Background: PCNA is a multifunctional component of DNA replication and repair machinery.
Results: A novel small molecule inhibitor of the PCNA protein-protein interaction inhibited DNA replication, induced DNA replication stress, and increased cisplatin-mediated DNA damage response in cells.
Conclusion: The biochemical PCNA inhibitor can inhibit PCNA functions essential for cells.
Significance: Inhibition of the PCNA protein-protein interaction can be a new strategy to sensitize cancer cells to chemotherapy.
Keywords: Anticancer Drug, Chemical Biology, Crystallography, DNA Repair, DNA Replication, Drug Design, Protein-Protein Interactions, PCNA, Small Molecule, Translesion Synthesis
Abstract
We have discovered that 3,3′,5-triiodothyronine (T3) inhibits binding of a PIP-box sequence peptide to proliferating cell nuclear antigen (PCNA) protein by competing for the same binding site, as evidenced by the co-crystal structure of the PCNA-T3 complex at 2.1 Å resolution. Based on this observation, we have designed a novel, non-peptide small molecule PCNA inhibitor, T2 amino alcohol (T2AA), a T3 derivative that lacks thyroid hormone activity. T2AA inhibited interaction of PCNA/PIP-box peptide with an IC50 of ∼1 μm and also PCNA and full-length p21 protein, the tightest PCNA ligand protein known to date. T2AA abolished interaction of PCNA and DNA polymerase δ in cellular chromatin. De novo DNA synthesis was inhibited by T2AA, and the cells were arrested in S-phase. T2AA inhibited growth of cancer cells with induction of early apoptosis. Concurrently, Chk1 and RPA32 in the chromatin are phosphorylated, suggesting that T2AA causes DNA replication stress by stalling DNA replication forks. T2AA significantly inhibited translesion DNA synthesis on a cisplatin-cross-linked template in cells. When cells were treated with a combination of cisplatin and T2AA, a significant increase in phospho(Ser139)histone H2AX induction and cell growth inhibition was observed.
Introduction
Numerous cancer drug discovery programs have targeted specific pathways mediated by oncogene products. Often these agents are not therapeutically efficacious in diverse cancers, presumably because tumors can be supported by multiple mechanisms (e.g. not “addictive” to the pathway targeted). This leads to failure of proof-of-concept validation in the cancer drug discovery programs. Such therapeutics have been proven successful for cancers if they are addictive to the pathway (1). However, there is a high risk of resistance based on mutation of the pathway components targeted, as seen in mutations of BCR-ABL by imatinib (2) and of SMO by vismodegib (3). In contrast, therapeutics targeting non-oncogenic mediators that ubiquitously support cancers, such as histone deacetylase, heat-shock proteins, ubiquitin ligase, spliceosome, and proteasome, have been successful (4).
PCNA3 is an essential DNA clamp loader, which acts as a scaffold protein that organizes numerous components for DNA replication, DNA damage repair, chromatin formation, and cell cycle progression (5). Several translesion DNA synthesis (TLS) DNA polymerases also interact with PCNA (6, 7) and engage in replication repair of DNA damage induced by chemotherapy agents (8), therefore promoting chemotherapy resistance. Given its diverse functions, PCNA is regarded as one of the essential non-oncogenic mediators supporting cancer growth (9); therefore, inhibitors of PCNA could be useful for cancer therapeutics. Although post-translational modifications, such as ubiquitination, emerged as important for PCNA functions (10), the majority of PCNA-interacting components bind to a cavity of PCNA with a short sequence motif called a PIP-box in a very similar manner (5). Thus, the PCNA/PIP-box interaction is absolutely essential for many PCNA functions, and consequently compounds inhibiting this interaction can be expected to inhibit PCNA functions. In this study, we have created a small molecular inhibitor of the PCNA/PIP-box interaction and investigated its pharmacological effects in cells from a chemotherapeutic viewpoint.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Cell Cultures
The following antibodies were used per the manufacturers' recommendation: anti-PCNA PC10 mouse mAb (Cell Signaling 2586), anti-Polδ3 rabbit (Sigma Prestige Antibodies HPA039627), anti-BrdU B44 mouse mAb (BD Biosciences Immunocytometry Systems 347580), anti-phospho(Ser33)RPA32 (replication protein A 32-kDa subunit) rabbit (Bethyl Laboratories A300-246A), anti-phospho(Ser345)Chk1 (checkpoint kinase 1) 133D3 rabbit (Cell Signaling 2348), Anti-phospho(Ser139)histone H2AX JBW301 mouse (Millipore 05-636), IgG-HRP-conjugated secondary antibodies (Cell Signaling), and Alexa Fluor-conjugated secondary antibodies (Invitrogen). Sources for plasmids in this study are given in the supplemental material. U2OS and HeLa cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. XP2OS(SV40) cells were cultured in RPMI1640 medium containing 10% FBS. All cells were maintained at 37 °C in a humidified, 5% carbon dioxide incubator.
