Background: The contribution of endometrial microRNAs (miRNAs) to female reproduction in rhesus monkey is unknown.
Results: Progesterone receptor is negatively modulated by miRNAs through non-conserved miRNA binding sites in the 3′-UTR.
Conclusion: miRNA regulation of endometrial receptivity is species-dependent.
Significance: Our study provides new insights into the species-biased molecular mechanisms underlying endometrial receptivity from the aspects of miRNA-mediated regulation.
Keywords: Development, MicroRNA, Mouse, Pregnancy, Uterus, Embryo Implantation, Rhesus Monkey, Uterus
Abstract
The establishment of endometrial receptivity is a prerequisite for successful pregnancy, which is controlled by a complex mechanism. MicroRNAs (miRNAs) are small non-coding RNAs that have emerged as important regulators of gene expression. However, the contribution of miRNAs in endometrial receptivity is still unknown. Here we used rhesus monkey as an animal model and compared the endometrial miRNA expression profiles during early-secretory (pre-receptive) phase and mid-secretory (receptive) phase by deep sequencing. A set of differentially expressed miRNAs were identified, 8 of which were selected and validated using quantitative RT-PCR. To facilitate the prediction of their target genes, the 3′-UTRome was also determined using tag sequencing of mRNA 3′-termini. Surprisingly, about 50% of the 10,677 genes expressed in the rhesus monkey endometrium exhibited alternative 3′-UTRs. Of special interest, the progesterone receptor (PGR) gene, which is necessary for endometrial receptivity, processes an ultra long 3′-UTR (∼10 kb) along with a short variant (∼2.5 kb). Evolutionary analysis showed that the 3′-UTR sequences of PGR are poorly conserved between primates and rodents, suggesting a species-biased miRNA binding pattern. We further demonstrated that PGR is a valid target of miR-96 in rhesus monkey and human but not in rodents, whereas the regulation of PGR by miR-375 is rhesus monkey-specific. Additionally, we found that miR-219–5p regulates PGR expression through a primate-specific long non-coding RNA immediately downstream of the PGR locus. Our study provides new insights into the molecular mechanisms underlying endometrial receptivity and presents intriguing species-specific regulatory roles of miRNAs.
Introduction
The establishment of endometrial receptivity to embryo implantation is a prerequisite for the success of human reproduction. Endometrial receptivity is strictly controlled by the ovarian steroid hormones, estrogen, and progesterone, through their nuclear receptors (1). Although both ovarian progesterone and estrogen are needed for embryo implantation in rats and mice (2), progesterone alone may be sufficient in some other species, such as rhesus monkeys and humans (3, 4). However, the molecular mechanism underlying progesterone regulation of endometrial receptivity is still poorly understood.
Recently, microRNAs (miRNAs)3 have emerged as important regulators of gene expression. miRNAs are a class of ∼22 nucleotides (nt) long non-coding RNAs processed from long primary transcripts via consecutive cleavage by two RNase III enzymes, Drosha and Dicer. Mature miRNAs recognize target mRNAs by imperfect base-pairing to complementary target sites, typically in the 3′-untranslated region (3′-UTR), leading to translational repression and/or mRNA degradation (5). In humans, more than 1,000 miRNAs have been discovered (miRBase Release 16). Since each miRNA is capable of targeting several hundred mRNAs, more than 60% of protein-coding genes are predicted to undergo direct miRNA regulation (6). Thus, miRNAs provide a widespread mechanism to control gene expression. Studies have indicated that miRNAs play an important role in the postnatal development of mouse uterus and oviduct (7–9). Uterine miRNAs are expressed in a steroid hormone-dependent manner in mice (10), and several groups have demonstrated that miRNA expression profiles change dynamically in the uterus during embryo implantation in mice and humans (11–14).
In this study, we aimed to study the contribution of miRNAs in endometrial receptivity using the rhesus monkey as an animal model. Because of ethical restrictions and experimental difficulties, it is not practical for direct analysis of embryo implantation process in humans (15, 16). Current studies heavily rely on mice. Mice are easy to maintain and handle, and reproduce quickly. Most importantly, the mouse genome supports targeted mutagenesis in specific genes by homologous recombination in embryonic stem (ES) cells, which allows genes to be altered efficiently and precisely (16). However, a recent study has indicated that null mutations in human and mouse orthologs frequently result in different phenotypes, suggesting that results from mouse models should be extrapolated to humans with caution (17). In particular, there are considerable differences in morphological changes, hormonal dependence, and molecular interactions between mice and humans during embryo implantation (18). Therefore, non-human primates are ideal models for studying endometrial receptivity and embryo implantation in humans.
Rhesus monkeys (Macaca mulatta) are the most frequently used laboratory non-human primates. They have a regular menstrual cycle (∼28 days), similar to women. However, the genomic resources for the rhesus monkey are relatively limited. The current list of rhesus monkey miRNAs reported in miRBase is still largely incomplete (comprising only 479 sequences, miRBase Release 16), most of which were identified by searching the rhesus monkey genome for human miRNA homologs (19). Given that microarray are restricted to detect known miRNA sequences and have some limitations in quantification due to nonspecific hybridization (20), we constructed two small RNA libraries from the endometrium samples collected on menstrual cycle days 15 (pre-receptive phase) and 21 (receptive phase) and subjected to direct deep sequencing. Deep sequencing measures the copy numbers of small RNA sequences, thus allowing for the discovery of novel miRNAs (20, 21). Additionally, most rhesus monkey genes have lacked annotated 3′-UTRs, making it hard to predict miRNA target genes. Although human 3′-UTRs can be used to predict homologous 3′-UTRs in the rhesus monkey, previous studies have indicated that alternative polyadenylation pattern of genes varies across tissues and developmental stages (22–24). Therefore, we also determined the 3′-UTRome using 3′-end tag sequencing of mRNA termini. Joint analysis of microRNome and 3′-UTRome enables us to characterize the role of miRNAs in endometrial receptivity. Through data mining followed by experimental validation, we found that the progesterone receptor gene (PGR) is negatively regulated by several miRNAs that are highly expressed in receptive endometrium in a species-specific manner. Our approach represents a novel strategy for regulatory and functional studies on miRNAs.
EXPERIMENTAL PROCEDURES
Tissue Collection
Healthy and mature female rhesus monkeys (Macaca mulatta) were housed singly in a controlled environment with a 12 h light/12 h dark cycle at the Fuzhou Non-human Primate Research Center (Fujian, China). All female rhesus monkeys were evaluated daily by visual examination of the perineum for menses, with the onset of menses defined as day 1 of the menstrual cycle. Female rhesus monkeys showing at least two consecutive menstrual cycles of ∼28 days were chosen for this study. Serum estrogen concentration was measured daily to screen estrogen peak. Both estrogen peak and the dating of menstrual cycle were used to determine the time of ovulation (25). Endometrial tissues were collected on day 15 (pre-receptive phase, n = 3) and day 21 (receptive phase, n = 3) of menstrual cycle by hysterectomy. Samples were flash-frozen in liquid nitrogen and stored at −80 °C for further analysis. This study design was approved by the Institutional Animal Care and Use Committee of Shantou University.
