Background: Engulfment of apoptotic cell is facilitated by Rac1-GTPase and phosphatidylinositol 3-kinase (PI3K). Beclin 1 is known to form PI3K complex.
Results: Beclin 1 was bound to Rac1, recruited to early phagocytotic cups, and required for apoptotic cell internalization.
Conclusion: Beclin 1 regulates apoptotic cell engulfment in cooperation with Rac1.
Significance: This report reveals a novel function of Beclin 1 in apoptotic cell clearance.
Keywords: Autophagy, Electron Microscopy (EM), Phagocytosis, Phosphatidylinositol 3-Kinase, Rac1
Abstract
Efficient apoptotic cell engulfment is important for both tissue homeostasis and immune response in mammals. In the present study, we report that Beclin 1 (a regulator of autophagy) is required for apoptotic cell engulfment. The engulfment process was largely abolished in Beclin 1 knock-out cells, and Beclin 1 knockdown significantly decreased apoptotic cell internalization in macrophage and fibroblast cell lines. Beclin 1 was recruited to the early phagocytic cup along with the generation of phosphatidylinositol 3-phosphate and Rac1, which regulates actin dynamics in lamellipodia. No lamellipodia were formed in Beclin 1 knock-out cells, and Beclin 1 knockdown completely inhibited the promotion of engulfment by ectopic expression of Rac1. Beclin 1 was co-immunoprecipitated with Rac1. These data indicate that Beclin 1 coordinates actin dynamics and membrane phospholipid synthesis to promote efficient apoptotic cell engulfment.
Introduction
Apoptosis is the process of programmed cell death that plays an important role in various biological events of all multicellular organisms, including development, maintenance of tissue homeostasis, and elimination of harmful cells. Rapid and efficient removal of apoptotic cells is essential to avoid the leakage of immunogenic contents from dying cells (1). Therefore, apoptotic cells are engulfed efficiently by professional phagocytes such as macrophages and monocytes or less efficiently by nonprofessional phagocytes such as fibroblasts and epithelial cells. Phagocytes engulf apoptotic cells but not healthy cells, indicating that phagocytes recognize a special signal from dying cells: the so-called “eat me” signal. Phosphatidylserine (PS)3 is considered to be the best candidate for an “eat me” signal, although various other molecules have been proposed. PS is exposed on the cell surface only when cells undergo apoptosis. It binds to the PS receptor directly (2, 3) or indirectly through bridging molecules such as milk fat globule EGF factor 8 or growth arrest-specific 6 so that the apoptotic cells are recognized by phagocytes (4, 5).
Once the apoptotic cells have been recognized, a cascade of intracellular molecules is activated in phagocytes, leading to rearrangement of the cytoskeleton for efficient engulfment. The molecules involved in apoptotic cell engulfment have been identified by genetic analysis of Caenorhabditis elegans. Evolutionarily conserved pathways that function upstream of CED-10, which corresponds to mammalian Rac1, a Rho family GTPase, have been described (6, 7). Various small GTPases such as Rac1, RhoA, and Rab5 are required for apoptotic cell engulfment (8). FRET analysis has revealed that Rac1 is activated in phagocytic cups only when a dead cell is being internalized, whereas Rab5 regulates the process by which phagosomes containing the dead cell fuse with lysosomes (9, 10).
Autophagy is a bulk degradation system that involves the transport of cytoplasmic components from autophagosomes to lysosomes, which recycle cellular materials. In the first step of autophagy, cytoplasmic constituents are sequestered by an isolation membrane to form an autophagosome. The process of autophagosome maturation after enclosure of the isolation membrane is topologically similar to that of phagosome maturation. Both autophagosomes and phagosomes undergo fusion with lysosomes, followed by acidification and degradation of their contents. Apart from VPS34, few molecules show overlap between the processes of autophagosome and phagosome maturation.
Beclin 1 is a coiled-coil protein that is well known as a regulator of autophagy in mammalian cells (11). Beclin 1 binds to Vps34/class III PI3K to generate phosphatidylinositol 3-phosphate (PI(3)P), which plays an important role in membrane trafficking (12). PI(3)P signaling is involved in the formation and maturation of both autophagosomes and phagosomes (13, 14). Vps34 also regulates apoptotic cell engulfment (15). Although it has been reported that Beclin 1 is involved in the clearance of apoptotic cells during embryonic development by regulating the expression of PS (16), whether or not Beclin 1 plays a role in apoptotic cell engulfment by phagocytes is unclear. In the present study, we report that Beclin 1 is involved in the clearance of apoptotic cells by regulating their engulfment by phagocytes in cooperation with Rac1 GTPase.
EXPERIMENTAL PROCEDURES
Cell Culture
ES cells were maintained in Glasgow minimum essential medium (Invitrogen) containing 15% FBS (Invitrogen) and 1,000 units/ml leukemia inhibitory factor (LIF) (Invitrogen). J774A1 cells, mouse embryonic fibroblasts (MEFs), Plat-E cells, and NIH-3T3 cells were maintained in DMEM (Nacalai Tesque) containing 10% FBS. Thymocytes were collected from C57BL/6 mice, treated with red blood cell lysis buffer (17 mm Tris-HCl, pH 7.5, and 144 mm NH4Cl), and maintained in RPMI medium (Nacalai Tesque) containing 10% FBS. Jurkat cells were maintained in RPMI medium containing 10% FBS.
Construction of Expression Plasmids
The full-length coding sequences for mouse Rac1, Rab5, and Beclin 1 were prepared by PCR from a mouse embryonic brain cDNA library and were inserted into the retroviral expression pLPC and pWzl vectors with an N-terminal Myc tag or HA tag. The cDNAs were verified by DNA sequencing. cDNAs for the dominant-negative forms of Rac1 (Rac1 T17N) and Rab5 (Rab5 S34N) and the constitutive active form of Rac1 (Rac1 Q61L) were prepared by PCR-mediated mutagenesis using a QuikChange Lightning site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The retroviral GFP fusion protein expression vector pMSCV-C-EGFP-zeo was constructed by replacing the puromycin resistance gene of pMSCV puro (Clontech) with Zeocin and subsequently cloning the enhanced GFP (EGFP) sequence into the multicloning site. The stop codon of mouse Beclin 1 cDNA was removed, and it was cloned into the pMSCV-C-EGFP-zeo vector. Two retroviral mCherry fusion expression vectors, pBabe-N-mCherry and pMSCV-N-mCherry, were constructed by cloning mCherry without the stop codon into the pBabe and pMSCV-Hygro (Clontech) vectors. The start codon was removed from cDNAs for mouse Rac1 and Rab5, which were then cloned into the pBabe-N-mCherry vector. The coding sequence of the FYVE zinc finger domain of mouse hepatocyte growth factor-regulated tyrosine kinase substrate was tandemly cloned into the pMSCV-N-mCherry-Hygro vector. The Rac1 biosensor, pBabe-mCherry-PBD, was made by replacing Ypet in pBabe-Ypet-PBD plasmid (Addgene plasmid 24533) (17) with mCherry.
