Wang F., Stieren E., Tong Q. (2005) SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 280, 13560–13567 [DOI] [PubMed] [Google Scholar]
- 27. Weindruch R., Walford R. L. (1988) The Retardation of Aging and Disease by Dietary Restriction, C.C. Thomas, Springfield, IL [Google Scholar]
- 28. Sohal R. S., Weindruch R. (1996) Oxidative stress, caloric restriction, and aging. Science 273, 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee C. K., Klopp R. G., Weindruch R., Prolla T. A. (1999) Gene expression profile of aging and its retardation by caloric restriction. Science 285, 1390–1393 [DOI] [PubMed] [Google Scholar]
- 30. Colman R. J., Anderson R. M., Johnson S. C., Kastman E. K., Kosmatka K. J., Beasley T. M., Allison D. B., Cruzen C., Simmons H. A., Kemnitz J. W., Weindruch R. (2009) Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neumann H., Hancock S. M., Buning R., Routh A., Chapman L., Somers J., Owen-Hughes T., van Noort J., Rhodes D., Chin J. W. (2009) A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell 36, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neumann H., Peak-Chew S. Y., Chin J. W. (2008) Genetically encoding Nϵ-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 [DOI] [PubMed] [Google Scholar]
- 33. Zerez C. R., Lee S. J., Tanaka K. R. (1987) Spectrophotometric determination of oxidized and reduced pyridine nucleotides in erythrocytes using a single extraction procedure. Anal. Biochem. 164, 367–373 [DOI] [PubMed] [Google Scholar]
- 34. Bell E. L., Emerling B. M., Ricoult S. J., Guarente L. (2011) SirT3 suppresses hypoxia-inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Di Matteo M. A., Loweth A. C., Thomas S., Mabley J. G., Morgan N. G., Thorpe J. R., Green I. C. (1997) Superoxide, nitric oxide, peroxynitrite, and cytokine combinations all cause functional impairment and morphological changes in rat islets of Langerhans and insulin secreting cell lines, but dictate cell death by different mechanisms. Apoptosis 2, 164–177 [DOI] [PubMed] [Google Scholar]
- 36. Kornberg A. (1955) Isocitric dehydrogenase of yeast (TPN). Methods Enzymol. 1, 3 [Google Scholar]
- 37. Albaugh B. N., Arnold K. M., Lee S., Denu J. M. (2011) Autoacetylation of the histone acetyltransferase Rtt109. J. Biol. Chem. 286, 24694–24701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vogel R., Wiesinger H., Hamprecht B., Dringen R. (1999) The regeneration of reduced glutathione in rat forebrain mitochondria identifies metabolic pathways providing the NADPH required. Neurosci. Lett. 275, 97–100 [DOI] [PubMed] [Google Scholar]
- 39. Barger J. L., Kayo T., Vann J. M., Arias E. B., Wang J., Hacker T. A., Wang Y., Raederstorff D., Morrow J. D., Leeuwenburgh C., Allison D. B., Saupe K. W., Cartee G. D., Weindruch R., Prolla T. A. (2008) A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One 3, e2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwer B., Eckersdorff M., Li Y., Silva J. C., Fermin D., Kurtev M. V., Giallourakis C., Comb M. J., Alt F. W., Lombard D. B. (2009) Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8, 604–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uhr M. L., Thompson V. W., Cleland W. W. (1974) The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. I. Initial velocity, substrate and product inhibition, and isotope exchange studies. J. Biol. Chem. 249, 2920–2927 [PubMed] [Google Scholar]
- 42. Peng Y., Zhong C., Huang W., Ding J. (2008) Structural studies of Saccharomyces cerevisiae mitochondrial NADP-dependent isocitrate dehydrogenase in different enzymatic states reveal substantial conformational changes during the catalytic reaction. Protein Sci. 17, 1542–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wagner G. R., Pride P. M., Babbey C. M., Payne R. M. (2012) Friedreich's ataxia reveals a mechanism for coordinate regulation of oxidative metabolism via feedback inhibition of the SIRT3 deacetylase. Hum. Mol. Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dröge W. (2002) Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95 [DOI] [PubMed] [Google Scholar]
- 45. Starkov A. A. (2008) The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N.Y. Acad. Sci. 1147, 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benhar M., Engelberg D., Levitzki A. (2002) ROS, stress-activated kinases, and stress signaling in cancer. EMBO Rep. 3, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pelicano H., Carney D., Huang P. (2004) ROS stress in cancer cells and therapeutic implications. Drug Resist. Updat. 7, 97–110 [DOI] [PubMed] [Google Scholar]
- 48. Yin F., Sancheti H., Cadenas E. (2012) Silencing of nicotinamide nucleotide transhydrogenase impairs cellular redox homeostasis and energy metabolism in PC12 cells. Biochim Biophys Acta 1817, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim H. S., Patel K., Muldoon-Jacobs K., Bisht K. S., Aykin-Burns N., Pennington J. D., van der Meer R., Nguyen P., Savage J., Owens K. M., Vassilopoulos A., Ozden O., Park S. H., Singh K. K., Abdulkadir S. A., Spitz D. R., Deng C. X., Gius D. (2010) SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Finley L. W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P. I., Cardoso S. M., Clish C. B., Pandolfi P. P., Haigis M. C. (2011) SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alhazzazi T. Y., Kamarajan P., Verdin E., Kapila Y. L. (2011) SIRT3 and cancer: tumor promoter or suppressor? Biochim. Biophys. Acta 1816, 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.