Deep Sequencing of miRNAs
Total RNAs of rhesus monkey endometrial tissues were extracted using TRIzol reagent (Invitrogen). Small RNAs in the size range of 18∼30 nt were purified from denaturing 15% polyacrylamide gel and ligated with adapters. After purification, small RNAs were reverse transcribed and amplified for 15 cycles. PCR products were purified and quantified for high-throughput sequencing with the Illumina Genome Analyzer IIx at Huada Genomics Institute Co. Ltd (Shenzhen, China).
A computational pipeline was used to process the sequencing data. All small RNA reads without perfect matches to the most proximal 8 nt of the 5′-adaptor sequences were removed. The adaptor-free reads were then queried against known miRNA and other non-coding RNAs obtained from miRbase release 16, piRNABank version 2.0 and fRNAdb version 3.4 using the Bowtie align tool version 0.9.9.1 (51). The miRNA expression profile was calculated by summing the number of reads that mapped uniquely to known miRNAs. Read counts were then normalized to reads per million in each dataset, and fold changes between datasets were calculated according to normalized read counts. When comparing two groups of profile differences, a two-sided z-test with Bonferroni multiple test correction was employed. The fold change values in miRNA expression between groups were also calculated. Differentially expressed miRNAs were chosen according to the criteria of a fold-change greater than 2 and a p value less than 0.001.
After all known miRNA and other non-coding RNA sequences were extracted from the two datasets, the remaining unique small RNAs were aligned to the rhesus monkey genome assembly (rheMac2) downloaded from the UCSC genome browser. MIREAP software version 0.2 (Huada Genomics Institute Co. Ltd.) was used to predict novel miRNA candidates under default settings. A novel miRNA was considered typical only when it fulfilled three criteria: 1) mature miRNA has to be supported by a minimum of five independent sequence reads originating from at least one small RNA library; 2) the secondary structure of the hairpin is steady, with the free energy of hybridization lower than −20 kcal/mol; and 3) hairpin is located in intergenic or intronic regions.
Tag Sequencing of mRNA 3′-Termini
DpnII-tag sequencing was performed according to the protocol of Digital Gene Expression Tag Profile Kit (Illumina). Briefly, total RNAs (1 μg) were incubated with oligo-dT beads to capture the polyadenylated RNA fraction. First-strand and second-strand cDNAs were synthesized while the RNA was bound to the beads, and the double stranded product was digested with restriction enzyme DpnII, which cuts ∼250 bp upstream of the messenger RNA poly(A) tail. The fragments that remained attached to the beads were ligated to an adapter, which contains a MmeI recognition site. The restriction enzyme MmeI was then used to cut 16 bp downstream from the recognition site, thus creating a 20 bp unique tag (including 4 bp of the DpnII recognition site). Then another adapter was ligated at the site of MmeI cleavage. The resulting sequences, containing adaptors on both 5′- and 3′-ends, were amplified using PCR and purified on PAGE gels. Finally, high throughput sequencing was carried out using the same platform as described above.
After sequencing, all of 20 bp tags were extracted and potentially erroneous tags (single copy tags or tags containing ambiguous bases “N”) were filtered out. Most rhesus monkey genes have lacked annotated 3′-UTRs. In order to identify 3′-UTR sequence, tags were mapped to the rhesus monkey genome assembly (rheMac2) using the Bowtie alignment tool. Only unique mapping tags were used for further analysis. Gene annotations for rhesus monkey were downloaded from ENSEMBL release 61, and the stop codon coordinates were determined in the genome. For a given gene, tags falling into regions from 1 kb upstream to 10 kb downstream of the stop codon were considered to support the evidence of transcriptional termination. The Polyadq program (52), which scans sequences for AATAAA or ATTAAA and downstream elements typical of mammalian poly(A) signals, was used to predict putative polyadenylation sites between the matched tag and the closest DpnII (GATC) site downstream from the tag. When more than one poly(A) signals were found, only the one with the highest score was chosen. The full-length of 3′-UTR sequences were defined as the region from the stop codon to the poly(A) signal site. In the end, quantitative analysis of these tags was performed using the same statistical framework as described in the section above.
Quantitative RT-PCR for miRNA and mRNA Quantification
Quantitative RT-PCR was carried out to validate the expression levels of miRNAs determined by deep sequencing. Total RNAs were extracted using TRIzol reagent (Invitrogen), digested with DNase I and reverse-transcribed into cDNA with PrimeScriptTM RT reagent kit (TaKaRa). Each reverse transcription reaction was performed in 10 μl reaction volume containing 0.1 μg of total RNAs, 2 μl of 5 × PrimeScript buffer, 250 fmol special stem-loop RT primers, and 0.5 μl of PrimeScript RT Enzyme Mix I. Reverse transcription conditions were as follows: 16 °C for 30 min, 60 cycles of 30 °C for 30 s, 42 °C for 30 s, 50 °C for 1 s, and 85 °C for 5 min. The reverse transcription products were amplified using a SYBR Premix Ex TaqTM kit (TaKaRa) on the Rotor-Gene 3000A system for 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 5 s and 72 °C for 8 s. U6 was amplified as a reference for normalization.
For mRNA and long non-coding RNA quantification, rhesus monkey GAPDH was selected as the internal control. For reverse transcription, 25 pmol of Oligo dT primers and 50 pmol of random 6-mer were used as RT primers in a 10 μl reaction system of PrimeScriptTM RT reagent kit according to the manufacturer's instructions. The cDNAs were amplified using the following PCR program: 95 °C for 10 s, 40 cycles of 95 °C for 5 s and 60 °C for 34 s. All primers used in this study were listed in supplemental Table S1.
Northern Blot
For the preparation of the probe for Northern blot, a 425 bp fragment of rhesus monkey PGR gene was amplified by RT-PCR. The derived fragment was cloned into pGEM-T plasmid (Promega) and verified by sequencing. The recombinant plasmid was amplified with the primers for T7 and SP6 to prepare templates for labeling sense or antisense probes, respectively. Digoxigenin (DIG)-labeled complementary RNA (cRNA) probes were transcribed in vitro using DIG RNA labeling kit (Roche).
Total RNAs were extracted from rhesus monkey endometrium, run on a 1% agarose gel in the presence of 2.2 m formaldehyde, transferred onto positively charged nylon membrane (Roche), and hybridized with the DIG-labeled RNA probes in DIG Easy Hyb (Roche) at 68 °C for 16 h. The membranes were washed twice for 5 min each at room temperature with 1 × SSC, 0.1% SDS, and then washed twice for 15 min each at 68 °C with 0.1 × SSC and 0.1% SDS. The hybridized bands were visualized using CDP-Star (Roche).
Western Blot
Cultured cells were collected directly in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, and 0.25% sodium deoxycholate). Protein concentration was measured with BCA Reagent kit (Applygen, Beijing, China). Samples were run on a 10% PAGE gel and transferred onto nitrocellulose membranes. After blocking in 5% nonfat dry milk in PBST (0.1% Tween 20 in PBS) for 1 h, membranes were incubated overnight at 4 °C with primary antibody for PGR (ab2764, Abcam) or β-Actin (ab8227, Abcam). After washed three times in washing buffer, membranes were incubated in matched second antibody conjugated with horseradish peroxidase. The signals were developed with ECL Chemiluminescent kit (Amersham Biosciences, Arlington Heights, IL).
