Background: Macrophage differentiation is accompanied by expression of unique extracellular matrix molecules.
Results: Monocyte-to-macrophage transition involves selective expression of serglycin, TSG-6, hyaluronan, and versican and the formation of inter-α-trypsin inhibitor and amyloid-like precursor protein complexes.
Conclusion: Differentiating macrophages synthesize and secrete novel ECM molecules.
Significance: These ECM secretory products likely play a role in macrophage differentiation and the pathogenesis of atherosclerosis.
Keywords: Atherosclerosis, Extracellular Matrix, Hyaluronate, Macrophages, Proteoglycan, Inter-α-trypsin Inhibitor Heavy Chain 2, TNF-stimulated Gene-6, Amyloid Precursor-like Protein 2, Hyaluronan, Versican
Abstract
Although monocyte- and macrophage-derived molecules are known to promote extracellular matrix (ECM) disruption and destabilization, it is less appreciated that they also synthesize molecules contributing to ECM formation, stabilization, and function. We have identified and characterized the synthesis of proteoglycans and related proteins, some not previously known to be associated with macrophages. Proteoglycan extracts of [35S]sulfate- and 35S-trans amino acid-radiolabeled culture media from THP-1 monocytes induced to differentiate by treatment with phorbol myristate acetate revealed three major proteins of ∼25, 90, and 100 kDa following chondroitin ABC lyase digestion. The 25-kDa protein was predominant for monocytes, whereas the 90- and 100-kDa proteins were predominant for macrophages. Tandem mass spectrometry identified (i) the 25-kDa core protein as serglycin, (ii) the 90-kDa core protein as inter-α-inhibitor heavy chain 2 (IαIHC2), and (iii) the 100-kDa core as amyloid precursor-like protein 2 (APLP2). Differentiation was also associated with (i) a >500-fold increase in mRNA for TNF-stimulated gene-6, an essential cofactor for heavy chain-mediated matrix stabilization; (ii) a >800-fold increase in mRNA for HAS2, which is responsible for hyaluronan synthesis; and (iii) a 3-fold increase in mRNA for versican, which interacts with hyaluronan. Biochemical evidence is also presented for an IαIHC2-APLP2 complex, and immunohistochemical staining of human atherosclerotic lesions demonstrates similar staining patterns for APLP2 and IαIHC2 with macrophages, whereas serglycin localizes to the underlying glycosaminoglycan-rich region. These findings indicate that macrophages synthesize many of the molecules participating in ECM formation and function, suggesting a novel role for these molecules in the differentiation of macrophages in the development of atherosclerosis.
Introduction
Much of the understanding of vascular wall proteoglycan structure and function has been derived from studies of arterial smooth muscle cell proteoglycans. In short, proteoglycans have a variety of important roles in the vascular wall, including (i) organization of extracellular matrix (ECM)2 structure and maintenance of viscoelastic properties; (ii) regulation of cell adhesion, migration, and proliferation; (iii) regulation of thrombosis and hemostasis; and (iv) regulation of lipoprotein metabolism and retention (1). The general consensus is that these phenomena contribute to macrophage foam cell formation and the development of atherosclerosis. However, there has been less attention paid to the possibility that the monocyte-to-macrophage transition involves the production of an ECM that contributes to events associated with the development of the atherosclerotic plaque.
The monocyte/macrophage is a critical cell in the pathogenesis of atherosclerosis (2–4) capable of secreting many factors, such as chemokines, cytokines, growth factors, and reactive oxygen species, which contribute to lesion development. Macrophages are also an important source of matrix-degrading proteinases as well as their activators and inhibitors, which contribute to plaque disruption and vascular remodeling. As such, it is widely appreciated that monocyte- and macrophage-derived molecules promote the progression and destabilization of atherosclerotic plaques.
It is less appreciated but well established that macrophages also are capable of secreting matrix molecules themselves. We (5) and others (6–12) have shown that macrophages synthesize and secrete proteoglycans. A common feature of both normal and neoplastic monocytes and macrophages is the ability to synthesize serglycin (7, 9), a proteoglycan known to be made by a variety of hematopoietic cells. Serglycin is essential for the maturation and function of macrophage secretory vesicles (10). Serglycin also is known to have an important protective role in the storage and secretion of tumor necrosis factor (TNF) (9) and perhaps other select macrophage secretory products as well.
In addition to serglycin, other proteoglycans also have been reported in monocytes and macrophages. THP-1 monocytes have been found to express mRNA for versican (11, 12), perlecan, syndecan, glypican-1, and thrombomodulin but not biglycan (13). Similarly, no mRNA for biglycan was detected in unstimulated thioglycolate-elicited peritoneal macrophages (14). However, peritoneal macrophages stimulated with proinflammatory cytokines express biglycan mRNA and protein, which then can function as both a signaling molecule and a proinflammatory factor (14). These important findings indicate that the state of activation of mononuclear cells influences proteoglycan expression and suggest that proteoglycan synthesis is an important component of the inflammatory response.
As monocyte-to-macrophage maturation is a critical process in atherosclerosis, we have been interested in understanding how differentiation and activation might influence proteoglycan production and the contribution that these molecules might make to the development of atherosclerosis. Our findings confirm the synthesis and secretion of serglycin by both THP-1 immature monocytes and mature macrophages. However, we also found an abundance of another proteoglycan species, amyloid precursor-like protein 2 (APLP2), as well as the proteoglycan-related molecule inter-α-inhibitor heavy chain 2 (IαIHC2), which is associated with the proteoglycan fraction of macrophage secretory products. Because of the critical role that the heavy chains of IαI have in the stabilization of hyaluronan-rich matrices (15–19), we also investigated changes in expression of other molecules involved in matrix stabilization, namely versican, TNF-stimulated gene-6 (TSG-6) (15, 20), and the hyaluronan synthases (HASs 1, 2, and 3). TSG-6 mediates the covalent transfer of IαI heavy chains onto hyaluronan (21, 22) and can also directly cross-link hyaluronan (23), thus having a central role as a matrix reorganizer, which may contribute to the pro- and/or anti-inflammatory properties of the ECM (24–26).
MATERIALS AND METHODS
Cell Culture
The human monocytic leukemia cell line THP-1 was obtained from the American Type Culture Collection (Manassas, VA) and maintained as described (27). Cells were cultured in RPMI 1640 medium (BioWhittaker, Walkersville, MD) supplemented with 10% (v/v) heat-inactivated FBS (HyClone, Logan, UT). THP-1 monocytes were treated for 24 h with 1.6 × 10−7 m phorbol myristate acetate (PMA) (Sigma) in fresh RPMI 1640 medium with 10% FBS. This medium was replaced by fresh RPMI 1640 medium with 10% FBS for another 24 h after which time the cells were considered fully differentiated into macrophages (28). For some experiments, monocytes were washed extensively with PBS prior to induction of differentiation with PMA in serum-free RPMI 1640 medium. For other experiments, monocytes were maintained in AIM V serum-free medium (Invitrogen). Monocytes and macrophages were metabolically labeled with either 50 μCi/ml [35S]SO4 or trans label ([35S]methionine/cysteine) (ICN Diagnostics, Costa Mesa, CA).
Proteoglycan Isolation
Labeled medium was harvested, and protease inhibitors (0.1 m 6-aminohexanoic acid, 5 mm benzamidine HCl, and 0.1 mm phenylmethylsulfonyl fluoride) were added (29). Medium was applied to DEAE-Sephacel minicolumns equilibrated in 8 m urea buffer, 0.25 m NaCl, 0.5% Triton X-100, and proteoglycans were eluted with 8 m urea buffer containing either 3 m NaCl, 0.5% Triton X-100 or a gradient of 0.25–1 m NaCl (29). This ion exchange chromatography step served to remove free radiolabel and concentrate proteoglycan samples.
Analysis of Molecular Size
35SO4-proteoglycan and trans-labeled core protein molecular sizes were characterized by SDS-PAGE under reducing conditions according to the procedure of Laemmli (30). Chondroitin and dermatan sulfate glycosaminoglycan chains were removed by digestion with 0.03 unit/ml chondroitin ABC lyase (ICN Diagnostics) in 0.3 m Tris-HCl, pH 8.0, 0.6 mg/ml bovine serum albumin, 18 mm sodium acetate with protease inhibitors (0.1 m 6-aminohexanoic acid and 5 mm benzamidine hydrochloride) for 3 h at 37 °C (29). Heparan sulfate glycosaminoglycan chains were removed by digestion with 20 units/ml heparinase I (Sigma) in 0.1 m Tris-HCl, pH 7.0, 10 mm calcium acetate, 18 mm sodium acetate with protease inhibitors for 3 h at 37 °C followed by the addition of 20 units/ml heparinase II (Sigma) and incubation overnight at 41 °C (29). The 35S-labeled intact proteoglycans and core proteins (after enzyme digestion) were visualized by phosphorimage analysis.
Analysis of Hydrodynamic Size
The hydrodynamic size of 35SO4-proteoglycans was analyzed on a Sepharose CL-2B (0.7 × 110-cm) molecular sieve column equilibrated in 8 m urea buffer with 0.5% Triton X-100 (29). Fractions of 1 ml were collected, and an aliquot of each fraction was assayed for radioactivity by liquid scintillation counting. The elution position of free 35SO4 was used as a marker for the total volume (Vt).
Glycosaminoglycan Isolation and Analysis
Glycosaminoglycan chains were released from proteoglycans by reductive β-elimination with 1 m sodium borohydride in 50 mm NaOH for 4 h at 45 °C (29). The reaction was terminated by neutralizing the sample with glacial acetic acid. Glycosaminoglycan chains then were applied to a Sepharose CL-6B column (0.7 × 63 cm) in 0.2 m Tris, pH 7.0 with 0.2 m NaCl for analysis of chain length by size exclusion chromatography (31). The elution position of free 35SO4 was used as a marker for the Vt.
Isolation of Proteoglycans from Agarose
35SO4- or trans-labeled proteoglycans were resolved further by 1.2% agarose gel electrophoresis and characterized. Approximately 1 × 104 dpm proteoglycans were applied to a single narrow lane of an agarose gel, and the remainder of ∼1 × 106 dpm was applied to a single wide lane on the same gel. The gel was electrophoresed for 7 h at 60 V at 4 °C. The narrow lane was sliced into 0.5-cm segments for scintillation counting and localization of the proteoglycan species. Based on this localization, proteoglycans were cut out of the wide lane in 0.5-cm fractions. Each agarose fraction was individually melted and diluted 10-fold with 8 m urea buffer containing 0.5% Triton X-100 and 0.25 m NaCl and reconcentrated on DEAE-Sephacel minicolumns, and proteoglycans were eluted with 8 m urea buffer containing 0.5% Triton X-100 and 3 m NaCl (32). These proteoglycans were then divided into two aliquots of ∼4 × 104 dpm; one aliquot was digested with chondroitin ABC lyase as described earlier for further analysis by SDS-PAGE.
Protein Identification
Core proteins of chondroitin ABC lyase-digested samples were separated by SDS-PAGE. Silver-stained bands were excised from the gel and diced, destained with 50% acetonitrile in 50 mm ammonium bicarbonate, shrunk using acetonitrile, and dried under vacuum centrifugation. The gel pieces were rehydrated in 30 μl of sequencing grade modified trypsin (Promega; 12.5 ng/ml in 50 mm ammonium bicarbonate) for 60 min on ice. The supernatant was replaced by 50 mm ammonium bicarbonate and incubated overnight at 37 °C. The supernatant was collected, and gels were extracted with 30 μl of 50% acetonitrile. Combined solutions were dried down to near completeness, and the volume was adjusted to ∼20 μl. The sample was then desalted on a C18 ZipTip (Millipore) and analyzed on a tandem matrix-assisted laser desorption ionization time of flight (MALDI-TOF/TOF) mass spectrometer (4700 MALDI-TOF/TOF Analyzer, Applied Biosystems, Foster City, CA). The tandem mass spectrum search was performed using the Mascot search engine against the Swiss-Prot database. Acceptance criteria for protein hits were set as a minimum of two unique peptides per protein and a probability-based Mowse score of >45 where Mowse stands for molecular weight search and allows assignment of a probability to each identified peptide of a given molecular weight.
