Background: Ku has a 5′-dRP/AP lyase activity that excises abasic sites near double-strand break ends.
Results: Ku excises only those abasic sites that would otherwise interfere with ligation.
Conclusion: This substrate specificity promotes NHEJ fidelity.
Significance: Tight control of damage excision is essential to nonhomologous end joining because this pathway typically cannot accurately replace excised DNA.
Keywords: DNA Damage, DNA Enzymes, DNA Recombination, DNA Repair, Protein DNA-Interaction, 5'-dRPase, AP Lyase, Ku, Nonhomologous End Joining, Abasic Site
Abstract
Nonhomologous end joining (NHEJ) is essential for efficient repair of chromosome breaks. However, the NHEJ ligation step is often obstructed by break-associated nucleotide damage, including base loss (abasic site or 5′-dRP/AP sites). Ku, a 5′-dRP/AP lyase, can excise such damage at ends in preparation for the ligation step. We show here that this activity is greatest if the abasic site is within a short 5′ overhang, when this activity is necessary and sufficient to prepare such termini for ligation. In contrast, Ku is less active near 3′ strand termini, where excision would leave a ligation-blocking α,β-unsaturated aldehyde. The Ku AP lyase activity is also strongly suppressed by as little as two paired bases 5′ of the abasic site. Importantly, in vitro end joining experiments show that abasic sites significantly embedded in double-stranded DNA do not block the NHEJ ligation step. Suppression of the excision activity of Ku in this context therefore is not essential for ligation and further helps NHEJ retain terminal sequence in junctions. We show that the DNA between the 5′ terminus and the abasic site can also be retained in junctions formed by cellular NHEJ, indicating that these sites are at least partly resistant to other abasic site-cleaving activities as well. High levels of the 5′-dRP/AP lyase activity of Ku are thus restricted to substrates where excision of an abasic site is required for ligation, a degree of specificity that promotes more accurate joining.
Introduction
Nonhomologous end joining (NHEJ)3 is a major pathway for resolving chromosome breaks (or double-strand breaks (DSBs)) in mammals; deficiency in NHEJ results in immunodeficiency (1), severe radiosensitivity (2), and premature replicative senescence (3, 4). NHEJ resolves chromosome breaks by ligating them together. In cells, chromosome breaks often have damage to flanking nucleotides (5) that can block the ligation step (6–9); thus this damage must be removed by end-processing enzymes for ligation to proceed. The ability to tightly couple such processing steps to the ligation step is probably the most important characteristic in distinguishing classical NHEJ from more error-prone and less efficient alternate end joining pathways. However, little is known regarding the specifics of how the context of nucleotide damage (what type of damage and where, relative to the DSB terminus) impacts the NHEJ ligation step.
We previously focused on how NHEJ resolves broken ends with flanking abasic sites, either at the 5′ terminus (5′-deoxyribose-phosphate, or 5′-dRP) or located between intact nucleotides (apurinic/apyrimidinic, or AP) (9). Abasic sites are associated with chromosome breaks generated by exposure to ionizing radiation (as well as other sources of reactive oxygen species) (5, 10) and aborted base excision repair (BER) (11–14) and associated with intermediates in immunoglobulin isotype class switch recombination (15, 16). Such lesions are a strong block to ligation when present near a DSB terminus and must be excised (7–9, 12). Abasic sites in this context can be specifically excised by a 5′-dRP/AP lyase activity that is part of the classically defined NHEJ pathway (9), similar to a step mediated by DNA polymerase β in base excision repair (BER)/single-stand break repair (17–20). 5′-dRP/AP lyases cleave such sites by forming a covalent intermediate between an active site nucleophile (typically a lysine ϵ-amine) and the abasic site 1′-carbon. Subsequent β-elimination results in cleavage of the phosphodiester bond 3′ of the abasic site, leaving behind a 5′-phosphate immediately downstream of the abasic site position that can now participate in ligation (21). Unless the AP site is at the 5′ terminus, cleavage also leaves behind a 3′ fragment terminus with an α,β-unsaturated aldehyde that, similar to a 3′-phosphoglycolate (6), will require additional processing before it can participate in ligation.
We further identified Ku as the primary source of this activity within NHEJ (9). Ku is a heterodimer of 70- and 80-kDa subunits and binds to DNA ends by threading them through a central channel or ring (22). Its primary role in NHEJ is to recognize broken chromosome ends and recruit other factors, including the NHEJ ligase, to promote end joining (23). We demonstrated that Ku also possesses 5′-dRP/AP lyase activity and employs this activity to excise abasic sites near DSB termini to help prepare such ends for ligation. Ku is uniquely effective in this role as extracts depleted of Ku no longer support efficient excision of near terminal abasic sites, and cellular NHEJ of such substrates is specifically reduced when cells are complemented with a 5′-dRP/AP lyase attenuated Ku mutant (9).
To help explain the advantages to having Ku uniquely fulfill this role, we characterized both the abasic site contexts in which the lyase of Ku is active, as well as those that block ligation. We describe significant restrictions that limit the activity of Ku to abasic sites within short 5′ overhangs and show how the limited spectrum of substrates of Ku correlates well with abasic site contexts that also block activity of the NHEJ ligase, both in vitro and in cells. Importantly, this restricted activity promotes more accurate NHEJ, both in vitro and in cells.