Retroviral Vector Production and Infection
Plat-E retroviral packaging cells (18) were transfected with the desired DNA plasmids. At 48, 60, and 72 h after transfection, the viral medium was collected and used to infect cells in the presence of polybrene (4 μg/ml). At 48 h after the last infection, the cells were selected with the desired drug and then were maintained in the selection medium.
In Vitro Phagocytosis Assay and Image Analysis
Mouse thymocytes and Jurkat cells were used as the targets. Thymocytes were stained with 5 μm 5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine (CMTMR) (Molecular Probes), treated with 10 μm dexamethasone (Sigma) for 3 h, washed thoroughly, and used for the phagocytosis assay. Jurkat cells were stained with 5 μm CMTMR or 2 μm CypHer5E (GE Healthcare Life Science), exposed to UV light (UV cross-linker, UVP) at 200 J/m2, incubated for 12 h, washed thoroughly, and used for the phagocytosis assay. J774A1 or NIH-3T3 cells were stained with 1 μm 5-chloromethylfluorescein diacetate (CMFDA) (Molecular Probes). The target cells (1.5 × 106/well) were added to CMFDA-labeled J774A1 or NIH-3T3 cells (3 × 105/well) in a 24-well plate and then incubated for 3 h at 37 °C. J774A1 or NIH-3T3 cells were washed thoroughly with DMEM, collected, and analyzed by flow cytometry using a FACSCalibur (BD Biosciences). The data were compared by nonpaired Student's t tests using Prism 5 software (GraphPad Software). All of the p values were two-tailed. p values of <0.05 were considered statistically significant for all experiments.
Fluorescence images were acquired with an inverted microscope (Olympus IX71) equipped with a cooled CCD camera (Olympus DP71) using 40× (0.85 NA) and 60× (1.2 NA) objective or with IN Cell Analyzer 1000 (GE Healthcare Life Science) equipped with a 20× objective. Staining of F-actin was performed using Alexa Fluor568 phalloidin (Molecular Probes) according to the manufacturer's instructions. The images were processed using Photoshop CS3 (Adobe). Time lapse images were acquired with an inverted microscope (Olympus IX81) equipped with a cooled CCD camera (Hamamatsu Photonics ORCA-R2) using 40× (0.95 NA) objective and were processed using MetaMorph software (Molecular Devices).
Construction of Retroviral shRNA Hairpin Expression Vectors
Synthetic oligonucleotides for shRNAs targeted for Beclin 1 and ATG5 were cloned into retroviral shRNA expression pLMP vector (19). The sequences of the shRNA targets were as follows: shBeclin1 #1, ACAGCTCCATTACTTACCA; shBeclin1 #2, ATACTGTGTGCGACGTGGA; and shATG5, GCATTATCCAATTGGTTTA.
Electron Microscopy
Electron microscopy was performed as reported previously (20). Briefly, the cells were fixed by a conventional method (1.5% paraformaldehyde and 3% glutaraldehyde in 0.1 m phosphate buffer, pH 7.3, followed by an aqueous solution of 1% OsO4). The fixed cells were embedded in Epon 812, after which thin sections (70–80 nm) were cut and stained with uranyl acetate and lead citrate for observation under a JEOL-1010 electron microscope (JEOL) at 80 kV.
Co-immunoprecipitation
Cell extracts were prepared from 293T cells that were transfected with HA-Beclin 1, Myc-Rac1, Myc-Rab5, and Myc-Cdc42 and incubated with an anti-HA antibody for 2 h at 4 °C. Immunoprecipitates were prepared by incubation with Dynabeads protein A (Invitrogen) and subjected to immunoblot with anti-Myc antibody.
Reagents and Antibodies
An antibody for Beclin 1 was purchased from BD Biosciences; anti-ATG5 and anti-ULK1 antibodies were obtained from Sigma; anti-active caspase 3 was from R & D Systems; anti-HA antibody was from Abcam; and anti-GAPDH and anti-Myc antibodies were purchased from Santa Cruz Biotechnology. Etoposide and 3-methyladenine were purchased from Sigma. siRNAs against Beclin 1 and ULK1 were purchased from Qiagen, and siRNA against ATG5 was obtained from Dharmacon.
RESULTS
Inhibition of Apoptotic Cell Engulfment by Beclin 1−/− ES Cells
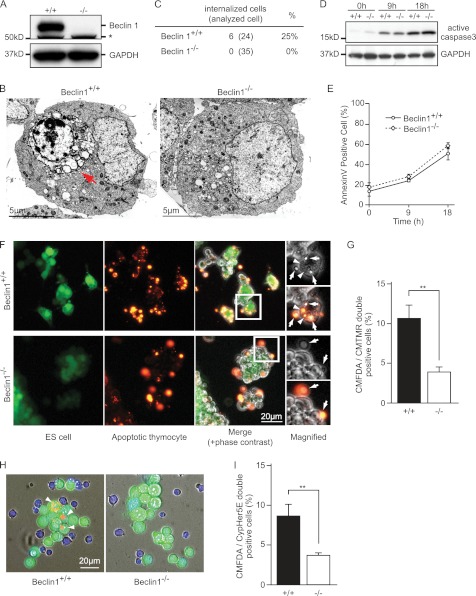
During morphological analysis of the apoptotic process of Beclin 1+/+ or Beclin 1−/− ES cells (21) (Fig. 1A), we found that apoptotic Beclin 1+/+ ES cells were engulfed by other ES cells, whereas Beclin 1−/− ES cell engulfment was uncommon (Fig. 1, B and C). There were no significant differences in the induction of apoptosis and surface exposure of PS between Beclin 1+/+ and Beclin 1−/− ES cells treated with etoposide, a reagent commonly used to induce apoptosis (Fig. 1, D and E). This observation led us to hypothesize that Beclin 1 regulates apoptotic cell engulfment.
FIGURE 1.