Immunoblot Analysis
Core proteins of chondroitin ABC lyase- or heparinase I- and II-digested samples were separated by SDS-PAGE on 4–12% gradient gels and transferred to nitrocellulose membranes at 20 V for 70 min in 40 mm Tris, 50 mm glycine with 20% methanol and 0.0375% SDS in a semidry electrophoretic transfer apparatus (Bio-Rad). Membranes were blocked with 2% calf serum in 50 mm Tris-buffered saline, pH 7.4 with 0.05% Tween 20 (TBST) and then incubated overnight at 4 °C with rabbit antiserum prepared against recombinant human versican (1:1,000 in blocking buffer) (kindly provided by Dr. R. LeBaron, San Antonio, TX) (33), a peptide sequence near the N terminus of human biglycan (1:1,000) (kindly provided by Dr. L. Fisher, National Institutes of Health) (34), a peptide from human decorin (1:1,000) (also provided by Dr. L. Fisher) (34), and serglycin (1:100) (Atlas Antibodies, Stockholm, Sweden). Nitrocellulose membranes were washed with 0.1% calf serum in TBST before incubation with alkaline phosphatase-conjugated goat anti-rabbit antiserum (1:2,000 dilution in TBST with 0.1% bovine serum albumin) (Roche Applied Science) for 1 h at room temperature. After washing, the membranes were developed by enhanced chemiluminescence (Pierce) and visualized by phosphorimage analysis.
Immunoprecipitation
Proteoglycans were isolated by ion exchange chromatography as described above, precipitated with 1.3% potassium acetate in 95% EtOH, and resuspended in PBS. Samples were precleared with recombinant Protein A beads (RepliGen, Waltham, MA) for 1 h at 4 °C to remove background binding. Precleared supernatants were divided into three equal aliquots. 1) The first aliquot was immunoprecipitated with anti-APLP2 (Calbiochem) overnight at 4 °C, 2) the second aliquot was incubated with rabbit IgG overnight at 4 °C and served as the nonspecific immunoprecipitation control, and 3) the third aliquot received no antibody and served as the total preimmunoprecipitation control. Aliquots 1 and 2 were incubated with Protein A beads for 1 h at 4 °C followed by three washes with PBS, 0.2% Tween and one wash with PBS. Beads were then digested with chondroitin ABC lyase as described above and boiled in 3× SDS-PAGE reducing sample buffer for 5 min. Aliquot 3 also was digested with chondroitin ABC lyase and boiled in sample buffer. Supernatants were loaded onto a 7% SDS-polyacrylamide gel for Western immunoblotting with MIM-7, an anti-peptide antiserum raised against the C-terminal 12-amino-acid peptide of human HC2 (35).
Quantitative Real Time Reverse Transcription-PCR
RNA was obtained from cell cultures using the Total RNA Isolation kit from Agilent Technologies (Wilmington, DE) according to the manufacturer's directions. cDNA was prepared by reverse transcription using random primers with the High Capacity cDNA Archive kit from the Applied Biosystems Division of PerkinElmer Life Sciences (ABI, Foster City, CA). PCR was carried out using an ABI Prism 7900HT Sequence Detection System and the TaqMan Fast Universal PCR Master Mix Reagents kit from ABI as directed by the manufacturer. The ABI Gene Expression Assays used are as follows: APLP2, Hs00920883_m1; ITIH2, Hs00158297_m1; serglycin, Hs01004159_m1; versican, Hs00171642_m1; HAS1, Hs00758053_m1; HAS2, Hs00193435_m1; HAS3, Hs00193436_m1; TSG-6, Hs00200180_m1; and 18 S, Hs99999901_s1. The ABI Gene Expression Assay forward and reverse primers are proprietary. A custom-designed primer set spanning the exon 10-11 junction and containing the sequence for the peptide IQPSGGTNINEALLR also was used for ITIH2 (forward, TGTGGAAGCAATGAAGACCA; reverse, GACGGAGTTGGGGTCTAACA). Custom-designed primers were used for microtubule-associated protein 1 light chain 3 (LC3) (forward, GCCGACCGCTGTAAGGAGGTAC; reverse, CTTGGTCTTGTCCAGGACGGGC) and cyclin D3 (forward, ATCGCCACGGGCAGCATTGG; reverse, CTGACAGGCCCGCAGGCAGT). For each group, assays were run in duplicate on RNA samples isolated from n = 2–3 different experiments from different passage numbers done at separate times. Normalized mRNA levels were then expressed as -fold of levels in control cells using the comparative Ct method (Applied Biosystems).
Histological and Immunohistochemical Staining
Serial 6-μm-thick tissue sections of human coronary artery obtained as described previously (36) were mounted on glass slides, deparaffinized, and then stained with Movat's pentachrome stain for morphology. Immunohistochemistry was performed on adjacent sections using rabbit polyclonal antisera raised against human APLP2 (1:200; Santa Cruz Biotechnology), IαIHC2 (1:200; Santa Cruz Biotechnology), or serglycin (1:100; Atlas Antibodies). Macrophages were detected using HAM-56 (1:100) (kindly provided by Dr. Allen Gown, University of Washington, Seattle, WA) (37). Single label immunohistochemistry was performed as described previously (38). Briefly, tissue sections were deparaffinized using xylene and rehydrated with graded alcohols. The slides were blocked with 3% H2O2 for 5 min, washed with PBS, incubated with the primary antibody for 60 min or overnight, and then washed again with PBS. A biotinylated anti-rabbit secondary antibody was then applied for 30 min followed by an avidin-biotin-peroxidase conjugate (ABC Elite, Vector Laboratories, Burlingame, CA) for 30 min. The standard peroxidase enzyme substrate 3,3′-diaminobenzidine was added without nickel chloride to yield a brown reaction product or with nickel chloride to yield a black reaction product. The slides were counterstained with methyl green to identify cell nuclei. Negative controls included substitution of primary antiserum or antibody with PBS, normal rabbit serum, or irrelevant monoclonal antibodies. The control for hyaluronan staining was predigestion of sections with Streptomyces hyaluronidases (39).
Statistical Analysis
The significance of differences in mean values was determined by the two-sample t test assuming unequal variances.
RESULTS
Total Proteoglycan Synthesis by THP-1 Monocytes and Macrophages
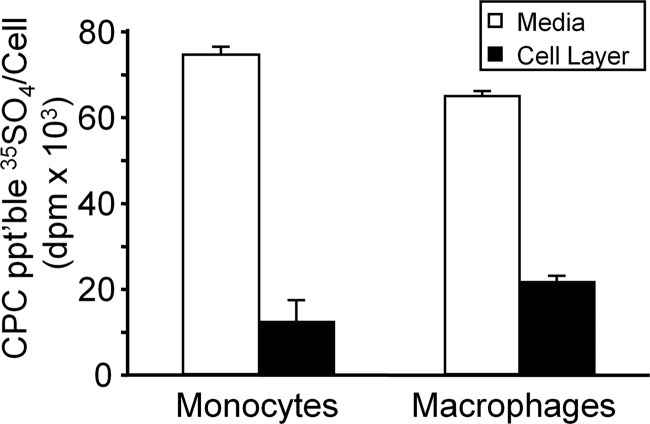
The distribution of newly synthesized proteoglycans secreted into the medium and associated with the cell layer (cell surface and intracellular) of monocyte and macrophage cultures was determined using cetylpyridinium chloride, a method for selective precipitation of anionic proteoglycans and glycosaminoglycans (40). After 24 h of metabolic labeling with [35S]SO4, there was no statistically significant difference in total (secreted + cell-associated) proteoglycan synthesis (cetylpyridinium chloride-precipitable 35SO4 per cell) between monocyte and macrophage cultures. For monocytes, 86.1 ± 0.7% of 35SO4-labeled material was secreted into the medium, and 13.9 ± 0.7% (n = 6) was associated with the cell layer. For macrophages, 74.9 ± 1.0% was secreted into the medium, and 25.1 ± 1.0% (n = 6) was associated with the cell layer (Fig. 1).
FIGURE 1.
Total proteoglycan synthesis by THP-1 monocytes and macrophages. THP-1 monocytes or PMA-differentiated macrophages were metabolically labeled with 35SO4. 35SO4-labeled proteoglycans secreted into the medium (open bars) or remaining associated with the cell layer (solid bars) were analyzed by cetylpyridinium chloride precipitation. Values are expressed as means ± S.D. (n = 6). CPC ppt'ble, cetylpyridinium chloride-precipitable. Error bars indicate S.D.
Macrophage Differentiation Is Accompanied by Increase in Proteoglycan Size Due to Changes in Glycosaminoglycan Chain and Protein Core Composition
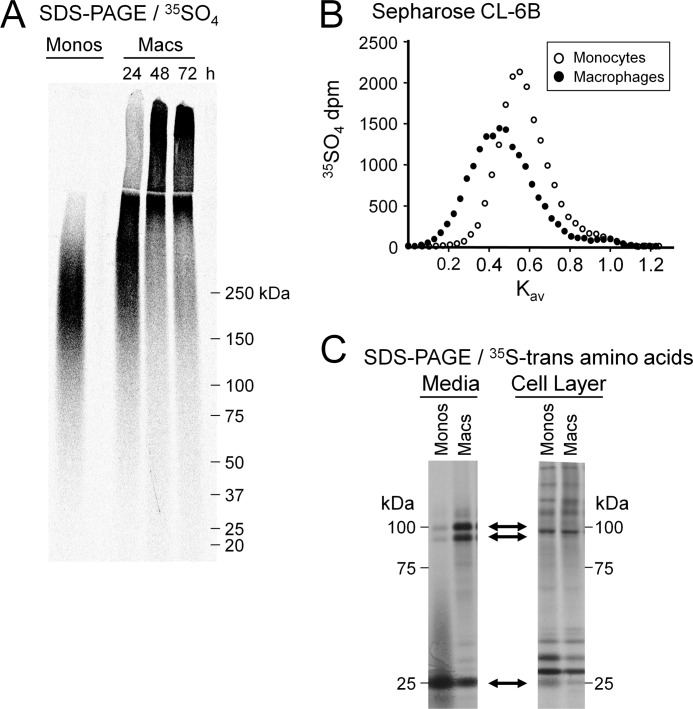
To evaluate the apparent molecular weight and hydrodynamic size of proteoglycans synthesized by monocytes and macrophages, 35SO4-proteoglycans were purified from the media by ion exchange chromatography with subsequent SDS-PAGE or size exclusion chromatography. Under reducing conditions on SDS-PAGE, monocyte 35SO4-proteoglycans migrated as a broad band as is characteristic of proteoglycans with an apparent molecular mass between ∼150 and 330 kDa (Fig. 2A). After induction of differentiation, proteoglycan size increased progressively with time. At 48 h after treatment with PMA, the cells were fully differentiated (41), and the apparent molecular mass of macrophage 35SO4-proteoglycans was greater than ∼250 kDa (Fig. 2A). Note that some of the macrophage proteoglycans are so large that they migrated only to the top of the resolving gel or remained in the stacking gel. Therefore, it is not possible to estimate the upper range of molecular mass for these molecules by SDS-PAGE. Size exclusion chromatography indicated an increase in hydrodynamic size of 35SO4-proteoglycans as monocytes differentiate into macrophages. Under dissociative conditions on Sepharose CL-2B, monocyte 35SO4-proteoglycans eluted as a single peak at Kav ∼0.67, whereas macrophage 35SO4-proteoglycans eluted at Kav ∼0.56 (p < 0.01) (data not shown). Thus, both methods indicate that the differentiation of monocytes into macrophages is associated with a dramatic increase in secreted proteoglycan size.