EXPERIMENTAL PROCEDURES
Proteins
Recombinant proteins were purified as described previously (9, 24). Briefly, Hi5 cells were infected with baculoviruses engineered to express hexahistidine-tagged Ku70 and Ku80 and XRCC4 and hexahistidine-tagged ligase IV, or hexahistidine-tagged XLF and proteins were purified using an FPLC with HisTrap affinity and MonoQ ion exchange columns (GE Healthcare).
Oligonucleotide Assays
Gel-purified oligonucleotides were obtained from Integrated DNA Technologies. Substrates with abasic sites near 5′ ends were radiolabeled at the 3′ end with [α-32P]cordycepin (PerkinElmer Life Sciences) and terminal deoxynucleotidyl transferase, whereas substrates with abasic sites near a 3′ end were labeled with γ-32P (PerkinElmer Life Sciences) and polynucleotide kinase (New England Biolabs). Labeled strands were then annealed to a complementary strand containing a terminal biotin-tetra-ethylene glycol modification, at either the 5′ or the 3′ terminus as appropriate to locate the biotin at the DNA end opposite to that of the abasic site (e.g. as illustrated in Fig. 1A). As described in Fig. 1A, oligonucleotide duplexes used in Figs. 1–3 are based on a common 40-bp core sequence, 5′-GAAATCAAACGTAAGTAGAATCCAAAGTCTCTTTCTTCCG-3′. With respect to substrates used in Fig. 1 the sequences 5′-P-GUG (substrates 1 and 2), 5′-OH-GUG (substrate 1NoP), 5′-P-GATCUG (substrate 3), 5′-OH-GATCUG (substrate 3NoP), 5′-P-GTGCATCUG (substrate 4), and 5′-P-GUGCATCTG (substrate 5) were appended 5′ of the core sequence above, and all were annealed to the complement of the 40-bp core. With respect to substrates used in Fig. 2, the sequences 3′-GUG (substrate 2) and 3′-GATCUG (substrate 3) were appended 3′ of the core strand complement. With respect to substrates used in Fig. 3, the strand complementary to the core was extended from the 3′ terminus, appending appropriate sequence complementary to 5′ overhangs to convert these substrates to fully complementary, blunt-ended substrates.
FIGURE 1.
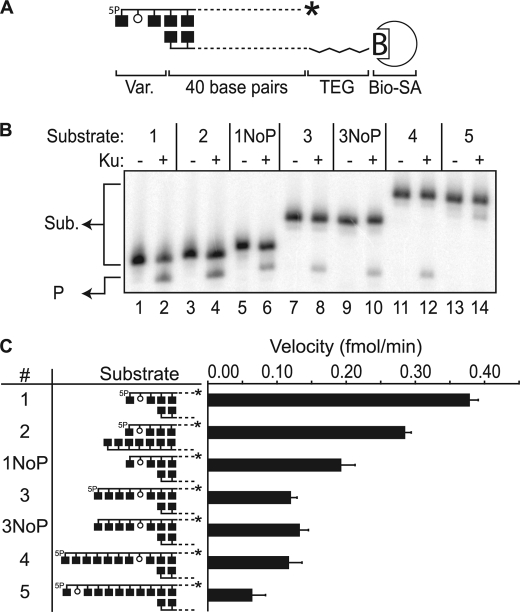
Activity on AP sites within 5′ overhangs. A, description of substrates with relative locations of radiolabel (*), 5′-phosphate (5P), abasic site (open circle), and biotin-streptavidin complex noted. Aligned boxes in opposing strands represent paired bases. Var., variable part; TEG, tetra-ethylene glycol. Bio-SA, biotin-streptavidin complex. B, representative denaturing PAGE analysis of reactions incubated for 15 min at 37 ºC with 5 nm Ku (+) and 1 nm substrates 1–5, with substrate 1–5 end structures varied as described in the graphics shown in panel C. Sub., substrates; P, product. C, average reaction velocities determined after 15 min. Standard deviations (error bars) are from the results of three independent experiments.
FIGURE 2.
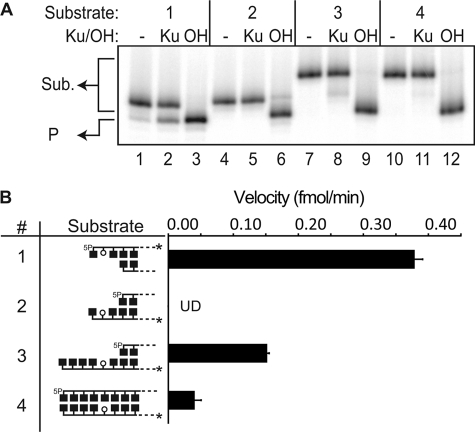
Activity on AP sites within 3′ overhangs. A, representative denaturing PAGE analysis of reactions incubated for 15 min at 37 ºC with 5 nm Ku (Ku) and 1 nm substrates 1–4 (reaction with 5′ overhang substrate 1 shown for comparative purposes), with substrate end structure varied as described in the graphics shown in panel B. OH, control reactions with substrates (Sub.) cleaved by alkali. Alkali cleavage generates a product (P) with a 3′-PO4 terminus with faster mobility than the 3′α,β-unsaturated aldehyde terminus generated by Ku. B, velocities for substrates 2, 3, and 4 were determined from the slope of reaction progression as measured after 15, 30, and 60 min. Velocity averages and standard deviations (error bars) are from the results of three independent experiments. Substrate 2 velocity was undetectable (UD). 5P, 5′-phosphate.
FIGURE 3.