Inhibition of apoptotic cell engulfment by Beclin 1−/− ES cells. A, immunoblot of Beclin 1 expression in ES cells. GAPDH was used as a loading control. *, nonspecific signal. B, EM observations of apoptotic cell engulfment by ES cells. Beclin 1+/+ and Beclin 1−/− ES cells were treated with 20 μm etoposide for 18 h. The cells were then harvested, concentrated by centrifugation, and processed for EM analysis. The red arrow indicates an internalized apoptotic cell. C, quantification of internalized cells in B. D, immunoblot of active caspase 3 in ES cells. Beclin 1+/+ and Beclin 1−/− ES cells were treated with 20 μm etoposide for the indicated times at 37 °C and then subjected to immunoblotting. GAPDH was used as a loading control. E, the level of exposure of “eat me” signal on apoptotic ES cells. Beclin 1+/+ and Beclin 1−/− ES cells were treated with 20 μm etoposide and subjected to the annexin V assay. Annexin V-bound cells were analyzed by flow cytometry. The graph shows the average values of three independent experiments and the S.D. (error bars). F, apoptotic cell engulfment by ES cells. Beclin 1+/+ and Beclin 1−/− ES cells were stained with CMFDA (green) and incubated with CMTMR-labeled apoptotic thymocytes (orange) for 3 h at 37 °C. The merged images include phase contrast images. The white boxes indicate the sites of the magnified images. The arrows indicate the attached apoptotic thymocytes. The arrowheads indicate the engulfed thymocytes. G, quantification of engulfment in F. The average levels of engulfment were obtained from three independent experiments. The bars indicate S.D. **, p < 0.01. H, engulfment assay using pH-sensitive CypHer5E dye. CMFDA-stained ES cells (green) were incubated with Cypher5E-labeled apoptotic Jurkat cells for 3 h at 37 °C. The images are merged DAPI (blue) and phase contrast images. ES cells are green, and apoptotic Jurkat cells are blue, whereas Jurkat cells that have been engulfed and transported to lysosomes are orange and are indicated by the arrowheads. I, quantification of the engulfed cells in H. The values represent the means ± S.D. (n = 3). **, p < 0.01).
To test this hypothesis, we set up a phagocytosis assay using Beclin 1 ES cells. CMFDA dye-labeled healthy ES cells (green) were incubated with CMTMR dye-labeled apoptotic thymocytes (orange) for 3 h, washed thoroughly, and observed under a fluorescence microscope. Wild-type ES cells occasionally engulfed apoptotic thymocytes (Fig. 1F, arrowheads). In contrast, Beclin 1−/− ES cells seldom engulfed dead thymocytes, even when they were bound to ES cells (Fig. 1F, arrows). We also quantified the extent of engulfment by measuring CMTMR-positive ES cells and observed that Beclin 1−/− ES cells performed significantly less engulfment than control ES cells (Fig. 1G).
To more easily distinguish ES cells internalizing apoptotic cells from those just bound to dying cells, we used a pH-sensitive dye (CypHer5E) that is nonfluorescent at neutral pH and shows increasing fluorescence under acidic conditions, such as those inside lysosomes. When CypHer5E-labeled apoptotic Jurkat cells were incubated with control ES cells, the engulfed Jurkat cells displayed red fluorescence after being transported to the lysosomes (Fig. 1H, left panel, arrowheads). In contrast, Beclin 1−/− ES cells showed significantly less acidification of internalized apoptotic cells (Fig. 1, H and I). On the basis of these findings, we conclude that Beclin 1 is required for the efficient apoptotic cell engulfment by ES cells.
Inhibition of Engulfment by Professional and Nonprofessional Phagocytes by Beclin 1 Knockdown
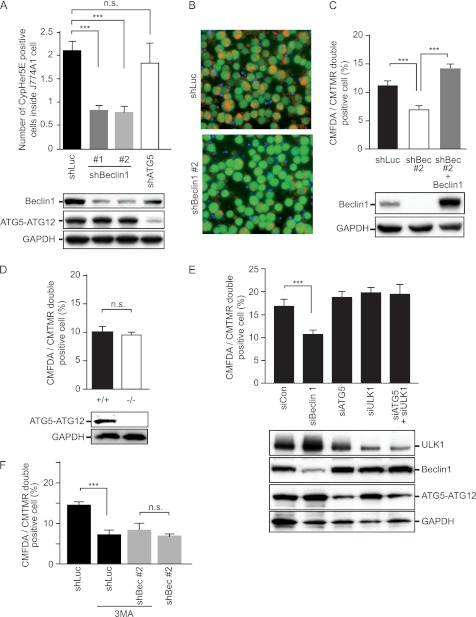
Next, we investigated the role of Beclin 1 in apoptotic cell engulfment by professional phagocytes. Beclin 1 was knocked down in a mouse macrophage cell line (J774A1) by retroviral introduction of Beclin 1 shRNA. J774A1 cells were then labeled with CMFDA and incubated with Cypher5E-labeled apoptotic Jurkat cells. Beclin 1 knockdown inhibited apoptotic cell engulfment (Fig. 2, A and B).
FIGURE 2.
Inhibition of engulfment by Beclin 1 knockdown in professional and nonprofessional phagocytes. A, effect of Beclin 1 knockdown on macrophages. CMFDA-stained J774A1 cells introduced with shRNAs for luciferase (shLuc, used as a nonsilencing control), Beclin 1 (shBeclin1 #1 and #2), and ATG5 (shATG5) were incubated with CypHer5E-stained apoptotic Jurkat cells for 3 h. Engulfment was assessed by CMFDA and CypHer5E double-positive cells quantified by flow cytometry. The values represent the means ± S.D. (n = 3). ***, p < 0.001; n.s., not significant. Efficiency of Beclin 1 and ATG5 knockdown was confirmed by immunoblotting, as shown in the bottom panel. GAPDH served as a loading control. B, representative images of the engulfment assay shown in A. J774A1 cells were introduced with the indicated shRNA. The images are merged CMFDA (green; J774A1 cells), DAPI (blue; not engulfed Jurkat cells), and CypHer5E (orange; engulfed Jurkat cells) images. C, effect of Beclin 1 knockdown on fibroblast cells. CMFDA-stained NIH-3T3 cells with the indicated shRNA were incubated with CMTMR-stained apoptotic Jurkat cells for 3 h at 37 °C. The expression of Beclin 1 was rescued with Beclin 1 cDNA (shBeclin1 #2 + Beclin 1). Engulfment was assessed by CMFDA and CMTMR double-positive cells quantified by flow cytometry. The values represent the means ± S.D. (n = 3). ***, p < 0.001). Beclin 1 knockdown and reintroduction was confirmed by immunoblotting, as shown in the bottom panel. D, engulfment by ATG5+/+ and Beclin 1−/− MEFs. CMFDA-stained MEFs were incubated with CMTMR-stained apoptotic Jurkat cells for 3 h at 37 °C, and the engulfment efficiency was quantified by flow cytometry. The values represent the means ± S.D. (n = 3). n.s., not significant. ATG5 knock-out was confirmed by immunoblotting, as shown in the bottom panel. E, effect of knockdown of autophagy-related genes in NIH-3T3 cells. CMFDA-stained NIH-3T3 cells transfected with the indicated siRNAs were subjected to the phagocytosis assay. The values represent the means ± S.D. (n = 3). ***, p < 0.001. The knockdown efficiency was confirmed by immunoblotting, as shown in the bottom panel. F, effect of the class III PI3K inhibitor on NIH-3T3 cells. CMFDA-stained NIH-3T3 cells with the indicated shRNAs were subjected to the phagocytosis assay with or without 10 mm 3MA. The values represent the means ± S.D. (n = 3). ***, p < 0.001; n.s., not significant.