FIGURE 2.
Monocyte-to-macrophage differentiation is accompanied by changes in proteoglycan synthesis. A, SDS-PAGE (3.5% stacking gel, 4–12% resolving gel) analysis of 35SO4-labeled proteoglycans secreted into the medium by monocytes (Monos) and macrophages (Macs). Gels were loaded with 40,000 dpm/lane and evaluated by phosphorimage analysis. The gel shown is representative of n = 6. B, size exclusion chromatography of the glycosaminoglycan chains from 35SO4-labeled proteoglycans secreted into the medium of monocytes and macrophages. Glycosaminoglycan chains were liberated by reductive alkaline β-elimination and analyzed on an analytical Sepharose CL-6B column. Columns were loaded with ∼10–15,000 dpm. The molecular weights of glycosaminoglycan chains were determined according to Wasteson et al. (31). The column profile shown is representative of n = 2. C, SDS-PAGE analysis of 35S-trans amino acid-labeled core proteins from both secreted and cell-associated proteoglycans. Core proteins were prepared by digestion of intact proteoglycans with chondroitin ABC lyase. Upper two arrows indicate 90- and 100-kDa core proteins present in the media but not in the cell layer; the lower arrow indicates 25-kDa core protein present in both the media and the cell layer.
The increase in proteoglycan size could be due in part to changes in glycosaminoglycan chain length or core protein composition. To evaluate whether there is an effect on glycosaminoglycan chain length, the 35SO4-labeled chains from monocyte and macrophage proteoglycans were released from their core proteins by reductive alkaline β-elimination and analyzed by size exclusion chromatography (Fig. 2B). On a Sepharose CL-6B column, the liberated glycosaminoglycan chains of monocyte proteoglycans eluted with a Kav of ∼0.57, corresponding to an apparent molecular mass of ∼14 kDa as determined according to Wasteson (31). The glycosaminoglycan chains of macrophage proteoglycans eluted with a Kav of ∼0.42, indicating a larger molecular mass of ∼32 kDa.
To evaluate whether there is also an effect on core protein composition, intact [35S]Met/Cys-labeled proteoglycans were treated with chondroitin ABC lyase for analysis by SDS-PAGE. As can be seen from Fig. 2C, chondroitin ABC lyase liberated three major core protein bands of ∼25, ∼90, and ∼100 kDa from both monocyte and macrophage proteoglycans. However, the relative abundance of these core proteins was strikingly different. For monocytes, the 25-kDa core was predominant, whereas the 90- and 100-kDa cores were minor. In contrast, for macrophages, the 90- and 100-kDa cores were predominant, whereas the 25-kDa core was less abundant. Several additional protein bands appeared to be secreted into the media during macrophage differentiation, but we chose to focus on the more abundant 25-, 90-, and 100-kDa proteins. Treatment of either monocyte or macrophage proteoglycans with heparinases I and II in addition to chondroitin ABC lyase did not reveal additional core protein bands (data not shown).
To examine whether monocyte-to-macrophage differentiation was associated with a change in proteoglycan compartmentalization, cell-associated core proteins also were examined (Fig. 2C). The 90- and 100-kDa cores were not present in the cell layer of either monocytes or macrophages. The 25-kDa core was present in the cell layer of both monocytes and macrophages. However, it was markedly less abundant in the cell layer than in the media, and as found in the media, the 25-kDa core was relatively more abundantly associated with the cell layer of monocytes than of macrophages. Several additional minor protein bands were associated with the cell layer of both monocytes and macrophages, but the expression levels of the majority of these did not differ between monocytes and macrophages. Thus, we chose to focus solely on the 25-, 90-, and 100-kDa proteins, each of which is largely secreted, and the difference in expression levels of these molecules is not due to changes in compartmentalization as monocytes mature into macrophages. Rather, macrophage differentiation is accompanied by an increase in proteoglycan size due to glycosaminoglycan chain elongation and changes in core protein composition.
Identification of Core Proteins
A combination of Western immunoblotting and mass spectrophotometric analysis was used to identify the core proteins associated with monocyte and macrophage proteoglycans. Based on its size, the 25-kDa protein was predicted to be the core protein serglycin, a proteoglycan that is important in the formation and function of macrophage secretory vesicles (9). This was confirmed by Western immunoblotting using an antibody against serglycin core protein (Fig. 3).
FIGURE 3.
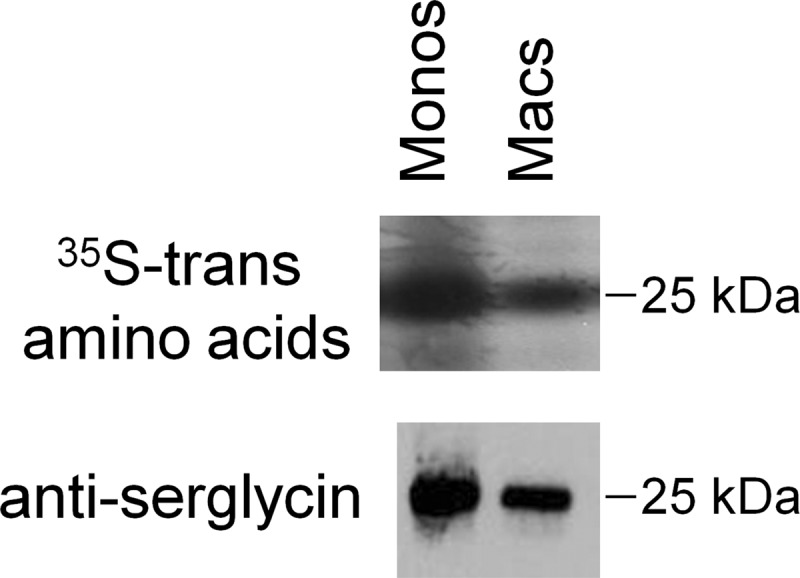
Identification of 25-kDa core protein as serglycin. Core proteins from monocytes (Monos) and macrophages (Macs) were evaluated by phosphorimage analysis (top panel) and Western immunoblotting (bottom panel) using antibodies specific for the core protein serglycin.
As the 90- and 100-kDa proteins did not match with the predicted sizes of the core proteins of other well described chondroitin sulfate proteoglycans (CSPGs) (i.e. versican, biglycan, and decorin), these bands were submitted for tandem mass spectrophotometric analysis (Table 1). For this analysis, the following criteria were used to identify proteins (42): (a) a high peptide identification score according to PeptideProphet (p > 0.95) (43), (b) a high protein identification score according to ProteinProphet (p > 0.90) (44), and (c) at least two peptides unique to the protein of interest detected in replicate analyses. Requiring at least two unique peptides with a high confidence score markedly decreases the false-positive rate of protein identification (45).
TABLE 1.
Protein identification by tandem mass spectrometry
Mowse, molecular weight search; CI, confidence interval.
| Band | Protein name | Swiss-Prot accession number | Peptide sequencesa | Amino acids | Mowse score | Total ion CI% |
|---|---|---|---|---|---|---|
| 1 | APLP2 | Q06481 | FVTPFK | 127–132 | 350 | 100 |
| WYFDLSK | 326–332 | |||||
| EWEEAELQAK | 411–420 | |||||
| QTLIQHFQAMVK | 428–439 | |||||
| HYQHVLAVDPEK | 510–521 | |||||
| SQVMTHLHVIEER | 527–539 | |||||
| ADMDQFTASISETPVDVR | 570–587 | |||||
| NKVDENMVIDETLDVK | 647–662 | |||||
| 2 | IαIHC2 | P19823 | VQFELHYQEVK | 177–187 | 65 | 100 |
| IQPSGGTNINEALLR | 380–394 | |||||
| FYNQVSTPLLR | 489–499 | |||||
| 3 | Serglycin | P10124 | GPMFELLPGESNK | 54–66 | 78 | 100 |
| LRTDLFPK | 70–77 |
a Individual peptide sequences and corresponding amino acid sequences are shown. Acceptance criteria are defined under “Materials and Methods” and “Results.”
By these criteria, the 100-kDa protein was identified as APLP2, a member of the amyloid precursor protein (APP) family (46). APLP2 is known to be synthesized both with and without a single chondroitin sulfate (CS) chain (47, 48); however, the specific functions of the chondroitin sulfate chain and of APLP2 itself are not yet known. The 90-kDa protein was identified as IαIHC2, which is one component of the IαI complex. IαIHC2 is itself not a proteoglycan but is covalently attached to the CS glycosaminoglycan chain of bikunin within the complex (16). The heavy chains of IαI (HC1 and HC2) are critical molecules in the stabilization of hyaluronan-rich matrices (16, 18). In addition, mass spectrophotometric analysis confirmed the Western blot identification of the 25-kDa protein as serglycin. Individual peptides and corresponding amino acid sequences based on Swiss-Prot database accession numbers are shown in Table 1. Thus, serglycin expression decreases but APLP2 and IαIHC2 expression increases as THP-1 monocytes mature into macrophages.
Expression of Serglycin, but Not APLP2 or IαIHC2, Is Regulated at Transcriptional Level
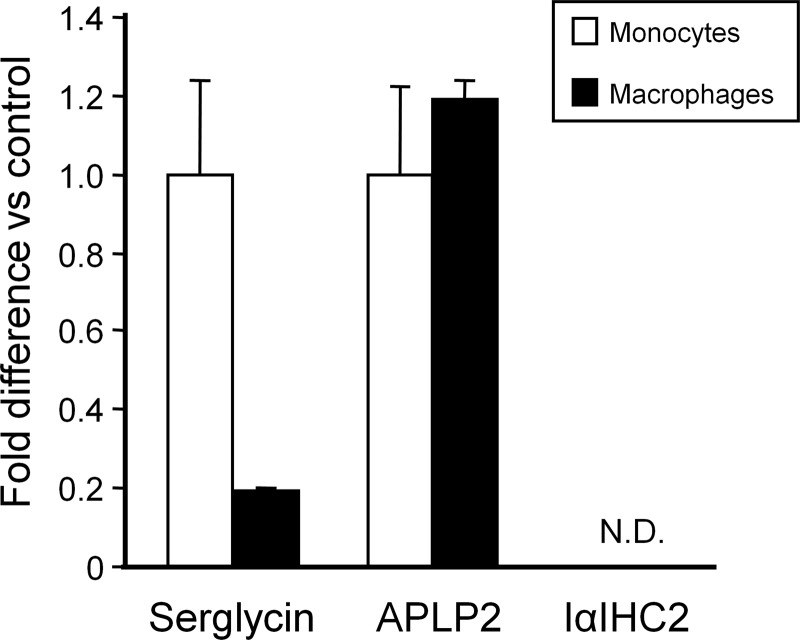
RT-PCR was performed to examine whether the changes in expression of these proteins associated with monocyte-to-macrophage differentiation are due to regulation at the transcriptional or post-translational level (Fig. 4). Consistent with the [35S]Met/Cys radiolabeling and Western immunoblot findings, serglycin mRNA expression was significantly higher (5.0-fold; p < 0.001) in monocytes than in fully differentiated macrophages. However, although APLP2 protein was more abundant in macrophages based on [35S]Met/Cys radiolabeling, there was no difference in mRNA expression between monocytes and macrophages. Furthermore, although IαIHC2 also was more abundant in macrophages based on [35S]Met/Cys radiolabeling, no signal for IαIHC2 mRNA could be detected even after 30 cycles of RT-PCR. The possibility that this surprising finding might indicate a splice variant form of IαIHC2 that could not be detected by the predesigned probe/primer set was considered. Therefore, a second probe/primer set corresponding to the coding region for one of the IαIHC2 peptide fragments (IQPSGGTNINEALLR) identified by mass spectrometry was custom-designed. Using this probe/primer set, no signal for IαIHC2 could be detected in THP-1 monocytes or macrophages at 2, 4, 8, 16, or 24 h as compared with HepG2 mRNA, a positive control for IαIHC2 (data not shown). These findings suggest that (i) serglycin expression is regulated at the transcriptional level but that (ii) APLP2 expression is regulated at the post-translational level and that (iii) although IαIHC2 is associated with macrophage-secreted proteoglycans it does not appear to be macrophage-derived.