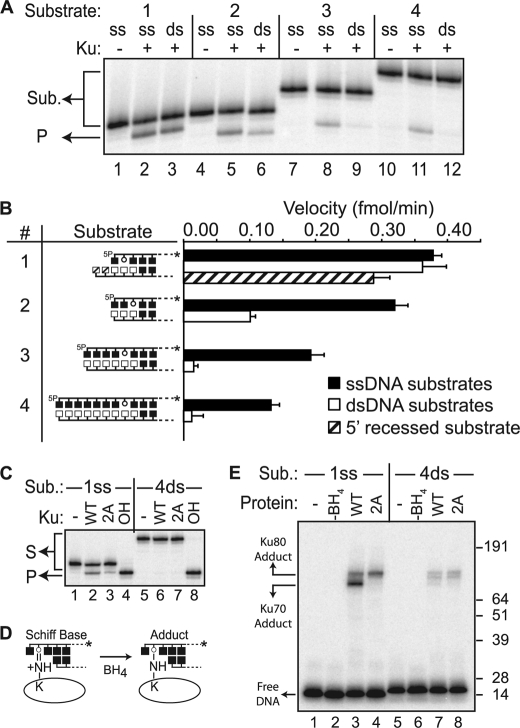
Activity on AP sites within double-stranded DNA. A, representative denaturing PAGE analysis of reactions incubated for 15 min at 37 ºC with 5 nm Ku (+) and 1 nm substrates 1–4, comparing activity on substrates (Sub.) where DNA 5′ of the abasic site is single-stranded (ss) (single-stranded versions of 1–4, as in Fig. 1) or double-stranded (ds) (double-stranded versions of 1–4). P, products. B, filled bars denote velocities on single-stranded versions (single-stranded DNA 5′ of the abasic site) of substrates 1–4, open bars denote velocities on double-stranded versions (double-stranded DNA 5′ of the abasic site) of substrates 1–4, and the hatched bar denotes velocity on a variant of substrate 1 with a 2-nucleotide 3′ overhang (5′ recessed, hatched bar). Average reaction velocities determined after 15 min and standard deviations (error bars) are from the results of three independent experiments. 5P, 5′-phosphate. C, representative denaturing PAGE analysis of reactions incubated as in panel A, using 5 nm wild type Ku heterodimer (WT) or a heterodimer of Ku70 with alanine substituted at Lys-31 and Lys-160 and WT Ku80 (2A). 1ss, substrate 1 with single-stranded overhang; 4ds, substrate 4 with double-stranded overhang; OH, alkali treatment. D, covalent intermediates in AP lyase reactions can be trapped by the addition of NaBH4 (BH4) at the start of reactions. E, SDS-PAGE analysis of trapped intermediates formed in reactions performed as in panel C, except with the addition of 5 mm NaBH4 at the start. Reactions were stopped after 10 min, and adducts were identified by SDS-PAGE.
Substrates were further prepared by preincubating them with 0.5 μm streptavidin (Pierce) and 1 unit of uracil DNA glycosylase (New England Biolabs) at 37 °C until control reactions with alkali confirmed complete deglycosylation. Reactions with 1 nm of prepared substrate in 25 mm NaPO4, pH 7.4, 0.1 mm EDTA, 150 mm KCl, and 1 mm dithiothreitol (DTT) were then mixed with 5 nm purified recombinant Ku and incubated at 37 °C for indicated times. Reactions were terminated by the addition of 200 mm NaBH4 and incubation on ice for 1 h. Products were then resolved by electrophoresis on a denaturing 12% urea-polyacrylamide gel, and the dried gel was analyzed by phosphorimaging (GE Healthcare).
The Schiff base intermediate formed between an abasic site and purified Ku was trapped by the addition of 5 mm NaBH4 immediately after adding Ku to the DNA substrate. Reactions were prepared under the same conditions as described above, incubated at 37 °C for 10 min, terminated by the addition of 8% sodium dodecyl sulfate (SDS), and heated to 95 °C for 5 min. Products were then resolved by electrophoresis on a 4–12% Bis-Tris SDS-polyacrylamide gel (Invitrogen), and the dried gel was analyzed by phosphorimaging (GE Healthcare).
NHEJ Assays
Substrates for NHEJ assays were made by amplification, in the presence of [α-32P]dATP (PerkinElmer Life Sciences), of a 300-bp fragment using primers containing a deoxyuridine at the appropriate position near a BsaI (New England Biolabs) site (to generate 4-nucleotide 5′ overhang end structures) or near or a SmuI (Fermentas) site (to generate 2-nucleotide 5′ overhangs). After digestion of the purified 300-bp fragment with BsaI or SmuI to make the appropriate end structures, deoxyuridines near the ends were converted to abasic sites by incubation with uracil DNA glycosylase and used directly (standard abasic sites) or after reduction with 100 mm NaBH4 for 30 min on ice and additional purification (QIAquick PCR cleanup; Qiagen).