On testing a mouse fibroblast cell line (NIH-3T3), we observed that Beclin 1 knockdown also inhibited engulfment by nonprofessional phagocytes. When CMFDA-labeled NIH-3T3 cells were incubated with CMTMR-labeled apoptotic Jurkat cells, NIH-3T3 cells expressing Beclin 1 shRNA showed less engulfment (Fig. 2C). To exclude an off target effect of Beclin 1 shRNA, Beclin 1 expression was rescued exogenously. The target sequence of Beclin 1 shRNA (shBeclin1 #2) was located in the 3′-UTR of Beclin 1 mRNA so that ectopic expression of Beclin 1 cDNA would not be affected. Complementation of Beclin 1 expression completely reversed the inhibition of engulfment seen with Beclin 1 shRNA (Fig. 2C). We also examined the effect of Beclin 1 overexpression by NIH-3T3 cells. As expected, Beclin 1 overexpression enhanced apoptotic cell engulfment (see Fig. 4A). Taken together, these results indicate that Beclin 1 is required for the efficient apoptotic cell engulfment by professional and nonprofessional phagocytes.
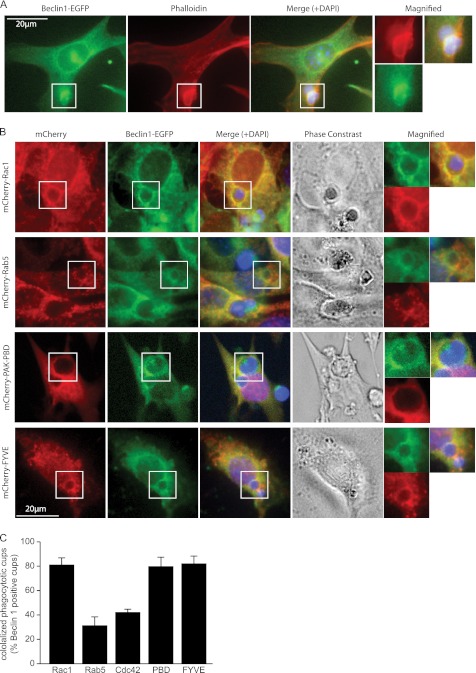
FIGURE 4.
Recruitment of Beclin 1 to the early phagocytic cup with PI3K and Rac1. A, localization of Beclin 1 during engulfment. NIH-3T3 cells introduced with EGFP-fused Beclin 1 were incubated with apoptotic Jurkat cells and stained with Alexa 568-phalloidin (red). Merged images also include the DAPI-stained images (blue). The open white boxes indicate the sites of the magnified images. B, co-localization of Beclin 1 with Rac1, Rab5, PAK-PBD, and FYVE at the phagocytic cup. EGFP-fused Beclin 1 and mCherry-fused Rac1, Rab5, PAK-PBD, and the FYVE domain were introduced into NIH-3T3 cells that were then incubated with apoptotic Jurkat cells for 3 h. C, quantification of co-localization in B. The bars represent the mean values from three independent experiments ± S.D.
This effect of Beclin 1 can be explained as either a direct effect on engulfment or a consequence of the inhibition of autophagy. To distinguish between these two possibilities, we assessed the effect of ATG5, another essential regulator of macroautophagy. As shown in Fig. 2A, ATG5 shRNA did not influence engulfment by J774A1 cells. Similar results were also observed on comparing ATG5−/− MEFs with ATG5+/+ MEFs (Fig. 2D). Recently, we discovered ATG5-independent alternative macroautophagy that is dependent on Beclin 1 and ULK1 (20) and examined the involvement of this alternative macroautophagy in apoptotic cell engulfment. Knockdown of ULK1, ATG5, or both had no effect on apoptotic cell internalization; however, only Beclin 1 single knockdown inhibited engulfment (Fig. 2E). Taken together, these data indicate that Beclin 1 and not ATG5-dependent or ATG5-independent alternative macroautophagy is required for engulfment by both professional and nonprofessional phagocytes.
Because Beclin 1 is part of the PI3K complex, we investigated the role of PI3K activity in apoptotic cell engulfment. We tested 3-methyladenine (3MA), a class III PI3K inhibitor, to study the requirement of PI3K activity for engulfment. As expected, 3MA inhibited apoptotic cell engulfment in NIH-3T3 cells. The inhibition of engulfment by 3MA was equivalent to that by Beclin 1 knockdown and had no additive effect on shBeclin1 (Fig. 2F), indicating that PI3K activity is required for engulfment in a Beclin 1-dependent manner.
Morphological Analysis of Apoptotic Cell Engulfment by ES Cells
We next investigated how Beclin 1 regulates apoptotic cell engulfment. The engulfment process can be divided into three steps: recognition, internalization, and degradation. To identify the step where Beclin 1 is active, we first analyzed the morphological changes of ES cells. Within 2 days of LIF removal, most ES cells became flat; therefore, it was easier to observe engulfment in detail by microscopy. Under these conditions, there was no significant difference in proliferation between Beclin 1+/+ and Beclin 1−/− cells.