FIGURE 4.
Expression of serglycin, but not APLP2 or IαIHC2, is regulated at transcriptional level. RT-PCR was performed for serglycin, APLP2, and IαIHC2 mRNA levels in both monocytes (open bars) and fully differentiated macrophages (solid bars). For each gene, macrophage mRNA levels are expressed relative to that in monocytes (control). No mRNA for IαIHC2 could be detected in either monocytes or macrophages (N.D.). Values represent means ± S.D. (n = 3). Error bars indicate S.D.
Expression of IαIHC2-related Molecules
Because of the critical role that IαIHC2 has in the stabilization of hyaluronan-rich matrices (19, 21, 22, 49, 50), the expression of other molecules implicated in this process, namely hyaluronan, TSG-6, and versican, also was examined using RT-PCR.
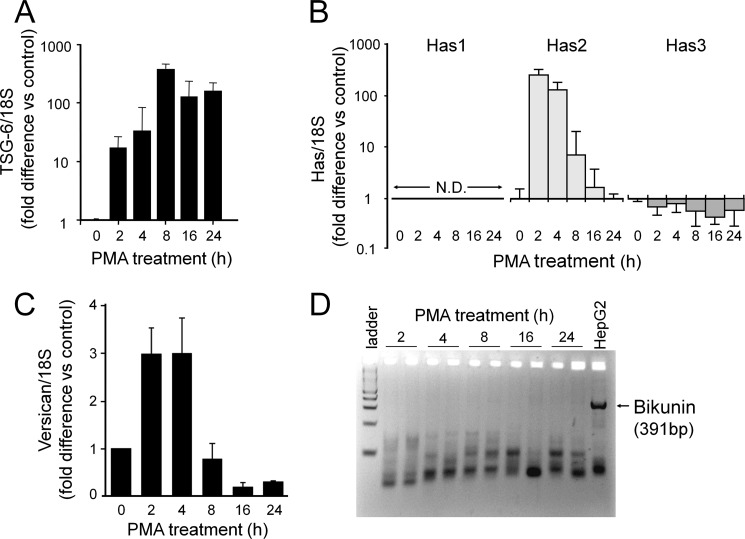
THP-1 monocytes (0 h of PMA treatment) expressed a very low copy number for TSG-6 mRNA (∼5 copies/10−5 18 S) that increased rapidly within 2 h (∼154 copies/10−5 18 S; p < 0.001; n = 3 versus monocytes) and was maximally elevated within 8 h of induction of differentiation (∼2,602 copies/10−5 18 S; p < 0.001) for a 520-fold increase (Fig. 5A). The increased expression of TSG-6 mRNA levels was sustained at least for 24 h after treatment with PMA by which time the cells were fully differentiated (∼1,668 copies/cell; p < 0.001).
FIGURE 5.
Monocyte-to-macrophage differentiation is accompanied by altered expression of other matrix-related molecules. RT-PCR was performed for TSG-6 (A); HASs 1, 2, and 3 (B); and versican (C) mRNA levels at the indicated times after induction of differentiation with PMA. For each gene, mRNA levels in monocytes (0 h of PMA treatment) represent control levels. Values represent means ± S.D. (n = 3). No mRNA for HAS1could be detected at any time (N.D.). D, standard PCR was performed for bikunin and evaluated on an agarose gel. Replicate samples were analyzed for each time point. mRNA from HepG2 cells was used as a positive control. Error bars indicate S.D.
Hyaluronan synthesis was investigated at the level of expression of the three isoforms of hyaluronan synthase, HASs 1–3 (Fig. 5B). HAS1 mRNA was essentially undetectable in either monocytes or macrophages (<1 copy/10−5 18 S). HAS2 mRNA also was present in extremely low abundance in monocytes (<0.1 copies/10−5 18 S) but increased markedly within 2 h of induction of differentiation (∼81 copies/10−5 18 S; p < 0.001; n = 3 versus monocytes) for an 810-fold increase. This increase was rapid and transient so that HAS2 mRNA was again undetectable in fully differentiated macrophages. HAS3 mRNA was present in THP-1 monocytes (∼71 copies/10−5 18 S), but its expression decreased upon induction of differentiation. The transient nature of the HAS2 response upon induction of differentiation with PMA suggests a role for autophagic endoplasmic reticulum stress activation in HAS stimulation (51). However, there were no substantial changes in mRNA levels of either LC3 or cyclin D3, two markers of endoplasmic stress activation (52). LC3 was present at ∼100 copies/10−5 18 S in monocytes and ∼180 copies/10−5 18 S in macrophages for only an ∼1.8-fold increase (data not shown); cyclin D3 was present at ∼650 copies/10−5 18 S in monocytes and decreased to ∼300 copies/10−5 18 S in macrophages (data not shown). Thus, at this point, it is not clear whether an endoplasmic reticulum stress response due to a hyperglycemic medium in these experiments contributes to the changes observed in hyaluronan synthesis observed during the differentiation of the macrophage.
Versican mRNA increased from ∼62 copies/10−5 18 S in monocytes to a maximum of ∼169 copies/10−5 18 S (p < 0.001; n = 3 versus monocytes) within 2 h of induction of differentiation for a 2.7-fold increase relative to control monocytes (Fig. 5C). This increase was rapid and transient so that versican mRNA was markedly down-regulated by the time the cells were fully differentiated (∼12 copies/10−5 18 S; p < 0.001; n = 3 versus monocytes).
These findings indicate that THP-1 macrophages have the capacity to synthesize TSG-6 and at least transiently hyaluronan and versican. Along with the mass spectrometry identification of IαIHC2 protein in the proteoglycan fraction of macrophages, these findings indicate that key molecules involved in hyaluronan matrix stabilization are associated with macrophages. However, as for IαIHC2, no bikunin mRNA could be detected in THP-1 macrophages as compared with HepG2 mRNA, a positive control for bikunin (Fig. 5D). Thus, the identification of radiolabeled IαIHC2 protein without finding mRNA for either IαIHC2 or bikunin (i.e. the CSPG to which this heavy chain is often covalently attached (31, 32)) is difficult to explain but may indicate covalent cross-linking to an unidentified radiolabeled protein.
Evidence for Novel IαIHC2 Complex Formation
It has been demonstrated that the heavy chains of IαI can form complexes with the chondroitin and dermatan sulfate chains on the core proteins of versican (53) and decorin (54), suggesting that the heavy chain can be linked to other CS-bearing core proteins as well. Three approaches were taken to investigate this further.
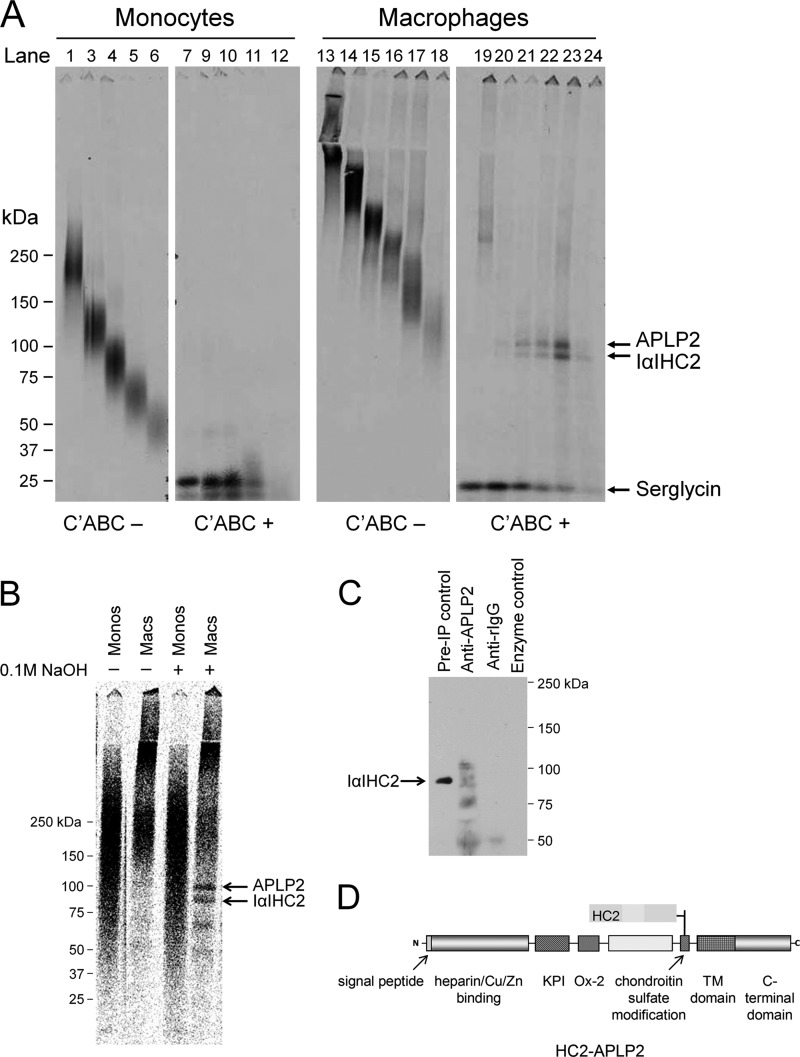
The broad banding pattern of proteoglycans on SDS-PAGE can be a reflection of the heterogeneity within a single species of proteoglycans and/or an indication of the presence of multiple distinct species of proteoglycans within a similar molecular weight range. Therefore, the first approach was to evaluate the size of the intact proteoglycan species associated with each individual core protein band using a combination of agarose gel electrophoresis and SDS-PAGE (Fig. 6A). For monocytes, the 25-kDa serglycin core protein liberated by chondroitin ABC lyase was found to be associated with every molecular mass fraction of intact proteoglycan (Fig. 6A, lanes 1–5, intact; lanes 6–10, digested). The 100-kDa APLP2 and 90-kDa IαIHC2 proteins could not be detected in association with any molecular mass fraction of intact proteoglycan by this approach. As these core proteins are in minor abundance among the monocyte proteoglycans (Fig. 2C), they might be difficult to detect in small fractions of secreted proteoglycans. For macrophages, the serglycin core protein was found to be associated with every molecular mass (>80 kDa) fraction of intact proteoglycan (Fig. 6A, lanes 11–16, intact; lanes 17–22, digested) but was the major core protein associated with the highest molecular mass fractions (>270 kDa) of intact proteoglycans (Fig. 6A, lanes 11–12, intact; lanes 17–18, digested). In contrast, the APLP2 and IαIHC2 proteins were found to be associated with the lower molecular mass fractions of intact proteoglycan (Fig. 6A, lanes 13–16, intact; lanes 19–22, digested). The finding of the APLP2 and IαIHC2 proteins in the same fractions of intact proteoglycans is consistent with the possibility that these molecules might be associated in the form of a complex within these fractions.