NHEJ assays were performed with 5 nm DNA substrate, 40 nm Ku, 80 nm XRCC4+Ligase IV complex, and 80 nm XLF and incubated for 30 min at 37 °C in 25 mm NaPO4, pH 7.4, 0.1 mm EDTA, 150 mm KCl, and 1 mm DTT, 10% (w/v) polyethylene glycol, and 25 ng of supercoiled plasmid DNA. Ligation was initiated by the addition of 10 mm MgCl2 and 0.1 mm ATP and incubated at 37 °C for another 10 min. Reactions were stopped by the addition of 10 mm EDTA and analyzed by native 5% PAGE. For the junction analysis shown in Fig. 4D, samples were purified (QIAquick PCR cleanup, Qiagen) and treated for 1 h either with 100 mm NaBH4 on ice or the abasic site-cleaving enzyme formamidopyrimidine glycosylase (fpg, New England Biolabs) at 37 °C. Treated samples were purified (MinElute QIAquick PCR cleanup, Qiagen) and digested with 25 units of HinfI (allows visualization of the many different concatemer head-to-tail ligation species, both linear and circular, as a single product of defined mobility), and the products were then resolved by denaturing electrophoresis on a 5% urea-polyacrylamide gel and visualized by phosphorimaging (GE Healthcare).
FIGURE 4.
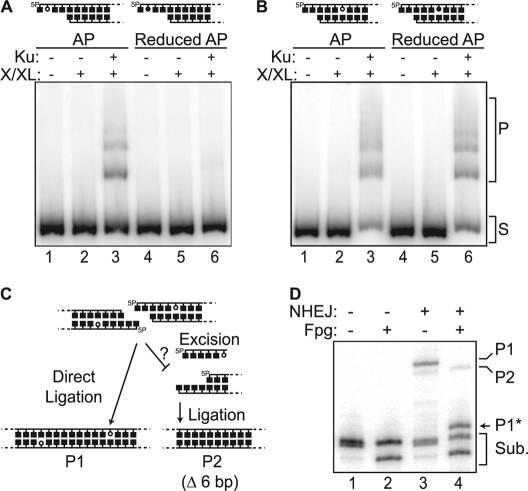
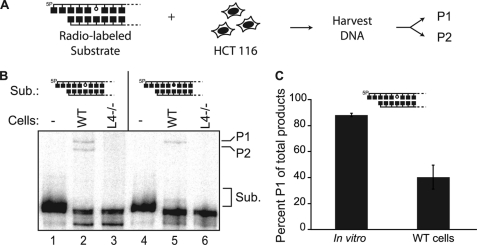
Joining of ends with near terminal abasic sites in vitro. A–D, 5 nm radiolabeled 300-bp substrates (S) were made with standard (open circle) or reduced (closed circle) AP sites located within a 5′ overhang (A) or embedded 3 bp in double-stranded DNA (B–D), and incubated with the core NHEJ factors (Ku and the ligase complex: XLF/Cernunnos, ligase IV, XRCC4 (X/LX)) as described under “Experimental Procedures.” A and B, ligation products (P) were analyzed by native PAGE. 5P, 5′-phosphate. C, the substrate described in panels A and B can be ligated in vitro directly, without prior excision (P1), or after excision at the abasic site (P2). D, denaturing PAGE analysis of head-to-tail ligation products generated in panel B, lane 3 after digestion with HinfI (allows visualization of the many different concatemer head-to-tail ligation species, both linear and circular, as a single product of defined mobility) and either further digested with fpg (+) at 37 ºC or stabilized with 100 mm NaBH4 (−). Sub., substrates. P1 is sensitive to fpg (P1*), as the AP site is incorporated in the junction.
For cellular assays, 100 ng of the 300-bp radiolabeled substrates used in in vitro experiments and 2 μg of pMAX-GFP supercoiled plasmid (carrier DNA) were introduced into one million HCT116 cells (25) by electroporation (Neon, MPK5000, Invitrogen) using a 10-μl chamber and a single 1530-V, 20-ms pulse. Flow cytometry of cells electroporated under these conditions and recovered after 5 h indicated that electroporation was efficient and did not have a major impact on cell integrity; 81% (± 4%) express green fluorescent protein from the carrier plasmid, and 88% (± 3.6%) exclude propidium iodide. Four replicate transfections were pooled and incubated at 37 °C in 1 ml of McCoy's 5A 1× media for 30 min. Cells were then washed three times with phosphate-buffered saline (PBS), resuspended in 200 μl of PBS, and treated with 100 mm NaBH4 for 10 min on ice. Cellular DNA was then harvested (QIAamp DNA mini kit, Qiagen; proteinase K step excluded to reduce processing time), and DNA samples were again treated with 100 mm NaBH4 on ice for 30 min. DNA was repurified (QIAquick PCR cleanup, Qiagen), treated with RNase A for 10 min at 37 °C, digested with HinfI for 1 h, and concentrated (MinElute QIAquick PCR cleanup, Qiagen) prior to electrophoresis on a 5% denaturing polyacrylamide gel (containing 20% formamide and 7 m urea) and analysis of the dried gel by phosphorimaging (GE Healthcare).
RESULTS
The Substrate Specificity of Ku
We applied a series of systematically varied abasic site-containing substrates to determine the substrate specificity of Ku. All substrates employ a common core double-stranded 40-bp-long DNA (Fig. 1A), which is sufficient for Ku to both load onto DNA ends and translocate internally (22). Because Ku loads on DNA ends asymmetrically, with the N terminus of its 70-kDa subunit oriented toward the broken end (26), we additionally blocked the end distal to the abasic site to ensure that asymmetry in loading onto broken ends appropriately contributes to the substrate specificity of Ku.