In Silico Analysis of miRNA Binding Sites
Rhesus monkey 3′-UTR sequences were scanned for perfect matches against the miRNA seed sequence (nucleotides 2∼7) and the identity of one upstream and one downstream flanking nucleotides were also considered for each seed match. In this study, three types of seed match as defined in the TargetScan 5.1 algorithm (53) were allowed: 8mer (exact match to positions 2–8 of the mature microRNA followed by an A), 7mer-m8 (exact match to positions 2–8 of the mature microRNA), and 7mer-1A (exact match to positions 2–7 of the mature microRNA followed by an A). The rhesus monkey 3′-UTR boundaries were used to acquire orthologous 3′-UTRs from the 46-genome vertebrate Multiz alignments from the UCSC genome browser. The conservation of target sites was studied in the aligned sequences of commonly used model organism for studying endometrial receptivity, including human, chimpanzee, rhesus monkey, mouse, and rat.
Dual-luciferase Activity Assay
For luciferase reporter experiments, the 3′-UTR segments of rhesus monkey harboring predicted miRNA binding sites were cloned into the XbaI-FseI restriction site downstream to the firefly luciferase reporter gene of pGL3 control vector (Promega). The miRNA-binding sites were site-directed mutated using the Quick Change site-directed mutagenesis kit (Stratagene). All the clones were confirmed by sequencing.
Rhesus monkey LLC-MK2 cells were cultured in DMEM Medium (Invitrogen) with 10% fetal bovine serum (Biochrom, Berlin, Germany). Human ECC-1 cells were maintained in RPMI 1640 Medium (Sigma) with 5% fetal bovine serum (Biochrom). Mouse TM3 cells were grown in DMEM/F12 Medium (Invitrogen) with 5% horse serum (Minghai, Lanzhou, China) and 2.5% fetal bovine serum (Biochrom). All cells were pre-plated in 24-well tissue culture plates at a concentration of 3 × 104 cells per well. Transfection was performed the next day using Lipofectamine 2000 (Invitrogen) with 40 ng of pRL-TK, 200 ng of pGL3 vector containing the desired 3′-UTR with or without site-directed mutations, and 50 ng of desired pre-miRNA, anti-miR inhibitor or negative control (Ambion). Cell lysate was collected and assayed 30 h after transfection. The activity of firefly and Renilla luciferase was measured using a Dual-luciferase reporter assay system (Promega). The plasmid pRL-TK containing the Renilla luciferase was used for normalization of transfection efficiency. Each experiment was run in triplicate and repeated at least three different times.
RESULTS
Global Analysis of miRNA Changes Associated with Endometrial Receptivity
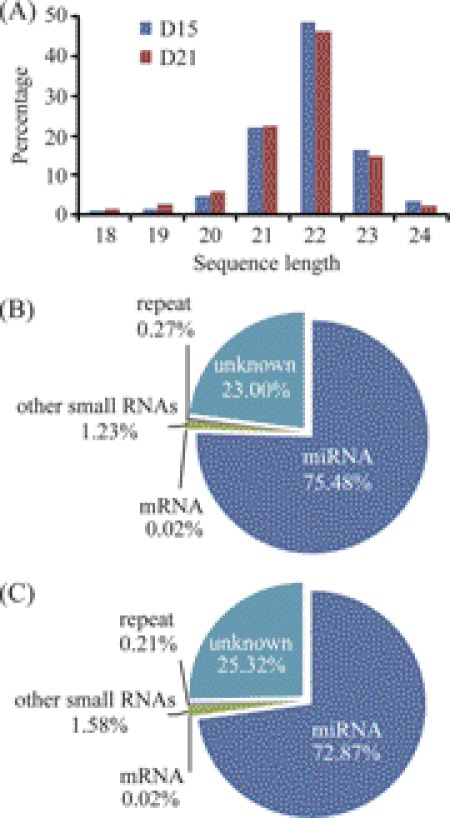
To address the issue of miRNAs associated with endometrial receptivity in rhesus monkey, endometrial tissues were collected on day 15 (D15, pre-receptive phase) and day 21 (D21, receptive phase) of the menstrual cycle. Through deep sequencing, we obtained 13,661,533 reads from day 15 and 12,296,254 reads from day 21, respectively. The raw data in this study were deposited in Gene Expression Omnibus database (GSE31041). Most of these reads were 18∼26 nt long with a peak around 22 nt (Fig. 1A), demonstrating that the small RNA libraries were highly enriched in mature miRNAs. Genomic mapping of these reads showed that the predominant RNA species (based on read counts) in both libraries were miRNAs, representing 75.48% of day 15 and 72.87% of day 21, respectively. There were small amounts of other non-coding RNAs (rRNAs, tRNAs, piRNAs, snRNAs, snoRNAs, etc.), mRNAs and genomic repeats, being no more than 2% of total reads (Fig. 1, B and C). From the unknown reads perfectly mapped to genome, we identified a low-level expression of 113 potential novel miRNAs (supplemental Table S2). These miRNA candidates deserve further investigation.
FIGURE 1.
Deep sequencing of small RNA libraries derived from the endometrium of rhesus monkey. A, length distribution of small RNAs. B and C, composition of small RNA species in the endometrium on days 15 and 21 of the menstrual cycle, respectively.
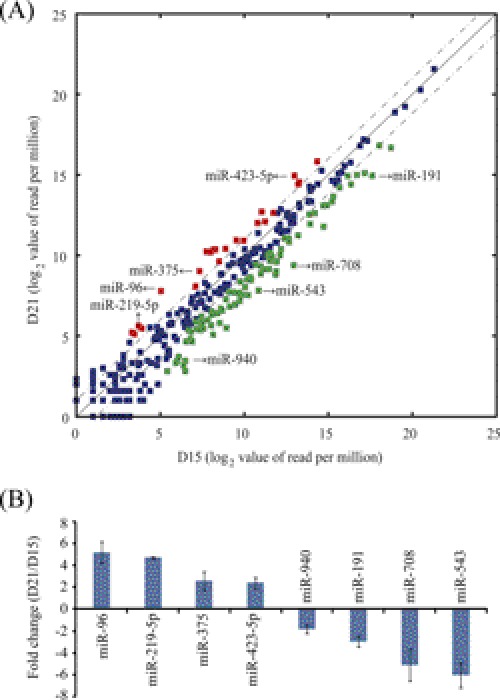
We then compared the relative endometrial miRNA abundance on days 15 and 21. Read counts for each library were normalized to reads per million. Differentially expressed miRNAs were identified according to their fold changes (>2-fold) and p values (<0.001). Compared with day 15, 85 miRNAs were down-regulated and 22 miRNAs were up-regulated on day 21 (Fig. 2A). To confirm the results from miRNA deep sequencing, 8 miRNAs were chosen for qRT-PCR validation according to their fold change or abundance index. The expression levels of these 8 miRNAs measured by qRT-PCR were similar to deep sequencing data (Fig. 2B).
FIGURE 2.
Identification of differentially expressed miRNAs. A, scatter plot represents miRNAs significantly changed at least 2-fold (p < 0.001). The RPM (reads per million) values are plotted in log2 scale. Dash lines indicate the 2-fold difference boundaries. Red dots indicate up-regulated miRNAs, and green dots represent down-regulated ones on day 21 compared with day 15. B, confirmation of 8 selected miRNAs by qRT-PCR analysis. Quantitative miRNA expression data were normalized to the expression levels of U6. Relative fold changes were shown as mean ± S.E.