FIGURE 6.
Evidence for IαIHC2-APLP2 complex formation. A, intact 35S-trans-labeled proteoglycans from monocytes or macrophages were isolated from agarose gels based on the migratory distance from the origin of the gel. Proteoglycans were then treated without (−) or with (+) chondroitin ABC (C'ABC) lyase and evaluated by SDS-PAGE. B, SDS-PAGE analysis of monocyte (Monos) and macrophage (Macs) proteoglycans treated without (−) or with (+) 0.1 m NaOH for 10 min at room temperature. C, SDS-PAGE analysis of macrophage proteoglycans immunoprecipitated (IP) using antibodies specific for APLP2 (lane 1). The unbound proteoglycans were subsequently digested with chondroitin ABC lyase and immunoprecipitated again using antibodies specific for APLP2 (lane 2). A significant amount of proteoglycans remained unbound even after two immunoprecipitations (lane 3). D, schematic of proposed IαIHC2-APLP2 complex. TM, transmembrane; KPI, Kunitz protease inhibitor domain.
The heavy chains of IαI are covalently linked to the CS chain of bikunin via an ester linkage (55), which can be cleaved by mild sodium hydroxide treatment (21). Thus, the second approach was to establish whether or not such a covalent ester linkage is present between IαIHC2 and the CS chain of APLP2. It is important to note that the traditional glycosaminoglycan attachment to core proteins (e.g. CS attached to bikunin) is also somewhat susceptible to mild sodium hydroxide treatment (56). No effect of sodium hydroxide on monocyte proteoglycans could be detected due to the overlap in molecular masses of the intact monocyte proteoglycans in the 90–100-kDa molecular mass range (Fig. 6B). Although mild sodium hydroxide treatment did not completely digest the intact macrophage proteoglycans (56), treatment did liberate both ∼100- and 90-kDa protein bands, corresponding to APLP2 and IαIHC2, respectively. This result is consistent with an ester bond linkage existing between IαIHC2 and the CS chain of APLP2 as well as the attachment of the CS chain to the APLP2 protein core. It is worth noting that sodium hydroxide treatment also generated two smaller bands of ∼75 and 60 kDa, which might be truncated forms of HC2 (35).
The third approach was to selectively co-isolate IαIHC2 and APLP2 from the macrophage proteoglycan population by immunoprecipitation. The total macrophage proteoglycan preparation was incubated with either anti-APLP2 or nonspecific rabbit IgG. Bound complexes were pulled down with Protein A beads, liberated with chondroitin ABC lyase, and analyzed by Western immunoblotting using MIM-7, an antiserum raised against the C-terminal region of HC2 (35). MIM-7 recognized a single species of ∼90 kDa, corresponding to HC2, in the preimmunoprecipitation control sample (Fig. 6C, lane 1). Importantly, MIM-7 also recognized a 90-kDa species in the APLP2-immunoprecipitated sample (Fig. 6C, lane 2) but not in the nonspecific IgG-immunoprecipitated sample (Fig. 6C, lane 3) or in the enzyme reaction mixture (Fig. 6C, lane 4). MIM-7 also recognized bands at ∼110, 75, and 50 kDa in the APLP2-immunoprecipitated sample. Previously, immunoreactive species of similar sizes (∼130, 75, and 55 kDa) were detected in cartilage extracts with MIM-7 as well as another HC2-specific antibody; the smaller bands likely indicate the presence of truncated forms of HC2 (35). Taken together, these three results support the existence of a novel IαIHC2-APLP2 complex (Fig. 6D).
Serglycin Glycosaminoglycan Chain Elongation Associated with Monocyte-to-Macrophage Differentiation
As seen in Fig. 6A, the intact proteoglycan size of serglycin increases as monocytes mature into macrophages. Serglycin secreted by monocytes ranges from ∼50 to 250 kDa, whereas serglycin secreted by macrophages is largely >250 kDa. In previous studies, we have found that such increases in overall size can be due to glycosaminoglycan chain elongation (57). To evaluate chain length, we electrophoresed 35SO4-labeled monocyte and macrophage proteoglycans on agarose, isolated the two largest molecular weight fractions corresponding to serglycin (see Fig. 6A, lanes 1 and 2 and lanes 11 and 12), and performed chain length analysis (Table 2). The glycosaminoglycan chains of monocyte serglycin fraction 1 eluted with a Kav of ∼0.40, whereas those of macrophage serglycin fraction 1 eluted at Kav ∼0.30. Similarly, the chains of monocyte serglycin fraction 2 eluted at Kav ∼0.42, and those of macrophage serglycin fraction 2 eluted at Kav ∼0.32. In short, macrophage serglycin chains were ∼50–60% longer than those of monocyte serglycin. Thus, the increase in overall size of serglycin associated with monocyte-to-macrophage differentiation is due at least in part to glycosaminoglycan chain elongation in the mature cells.
TABLE 2.
Glycosaminoglycan chain length analysis of monocyte versus macrophage serglycin
| Elution position, Kav | Molecular sizea | |
|---|---|---|
| kDa | ||
| Monocytes, fraction 1 | 0.40 | 34 |
| Macrophages, fraction 1 | 0.30 | 55 |
| Monocytes, fraction 2 | 0.42 | 32 |
| Macrophages, fraction 2 | 0.32 | 50 |
a Apparent molecular size was determined from the elution position on Sepharose CL-6B according to the method of Wasteson (31).
Localization of IαIHC2 and APLP2 in Human Atherosclerotic Lesions
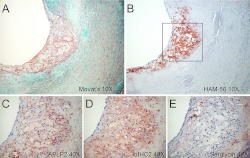
To establish the potential relevance of these cell culture findings to an in vivo situation characterized by an abundance of mature macrophages, we performed an immunohistochemical analysis of human atherosclerotic lesions (Fig. 7). Movat's pentachrome (Fig. 7A) and HAM-56 (Fig. 7B) staining of an aortic lesion indicates a thin fibrous cap overlying abundant macrophage foam cells and an underlying glycosaminoglycan-rich region. APLP2 (Fig. 7C) and IαIHC2 (Fig. 7D) have similar staining patterns. Both are predominantly found in macrophage-rich regions with a minor amount associated with the underlying glycosaminoglycan-rich region. In contrast, serglycin (Fig. 7E) is found primarily in the glycosaminoglycan-rich region and not associated with macrophages.
FIGURE 7.
Presence of APLP2 and IαIHC2 within macrophage-rich regions of atherosclerotic lesions. Shown is a section of human aorta stained with Movat's pentachrome (A) to identify cells (pink), elastin (black), glycosaminoglycans (blue), and collagen (yellow); HAM-56 to identify macrophages (B); anti-APLP2 (C); anti-IαIHC2 (D); or anti-serglycin (E).
DISCUSSION
In the present study, we have evaluated the proteoglycan changes associated with monocyte-to-macrophage differentiation. We confirm that serglycin is the primary proteoglycan species secreted by monocytes and that it is also secreted by mature macrophages as has been reported (9, 10). We also have identified APLP2, a less well known proteoglycan, and IαIHC2, a proteoglycan-associated molecule, in the proteoglycan fraction of mature macrophages. Furthermore, we found other proteoglycan and related genes to be markedly up-regulated during macrophage differentiation. These include versican, HAS2, and TSG-6. These findings add to the existing body of knowledge regarding proteoglycans and related molecules synthesized by monocytes and macrophages.
Serglycin is a proteoglycan known to be made by a variety of hematopoietic cells (including mast cells (58), monocytes (59), macrophages (7, 9), T-lymphocytes (60, 61), neutrophils (62), and platelets (63–65)) and to have a vital function in the formation of secretory vesicles and storage granules within these cells (9, 61, 65, 66). Two distinctive structural features confer added functionality. First, the core protein serglycin has a high density of Ser-Gly repeats in the glycosaminoglycan attachment region, making it protease-resistant. Second, the core protein carries up to 24 glycosaminoglycan chains, giving serglycin a high degree of negative charge. We found that the transition of serglycin to an overall larger molecular size as monocytes mature into macrophages is due at least in part to glycosaminoglycan chain elongation. In previous studies, we have found that such increases in chain length serve to enhance the negative charge-dependent ligand binding characteristics of proteoglycans (57, 67). Thus, structural features of both the core protein and glycosaminoglycan chains likely contribute to the ability of serglycin to bind a variety of molecules (e.g. cytokines, growth factors, and proteases) and to protect them from proteolytic degradation. In the specific case of TNF, it is suggested that serglycin controls secretion of this cytokine to prevent the release of excessive amounts (9). It is likely that serglycin has an important protective role in the storage and transport of other binding proteins as well. It also is likely that macrophage-derived serglycin will have an altered and perhaps enhanced role in this process as compared with monocyte-derived serglycin.
Given that serglycin has been shown to bind to and control secretion of TNF (9), it is particularly interesting that we found one of the TNF-inducible genes, TSG-6, to be dramatically up-regulated upon initiation of monocyte-to-macrophage differentiation. We suggest two possibilities by which serglycin and TSG-6 might interact. The first possibility involves an indirect interaction. Monocyte-to-macrophage differentiation has been shown to induce TNF secretion (68). The enhanced serglycin-mediated transport of TNF from the secretory vesicles and subsequent release of the cytokine in the extracellular environment in this response could in turn stimulate the macrophage to increase synthesis and secretion of TSG-6 as we report here. The second possibility involves a direct interaction. TSG-6 might itself be a serglycin-binding protein in which case serglycin might control transport as well as secretion of this molecule. Although this has not yet been examined, TSG-6 is known to bind chondroitin 4-sulfate (69, 70) and heparin/heparin sulfate (71).
TSG-6 is not constitutively present in most healthy adult tissues (with the exception of bone marrow and epidermis (72, 73)), but its expression is up-regulated in response to a variety of inflammatory mediators in a number of pathological and physiological contexts (20, 74, 75). The interactions of TSG-6 with a variety of ligands contribute to its multifunctionality (74). Of particular relevance are (i) the potent anti-inflammatory properties of TSG-6 (76–81) and (ii) the important role of TSG-6 in ECM remodeling (18, 22, 72, 82). These functions are mediated by the abilities of TSG-6 to inhibit neutrophil migration (76, 79–81, 83) and plasmin activity (and thus matrix metalloproteinase activation) (71, 76, 77, 80), to promote expression of negative modulators of inflammation (78), and to form cross-linked hyaluronan networks (i.e. either via the formation of covalent complexes between hyaluronan and IαI and pre-α-inhibitor heavy chains (18, 21, 50, 84) or through direct TSG-6-mediated hyaluronan cross-linking (23, 26) where these might induce anti-inflammatory signals (e.g. in macrophages (85)).
Versican is another monocyte/macrophage gene that is up-regulated in a number of disease states, such as myocardial infarction (86), coronary stenosis (87), autoimmunity (88, 89), and in response to proinflammatory stimulants, such as LPS (90) and hypoxia (11, 12). In addition, versican has been identified as a gene that is differentially expressed in M1 macrophages as opposed to M2 macrophages as they differentiate from monocytes (91). Thus, our finding of up-regulation of TSG-6 and versican expression suggests that both an anti-inflammatory response and matrix remodeling are features characteristic of monocyte-to-macrophage differentiation.
APLP2 is known to be secreted in both a CSPG and a non-proteoglycan form by a variety of cell types (48), but to our knowledge, it has not been described as a product of monocytes and macrophages. However, a related family member, APP, has been demonstrated on circulating monocytes with increased expression in macrophage colony stimulating factor-treated monocytes (92). APLP2, APP, and a third family member, APLP1, all have a high degree of amino acid and domain homology (46). The most notable difference between APLP2 and APP is APLP2's lack of the Aβ sequence. Thus, upon proteolytic processing, APLP2 does not give rise to the Aβ polypeptide. Apparently, APLP2 does have important non-redundant functions with APP that might be related to cell adhesion, migration and wound healing (93, 94).