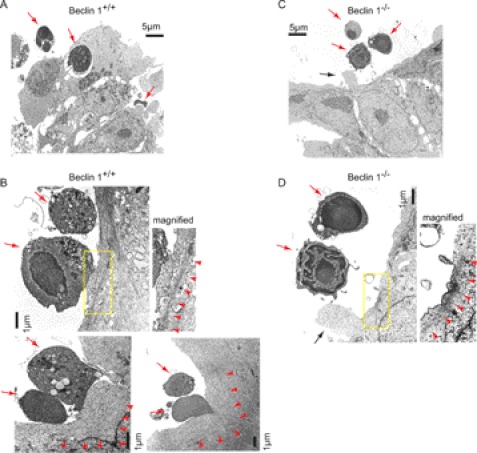
When LIF-free ES cells were incubated with apoptotic thymocytes, Beclin 1+/+ ES cells efficiently engulfed the apoptotic thymocytes (Fig. 3A) in a similar way to ES cells cultured with LIF (Fig. 1). In contrast, very little engulfment was seen with Beclin 1−/− ES cells (Fig. 3C). Morphological analysis revealed that engulfment occurred at the site of lamellipodia lined by polymerized actin filaments in Beclin 1+/+ cells (Fig. 3B, arrowheads). After the plasma membrane of an ES cell came into close contact with an apoptotic cell, the contact surface expanded, and lamellipodia were formed that later encircled the target cell (Fig. 3B).
FIGURE 3.
Morphological analysis of apoptotic cell engulfment by ES cells. EM observation of apoptotic thymocyte engulfment by Beclin 1+/+ (A and B) and Beclin 1−/− (C and D) ES cells. ES cells cultured without LIF were incubated with apoptotic thymocytes for 3 h and subjected to EM analysis. The red arrows indicate apoptotic thymocytes. The yellow boxes indicate the sites of the magnified images. The red arrowheads indicate polymerized actin filaments. The black arrows indicate filopodia.
In contrast, Beclin 1−/− ES cells failed to form lamellipodia and increase the surface in contact with the apoptotic cells (Fig. 3D). Beclin 1−/− cells still formed polymerized actin filaments adjacent to apoptotic cells (Fig. 3D, red arrowheads), as well as small ruffles and filopodia (Fig. 3D, black arrow), suggesting that the apoptotic cells were recognized and that engulfment was initiated, but the formation of lamellipodia and engulfment did not occur.
Recruitment of Beclin 1 to Early Phagocytic Cup with PI3K and Rac1
To investigate the role of Beclin 1 in apoptotic cell internalization, we examined its localization during the engulfment process. NIH-3T3 cells were transfected with EGFP-fused Beclin 1 (Beclin 1-EGFP) and incubated with apoptotic Jurkat cells. Interestingly, Beclin 1 accumulated around engulfed apoptotic cells, and its localization coincided with that of actin filaments detected by Alexa 568-phalloidin (Fig. 4A). The engulfment process is known to be initiated by actin filament rearrangement that can be visualized with phalloidin. Therefore, these findings indicate that Beclin 1 was recruited to the early phagocytic cup.
Because the polymerization of actin filaments is regulated by Rac1, we next examined the localization of Beclin 1 and Rac1 to determine whether Beclin 1 cooperated with Rac1. NIH-3T3 cells were co-introduced with Beclin 1-EGFP and mCherry-fused Rac1 (mCherry-Rac1) and then incubated with apoptotic Jurkat cells. As previously reported (9), Rac1 was observed around the early phagocytic cup, whereas Beclin 1 also accumulated on the phagocytic cup and co-localized with Rac1 (Fig. 4, B and C). In contrast, there was little Beclin 1 co-localization with other small GTPases such as Rab5 and Cdc42 (Fig. 4, B and C). We also investigated whether Beclin 1 co-localized with an active form of Rac1. For this purpose, we used the mCherry-fused p21-binding domain (PBD) of p21-activated kinase 1 (PAK1) because PAK-PBD binds to Rac1 only when Rac1 is activated as a GTP-bound form (22). We found that mCherry-PAK-PBD was accumulated at the early phagocytic cups and co-localized with Beclin 1 (Fig. 4, B and C).
It has been reported that PI(3)P is generated by the VPS34-PI3K complex on the phagosome membrane during apoptotic cell engulfment (15). The FYVE zinc-finger domain binds to PI(3)P. Therefore, a fluorescent protein-fused FYVE domain is widely used as a marker for PI(3)P. Accordingly, we examined the co-localization of an mCherry-fused FYVE domain (mCherry-FYVE) with Beclin 1. mCherry-FYVE accumulation was observed around the phagocytic cups, and there was prominent co-localization with Beclin 1 (Fig. 4, B and C), indicating that Beclin 1 formed a complex with active PI3K in the early phagocytic cup in cooperation with active Rac1.
Regulation of Engulfment by Beclin 1 in Cooperation with Rac1
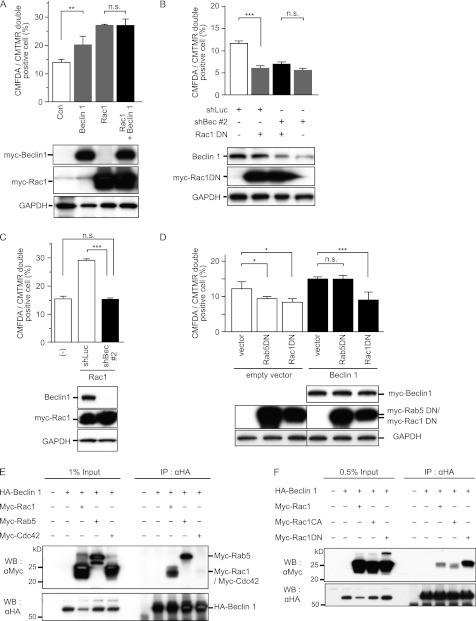
Next, we examined the functional relationship between Beclin 1 and Rac1 during apoptotic cell engulfment. It has been reported that Rac1 is involved in apoptotic cell internalization and that ectopic expression of Rac1 enhances engulfment by NIH-3T3 cells. As reported previously (8), engulfment by NIH-3T3 cells was increased by Rac1 overexpression (Fig. 5A). Beclin 1 overexpression also increased apoptotic cell engulfment but did not show any additive effect on the engulfment promoted by Rac1 (Fig. 5A). Furthermore, as reported previously (8), ectopic expression of the dominant-negative form of Rac1 (Rac1DN) inhibited apoptotic cell engulfment (Fig. 5B), whereas Rac1DN had no additive inhibitory effect on engulfment by Beclin 1 knockdown cells (Fig. 5B). These epistasis results suggest that Beclin 1 and Rac1 are involved in the same molecular pathway for regulating apoptotic cell engulfment.