The Landscape of Endometrial 3′-UTRome
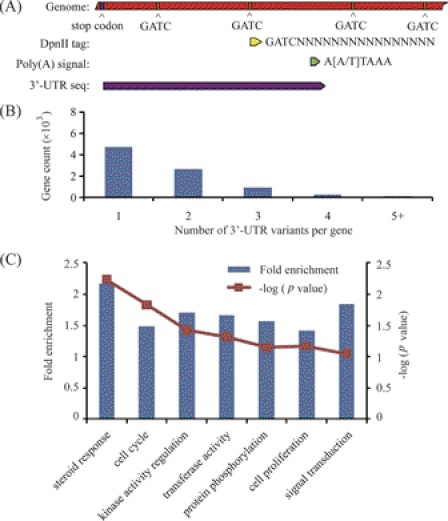
MiRNAs regulate the stability and translation of target mRNAs through complimentary binding sites in their 3′-UTRs. However, the gene sequence resources for rhesus monkey are relatively limited and most of the genes have lacked annotated 3′-UTRs. To predict the target genes of miRNAs, we analyzed the 3′-UTRome of rhesus monkey endometrium through tag sequencing of mRNA 3′-termini. The tags were generated from the 3′ most recognition site of restriction enzyme DpnII, which recognizes GATC, proving transcriptional termination information for each mRNA. The polyadenylation site, which locates between the DpnII-tag and the next GATC site, was predicted using the polydq algorithm. As most 3′-UTRs have no introns (23, 24), the 3′-UTR sequences were extracted from the stop codon to the poly(A) signal site in the genome (illustrated in Fig. 3A). Using this method, we defined 20,149 distinct 3′-UTRs in rhesus monkey for 10,677 protein-coding genes. About 50% of the genes expressed in the rhesus monkey endometrium exhibited alternative 3′-UTRs (Fig. 3B). By counting the 3′-UTR sequence reads for each gene, we measured the relative expression levels of genes in the rhesus monkey endometrium. In comparison with day 15, there were 487 genes up-regulated and 453 genes down-regulated at least 2-fold (p < 0.001) on day 21 (supplemental Table S3). Based on Gene Ontology analysis, the genes involved in steroid response were highly enriched (Fig. 3C), in agreement with the idea that steroid hormones are key regulators of endometrial receptivity (26, 27).
FIGURE 3.
The 3′-UTRome of the rhesus monkey endometrium. A, 3′-UTRome discovery pipeline. B, distribution of 3′-UTR variants per gene. C, functional clustering and enrichment analysis of differentially expressed genes identified by tag sequencing. DAVID online tools were used for enrichment analysis. Genes were classified according to biological processes.
Characterization of Progesterone Receptor 3′-UTR
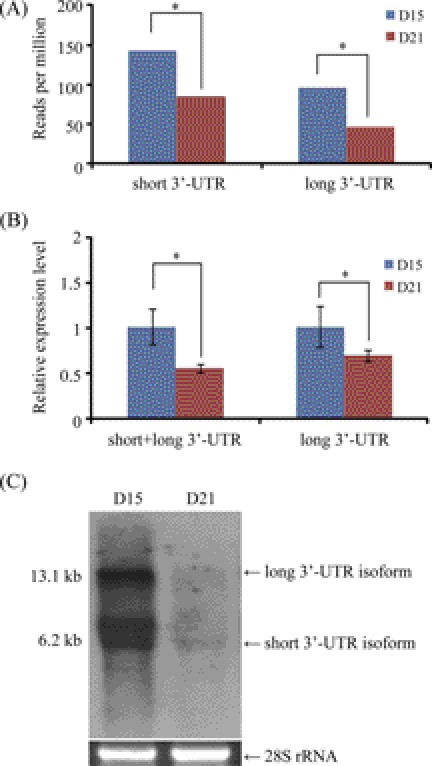
Among these steroid response-related genes, we focused on the progesterone receptor (PGR), because PGR is a key component of progesterone signaling and is necessary for embryo implantation (28, 29). Interestingly, we identified two tags falling into the 3′-UTR region of PGR. One is 5′-GATCATGGATTTTAACGGTA-3′, which represents a long 3′-UTR of 9,398 nt in length. The other is 5′-GATCTCTGTCTGCAATGTAG-3′, corresponding to a short 3′-UTR of 2,482 nt. According to tag abundance, both long and short 3′-UTRs were significantly decreased on day 21 compared with day 15 (Fig. 4A). The existence of these 2 isoforms was confirmed by qRT-PCR (Fig. 4B) and Northern blot (Fig. 4C).
FIGURE 4.
The 3′-UTR variants of PGR. A, expression of long and short 3′-UTR variants revealed by tag sequencing. B, real-time RT-PCR quantification of PGR 3′-UTR variants. Primers were designed in the region shared by short and long 3′-UTR variants, as well as the region specific to the long 3′-UTR variant alone. *, p < 0.05. C, confirmation of PGR mRNA in the endometrium of rhesus monkey by Northern blot hybridization. The 28 S rRNA was used as RNA loading control.
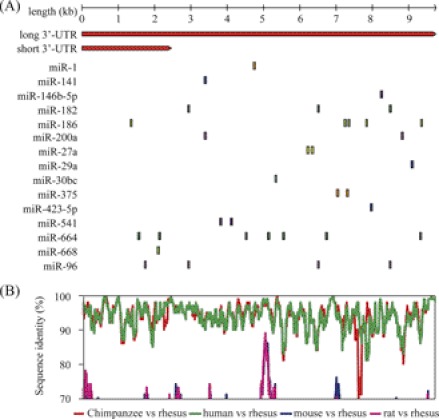
The coding sequence (CDS) of PGR is conserved among mammals (30). To further address the question of whether the 3′-UTR sequences of PGR are also conserved, we used the BLAST tool to search the gene database for matches. Close matches were found only in the human but not in other mammals. The long 3′-UTR is homologous to human PGR (NM_000926.4). As the 3′-UTR annotation for genes may be incomplete, we next retrieved the sequences corresponding to the 3′-UTRs of PGR aligned by Multiz against 5 representative mammals (rhesus monkey, human, chimpanzee, mouse, and rat) from the UCSC Genome Browser database. Pair-wise conservation profile was obtained and identity score was calculated in a 100 bp sliding window. Using a cutoff of 70% identity, we found that the 3′-UTR of PGR is conserved in primates, but not in rodents (Fig. 5B), suggesting a species-biased miRNA binding pattern.
FIGURE 5.
Prediction of miRNAs targeting PGR. A, schematic diagram of potential binding sites of miRNAs in the PGR 3′-UTR. A total of 34 potential sites corresponding to 15 unique miRNAs were identified. B, conservation of the 3′-UTR sequence. Aligned sequences corresponding to the 3′-UTR of rhesus monkey PGR gene were retrieved from the UCSC Genome Browser database. Pairwise conservation profile was obtained, and identity score was calculated in a 100 nt sliding window. The identity score cut-off value was set as 70%.
Because miRNAs are negative regulators of gene expression, we hypothesized that miRNAs up-regulated in the receptive endometrium may involve in the down-regulation of PGR. We screened the 3′-UTR sequence of PGR for miRNA binding sites using the seed-match algorithm. A total of 34 potential sites corresponding to 15 unique miRNAs were identified (Fig. 5A). Among these sites, 7 sites were rhesus monkey-specific, and 26 sites were conserved only in primates. The miR-1 site was the only one conserved between primates and rodents. However, the 3′-UTR of PGR is only 3,473 bp long (RefSeq: NM_008829.2, in agreement with Northern blot results from a previous study (31)) and the miR-1 binding site was out of the 3′-UTR range in the mouse. Thus, it is not a conserved site in the mRNA level between primates and rodents.