We found a dramatic increase in APLP2 protein in the proteoglycan fraction of macrophages as compared with that of monocytes. However, we also found similarly high copy numbers for APLP2 mRNA in both monocytes and macrophages with no increase in APLP2 mRNA levels associated with macrophage differentiation. This suggests that monocytes have the capacity to make the non-proteoglycan form of APLP2, whereas macrophages make the CSPG form. Thus, the dramatic increase in APLP2 protein associated with macrophages is likely due to changes in glycosaminoglycan chain addition rather than changes in core protein synthesis. To date, little is known regarding what factors control addition of the CS chain onto the APLP2 core protein. However, it has similarly been suggested that chain addition might be related to differentiation in neuronal cells (48).
IαIHC2, although not a proteoglycan itself, is one component of the IαI proteoglycan complex. The IαI family consists of three protein complexes resulting from different arrangements of heavy chains (HC1, HC2, and HC3) covalently attached to the CS moiety of bikunin (15). A major function of IαI is to stabilize hyaluronan-rich matrices (18, 54); therefore, IαI has important roles in ECM structure and essential roles in normal physiological processes (e.g. ovulation (95)) as well as in pathological conditions (e.g. rheumatoid arthritis (96) and inflammatory bowel disease (97, 98)). The ability of IαI to form complexes with hyaluronan is due to the covalent transfer of heavy chains from IαI to hyaluronan (16, 21). Notably, TSG-6 is a critical molecule for such heavy chain transfer to occur (21, 22).
Our finding of IαIHC2 protein, but not IαIHC2 mRNA or bikunin mRNA, associated with mature macrophages was unexpected. The inability to detect IαIHC2 mRNA suggests that the protein is not macrophage-derived. More likely, IαIHC2 must be exogenously derived from the serum. It is imperative to emphasize that the mass spectrophotometric identification of IαIHC2 was based on the identification of three peptide fragments, which did match the sequence of this human protein with a high degree of confidence. Tandem mass spectrometry identification was performed three times on three separate batches of THP-1 cells. On no occasion was another protein detected in the 90-kDa band. We are aware that lack of detection of another protein by mass spectrometry does not definitively rule out the possibility of a co-migrating protein. We also have performed Western immunoblotting using MIM-7, a highly specific anti-peptide antiserum raised against the C-terminal 12-amino acid peptide of human HC2 (35). MIM-7 recognizes the 90-kDa band identified as IαIHC2 by tandem mass spectrometry (data not shown). Two additional factors were taken into consideration. First, there is a high degree of similarity between the sequences for human and bovine IαIHC2 (NCBI Reference Sequence NP_001091485.1). IαIHC2 and the three peptide fragments we identified also match the bovine sequence. Second, IαIHC2 has to date only been reported to be synthesized by hepatocytes and chondrocytes. All factors considered, we make the cautious interpretation that IαIHC2 is serum-derived. The inability to detect bikunin message in macrophages not only is further support for this possibility but also suggests that IαIHC2 might be linked to a different core protein. This is not unprecedented as IαIHC3, another heavy chain component of the IαI complex, can be linked both to the CS chains of decorin and to free CS chains (54). In addition, truncated heavy chains have been found to be attached to a CSPG other than bikunin in extracts from normal and osteoarthritic cartilage (35). Thus, heavy chains could be linked to the CS chains of macrophage proteoglycans as well.
The possibility that macrophage-secreted proteoglycans might form a complex with other molecules also is not unprecedented. In THP-1 macrophages, a fraction of the secreted matrix metalloproteinase 9 has been found to be covalently linked via disulfide bonds to the core protein serglycin (8, 99). We, however, were not able to detect matrix metalloproteinase 9 in the proteoglycan fraction of THP-1 macrophages (data not shown). It also has been suggested that serglycin could form a complex with versican and another unidentified core protein secreted by interleukin-1-stimulated arterial smooth muscle cells (100).
In short, our data support the formation of a complex between IαIHC2 and APLP2. Regardless of the source of IαIHC2, its presence as a complex bound to APLP2 is a novel finding. Although we have not yet quantified the amounts of free APLP2 relative to IαIHC2-bound APLP2, it is likely that the increased expression of APLP2 could impact the amount of IαIHC2 recruited into the macrophage milieu. We propose that IαIHC2 protein, carried on serum bikunin (e.g. in the context of IαI), is transferred onto macrophage-derived APLP2 and that this transfer is mediated by TSG-6, which is induced during monocyte-to-macrophage differentiation. The simultaneous temporal up-regulation of other essential molecules involved in matrix stabilization (HAS2 and versican) suggests that macrophages temporarily produce a novel, stabilized matrix, which may be essential to the maturation process. Our immunohistochemistry results showing that both are localized in the same region in macrophage-rich regions in atherosclerotic plaques also suggests, but does not prove, that these components exist together in a complex.
It should be noted that these studies were conducted using THP-1 monocytes and PMA-induced macrophages. The similarities and differences between this human monocytic leukemia cell line and normal human peripheral blood monocyte-derived macrophages have been evaluated (28, 101). Although these cells undergo comparable morphological changes and expression of differentiation markers (e.g. apolipoprotein E, scavenger receptor type-A, and lipoprotein lipase), differences in expression of a variety of other proteins have been identified. Thus, to validate our in vitro findings using THP-1 cells, we performed an immunohistochemical analysis of human atherosclerotic lesions, a condition characterized by an abundance of mature macrophages. The co-existence of APLP2 and IαIHC2 in macrophage-rich regions of human lesions confirms that these molecules are present in atherosclerosis and suggests that these molecules have a role in the ECM changes that occur during the development of this disease. How these molecules contribute to the development of atherosclerosis and whether they contribute to pro- or anti-inflammatory processes remain to be determined. It will also be of interest to evaluate whether these molecules are associated with each other in other inflammatory diseases characterized by macrophage development as well.
Acknowledgment
We thank Dr. Virginia M. Green for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL018645 and HL098067 (to T. N. W.) and HL092969 (to A. C. and T. N. W.). This work was also supported by Arthritis Research UK Grants 18472 and 19489 (to A. J. D.).
- ECM
- extracellular matrix
- APLP2
- amyloid precursor-like protein 2
- IαI
- inter-α-trypsin inhibitor
- IαIHC
- inter-α-trypsin inhibitor heavy chain
- TSG-6
- TNF-stimulated gene-6
- HAS
- hyaluronan synthases
- PMA
- phorbol myristate acetate
- CSPG
- chondroitin sulfate proteoglycan
- APP
- amyloid precursor protein
- CS
- chondroitin sulfate
- HC
- heavy chain
- LC3
- microtubule-associated protein 1 light chain 3.
REFERENCES
- 1. Wight T. (2005) in Atherothrombosis and Coronary Artery Disease (Fuster V., Topol E., Nabel E., eds) pp. 421–437, Lippincott Williams and Wilkins, Philadelphia [Google Scholar]
- 2. Libby P., Geng Y. J., Aikawa M., Schoenbeck U., Mach F., Clinton S. K., Sukhova G. K., Lee R. T. (1996) Macrophages and atherosclerotic plaque stability. Curr. Opin. Lipidol. 7, 330–335 [DOI] [PubMed] [Google Scholar]
- 3. Ross R. (1999) Atherosclerosis—an inflammatory disease. N. Engl. J. Med. 340, 115–126 [DOI] [PubMed] [Google Scholar]
- 4. Shibata N., Glass C. K. (2009) Regulation of macrophage function in inflammation and atherosclerosis. J. Lipid Res. 50, (suppl.) S277–S281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang M. Y., Olin K. L., Tsoi C., Wight T. N., Chait A. (1998) Human monocyte-derived macrophages secrete two forms of proteoglycan-macrophage colony-stimulating factor that differ in their ability to bind low density lipoproteins. J. Biol. Chem. 273, 15985–15992 [DOI] [PubMed] [Google Scholar]
- 6. Uhlin-Hansen L., Eskeland T., Kolset S. (1989) Modulation of the expression of chondroitin sulfate proteoglycan in stimulated human monocytes. J. Biol. Chem. 264, 14916–14922 [PubMed] [Google Scholar]
- 7. Uhlin-Hansen L., Wik T., Kjellén L., Berg E., Forsdahl F., Kolset S. O. (1993) Proteoglycan metabolism in normal and inflammatory human macrophages. Blood 82, 2880–2889 [PubMed] [Google Scholar]
- 8. Winberg J. O., Kolset S. O., Berg E., Uhlin-Hansen L. (2000) Macrophages secrete matrix metalloproteinase 9 covalently linked to the core protein of chondroitin sulphate proteoglycans. J. Mol. Biol. 304, 669–680 [DOI] [PubMed] [Google Scholar]
- 9. Zernichow L., Abrink M., Hallgren J., Grujic M., Pejler G., Kolset S. O. (2006) Serglycin is the major secreted proteoglycan in macrophages and has a role in the regulation of macrophage tumor necrosis factor-α secretion in response to lipopolysaccharide. J. Biol. Chem. 281, 26792–26801 [DOI] [PubMed] [Google Scholar]
- 10. Kolset S. O., Zernichow L. (2008) Serglycin and secretion in human monocytes. Glycoconj. J. 25, 305–311 [DOI] [PubMed] [Google Scholar]
- 11. Asplund A., Stillemark-Billton P., Larsson E., Rydberg E. K., Moses J., Hultén L. M., Fagerberg B., Camejo G., Bondjers G. (2010) Hypoxic regulation of secreted proteoglycans in macrophages. Glycobiology 20, 33–40 [DOI] [PubMed] [Google Scholar]
- 12. Asplund A., Fridén V., Stillemark-Billton P., Camejo G., Bondjers G. (2011) Macrophages exposed to hypoxia secrete proteoglycans for which LDL has higher affinity. Atherosclerosis 215, 77–81 [DOI] [PubMed] [Google Scholar]
- 13. Makatsori E., Lamari F. N., Theocharis A. D., Anagnostides S., Hjerpe A., Tsegenidis T., Karamanos N. K. (2003) Large matrix proteoglycans, versican and perlecan, are expressed and secreted by human leukemic monocytes. Anticancer Res. 23, 3303–3309 [PubMed] [Google Scholar]
- 14. Schaefer L., Babelova A., Kiss E., Hausser H. J., Baliova M., Krzyzankova M., Marsche G., Young M. F., Mihalik D., Götte M., Malle E., Schaefer R. M., Gröne H. J. (2005) The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J. Clin. Investig. 115, 2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bost F., Diarra-Mehrpour M., Martin J. P. (1998) Inter-α-trypsin inhibitor proteoglycan family—a group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 252, 339–346 [DOI] [PubMed] [Google Scholar]