FIGURE 5.
Regulation of engulfment by Beclin 1 in cooperation with Rac1. A, effect of Beclin 1 and Rac1 overexpression on engulfment. CMFDA-stained NIH-3T3 cells introduced with Myc-tagged Beclin 1, Rac1, or both were subjected to the phagocytosis assay. Engulfment of CMTMR-labeled apoptotic Jurkat cells was quantified by flow cytometry. The values represent the means ± S.D. (n = 3). **, p < 0.01; n.s., not significant. The expression of ectopic Beclin 1 and Rac1 was confirmed by immunoblotting, as shown in the bottom panel. GAPDH served as a loading control. Con, control. B, effect of Beclin 1 knockdown and the dominant-negative form of Rac1 (Rac1DN) on engulfment. After introduction of shBeclin1 #2 (shBec #2), Myc-tagged Rac1DN, or both, NIH-3T3 cells were subjected to the phagocytosis assay. Luciferase shRNA (shLuc) was used as a nonsilencing control. The values represent the means ± S.D. (n = 3). ***, p < 0.001; n.s., not significant. The efficiency of Beclin 1 knockdown and ectopic expression of Rac1DN were confirmed by immunoblotting, as shown in the bottom panel. C, effect of Beclin 1 knockdown on cells with Rac1overexpression. shBeclin1 #2 was introduced into NIH-3T3 cells overexpressing Rac1, and the cells were then subjected to the phagocytosis assay. The values represent the means ± S.D. (n = 3). ***, p < 0.001; n.s., not significant. Knockdown of Beclin 1 and ectopic expression of Rac1 were confirmed by immunoblotting, as shown in the bottom panel. D, effect of Rac1DN on cells with Beclin 1 overexpression. Dominant-negative forms of Rab5 (Rab5DN) and Rac1 (Rac1DN) were introduced into NIH-3T3 cells overexpressing Beclin 1, and the phagocytosis assay was performed. The values represent the means ± S.D. (n = 3). *, p < 0.05; ***, p < 0.001; n.s., not significant. Expression of ectopic Beclin 1, Rab5DN, or Rac1DN was confirmed by immunoblotting, as shown in the bottom panel. E, co-immunoprecipitation of Beclin 1 and small GTPases. 293T cells were transfected with HA-tagged Beclin 1 (HA-Beclin 1) alone or co-transfected with HA-Beclin 1 and the indicated Myc-tagged small GTPases and then subjected to co-immunoprecipitation experiments with anti-HA antibody. Immunoprecipitates were then subjected to immunoblotting with anti-Myc antibody. F, co-immunoprecipitation of Beclin 1 and Rac1. 293T cells were transfected with HA-Beclin 1 alone or co-transfected with HA-Beclin 1 and Myc-tagged Rac1, the constitutive active form of Rac1 (Rac1CA), or Rac1DN and then subjected to co-immunoprecipitation experiments with anti-HA antibody. Immunoprecipitates were then subjected to immunoblotting with anti-Myc antibody. Equivalence of Beclin 1 immunoprecipitation was confirmed by immunoblotting with anti-HA antibody, as shown in the bottom panel. WB, Western blot; IP, immunoprecipitation.
We also tested the effect of the combination of Beclin 1 inhibition and Rac1 overexpression on apoptotic cell engulfment. Rac1-induced increase in engulfment was totally inhibited by co-introduction of Beclin 1 shRNA (Fig. 5C). Moreover, Beclin 1 overexpression increased engulfment, whereas ectopic expression of Rac1DN inhibited this enhancement (Fig. 5D). Rab5 has also been reported to act during apoptotic cell engulfment by being involved in phagosome maturation that occurs downstream of internalization (8, 15). Unlike Rac1DN, Rab5DN did not inhibit the engulfment increased by Beclin 1 (Fig. 5D). This result was compatible with the finding of limited co-localization of Beclin 1 with Rab5 during apoptotic cell engulfment (Fig. 4, B and C) and suggested that Beclin 1 regulates the engulfment at an upstream step of Rab5 such as apoptotic cell internalization.
Beclin 1 could bind to Rac1 when transiently overexpressed in 293T cells (Fig. 5, E and F). Beclin 1 was co-immunoprecipitated with Rac1 when both Beclin 1 and Rac1 were overexpressed in 293T cells (Fig. 5E); however, it did not bind to Cdc42, another Rho family GTPase, suggesting that Beclin 1 specifically binds to Rac1. Beclin 1 was bound to both active and inactive forms of Rac1 (Fig. 5F). The interaction between Beclin 1 and active Rac1 was less stable (Fig. 5F) and could not be observed at endogenous levels (data not shown), suggesting that the complex of Beclin 1 and an active form of Rac1 is transient. Beclin 1 was also bound to Rab5, which was previously reported to interact with Beclin 1 to form autophagosomes (23). These data indicate that Beclin 1 contributes to apoptotic cell engulfment in cooperation with the Rac1 complex.
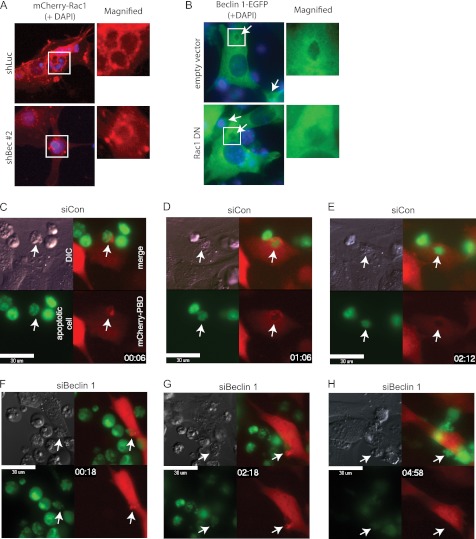
We next investigated how Beclin 1 cooperates with Rac1. During apoptotic cell engulfment, both Beclin 1 and Rac1 were recruited to the early phagocytic cup. Rac1 was still recruited to the early phagocytic cup in NIH-3T3 cells when Beclin 1 was knocked down (Fig. 6, A and F, and supplemental Movie S2), indicating that the initiation of Rac1 activation was independent of Beclin 1. This was consistent with the finding of actin filament polymerization in Beclin 1−/− ES cells (Fig. 3D). However, the recruitment of Beclin 1 was largely diminished by co-introduction of Rac1DN (Fig. 6B), suggesting that Rac1 activation is indispensable to Beclin 1.