MiRNA Regulation on PGR through Non-conserved miRNA Binding Sites in the 3′-UTR
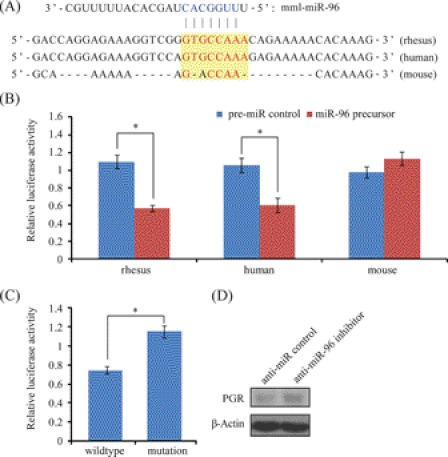
MiR-96 is highly conserved among mammals (32). It was up-regulated in the rhesus monkey endometrium on day 21 compared with day 15. According to the bioinformatic predictions, there were 4 potential miR-96 binding sites in the 3′-UTR of PGR. Luciferase reporter assay was used to validate these sites. The 3′-UTR fragments of rhesus monkey PGR spanning each miR-96 binding site were cloned downstream to a firefly luciferase gene. In LLC-MK2 cells, a significant decrease of firefly luciferase activity was observed in the forth construct, containing the binding site located at position 8,817∼8,825 (364 bp) in the 3′-UTR of PGR (Fig. 6, A and B). To further test the direct binding of this site to miR-96, we introduced point mutations into the seed binding sequence. The reduction of luciferase reporter was eliminated by mutations (Fig. 6C). Then the orthologous target sites in other species were retrieved from UCSC Multiz genome alignment. In the human, this site is the same as rhesus monkey, whereas 3 mismatches were found in the mouse. We tested the responsiveness of the human and mouse sequences to miR-96 with luciferase reporter assay. The 3′-UTR fragment homologs from human and mouse were cloned and assayed luciferase activity. As expected, miR-96 mediated regulation on PGR 3′-UTR was retained in humans, but lost in mice (Fig. 6B). Finally, we checked miR-96 regulation of endogenous PGR protein expression using antisense inhibitor. In rhesus LLC-MK2 cells, PGR-A protein level was induced upon anti-miR-96 inhibitor treatment (Fig. 6D). Collectively, our results indicate that PGR is a valid target of miR-96 in rhesus monkey and human, but not in rodents.
FIGURE 6.
MiR-96 regulation on PGR. A, sequences of predicted miR-96 binding site in the 3′-UTR of PGR. Sequence conservation was compared among rhesus monkey, human, and mouse. The seed sequence of miR-96 was colored in blue and the seed-complementary sequence in the 3′-UTR of PGR is indicated in red. B, target validation by luciferase assay in three different species. The 3′-UTR fragments derived from rhesus monkey, human, and mouse were cloned and subjected to luciferase assay in rhesus LLC-MK2 cells, human ECC-1 cells and mouse TM3 cells, respectively. C, point mutation analysis of miR-96 seed binding sequence. The seed binding site (GTGCCAAA) was mutated into GACCAACA (to mimic the seed binding sequence in mouse). Cells were co-transfected with miR-96 precursor. D, Western blot analysis of PGR-A protein expression after a transfection of anti-miR-96 inhibitor or negative control in rhesus LLC-MK2 cells.
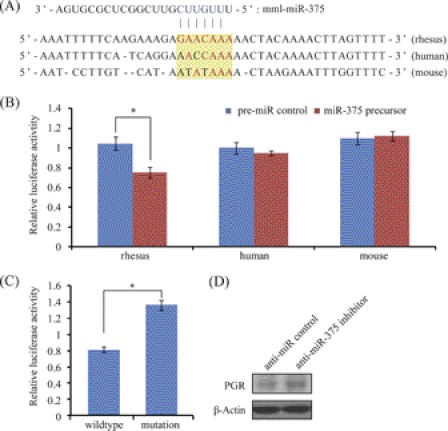
Like miR-96, miR-375 is conserved in most mammals (33). It was also up-regulated in the receptive endometrium in our study. MiR-375 was predicted to target PGR on 2 sites. Using luciferase reporter assay, we showed that only the second site was functional. This site was located at position 7,309∼7,315 (570 bp) in the 3′-UTR of PGR (Fig. 7, A and B). Introduction of point mutations into the predicted seed binding sequence abrogated miR-375 repression of luciferase reporter (Fig. 7C). Evolutionary analysis showed that the binding site is rhesus monkey-specific. There were 2∼3 mismatches in human and mouse. Further luciferase reporter assay showed that PGR 3′-UTR fragments derived from human and mouse escaped miR-375 regulation (Fig. 7B). Consistently, anti-miR-375 inhibitor was able to elevate endogenous PGR-A protein level in rhesus LLC-MK2 cells (Fig. 7D). Thus, the miR-375 regulation on PGR was rhesus monkey-specific.
FIGURE 7.
MiR-375 regulation on PGR. A, sequences of predicted miR-375 binding site in the 3′-UTR of PGR. B, target validation by luciferase assay in three different species. C, point mutation analysis of miR-375 seed binding sequence. The seed binding site (GAACAAA) was mutated into AACCAAA (to mimic the seed binding sequence in human). Cells were co-transfected with miR-375 precursor. D, Western blot analysis of PGR-A protein expression after a transfection of anti-miR-375 inhibitor or negative control in rhesus LLC-MK2 cells.
MiR-219–5p Regulation on a Long Non-coding RNA Immediately Downstream of the PGR Locus
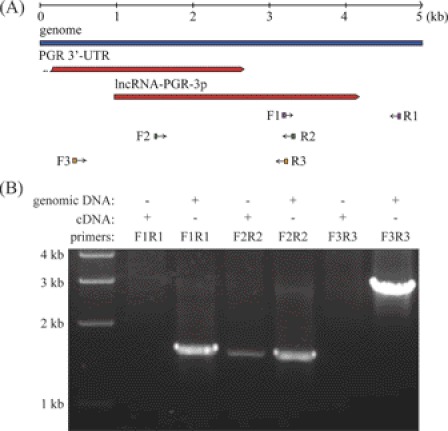
In our sequencing data, we found a tag (5′-GATCATTTGGCCATGAAAAT-3′) from an unknown transcript. This tag was located in the intergenic region immediately downstream of PGR (Fig. 8A). Using RT-PCR and primer sets spanning the upstream and downstream of this transcript, we confirmed that this transcript was an independent transcript rather than another PGR variant generated by 3′-UTR extension (Fig. 8B). This transcript was classified as a long non-coding RNA (lncRNA) using the Coding Potential Calculator (CPC) tool version 0.9r2 (54). By searching the transcript data base, we found that it was highly homologous to a human non-coding RNA, which was characterized as a cis-regulator of PGR (34). No matches were found in non-primate mammals, implying that this long non-coding RNA was primate-specific. We designated this lncRNA as lncRNA-PGR-3p. Like PGR, lncRNA-PGR-3p was also down-regulated on day 21 compared with day 15.