- 16. Zhuo L., Hascall V. C., Kimata K. (2004) Inter-α-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J. Biol. Chem. 279, 38079–38082 [DOI] [PubMed] [Google Scholar]
- 17. Milner C. M., Tongsoongnoen W., Rugg M. S., Day A. J. (2007) The molecular basis of inter-α-inhibitor heavy chain transfer on to hyaluronan. Biochem. Soc. Trans. 35, 672–676 [DOI] [PubMed] [Google Scholar]
- 18. Day A. J., de la Motte C. A. (2005) Hyaluronan cross-linking: a protective mechanism in inflammation? Trends Immunol. 26, 637–643 [DOI] [PubMed] [Google Scholar]
- 19. Yingsung W., Zhuo L., Morgelin M., Yoneda M., Kida D., Watanabe H., Ishiguro N., Iwata H., Kimata K. (2003) Molecular heterogeneity of the SHAP-hyaluronan complex. Isolation and characterization of the complex in synovial fluid from patients with rheumatoid arthritis. J. Biol. Chem. 278, 32710–32718 [DOI] [PubMed] [Google Scholar]
- 20. Milner C. M., Higman V. A., Day A. J. (2006) TSG-6: a pluripotent inflammatory mediator? Biochem. Soc. Trans. 34, 446–450 [DOI] [PubMed] [Google Scholar]
- 21. Rugg M. S., Willis A. C., Mukhopadhyay D., Hascall V. C., Fries E., Fülöp C., Milner C. M., Day A. J. (2005) Characterization of complexes formed between TSG-6 and inter-α-inhibitor that act as intermediates in the covalent transfer of heavy chains onto hyaluronan. J. Biol. Chem. 280, 25674–25686 [DOI] [PubMed] [Google Scholar]
- 22. Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R. V., Rugg M. S., Day A. J., Salustri A., Hascall V. C., Glant T. T., Mikecz K. (2003) Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development 130, 2253–2261 [DOI] [PubMed] [Google Scholar]
- 23. Baranova N. S., Nilebäck E., Haller F. M., Briggs D. C., Svedhem S., Day A. J., Richter R. P. (2011) The inflammation-associated protein TSG-6 cross-links hyaluronan via hyaluronan-induced TSG-6 oligomers. J. Biol. Chem. 286, 25675–25686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhuo L., Kanamori A., Kannagi R., Itano N., Wu J., Hamaguchi M., Ishiguro N., Kimata K. (2006) SHAP potentiates the CD44-mediated leukocyte adhesion to the hyaluronan substratum. J. Biol. Chem. 281, 20303–20314 [DOI] [PubMed] [Google Scholar]
- 25. He H., Li W., Tseng D. Y., Zhang S., Chen S. Y., Day A. J., Tseng S. C. (2009) Biochemical characterization and function of complexes formed by hyaluronan and the heavy chains of inter-α-inhibitor (HC*HA) purified from extracts of human amniotic membrane. J. Biol. Chem. 284, 20136–20146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lesley J., Gál I., Mahoney D. J., Cordell M. R., Rugg M. S., Hyman R., Day A. J., Mikecz K. (2004) TSG-6 modulates the interaction between hyaluronan and cell surface CD44. J. Biol. Chem. 279, 25745–25754 [DOI] [PubMed] [Google Scholar]
- 27. Auwerx J. H., Deeb S., Brunzell J. D., Peng R., Chait A. (1988) Transcriptional activation of the lipoprotein lipase and apolipoprotein E genes accompanies differentiation in some human macrophage-like cell lines. Biochemistry 27, 2651–2655 [DOI] [PubMed] [Google Scholar]
- 28. Auwerx J. (1991) The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia 47, 22–31 [DOI] [PubMed] [Google Scholar]
- 29. Schönherr E., Järveläinen H. T., Sandell L. J., Wight T. N. (1991) Effects of platelet-derived growth factor and transforming growth factor-β1 on the synthesis of a large versican-like chondroitin sulfate proteoglycan by arterial smooth muscle cells. J. Biol. Chem. 266, 17640–17647 [PubMed] [Google Scholar]
- 30. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31. Wasteson A. (1971) A method for the determination of the molecular weight and molecular weight distribution of chondroitin sulfate. J. Chromatogr. 59, 87–97 [DOI] [PubMed] [Google Scholar]
- 32. Schönherr E., Kinsella M. G., Wight T. N. (1997) Genistein selectively inhibits platelet-derived growth factor stimulated versican biosynthesis in monkey arterial smooth muscle cells. Arch. Biochem. Biophys. 339, 353–361 [DOI] [PubMed] [Google Scholar]
- 33. Lebaron R. G. (1996) Versican. Perspect. Dev. Neurobiol. 3, 261–271 [PubMed] [Google Scholar]
- 34. Fleischmajer R., Fisher L. W., MacDonald E. D., Jacobs L., Jr., Perlish J. S., Termine J. D. (1991) Decorin interacts with fibrillar collagen of embryonic and adult human skin. J. Struct. Biol. 106, 82–90 [DOI] [PubMed] [Google Scholar]
- 35. Yoshihara Y., Plaas A., Osborn B., Margulis A., Nelson F., Stewart M., Rugg M. S., Milner C. M., Day A. J., Nemoto K., Sandy J. D. (2008) Superficial zone chondrocytes in normal and osteoarthritic human articular cartilages synthesize novel truncated forms of inter-α-trypsin inhibitor heavy chains which are attached to a chondroitin sulfate proteoglycan other than bikunin. Osteoarthritis Cartilage 16, 1343–1355 [DOI] [PubMed] [Google Scholar]
- 36. Stewart C. R., Haw A., 3rd, Lopez R., McDonald T. O., Callaghan J. M., McConville M. J., Moore K. J., Howlett G. J., O'Brien K. D. (2007) Serum amyloid P colocalizes with apolipoproteins in human atheroma: functional implications. J. Lipid Res. 48, 2162–2171 [DOI] [PubMed] [Google Scholar]
- 37. Gown A. M., Tsukada T., Ross R. (1986) Human atherosclerosis. II. Immunocytochemical analysis of the cellular composition of human atherosclerotic lesions. Am. J. Pathol. 125, 191–207 [PMC free article] [PubMed] [Google Scholar]
- 38. McDonald T. O., Gerrity R. G., Jen C., Chen H. J., Wark K., Wight T. N., Chait A., O'Brien K. D. (2007) Diabetes and arterial extracellular matrix changes in a porcine model of atherosclerosis. J. Histochem. Cytochem. 55, 1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Riessen R., Wight T. N., Pastore C., Henley C., Isner J. M. (1996) Distribution of hyaluronan during extracellular matrix remodeling in human restenotic arteries and balloon-injured rat carotid arteries. Circulation 93, 1141–1147 [DOI] [PubMed] [Google Scholar]
- 40. Wasteson A., Uthne K., Westermark B. (1973) A novel assay for the biosynthesis of sulfated polysaccharide and its application to studies on the effects of somatomedin on cultured cells. Biochem. J. 136, 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edelson P. J., Cohn Z. A. (1976) in In Vitro Methods in Cell-mediated and Tumor Immunity (Bloom B. R., David J. R., eds) pp. 333–340, Academic Press, New York [Google Scholar]
- 42. Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., Chea H., Knopp R. H., Brunzell J., Geary R., Chait A., Zhao X. Q., Elkon K., Marcovina S., Ridker P., Oram J. F., Heinecke J. W. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 117, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 44. Yan W., Lee H., Deutsch E. W., Lazaro C. A., Tang W., Chen E., Fausto N., Katze M. G., Aebersold R. (2004) A dataset of human liver proteins identified by protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. Mol. Cell. Proteomics 3, 1039–1041 [DOI] [PubMed] [Google Scholar]
- 45. Resing K. A., Ahn N. G. (2005) Proteomics strategies for protein identification. FEBS Lett. 579, 885–889 [DOI] [PubMed] [Google Scholar]
- 46. Walsh D. M., Minogue A. M., Sala Frigerio C., Fadeeva J. V., Wasco W., Selkoe D. J. (2007) The APP family of proteins: similarities and differences. Biochem. Soc. Trans. 35, 416–420 [DOI] [PubMed] [Google Scholar]
- 47. Thinakaran G., Sisodia S. (1994) Amyloid precursor-like protein 2 (APLP2) is modified by the addition of chondroitin sulfate glycosaminoglycan at a single site. J. Biol. Chem. 269, 22099–22104 [PubMed] [Google Scholar]
- 48. Pangalos M. N., Shioi J., Robakis N. K. (1995) Expression of the chondroitin sulfate proteoglycans of amyloid precursor (appican) and amyloid precursor-like protein 2. J. Neurochem. 65, 762–769 [DOI] [PubMed] [Google Scholar]
- 49. Zhuo L., Yoneda M., Zhao M., Yingsung W., Yoshida N., Kitagawa Y., Kawamura K., Suzuki T., Kimata K. (2001) Defect in SHAP-hyaluronan complex causes severe female infertility. A study by inactivation of the bikunin gene in mice. J. Biol. Chem. 276, 7693–7696 [DOI] [PubMed] [Google Scholar]
- 50. Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Lorentzen K. A., Wisniewski H. G., Thøgersen I. B., Enghild J. J. (2008) The transfer of heavy chains from bikunin proteins to hyaluronan requires both TSG-6 and HC2. J. Biol. Chem. 283, 18530–18537 [DOI] [PubMed] [Google Scholar]
- 51. Wang A., Hascall V. C. (2009) Hyperglycemia, intracellular hyaluronan synthesis, cyclin D3 and autophagy. Autophagy 5, 864–865 [DOI] [PubMed] [Google Scholar]
- 52. Wang A., de la Motte C., Lauer M., Hascall V. (2011) Hyaluronan matrices in pathobiological processes. FEBS J. 278, 1412–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eriksen G. V., Carlstedt I., Mörgelin M., Uldbjerg N., Malmström A. (1999) Isolation and characterization of proteoglycans from human follicular fluid. Biochem. J. 340, 613–620 [PMC free article] [PubMed] [Google Scholar]
- 54. Kaczmarczyk A., Thuveson M., Fries E. (2002) Intracellular coupling of the heavy chain of pre-α-inhibitor to chondroitin sulfate. J. Biol. Chem. 277, 13578–13582 [DOI] [PubMed] [Google Scholar]
- 55. Enghild J. J., Salvesen G., Thøgersen I. B., Valnickova Z., Pizzo S. V., Hefta S. A. (1993) Presence of the protein-glycosaminoglycan-protein covalent cross-link in the inter-α-inhibitor-related proteinase inhibitor heavy chain 2/bikunin. J. Biol. Chem. 268, 8711–8716 [PubMed] [Google Scholar]
- 56. Forteza R., Casalino-Matsuda S. M., Monzon M. E., Fries E., Rugg M. S., Milner C. M., Day A. J. (2007) TSG-6 potentiates the antitissue kallikrein activity of inter-α-inhibitor through bikunin release. Am. J. Respir. Cell Mol. Biol. 36, 20–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang M. Y., Potter-Perigo S., Tsoi C., Chait A., Wight T. N. (2000) Oxidized low density lipoproteins regulate synthesis of monkey aortic smooth muscle cell proteoglycans that have enhanced native low density lipoprotein binding properties. J. Biol. Chem. 275, 4766–4773 [DOI] [PubMed] [Google Scholar]
- 58. Tantravahi R. V., Stevens R. L., Austen K. F., Weis J. H. (1986) A single gene in mast cells encodes the core peptides of heparin and chondroitin sulfate proteoglycans. Proc. Natl. Acad. Sci. U.S.A. 83, 9207–9210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kolset S. O., Mann D. M., Uhlin-Hansen L., Winberg J. O., Ruoslahti E. (1996) Serglycin-binding proteins in activated macrophages and platelets. J. Leukoc. Biol. 59, 545–554 [DOI] [PubMed] [Google Scholar]