FIGURE 6.
Regulation of apoptotic cell internalization by Beclin 1. A, localization of Rac1 in a Beclin 1 knockdown cell. shBeclin1 #2 was introduced into NIH-3T3 cells with mCherry-Rac1 expression. The cells were then incubated with apoptotic Jurkat cells for 3 h. shLuc was used as a control. The images were merged with DAPI-stained images (blue). The open white boxes indicate the sites of the magnified images. B, localization of Beclin 1 in cells with ectopic Rac1DN. Rac1DN was introduced into NIH-3T3 cells with Beclin 1-EGFP expression. The cells were then incubated with apoptotic Jurkat cells for 3 h. The empty vector was used as a control. The images were merged with DAPI-stained images (blue). The open white boxes indicate the sites of the magnified images. The arrows indicate the phagocytic cups. C–H, time lapse images of apoptotic cell engulfment by Beclin 1 knockdown cells. NIH-3T3 cells with mCherry-PAK-PBD expression were transfected with nonsilencing control siRNA (siCon) (C–E) or siRNA for Beclin 1 (siBeclin 1) (F–H) and then incubated with CMFDA dye-labeled apoptotic Jurkat cells (green). The images are merged mCherry (red; NIH-3T3 cells) and CFMDA (green; apoptotic Jurkat cells) images. The embedded time indicates the time after the addition of apoptotic cells. The arrows indicate the site of engulfment. DIC, differential interference contrast. Bars, 30 μm.
To confirm the aforementioned findings, we performed time lapse analysis using NIH-3T3 cells expressing mCherry-PAK-PBD. First, mCherry-PAK-PBD was accumulated at the site of contact with apoptotic cells when NIH-3T3 cells transfected with nonsilencing control siRNA (siCon) were incubated with CMFDA dye-labeled apoptotic Jurkat cells (Fig. 6C, arrows). mCherry-PAK-PBD encircled the target apoptotic cell to form the phagocytic cup within 1 h (Fig. 6D). Apoptotic cell internalization was completed in 2 h, as judged by the loss of contrast of the apoptotic cell outline in differential interference contrast images (Fig. 6E and supplemental Movie S1). In contrast, Beclin 1 knockdown cells failed to circularize mCherry-PAK-PBD surrounding the apoptotic cells (Fig. 6, G and H), whereas mCherry-PAK-PBD was initially accumulated at the site of contact (Fig. 6F), and filopodia and small raffles with enriched mCherry-PAK-PBD were repeatedly formed near the target cells (Fig. 6G and supplemental Movie S2). Beclin 1 knockdown cells failed to complete engulfment and finally released the target cells (Fig. 6H). Taken together, these observations indicate that Beclin 1 is recruited by active Rac1 at the site of initiation of engulfment and promotes the proper formation of lamellipodia in cooperation with Rac1 to facilitate apoptotic cell internalization.
DISCUSSION
In the present study, we showed that Beclin 1, a well known regulator of autophagy, plays another role in apoptotic cell engulfment. Beclin 1 knock-out or knockdown led to decreased engulfment by macrophages, fibroblasts, and ES cells. Previously, Qu et al. (16) reported that Beclin 1 is required for the clearance of apoptotic cells during embryonic development. They showed that the macroautophagy mechanism is required to expose an “eat me” signal or secrete a “find me” signal by maintaining the energy level of cells that are scheduled to die during embryonic development. Our findings demonstrated another role of Beclin 1 in the clearance of apoptotic cells. Even when Beclin 1 did not participate in the exposure of “eat me” signals on dead cells, it regulated the machinery involved in apoptotic cell internalization by phagocytes. The role of Beclin 1 in regulating engulfment by phagocytes is considered to be independent of its function in autophagy because other essential autophagy genes, ATG5 and ULK1, which are needed for the conventional and the recently discovered ATG5-independent alternative macroautophagy, respectively, were not required for the occurrence of engulfment. Indeed, a nonautophagy role of Beclin 1 was suggested previously; however, how Beclin 1 coordinates multiple cellular functions as a key component of the Vps34 complex remains unclear (12).
It has been reported that apoptotic cells are engulfed by phagocytes at the site of their lamellipodia (9). Our EM observations of ES cells revealed findings consistent with such reports. The formation of lamellipodia is closely linked to reorganization of actin filaments. It appeared that coordination between actin filament polymerization and the formation of lamellipodia was impaired in Beclin 1−/− cells because polymerized actin fibers were observed in these cells even though the lamellipodia failed to form. Our time lapse imaging strengthened these findings. Beclin 1 knockdown cells could initiate the activation of Rac1 at the site of contact with apoptotic cells and form small ruffles and filopodia to internalize the target cells; however, they failed to induce the proper formation of the phagocytic cup and did not accomplish apoptotic cell internalization.
Rho family GTPases and PI3K are required for apoptotic cell engulfment (8, 24). The PI3K product accumulates in the early phagocyte cup, and PI3K activity is required for efficient phagocytosis by macrophages (25, 26), whereas PI3K inhibitors block the clearance of apoptotic cells both in vitro and in vivo (24, 27). Beclin 1 is known to bind with VPS34 and form the class III PI3K complex, making it a good candidate for the recruitment of active PI3K to the lamellipodia. These reports support our fluorescent imaging observations, which showed that Beclin 1, Rac1, and a PI(3)P marker (FYVE domain) accumulated together in the early phagocytic cup.
VPS34 is known to cooperate with Rab5 in the early endosome of yeasts and mammals, and it has also been reported to interact with Rab5 during phagosome maturation for apoptotic cell engulfment (15). Our finding that the PI3K complex containing Beclin 1 cooperated with Rac1 but not with Rab5 during apoptotic cell engulfment did not support this report. However, our findings are not inconsistent if Beclin 1 also regulates the phagosome maturation process because Rab5 was bound to Beclin 1 in our co-immunoprecipitation experiment. Indeed, Martinez et al. (29) recently reported that Beclin 1 is required for the phagosome maturation to degrade engulfed apoptotic cells. They also showed that other autophagy-related genes, such as ATG5 and ATG7, participate in phagosome maturation, whereas we showed that ATG5 is not involved in apoptotic cell internalization.