FIGURE 8.
Characterization of a long non-coding RNA, lncRNA-PGR-3p. A, genomic location of lncRNA-PGR-3p and primers used for RT-PCR. B, agarose gel analysis of RT-PCR amplification products. The amplification of genomic DNA was included as a positive control for primer design. RT-PCR with primers F2 and R2 revealed a novel transcript immediately downstream of the PGR gene locus. No transcript was detected from primers F1 and R1 or from primers F3 and R3, even though these primer sets could amplify genomic DNA efficiently.
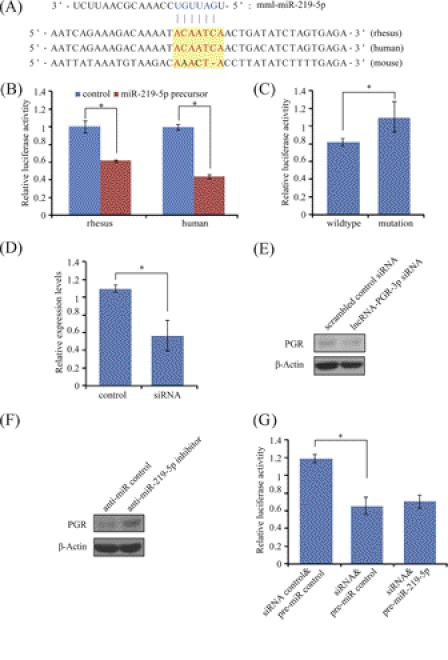
Previous studies have indicated that lncRNAs are valid targets for miRNAs (35–37). We then predicted the miRNAs that have potential binding sites on lncRNA-PGR-3p. A candidate, miR-219–5p, one of the up-regulated miRNAs in the receptive endometrium, was selected (Fig. 9A). MiR-219–5p had no predicted binding sites in the 5′-UTR, CDS, or 3′-UTR of PGR. In order to examine the interaction between lncRNA-PGR-3p and miR-219–5p, we cloned a 485 bp fragment of lncRNA-PGR-3p containing the putative miRNA binding site into the luciferase reporter vector. Luciferase assay and following point mutation analysis confirmed that lncRNA-PGR-3p was genuinely targeted by miR-219–5p in primates (Fig. 9, B and C).
FIGURE 9.
MiR-219–5p regulation on PGR via lncRNA-PGR-3p. A, sequences of predicted miR-219–5p binding site in the lncRNA-PGR-3p non-coding RNA. B, target validation by luciferase assay in rhesus monkey and human. The 3′-UTR fragments derived from rhesus monkey and human were cloned and subjected to luciferase assay in rhesus LLC-MK2 cells and human ECC-1 cells, respectively. C, point mutation analysis of miR-219–5p seed binding sequence. The seed binding site (ACAATCA) was mutated into AAACTAC. Cells were co-transfected with miR-219–5p precursor. D, efficiency of lncRNA-PGR-3p RNAi on lncRNA-PGR-3p RNA level determined by qRT-PCR. The siRNA sequence designed for rhesus lncRNA-PGR-3p was 5′-GCUGUAAGUUUGAGUUGAUdTdT-3′. E, Western blot analysis of PGR-A protein expression after rhesus LLC-MK2 cells were treated with lncRNA-PGR-3p siRNA. F, level of endogenous PGR-A protein measured by Western blot after rhesus LLC-MK2 cells were transfected with the anti-miR-219-5p inhibitor or control. G, indirect regulation of PGR by miR-219-5p via lncRNA-PGR-3p. When rhesus LLC-MK2 cells were co-transfected with lncRNA-PGR-3p siRNA, miR-219–5p down-regulation on PGR luciferase activity was abrogated.
To test whether lncRNA-PGR-3p could regulate PGR expression in rhesus LLC-MK2 cells, lncRNA-PGR-3p RNAi was performed. After cells were transfected with designed siRNA, lncRNA-PGR-3p expression level was confirmed by qRT-PCR (Fig. 9D). With lncRNA-PGR-3p expression level reduced to about 60% by RNAi, PGR-A protein expression was repressed (Fig. 9E), suggesting that lncRNA-PGR-3p was a positive regulator of PGR in rhesus LLC-MK2 cells.
Because lncRNA-PGR-3p is a target of miR-219–5p and also a regulator of PGR, we assumed that miR-219–5p may exert its regulation on PGR expression through lncRNA-PGR-3p. To confirm this, LLC-MK2 cells were transfected with the miR-219–5p antisense inhibitor. Compared with the control, the level of PGR-A protein was up-regulated after cells were transfected with anti-miR-219-5p inhibitor (Fig. 9F). Additionally, miR-219-5p down-regulation on PGR luciferase activity was abrogated when rhesus LLC-MK2 cells were co-transfected with lncRNA-PGR-3p siRNA (Fig. 9G). Our data indicated that miR-219-5p is potentially an indirect regulator of PGR expression through lncRNA-PGR-3p.
DISCUSSION
In this study, deep sequencing was applied to determine the miRNA expression profiles in the endometrium of rhesus monkey. We detected 325 known miRNAs along with 113 potential novel miRNAs that had not been reported in any species before. Although 89.3% of known miRNAs are conserved in primates and rodents, 11.5% of potential novel miRNAs are conserved according to genomic sequence alignments. Most of them are primate-specific. All these novel miRNAs were expressed at very low levels in the endometrium. Therefore, it seems that the conserved known miRNAs are the major players in the endometrium, and the roles of novel miRNAs were not pursued further.
In this study, two PGR variants with different 3′-UTR sequences were identified in the endometrium of rhesus monkey. Both of them were significantly down-regulated in receptive endometrium. These two variants were different from the well known PGR-A and PGR-B variants that result from the usage of different transcription start sites (38, 39). Previous studies have shown that alternative 3′-UTRs in varying length caused by alternative polyadenylation are frequent in mammals, often differentially expressed in a tissue- and phase-specific manner (22). The long and short 3′-UTRs are different substrates for miRNA regulation. Genes that prefer to increase the relative abundance of short 3′-UTR isoforms in receptive endometrium might be due to the need to escape miRNA regulation through evolutional selection when particularly high levels of gene products are needed. On the other hand, genes that choose to express a higher relative abundance of long 3′-UTR isoforms might provide additional regulatory interfaces that mediate the interactions with miRNAs at selected spatiotemporal coordinates. The existence of isoform preferences adds another layer of complexity in miRNA-mediated post-transcriptional regulation.
Using bioinformatic tools, we found that the 8 qRT-PCR-validated miRNAs in this study potentially targeted 492 genes. Functional classification analysis indicated that genes responsible for steroid response were highly enriched (data not shown). Among these steroid response genes, we focused on the PGR gene, a key component of the progesterone signaling. It has been established that PGR is indispensable for endometrial receptivity and embryo implantation (28, 29). Paradoxically, PGR is repressed in receptive endometrium in most mammals. In humans, the retention of PGR expression in endometrial epithelium is associated with embryo implantation failure. Loss of the PGR in the luminal epithelium is concomitant with the decrease of mucin glycoprotein 1 (MUC-1), an inhibitor of blastocyst implantation (40, 41). It is notable that MUC-1 remains abundant at the luminal epithelial surface during the receptive phase in humans, in contrast to other primates (42). This phenomenon is thought to be an evolutionary advantage to sort out defective embryos prior to implantation by the continued MUC-1 barrier. The loss of epithelial PGR also provides the opportunity for stromal cells to exercise a paracrine-mediated effect on epithelial αvβ3 integrin expression (43). However, little is known about how PGR is down-regulated. It has been reported that miR-26a and miR-181a target PGR and repress PGR expression in human MCF7 cells (44). However, no change in miR-26a or miR-181a expression was observed in the receptive endometrium in this study, demonstrating the down-regulation of PGR is not linked to miR-26a or miR-181a. In this study, we showed that both miR-96 and miR-375 act as direct negative regulators on the long 3′-UTR PGR isoform. These miRNAs may fine-tune the dosage of PGR signaling and contribute to endometrial receptivity. Noticeably, the short 3′-UTR PGR isoform escapes the regulation by these two miRNAs.