We previously reported that Ku is active in cleaving abasic sites located within short (<4 nucleotides) 5′ overhangs (9). Here we varied the length of this overhang and the position of the abasic site within that overhang: first increasing the distance of single-stranded DNA 5′ of the abasic site (Fig. 1, B and C, Substrates 1, 3, and 4) and then increasing the distance between the abasic site and the duplex region of the substrate (Fig. 1, B and C, Substrate 5). The substrate with an abasic site farthest away from the duplex (7 nucleotides) was least active, indicating an important role for nearby double-stranded DNA (Fig. 1, B and C Substrate 5). A 5′-phosphate also contributes significantly (at least for abasic sites near duplex termini) as lyase activity is reduced 2-fold in its absence (Fig. 1C, compare Substrates 1 and 1NoP). Notably, placing the 5′-phosphate and strand terminus 4 and 7 nucleotides away from the abasic site (Fig. 1C, Substrates 3 and 4) resulted in only a slightly greater reduction in activity, and activity on substrate 3 was no longer affected by additional omission of the 5′-phosphate (Fig. 1C, compare Substrates 3 and 3NoP). In sum, the AP lyase activity of Ku on 5′ overhangs is constrained mostly by its preference that the abasic site is close to the end of double-stranded DNA (Fig. 1C, compare Substrates 1 and 5). Within this context, Ku is stimulated when the abasic site is also within 1–2 nucleotides of a strand-terminating 5′-phosphate (Fig. 1C, compare Substrate 1 with Substrates 2–4).
Is the AP lyase of Ku similarly active on abasic sites in 3′ overhangs (i.e. in the bottom strand)? AP lyase activity near 3′ termini would fail to “clean” termini in preparation for ligation, instead leaving behind a 3′-α,β-unsaturated aldehyde residue similar to a 3′-phosphoglycolate (6). Strikingly, Ku has no significant AP lyase activity on the 3′ overhang context most comparable with its optimum 5′ overhang substrate (Fig. 2; compare Substrates 1 and 2), although its activity is somewhat restored with additional 3′-flanking DNA (Fig. 2, Substrates 3 and 4). Therefore, the AP lyase activity of Ku is reduced in a context where its activity alone would be insufficient to allow joining.
Given that Ku is more active near 5′ termini, is it important that the abasic site be located within an overhang, or is Ku equally active on abasic sites near 5′ termini in duplex DNA? To address this question, we assessed AP lyase activity on substrates with abasic sites embedded in double-stranded DNA increasingly distal from the 5′ terminus (1, 2, 4, and 7 base pairs). To isolate the contribution of flanking double-stranded DNA, we directly compared the activity of Ku on each of these substrates in pairwise fashion with the substrate with an abasic site positioned the same distance relative to the 5′ terminus, but within 5′ single-stranded overhangs (Fig. 3A, ss versus ds). A single complementary base pair preceding the abasic site has no significant impact (such a substrate presumably “breathes” sufficiently and therefore should not be considered double-stranded), nor did a slight extension of the bottom strand (preserving a single 5′ complementary base pair) (Fig. 3B, Substrate 1). However, embedding the abasic site another complementary base pair reduces activity 3.2-fold, relative to the cognate substrate in an ssDNA overhang (Fig. 3, Substrate 2), and four or more complementary bases preceding the abasic site resulted in a greater than 10-fold reduction (Fig. 3A, Substrates 3 and 4).
We previously characterized a Ku heterodimer with a mutated Ku70 subunit, Ku70 with Lys-31, Lys-160, and Lys-164 substituted with alanine, that had diminished 5′-dRP/AP lyase activity, but normal DNA binding (9). We show here that substitution of Lys-31 and Lys-160 alone (Ku70 2A) is sufficient to reduce activity on 5′ overhang substrates 4-fold (Fig. 3C). As with previous work using the 3A mutant, trapping of the 5′-dRP/AP lyase covalent reaction intermediate with borohydride (Fig. 3D) shows that the Ku70 2A mutant no longer has significant ability to form this covalent intermediate. Additionally, adduct formation with Ku80 is slightly stimulated, arguing that residual activity of the Ku70 2A mutant heterodimer is due to partial compensation by sites in Ku80 (9) (Fig. 3E, lanes 3 and 4). Strikingly, the Ku70 subunit of a wild type heterodimer is also unable to efficiently form a covalent intermediate when the abasic site is embedded in double-stranded DNA, and even the ability of Ku80 to form adducts is reduced (Fig. 3E, lane 7). The 5′-dRP/AP lyase activity of Ku is thus strongly reduced by significant double-stranded DNA 5′ of the abasic site, and this reduction is due to an inability of the 5′-dRP/AP lyase active site of Ku (primarily defined by Ku70 Lys-31 and Lys-160) to effectively engage these substrates.
The 5′-dRP/AP Lyase Activity of Ku and the NHEJ Ligation Step
Very low AP lyase activity when abasic sites are embedded significantly (>2 bp) within double-stranded DNA near a DSB suggests that NHEJ may not be able to repair such breaks if an abasic site in this context remains a block to ligation. We therefore made 300-bp radiolabeled substrates with either standard abasic sites or chemically reduced abasic sites (the latter of which are resistant to lyase activity) and assessed whether they could be joined by Ku and the NHEJ ligase complex (XRCC4, XLF, ligase IV) in vitro. The substrates were further designed such that they could join directly, without processing (Fig. 4C, P1), or after precise excision of abasic sites reveals complementary overhangs (Fig. 4C, P2). As shown previously, Ku can and must excise abasic sites within 5′ overhangs; a reduced abasic site in this context blocks ligation (Fig. 4A, lane 6) (9), and ligation products formed with standard abasic sites were fully resistant to digestion by the AP lyase fpg or alkali treatment (supplemental Fig. 1) (9). In contrast, reduction of an abasic site embedded 3 bp from the end has no significant impact on ligation (Fig. 4B, compare lanes 3 and 6). It is therefore not necessary to excise abasic sites in the latter context. To address whether excision by Ku nevertheless precedes ligation when the abasic site is not reduced, we analyzed the structure of the ligation products. We observed that the majority of products (Fig. 4D, lane 3, P1; 88.1% (± 1.4%) of ligation) are sensitive to digestion with fpg, indicative of ligation without prior excision. A 6-bp-smaller and fpg-insensitive product indicative of sequential excision and ligation (Fig. 4C, P2) accounts for only 12% of total ligation product. Therefore, AP lyase activity is not only dispensable for ligation of a substrate containing an abasic site embedded 3 bp into double-stranded DNA, but also is so low in this context that ligation generally proceeds without excision of the abasic site, resulting in preservation of the terminal sequence.