- 60. Elliott J. F., Miller C. L., Pohajdak B., Talbot D., Helgason C. D., Bleackley R. C., Paetkau V. (1993) Induction of a proteoglycan core protein mRNA in mouse T lymphocytes. Mol. Immunol. 30, 749–754 [DOI] [PubMed] [Google Scholar]
- 61. Grujic M., Braga T., Lukinius A., Eloranta M. L., Knight S. D., Pejler G., Abrink M. (2005) Serglycin-deficient cytotoxic T lymphocytes display defective secretory granule maturation and granzyme B storage. J. Biol. Chem. 280, 33411–33418 [DOI] [PubMed] [Google Scholar]
- 62. Niemann C. U., Cowland J. B., Klausen P., Askaa J., Calafat J., Borregaard N. (2004) Localization of serglycin in human neutrophil granulocytes and their precursors. J. Leukoc. Biol. 76, 406–415 [DOI] [PubMed] [Google Scholar]
- 63. Alliel P. M., Périn J. P., Maillet P., Bonnet F., Rosa J. P., Jollès P. (1988) Complete amino acid sequence of a human platelet proteoglycan. FEBS Lett. 236, 123–126 [DOI] [PubMed] [Google Scholar]
- 64. Périn J. P., Bonnet F., Maillet P., Jollès P. (1988) Characterization and N-terminal sequence of human platelet proteoglycan. Biochem. J. 255, 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Woulfe D. S., Lilliendahl J. K., August S., Rauova L., Kowalska M. A., Abrink M., Pejler G., White J. G., Schick B. P. (2008) Serglycin proteoglycan deletion induces defects in platelet aggregation and thrombus formation in mice. Blood 111, 3458–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abrink M., Grujic M., Pejler G. (2004) Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 279, 40897–40905 [DOI] [PubMed] [Google Scholar]
- 67. Chang M. Y., Han C. Y., Wight T. N., Chait A. (2006) Antioxidants inhibit the ability of lysophosphatidylcholine to regulate proteoglycan synthesis. Arterioscler. Thromb. Vasc. Biol. 26, 494–500 [DOI] [PubMed] [Google Scholar]
- 68. Willeaume V., Kruys V., Mijatovic T., Huez G. (1995) Tumor necrosis factor-alpha production induced by viruses and by lipopolysaccharides in macrophages: similarities and differences. J. Inflamm. 46, 1–12 [PubMed] [Google Scholar]
- 69. Parkar A. A., Day A. J. (1997) Overlapping sites on the Link module of human TSG-6 mediate binding to hyaluronan and chrondroitin-4-sulphate. FEBS Lett. 410, 413–417 [DOI] [PubMed] [Google Scholar]
- 70. Heng B. C., Gribbon P. M., Day A. J., Hardingham T. E. (2008) Hyaluronan binding to link module of TSG-6 and to G1 domain of aggrecan is differently regulated by pH. J. Biol. Chem. 283, 32294–32301 [DOI] [PubMed] [Google Scholar]
- 71. Mahoney D. J., Mulloy B., Forster M. J., Blundell C. D., Fries E., Milner C. M., Day A. J. (2005) Characterization of the interaction between tumor necrosis factor-stimulated gene-6 and heparin: implications for the inhibition of plasmin in extracellular matrix microenvironments. J. Biol. Chem. 280, 27044–27055 [DOI] [PubMed] [Google Scholar]
- 72. Mahoney D. J., Mikecz K., Ali T., Mabilleau G., Benayahu D., Plaas A., Milner C. M., Day A. J., Sabokbar A. (2008) TSG-6 regulates bone remodeling through inhibition of osteoblastogenesis and osteoclast activation. J. Biol. Chem. 283, 25952–25962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tan K. T., McGrouther D. A., Day A. J., Milner C. M., Bayat A. (2011) Characterization of hyaluronan and TSG-6 in skin scarring: differential distribution in keloid scars, normal scars and unscarred skin. J. Eur. Acad. Dermatol. Venereol. 25, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Milner C. M., Day A. J. (2003) TSG-6: a multifunctional protein associated with inflammation. J. Cell Sci. 116, 1863–1873 [DOI] [PubMed] [Google Scholar]
- 75. Mahoney D. J., Swales C., Athanasou N. A., Bombardieri M., Pitzalis C., Kliskey K., Sharif M., Day A. J., Milner C. M., Sabokbar A. (2011) TSG-6 inhibits osteoclast activity via an autocrine mechanism and is functionally synergistic with osteoprotegerin. Arthritis Rheum. 63, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 76. Wisniewski H. G., Hua J. C., Poppers D. M., Naime D., Vilcek J., Cronstein B. N. (1996) TNF/IL-1-inducible protein TSG-6 potentiates plasmin inhibition by inter-α-inhibitor and exerts a strong anti-inflammatory effect in vivo. J. Immunol. 156, 1609–1615 [PubMed] [Google Scholar]
- 77. Wisniewski H. G., Vilcek J. (1997) TSG-6: an IL-1/TNF-inducible protein with anti-inflammatory activity. Cytokine Growth Factor Rev. 8, 143–156 [DOI] [PubMed] [Google Scholar]
- 78. Mindrescu C., Le J., Wisniewski H. G., Vilcek J. (2005) Up-regulation of cyclooxygenase-2 expression by TSG-6 protein in macrophage cell line. Biochem. Biophys. Res. Commun. 330, 737–745 [DOI] [PubMed] [Google Scholar]
- 79. Getting S. J., Mahoney D. J., Cao T., Rugg M. S., Fries E., Milner C. M., Perretti M., Day A. J. (2002) The link module from human TSG-6 inhibits neutrophil migration in a hyaluronan- and inter-α-inhibitor-independent manner. J. Biol. Chem. 277, 51068–51076 [DOI] [PubMed] [Google Scholar]
- 80. Lee R. H., Pulin A. A., Seo M. J., Kota D. J., Ylostalo J., Larson B. L., Semprun-Prieto L., Delafontaine P., Prockop D. J. (2009) Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell 5, 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Oh J. Y., Roddy G. W., Choi H., Lee R. H., Ylöstalo J. H., Rosa R. H., Jr., Prockop D. J. (2010) Anti-inflammatory protein TSG-6 reduces inflammatory damage to the cornea following chemical and mechanical injury. Proc. Natl. Acad. Sci. U.S.A. 107, 16875–16880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kuznetsova S. A., Mahoney D. J., Martin-Manso G., Ali T., Nentwich H. A., Sipes J. M., Zeng B., Vogel T., Day A. J., Roberts D. D. (2008) TSG-6 binds via its CUB_C domain to the cell-binding domain of fibronectin and increases fibronectin matrix assembly. Matrix Biol. 27, 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Szántó S., Bárdos T., Gál I., Glant T. T., Mikecz K. (2004) Enhanced neutrophil extravasation and rapid progression of proteoglycan-induced arthritis in TSG-6-knockout mice. Arthritis Rheum. 50, 3012–3022 [DOI] [PubMed] [Google Scholar]
- 84. Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Kristensen T., Wisniewski H. G., Thøgersen I. B., Enghild J. J. (2008) TSG-6 transfers proteins between glycosaminoglycans via a Ser28-mediated covalent catalytic mechanism. J. Biol. Chem. 283, 33919–33926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Choi H., Lee R. H., Bazhanov N., Oh J. Y., Prockop D. J. (2011) Anti-inflammatory protein TSG-6 secreted by activated MSCs attenuates zymosan-induced mouse peritonitis by decreasing TLR2/NF-κB signaling in resident macrophages. Blood 118, 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Toeda K., Nakamura K., Hirohata S., Hatipoglu O. F., Demircan K., Yamawaki H., Ogawa H., Kusachi S., Shiratori Y., Ninomiya Y. (2005) Versican is induced in infiltrating monocytes in myocardial infarction. Mol. Cell. Biochem. 280, 47–56 [DOI] [PubMed] [Google Scholar]
- 87. Wingrove J. A., Daniels S. E., Sehnert A. J., Tingley W., Elashoff M. R., Rosenberg S., Buellesfeld L., Grube E., Newby L. K., Ginsburg G. S., Kraus W. E. (2008) Correlation of peripheral-blood gene expression with the extent of coronary artery stenosis. Circ. Cardiovasc. Genet. 1, 31–38 [DOI] [PubMed] [Google Scholar]
- 88. Olsen N. J., Moore J. H., Aune T. M. (2004) Gene expression signatures for autoimmune disease in peripheral blood mononuclear cells. Arthritis Res. Ther. 6, 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shou J., Bull C. M., Li L., Qian H. R., Wei T., Luo S., Perkins D., Solenberg P. J., Tan S. L., Chen X. Y., Roehm N. W., Wolos J. A., Onyia J. E. (2006) Identification of blood biomarkers of rheumatoid arthritis by transcript profiling of peripheral blood mononuclear cells from the rat collagen-induced arthritis model. Arthritis. Res. Ther. 8, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lang R., Patel D., Morris J. J., Rutschman R. L., Murray P. J. (2002) Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 169, 2253–2263 [DOI] [PubMed] [Google Scholar]
- 91. Martinez F. O., Gordon S., Locati M., Mantovani A. (2006) Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]
- 92. Vehmas A., Lieu J., Pardo C. A., McArthur J. C., Gartner S. (2004) Amyloid precursor protein expression in circulating monocytes and brain macrophages from patients with HIV-associated cognitive impairment. J. Neuroimmunol. 157, 99–110 [DOI] [PubMed] [Google Scholar]
- 93. Coulson E. J., Paliga K., Beyreuther K., Masters C. L. (2000) What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem. Int. 36, 175–184 [DOI] [PubMed] [Google Scholar]
- 94. Li Q. X., Fuller S. J., Beyreuther K., Masters C. L. (1999) The amyloid precursor protein of Alzheimer disease in human brain and blood. J. Leukoc. Biol. 66, 567–574 [DOI] [PubMed] [Google Scholar]
- 95. Russell D. L., Salustri A. (2006) Extracellular matrix of the cumulus-oocyte complex. Semin. Reprod. Med. 24, 217–227 [DOI] [PubMed] [Google Scholar]
- 96. Yoshida M., Sai S., Marumo K., Tanaka T., Itano N., Kimata K., Fujii K. (2004) Expression analysis of three isoforms of hyaluronan synthase and hyaluronidase in the synovium of knees in osteoarthritis and rheumatoid arthritis by quantitative real-time reverse transcriptase polymerase chain reaction. Arthritis Res. Ther. 6, R514–R520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. de La Motte C. A., Hascall V. C., Calabro A., Yen-Lieberman B., Strong S. A. (1999) Mononuclear leukocytes preferentially bind via CD44 to hyaluronan on human intestinal mucosal smooth muscle cells after virus infection or treatment with poly(I·C). J. Biol. Chem. 274, 30747–30755 [DOI] [PubMed] [Google Scholar]
- 98. de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., Strong S. A. (2003) Mononuclear leukocytes bind to specific hyaluronan structures on colon mucosal smooth muscle cells treated with polyinosinic acid:polycytidylic acid: inter-α-trypsin inhibitor is crucial to structure and function. Am. J. Pathol. 163, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Winberg J. O., Berg E., Kolset S. O., Uhlin-Hansen L. (2003) Calcium-induced activation and truncation of promatrix metalloproteinase-9 linked to the core protein of chondroitin sulfate proteoglycans. Eur. J. Biochem. 270, 3996–4007 [DOI] [PubMed] [Google Scholar]
- 100. Lemire J. M., Chan C. K., Bressler S., Miller J., LeBaron R. G., Wight T. N. (2007) Interleukin-1β selectively decreases the synthesis of versican by arterial smooth muscle cells. J. Cell. Biochem. 101, 753–766 [DOI] [PubMed] [Google Scholar]
- 101. Kohro T., Tanaka T., Murakami T., Wada Y., Aburatani H., Hamakubo T., Kodama T. (2004) A comparison of differences in the gene expression profiles of phorbol 12-myristate 13-acetate differentiated THP-1 cells and human monocyte-derived macrophage. J. Atheroscler. Thromb. 11, 88–97 [DOI] [PubMed] [Google Scholar]