Despite the evolutionary conservation of most miRNAs, the 3′-UTR sequences of PGR are poorly conserved between primates and rodents. Consequently, all predicted miRNA binding sites in the 3′-UTR of PGR were rhesus monkey-specific or only conserved in primates. However, the lack of conservation does not necessarily mean lack of function. Using luciferase assay, we confirmed that PGR is a primate-specific target of miR-96 and a rhesus monkey-specific target of miR-375, respectively. Previously, Rosa et al. have shown that the miR-430/427/302 family of miRNAs have a distinct effect on Nodal signaling during mesendoderm differentiation in African frog and human (45), indicating a species-specific bias of miRNA target selection. To date, most algorithms for target prediction are limited to predicting evolutionarily conserved candidate target sites, while non-conserved targets are completely missed. It is reported that evolutionary conserved sites are more reliable than non-conserved ones, based on the hypothesis that the 3′-UTR sequences are under selective pressure to maintain miRNA binding sites, and these sites should be more conserved than random sequences (46). In this study, we tested 6 poorly conserved miRNA binding sites (4 for miR-96 and 2 for miR-375) using luciferase assay, 2 of which were proved functional. The false positive rate was 67%, higher than that for conserved miRNA-binding site (22∼31%) (47). In the whole 3′-UTRome scale, more than 75% of predicted miRNA binding sites were poorly conserved. Therefore, the number of functional poorly conserved sites is estimated to be very large, probably close to that of conserved sites. We believe that these widespread poorly conserved miRNA binding sites may help shape the species-specific characteristics of endometrial receptivity in the rhesus monkey and the primate order as well.
Recently, there are an increasing number of reports describing mRNA-like lncRNAs (48). Studies have indicated that some of these lncRNAs also harbor miRNA binding sites, just like protein-coding mRNAs (35–37). In this study, we identified an lncRNA, lncRNA-PGR-3p, immediately downstream of PGR. It was highly homologous to a known non-coding RNA in human, which was found to modulate the expression of PGR upon exogenous siRNA delivery (34). We discovered that lncRNA-PGR-3p was directly targeted by miR-219-5p. In rhesus LLC-MK2 cells, miR-219-5p was capable to repress PGR expression efficiently via lncRNA-PGR-3p. Because no other protein coding genes are affected, the regulation of PGR by miR-219-5p is highly specific. The regulation of PGR by miR-219-5p is species-dependent, as the sequence of lncRNA-PGR-3p is primate-specific. Actually, most lncRNAs are as poorly conserved as the intronic regions and less conserved than the 3′-UTR regions of protein coding genes (49, 50). MiRNA regulation through lncRNAs may represent an additional mechanism for species-specific miRNA target recognition.
Altogether, we defined the microRNome and 3′-UTRome in the receptive endometrium of rhesus monkey in this study. Through data mining, we revealed that the PGR gene expression is negatively modulated by miRNAs in a species-specific manner. Our study provides new insights into the species-biased molecular mechanisms underlying endometrial receptivity from the aspects of miRNA-mediated regulation.
Supplementary Material
This work was supported by the National Basic Research Program of China (2011CB944402) and the National Natural Science Foundation of China (30930013 and 31071276).

This article contains supplemental Tables S1–S3.
- miRNA
- microRNA
- IVF
- in vitro fertilization
- 3′-UTR
- 3′-untranslated region
- PGR
- progesterone receptor
- lncRNA
- long non-coding RNA.
REFERENCES
- 1. O'Malley B. W., Tsai M. J. (1992) Molecular pathways of steroid receptor action. Biol. Reprod. 46, 163–167 [DOI] [PubMed] [Google Scholar]
- 2. Gidley-Baird A. A. (1981) Endocrine control of implantation and delayed implantation in rats and mice. J. Reprod. Fertil. Suppl. 29, 97–109 [PubMed] [Google Scholar]
- 3. de Ziegler D., Bergeron C., Cornel C., Medalie D. A., Massai M. R., Milgrom E., Frydman R., Bouchard P. (1992) Effects of luteal estradiol on the secretory transformation of human endometrium and plasma gonadotropins. J. Clin. Endocrinol. Metab. 74, 322–331 [DOI] [PubMed] [Google Scholar]
- 4. Ghosh D., De P., Sengupta J. (1994) Luteal phase ovarian oestrogen is not essential for implantation and maintenance of pregnancy from surrogate embryo transfer in the rhesus monkey. Hum. Reprod. 9, 629–637 [DOI] [PubMed] [Google Scholar]
- 5. Nilsen T. W. (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 23, 243–249 [DOI] [PubMed] [Google Scholar]
- 6. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gonzalez G., Behringer R. R. (2009) Dicer is required for female reproductive tract development and fertility in the mouse. Mol. Reprod. Dev. 76, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagaraja A. K., Andreu-Vieyra C., Franco H. L., Ma L., Chen R., Han D. Y., Zhu H., Agno J. E., Gunaratne P. H., DeMayo F. J., Matzuk M. M. (2008) Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol. Endocrinol. 22, 2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong X., Luense L. J., McGinnis L. K., Nothnick W. B., Christenson L. K. (2008) Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 149, 6207–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nothnick W. B., Healy C. (2010) Estrogen induces distinct patterns of microRNA expression within the mouse uterus. Reprod. Sci. 17, 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qian K., Hu L., Chen H., Li H., Liu N., Li Y., Ai J., Zhu G., Tang Z., Zhang H. (2009) Hsa-miR-222 is involved in differentiation of endometrial stromal cells in vitro. Endocrinology 150, 4734–4743 [DOI] [PubMed] [Google Scholar]
- 12. Kuokkanen S., Chen B., Ojalvo L., Benard L., Santoro N., Pollard J. W. (2010) Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol. Reprod. 82, 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu S. J., Ren G., Liu J. L., Zhao Z. A., Yu Y. S., Su R. W., Ma X. H., Ni H., Lei W., Yang Z. M. (2008) MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 283, 23473–23484 [DOI] [PubMed] [Google Scholar]
- 14. Chakrabarty A., Tranguch S., Daikoku T., Jensen K., Furneaux H., Dey S. K. (2007) MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. U.S.A. 104, 15144–15149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dey S. K., Lim H., Das S. K., Reese J., Paria B. C., Daikoku T., Wang H. (2004) Molecular cues to implantation. Endocr. Rev. 25, 341–373 [DOI] [PubMed] [Google Scholar]
- 16. Wang H., Dey S. K. (2006) Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 7, 185–199 [DOI] [PubMed] [Google Scholar]
- 17. Liao B. Y., Zhang J. (2008) Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc. Natl. Acad. Sci. U.S.A. 105, 6987–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee K. Y., DeMayo F. J. (2004) Animal models of implantation. Reproduction 128, 679–695 [DOI] [PubMed] [Google Scholar]