In cells, a variety of abasic site-cleaving enzymes could act on the substrates described above. We therefore addressed how the same four NHEJ substrates used above are joined in cells by introducing them into wild type HCT116 cells, as well as a variant of this cell line made deficient in the ligase required for NHEJ (ligase IV−/−) by gene targeting (Fig. 5A) (25). Joined products were detectable only in wild type cells, confirming the dependence of this assay on classically defined NHEJ (Fig. 5B). We then determined that an abasic site within a 5′ overhang must be excised by a 5′-dRP/AP lyase for efficient NHEJ in wild type cells as we detect a defined product in this direct assay only if the abasic site is not reduced (supplemental Fig. 1C). This is consistent with previous analyses indicating that a reduced abasic site in this context made NHEJ both less accurate and 20-fold less efficient, relative to NHEJ with the same substrate containing a standard abasic site (9). By comparison, reduction of an abasic site embedded 3 bp in double-stranded DNA flanking the break does not block cellular NHEJ (Fig. 5B, lane 5), as also observed in vitro. An abasic site in this context is thus not a barrier to the NHEJ ligation step, and such ends can be joined without excision. However, when using a standard abasic site, 40% (± 9%) of cellular ligation products were consistent with direct ligation (Fig. 5B, lane 2, and 5C), a smaller fraction than observed in vitro (Figs. 4D, lane 3, and 5C). Abasic sites embedded in duplex DNA are thus imperfectly protected from excision during cellular NHEJ, possibly through activity of other abasic site-cleaving activities, as discussed below.
FIGURE 5.
Joining of ends with near-terminal abasic sites in cells. A, substrates described in the legend for Fig. 4 were introduced by electroporation into HCT116 cells, and recovered DNA was analyzed after HinfI digestion by denaturing PAGE as in Fig. 4D. 5P, 5′-phosphate; P1, ligation without prior excision; P2, ligation after excision at the abasic site. B, recovered DNA was analyzed either without electroporation (−) or after electroporation of substrate (Sub.) into standard (WT) or ligase IV-defective (L4−/−) HCT116 cells. C, quantification of proportion of directly ligated product (P1) of the total defined ligation product (P1/(P1+P2)) generated by NHEJ in vitro (from Fig. 4D) or in cells (from Fig. 5B). Averages and standard deviations (error bars) are from results of three independent experiments.
DISCUSSION
We have shown that the 5′-dRP/AP lyase of Ku is most active on abasic sites in single-stranded DNA, within 2–3 nucleotides from both a 5′-PO4 terminus and a double-strand/single-strand DNA transition. Ku is less active on abasic sites near 3′ strand termini and is nearly inactive on abasic sites significantly embedded (>3 bp) in double-stranded DNA. Comparison of the substrate spectrum of Ku with that of other dRP/AP lyases indicates that there is little overlap. The primary lyase implicated in single-strand break repair, DNA polymerase β, is relatively inactive on abasic sites within 5′ ssDNA overhangs (the optimum substrate of Ku) (9), but it is robustly active on 5′-dRP sites embedded in extended duplex DNA. Notably, the endonuclease VIII-like (Neil1, Neil2) enzymes also cleave abasic sites within single-stranded DNA (27), including the 5′ overhang substrates described here.4 However, Neil1/Neil2 expression is highest in S phase (28), and they interact specifically with replisome components (29); thus the biological targets for Neil1/Neil2 activity are thought to be primarily abasic sites within ssDNA bubbles generated during replication (28). This may help explain previous work showing that, among the abasic cleaving activities in mammalian cell extracts (including Neil1/Neil2), Ku easily accounts for the majority of activity on abasic sites near DSB 5′ termini (9). In general, the substrate specificity of Ku is consistent with a fairly clear division of labor for the principle dRP/AP lyases.
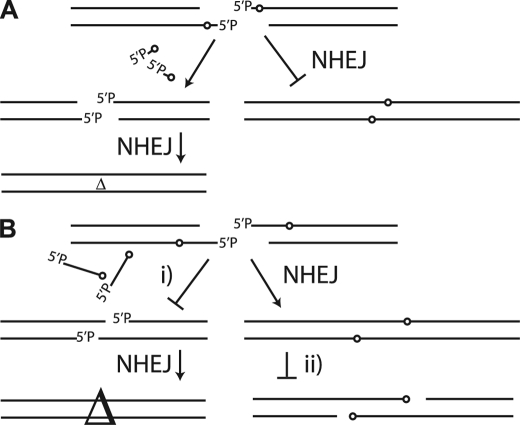
We suggest that there are significant advantages to this division of labor for NHEJ. Ku is most active in contexts where the abasic site blocks activity of the ligase required for NHEJ (Fig. 6A), but is relatively inactive in contexts where the abasic site does not interfere with activity of the NHEJ ligase (Fig. 6B). In other words, Ku cuts when it must, and does not cut when it does not have to. The latter restriction promotes retention of DNA between the abasic site and the DSB terminus in ligation products, effectively reducing deletion associated with NHEJ of damaged ends (Fig. 6B, step i). Specifically, 88% of in vitro NHEJ reaction products are P1 (ligation without excision), a joined product that is 6 bp larger than P2 (excision before ligation), because it retains sequence between the 5′ terminus and the abasic site (Fig. 4C).