It is also known that the Beclin 1-VPS34 complex changes its binding partner and forms various complexes for precise regulation of autophagy pathways (12, 28). Thus, the VPS34 complex might also form various other complexes and shift its functional location during apoptotic cell engulfment. Kinchen et al. (15) reported that dynamin is involved in the interaction of VPS34 with Rab5 during phagosome maturation. Therefore, it would be interesting to investigate the role of VPS34 in regulating apoptotic cell engulfment. Because the interaction is thought to be transient with rapid changes of complexes, spatiotemporal analysis using live cell imaging with the FRET technique would be required.
Supplementary Material
Acknowledgments
We thank Masato Tanaka for the advice of the phagocytosis experiment, Scott Lowe and Klaus Hahn for providing us with the plasmids, and Hirofumi Yamaguchi for the construction of the fluorescent protein fusion vectors.
This work was supported in part by the Program for Improvement of Research Environment for Young Researchers from Special Coordination Funds for Promoting Science and Technology commissioned by the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported by a Grant-in-Aid for Scientific Research on Innovative Areas, Grant-in-Aid for Scientific Research (S), a Grant-in-Aid for Challenging Exploratory Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a grant for the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation. This work was also supported by grants from the Takeda Science Foundation, the Uehara Memorial Foundation, the Sumitomo Foundation, and the Naito Foundation.

This article contains supplemental Movies S1 and S1.
- PS
- phosphatidylserine
- PI(3)P
- phosphatidylinositol 3-phosphate
- LIF
- leukemia inhibitory factor
- MEF
- mouse embryonic fibroblast
- EGFP
- enhanced GFP
- CMTMR
- 5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine
- CMFDA
- 5-chloromethylfluorescein diacetate
- PBD
- p21-binding domain
- PAK
- p21-activated kinase
- shBeclin1
- shRNA for Beclin 1
- DN
- dominant-negative.
REFERENCES
- 1. Nagata S., Hanayama R., Kawane K. (2010) Autoimmunity and the clearance of dead cells. Cell 140, 619–630 [DOI] [PubMed] [Google Scholar]
- 2. Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., Nagata S. (2007) Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 [DOI] [PubMed] [Google Scholar]
- 3. Kobayashi N., Karisola P., Peña-Cruz V., Dorfman D. M., Jinushi M., Umetsu S. E., Butte M. J., Nagumo H., Chernova I., Zhu B., Sharpe A. H., Ito S., Dranoff G., Kaplan G. G., Casasnovas J. M., Umetsu D. T., Dekruyff R. H., Freeman G. J. (2007) TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27, 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanayama R., Tanaka M., Miwa K., Shinohara A., Iwamatsu A., Nagata S. (2002) Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 [DOI] [PubMed] [Google Scholar]
- 5. Nakano T., Ishimoto Y., Kishino J., Umeda M., Inoue K., Nagata K., Ohashi K., Mizuno K., Arita H. (1997) Cell adhesion to phosphatidylserine mediated by a product of growth arrest-specific gene 6. J. Biol. Chem. 272, 29411–29414 [DOI] [PubMed] [Google Scholar]
- 6. Reddien P. W., Horvitz H. R. (2004) The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20, 193–221 [DOI] [PubMed] [Google Scholar]
- 7. Mangahas P. M., Zhou Z. (2005) Clearance of apoptotic cells in Caenorhabditis elegans. Semin. Cell Dev. Biol. 16, 295–306 [DOI] [PubMed] [Google Scholar]
- 8. Nakaya M., Tanaka M., Okabe Y., Hanayama R., Nagata S. (2006) Opposite effects of Rho family GTPases on engulfment of apoptotic cells by macrophages. J. Biol. Chem. 281, 8836–8842 [DOI] [PubMed] [Google Scholar]
- 9. Nakaya M., Kitano M., Matsuda M., Nagata S. (2008) Spatiotemporal activation of Rac1 for engulfment of apoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 105, 9198–9203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitano M., Nakaya M., Nakamura T., Nagata S., Matsuda M. (2008) Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453, 241–245 [DOI] [PubMed] [Google Scholar]
- 11. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 12. Funderburk S. F., Wang Q. J., Yue Z. (2010) The Beclin 1-VPS34 complex. At the crossroads of autophagy and beyond. Trends Cell Biol. 20, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindmo K., Stenmark H. (2006) Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 14. Backer J. M. (2008) The regulation and function of Class III PI3Ks. Novel roles for Vps34. Biochem. J. 410, 1–17 [DOI] [PubMed] [Google Scholar]
- 15. Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O., Ravichandran K. S. (2008) A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R. N., Gilpin C., Levine B. (2007) Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 [DOI] [PubMed] [Google Scholar]
- 17. Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009) Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morita S., Kojima T., Kitamura T. (2000) Plat-E. An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 19. Dickins R. A., Hemann M. T., Zilfou J. T., Simpson D. R., Ibarra I., Hannon G. J., Lowe S. W. (2005) Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 [DOI] [PubMed] [Google Scholar]
- 20. Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 [DOI] [PubMed] [Google Scholar]
- 21. Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kraynov V. S., Chamberlain C., Bokoch G. M., Schwartz M. A., Slabaugh S., Hahn K. M. (2000) Localized Rac activation dynamics visualized in living cells. Science 290, 333–337 [DOI] [PubMed] [Google Scholar]
- 23. Ravikumar B., Imarisio S., Sarkar S., O'Kane C. J., Rubinsztein D. C. (2008) Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 121, 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leverrier Y., Ridley A. J. (2001) Requirement for Rho GTPases and PI 3-kinases during apoptotic cell phagocytosis by macrophages. Curr. Biol. 11, 195–199 [DOI] [PubMed] [Google Scholar]
- 25. Marshall J. G., Booth J. W., Stambolic V., Mak T., Balla T., Schreiber A. D., Meyer T., Grinstein S. (2001) Restricted accumulation of phosphatidylinositol 3-kinase products in a plasmalemmal subdomain during Fcγ receptor-mediated phagocytosis. J. Cell Biol. 153, 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamen L. A., Levinsohn J., Swanson J. A. (2007) Differential association of phosphatidylinositol 3-kinase, SHIP-1, and PTEN with forming phagosomes. Mol. Biol. Cell 18, 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mellén M. A., de la Rosa E. J., Boya P. (2008) The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 15, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 28. He C., Levine B. (2010) The Beclin 1 interactome. Curr. Opin. Cell Biol. 22, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez J., Almendinger J., Oberst A., Ness R., Dillon C. P., Fitzgerald P., Hengartner M. O., Green D. R. (2011) Microtubule-associated protein 1 light chain 3α (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. U.S.A. 108, 17396–17401 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.