- 19. Brameier M. (2010) Genome-wide comparative analysis of microRNAs in three non-human primates. BMC Res. Notes 3, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Creighton C. J., Reid J. G., Gunaratne P. H. (2009) Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 10, 490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Creighton C. J., Benham A. L., Zhu H., Khan M. F., Reid J. G., Nagaraja A. K., Fountain M. D., Dziadek O., Han D., Ma L., Kim J., Hawkins S. M., Anderson M. L., Matzuk M. M., Gunaratne P. H. (2010) Discovery of novel microRNAs in female reproductive tract using next generation sequencing. PLoS One 5, e9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji Z., Lee J. Y., Pan Z., Jiang B., Tian B. (2009) Progressive lengthening of 3′-untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T., Mackowiak S. D., Mis E., Zegar C., Gutwein M. R., Khivansara V., Attie O., Chen K., Salehi-Ashtiani K., Vidal M., Harkins T. T., Bouffard P., Suzuki Y., Sugano S., Kohara Y., Rajewsky N., Piano F., Gunsalus K. C., Kim J. K. (2010) The landscape of C. elegans 3′-UTRs. Science 329, 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P. (2011) Formation, regulation, and evolution of Caenorhabditis elegans 3′-UTRs. Nature 469, 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pauerstein C. J., Eddy C. A., Croxatto H. D., Hess R., Siler-Khodr T. M., Croxatto H. B. (1978) Temporal relationships of estrogen, progesterone, and luteinizing hormone levels to ovulation in women and infrahuman primates. Am. J. Obstet. Gynecol. 130, 876–886 [DOI] [PubMed] [Google Scholar]
- 26. Critchley H. O., Saunders P. T. (2009) Hormone receptor dynamics in a receptive human endometrium. Reprod. Sci. 16, 191–199 [DOI] [PubMed] [Google Scholar]
- 27. Spencer T. E., Bazer F. W. (2002) Biology of progesterone action during pregnancy recognition and maintenance of pregnancy. Front. Biosci. 7, d1879–1898 [DOI] [PubMed] [Google Scholar]
- 28. Conneely O. M., Mulac-Jericevic B., Lydon J. P., De Mayo F. J. (2001) Reproductive functions of the progesterone receptor isoforms: lessons from knock-out mice. Mol. Cell Endocrinol. 179, 97–103 [DOI] [PubMed] [Google Scholar]
- 29. Ho P. C. (2001) Mifepristone: a potential postcoital contraceptive. Expert Opin. Pharmacother. 2, 1383–1388 [DOI] [PubMed] [Google Scholar]
- 30. Thornton J. W. (2001) Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. U.S.A. 98, 5671–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tan J., Paria B. C., Dey S. K., Das S. K. (1999) Differential uterine expression of estrogen and progesterone receptors correlates with uterine preparation for implantation and decidualization in the mouse. Endocrinology 140, 5310–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jensen K. P., Covault J. (2011) Human miR-1271 is a miR-96 paralog with distinct non-conserved brain expression pattern. Nucleic Acids Res. 39, 701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kloosterman W. P., Lagendijk A. K., Ketting R. F., Moulton J. D., Plasterk R. H. (2007) Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 5, e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yue X., Schwartz J. C., Chu Y., Younger S. T., Gagnon K. T., Elbashir S., Janowski B. A., Corey D. R. (2010) Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat. Chem. Biol. 6, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. (2010) CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38, 5366–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forrest A. R., Kanamori-Katayama M., Tomaru Y., Lassmann T., Ninomiya N., Takahashi Y., de Hoon M. J., Kubosaki A., Kaiho A., Suzuki M., Yasuda J., Kawai J., Hayashizaki Y., Hume D. A., Suzuki H. (2010) Induction of microRNAs, mir-155, mir-222, mir-424, and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 24, 460–466 [DOI] [PubMed] [Google Scholar]
- 37. Poliseno L., Salmena L., Zhang J., Carver B., Haveman W. J., Pandolfi P. P. (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meyer M. E., Quirin-Stricker C., Lerouge T., Bocquel M. T., Gronemeyer H. (1992) A limiting factor mediates the differential activation of promoters by the human progesterone receptor isoforms. J. Biol. Chem. 267, 10882–10887 [PubMed] [Google Scholar]
- 39. Wen D. X., Xu Y. F., Mais D. E., Goldman M. E., McDonnell D. P. (1994) The A and B isoforms of the human progesterone receptor operate through distinct signaling pathways within target cells. Mol. Cell Biol. 14, 8356–8364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Horne A. W., Lalani E. N., Margara R. A., Ryder T. A., Mobberley M. A., White J. O. (2005) The expression pattern of MUC1 glycoforms and other biomarkers of endometrial receptivity in fertile and infertile women. Mol. Reprod. Dev. 72, 216–229 [DOI] [PubMed] [Google Scholar]
- 41. Julian J., Enders A. C., Fazleabas A. T., Carson D. D. (2005) Compartmental distinctions in uterine Muc-1 expression during early pregnancy in cynomolgous macaque (Macaca fascicularis) and baboon (Papio anubis). Hum. Reprod. 20, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 42. Aplin J. D. (1999) MUC-1 glycosylation in endometrium: possible roles of the apical glycocalyx at implantation. Hum. Reprod. 14, (Suppl. 2), 17–25 [DOI] [PubMed] [Google Scholar]
- 43. Palomino W. A., Fuentes A., González R. R., Gabler F., Boric M. A., Vega M., Devoto L. (2005) Differential expression of endometrial integrins and progesterone receptor during the window of implantation in normo-ovulatory women treated with clomiphene citrate. Fertil. Steril. 83, 587–593 [DOI] [PubMed] [Google Scholar]
- 44. Maillot G., Lacroix-Triki M., Pierredon S., Gratadou L., Schmidt S., Bènés V., Roché H., Dalenc F., Auboeuf D., Millevoi S., Vagner S. (2009) Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 69, 8332–8340 [DOI] [PubMed] [Google Scholar]
- 45. Rosa A., Spagnoli F. M., Brivanlou A. H. (2009) The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517–527 [DOI] [PubMed] [Google Scholar]
- 46. Rajewsky N. (2006) MicroRNA target predictions in animals. Nat. Genet. 38, (suppl.), S8–S13 [DOI] [PubMed] [Google Scholar]
- 47. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 48. Alexander R. P., Fang G., Rozowsky J., Snyder M., Gerstein M. B. (2010) Annotating non-coding regions of the genome. Nat. Rev. Genet. 11, 559–571 [DOI] [PubMed] [Google Scholar]
- 49. Pang K. C., Frith M. C., Mattick J. S. (2006) Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22, 1–5 [DOI] [PubMed] [Google Scholar]
- 50. Marques A. C., Ponting C. P. (2009) Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 10, R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tabaska J. E., Zhang M. Q. (1999) Detection of polyadenylation signals in human DNA sequences. Gene 231, 77–86 [DOI] [PubMed] [Google Scholar]
- 53. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 54. Kong L., Zhang Y., Ye Z. Q., Liu X. Q., Zhao S. Q., Wei L., Gao G. (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.