FIGURE 6.
Role of substrate specificity in protecting damaged ends. A, abasic sites, and possibly other types of damage as well, block ligation activity when within 5′ overhangs and must be excised for NHEJ completion. 5P, 5′-phosphate. B, abasic sites in adjacent flanking DNA do not block the activity of the NHEJ ligase, and if not excised before ligation by either Ku or BER enzymes (step i), junctions possess less deleted sequence (△; symbol size is proportional to amount of deletion). Reduced activity from Ku and BER enzymes on the same abasic sites after joining (step ii) may also reduce the chance of recleavage of the product of NHEJ.
This restriction of the activity of Ku also influences cellular NHEJ as we can still detect full-length product, although at lower frequency than observed in vitro (Fig. 5C). By comparison, we cannot detect the comparable product when the abasic site is within the single-stranded overhang (supplemental Fig. 1). With respect to abasic sites flanked by double-stranded DNA, activity of cellular BER enzymes (e.g. DNA polymerase β or AP endonuclease 1 (APE1)), although reduced near a DSB terminus (7–9), could act prior to NHEJ and account for differences between our in vitro and cellular results. The division of labor between NHEJ and BER may thus not be perfect, at least in this specific context (abasic sites embedded 3 bp from the end of double-stranded DNA). Alternatively, abasic sites in either the NHEJ substrate or the product may not be completely stable during transfection and sample processing.
Assuming that Ku remains bound at least transiently to products of joining, reduced activity of Ku on double-stranded DNA also lowers the chance that Ku would act after joining, potentially recleaving junctions (Fig. 6B, step ii). This is similar to the apparent discouragement of simultaneous action of base excision repair on nearby, opposite strand damaged sites within a damage cluster, a mechanism that helps reduce generation of DSBs (aborted base excision repair) (11–13). Indeed, Ku has been reported to inhibit AP site-cleaving activity when the AP site is embedded in double-stranded DNA (30, 31). The persistence of Ku near the product of recently joined ends with residual damage may thus help discourage recleavage of the chromosome by an aborted BER reaction.
The ability of Ku to specifically recognize chromosome breaks relies on its ability to thread double-stranded DNA through a central channel, or ring. We initially anticipated that the 5′-dRP/AP lyase active site of Ku would be located within this ring, given its extensive interface with DNA. However, mutagenesis suggests a location of the active site within the N-terminal domain of Ku70 (9) (Fig. 3, C and D). This location, nearby but distinct from the ring that wraps around double-stranded DNA, is consistent with the specificity of Ku for abasic sites in ssDNA near an ssDNA/dsDNA transition (overhangs near a DSB terminus). Location within the N terminus of Ku70 also orients the active site on the appropriate side of the ring, given that Ku binds asymmetrically with the Ku70 N terminus toward the broken end (26). As noted above, the spectrum of substrates conferred by this active site location is far more useful than would likely be possible were the active site located within the ring, where cleavage activity would be restricted to double-stranded DNA.
NHEJ and BER/single-stand break repair must process similarly damaged termini in repair of double-strand and single-strand breaks, respectively. Not surprisingly, there are several examples where these pathways use the same enzyme for this function (e.g. polynucleotide kinase/phosphatase) (32). With regard to abasic sites, however, the two pathways employ different enzymes: Ku versus APE1/DNA polymerase β. The mechanism by which Ku recognizes double-strand breaks (and the consequent unparalleled specificity and affinity that Ku has for DSB termini) might be reason enough for this division of labor, but the additional restrictions on the activity of Ku described here suggest other significant advantages.
Supplementary Material
Acknowledgments
We thank Crystal Waters and Kenjiro Asagoshi for critical reading of the manuscript and Kenjiro Asagoshi for analysis of HCT116 transfections by flow cytometry.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA 84442 (to D. A. R.) and RO1 GM088351 and RO1 CA154461 (to E. A. H.).

This article contains supplemental Fig. 1.
N. T. Strande and D. A. Ramsden, unpublished observations.
- NHEJ
- nonhomologous end joining
- 5′-dRP
- 5′-deoxyribose 5′-phosphate
- AP
- apurinic/apyrimidinic
- DSB
- double-strand break
- BER
- base excision repair
- XRCC4
- X-ray cross-complementary gene 4
- XLF
- XRCC4-like factor
- fpg
- formamidopyrimidine glycosylase
- Neil1/Neil2
- endonuclease VIII-like
- APE1
- AP endonuclease 1
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Rooney S., Chaudhuri J., Alt F. W. (2004) The role of the nonhomologous end joining pathway in lymphocyte development. Immunol. Rev. 200, 115–131 [DOI] [PubMed] [Google Scholar]
- 2. Jeggo P. A. (1990) Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat. Res. 239, 1–16 [DOI] [PubMed] [Google Scholar]
- 3. Sekiguchi J. M., Gao Y., Gu Y., Frank K., Sun Y., Chaudhuri J., Zhu C., Cheng H. L., Manis J., Ferguson D., Davidson L., Greenberg M. E., Alt F. W. (1999) Nonhomologous end joining proteins are required for V(D)J recombination, normal growth, and neurogenesis. Cold Spring Harbor Symp. Quant. Biol. 64, 169–181 [DOI] [PubMed] [Google Scholar]
- 4. Vogel H., Lim D. S., Karsenty G., Finegold M., Hasty P. (1999) Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. U.S.A. 96, 10770–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ward J. F. (1994) The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat Biol. 66, 427–432 [DOI] [PubMed] [Google Scholar]
- 6. Chen S., Inamdar K. V., Pfeiffer P., Feldmann E., Hannah M. F., Yu Y., Lee J. W., Zhou T., Lees-Miller S. P., Povirk L. F. (2001) Accurate in vitro end joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 276, 24323–24330 [DOI] [PubMed] [Google Scholar]
- 7. Datta K., Purkayastha S., Neumann R. D., Pastwa E., Winters T. A. (2011) Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiation Res. 175, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dobbs T. A., Palmer P., Maniou Z., Lomax M. E., O'Neill P. (2008) Interplay of two major repair pathways in the processing of complex double-strand DNA breaks. DNA Repair 7, 1372–1383 [DOI] [PubMed] [Google Scholar]
- 9. Roberts S. A., Strande N., Burkhalter M. D., Strom C., Havener J. M., Hasty P., Ramsden D. A. (2010) Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 464, 1214–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pogozelski W. K., Tullius T. D. (1998) Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 98, 1089–1108 [DOI] [PubMed] [Google Scholar]
- 11. Ma W., Resnick M. A., Gordenin D. A. (2008) Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 36, 1836–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malyarchuk S., Castore R., Harrison L. (2008) DNA repair of clustered lesions in mammalian cells: involvement of nonhomologous end joining. Nucleic Acids Res. 36, 4872–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malyarchuk S., Castore R., Harrison L. (2009) Apex1 can cleave complex clustered DNA lesions in cells. DNA Repair 8, 1343–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang N., Galick H., Wallace S. S. (2004) Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double-strand breaks. DNA Repair 3, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 15. Di Noia J. M., Williams G. T., Chan D. T., Buerstedde J. M., Baldwin G. S., Neuberger M. S. (2007) Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 204, 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rada C., Di Noia J. M., Neuberger M. S. (2004) Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16, 163–171 [DOI] [PubMed] [Google Scholar]
- 17. Matsumoto Y., Kim K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 269, 699–702 [DOI] [PubMed] [Google Scholar]
- 18. Prasad R., Beard W. A., Strauss P. R., Wilson S. H. (1998) Human DNA polymerase β deoxyribose phosphate lyase: substrate specificity and catalytic mechanism. J. Biol. Chem. 273, 15263–15270 [DOI] [PubMed] [Google Scholar]
- 19. Sobol R. W., Prasad R., Evenski A., Baker A., Yang X. P., Horton J. K., Wilson S. H. (2000) The lyase activity of the DNA repair protein β-polymerase protects from DNA damage-induced cytotoxicity. Nature 405, 807–810 [DOI] [PubMed] [Google Scholar]
- 20. Allinson S. L., Dianova I. I., Dianov G. L. (2001) DNA polymerase β is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J. 20, 6919–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piersen C. E., McCullough A. K., Lloyd R. S. (2000) AP lyases and dRPases: commonality of mechanism. Mutat. Res. 459, 43–53 [DOI] [PubMed] [Google Scholar]
- 22. Dynan W. S., Yoo S. (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26, 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Downs J. A., Jackson S. P. (2004) A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5, 367–378 [DOI] [PubMed] [Google Scholar]
- 24. Nick McElhinny S. A., Snowden C. M., McCarville J., Ramsden D. A. (2000) Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell Biol. 20, 2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fattah F., Lee E. H., Weisensel N., Wang Y., Lichter N., Hendrickson E. A. (2010) Ku regulates the nonhomologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 6, e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoo S., Kimzey A., Dynan W. S. (1999) Photocross-linking of an oriented DNA repair complex: Ku bound at a single DNA end. J. Biol. Chem. 274, 20034–20039 [DOI] [PubMed] [Google Scholar]
- 27. Dou H., Mitra S., Hazra T. K. (2003) Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684 [DOI] [PubMed] [Google Scholar]
- 28. Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., Mitra S. (2002) Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 3523–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dou H., Theriot C. A., Das A., Hegde M. L., Matsumoto Y., Boldogh I., Hazra T. K., Bhakat K. K., Mitra S. (2008) Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen: the potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 283, 3130–3140 [DOI] [PubMed] [Google Scholar]
- 30. Hashimoto M., Donald C. D., Yannone S. M., Chen D. J., Roy R., Kow Y. W. (2001) A possible role of Ku in mediating sequential repair of closely opposed lesions. J. Biol. Chem. 276, 12827–12831 [DOI] [PubMed] [Google Scholar]
- 31. Ilina E. S., Lavrik O. I., Khodyreva S. N. (2008) Ku antigen interacts with abasic sites. Biochim. Biophys. Acta 1784, 1777–1785 [DOI] [PubMed] [Google Scholar]
- 32. Caldecott K. W. (2008) Single-strand break repair and genetic disease. Nat. Rev. Genet. 9, 619–631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.