High-throughput Screening and Fluorescence Polarization Assay
Screening for PCNA inhibitors was carried out using a fluorescence polarization (FP) assay in a solution consisting of 100 nm PCNA protein and 10 nm N-terminal 5-carboxyfluorescein-labeled PL-peptide (SAVLQKKITDYFHPKK) (11) in FP buffer (35 mm HEPES, pH 7.4, 10% glycerol, and 0.01% Triton X-100) for 38,035 compounds. Briefly, 20 μl of the assay solution was transferred into each well of a black 384-well plate using Wellmate (Matrix). Twenty nanoliters of the test compounds in DMSO solution were pin-transferred (V&P Scientific) by Biomek (Beckman Coulter) into the PCNA-PL solution in triplicate to give a final drug concentration of 10 μm in each well. The negative control in each plate was DMSO, whereas the positive control was unlabeled PL-peptide as a self-competitor. After 30 min, the fluorescence was read using an EnVision plate reader (PerkinElmer Life Sciences) with 485-nm excitation (20-nm bandpass) and 535-nm emission (20-nm bandpass) filters and the supplied 510-nm dichroic mirror. FP values were calculated in millipolarization units. All data processing was performed using in-house programs in the Pipeline Pilot (Accelrys, version 7.0.1). The quality of the primary screen was assessed by Z-prime and other screening quality metrics. 91 compounds (0.24% hit rate) showing >40% decrease in fluorescence polarization were further validated by establishing dose response (10 μm to 0.5 μm final concentration) in triplicate using a freshly prepared batch of each compound in at least two different FP buffers. Among compounds giving regenerative saturating sigmoidal dose-response curves, T3 was selected for validation in this study.
Crystallization and Structure Determination of PCNA in Complex with T3
Human PCNA was overexpressed and purified by a procedure similar to that previously reported (12). Crystals of PCNA-T3 complex were obtained by sitting-drop vapor diffusion methods by mixing of equal volumes of protein solution (9 mg/ml PCNA, 1 mm T3, 9 mm HEPES-NaOH, pH 7.4, 90 mm NaCl, and 10% DMSO) and a reservoir solution (100 mm sodium cacodylate, pH 6.5, 200 mm NaCl, and 2.0 m ammonium sulfate). Cubic crystals were obtained in several days at 293 K. Prior to x-ray diffraction experiments, crystals were transferred to the reservoir solution containing 20% ethylene glycol for cryoprotection. X-ray diffraction data were collected under N2 gas steam (100 K) at Photon Factory beamline BL-5A. Diffraction data were processed using the program HKL2000. The structure of the PCNA-T3 complex was solved by the molecular replacement method with the program MOLREP. Model building was performed with the program COOT. Structure refinement was performed with the programs CNS and REFMAC. The geometries of the final structure were validated with the program PROCHECK. Data collection and refinement statistics are given in Table 1. References for the programs are listed in the supplemental material.
TABLE 1.
Protein crystallography data collection and refinement statistics
Values in parentheses are for the highest resolution shell. Ramachandran statistics indicate the fraction of residues in the most favored/allowed/disallowed regions, respectively.
| Data collection | |
| Space group | I213 |
| a = b = c (Å) | 143.1 |
| Resolution range (Å) | 50.0–2.10 (2.18–2.10) |
| Observed reflections | 164,205 |
| Unique reflections | 28,025 |
| Rmerge | 0.066 (0.392) |
| Completeness (%) | 98.0 (92.9) |
| 〈I〉/σ〈I〉 | 11.0 (2.0) |
| Refinement | |
| Resolution range (Å) | 20.0–2.10 |
| R/Rfree | 0.205/0.252 |
| Root mean square bond distances (Å)/angles (degrees) | 0.011/1.331 |
| Ramachandran statistics (%) | 95.1/4.9/0 |
| Protein Data Bank code | 3VKX |
Preparation of T2 Amino Alcohol (T2AA) Compound
To a mixture of dry tetrahydrofuran (5.5 ml), dry 1,4-dioxane (5.0 ml), and 2 m lithium borohydride tetrahydrofuran solution (3.0 ml), chlorotrimethylsilane (1.53 ml) was added in dropwise manner with cooling by an ice bath under nitrogen. The mixture was stirred at room temperature for 15 min and cooled again in an ice bath, and 3,5-diiodothyronine (Toronto Research Chemicals) (1.05 g) was added at once. The mixture was stirred overnight, allowing it to reach the ambient temperature. The mixture has been heterogeneous throughout, in which the absence of 3,5-diiodothyronine was ensured by TLC and LC-MS. The reaction mixture was carefully added to ice water, adjusted to pH >9, and extracted by ethyl acetate. After drying the organic phase over anhydrous sodium sulfate and evaporation under reduced pressure, the crude material was solidified and successively washed with a mixture of n-hexane and diethyl ether (4:1) to isolate T2AA as white crystals (0.91 g, 89% yield), which were further purified by recrystallization from a mixture of diethyl ether and 1,4-dioxane (8:1) and dried in vacuo. 1H NMR (400 MHz, DMSO) δ 9.08 (broad s, 1H), 7.76 (s, 2H), 6.66 (d, J = 8.0 Hz, 2H), 6.52 (d, J = 8.0 Hz, 2H), 4.61 (broad s, 1H), 3.32–3.24 (m, 2H), 2.85 - 2.78 (m, 1H), 2.65 (dd, J = 13.4, 5.0 Hz, 1H), 2.35 (dd, J = 13.3, 8.2 Hz, 1H), 1.48 (broad s, 2H). MS (ESI+) m/z, 511.79.
Preparation of Recombinant Proteins and TR Reporter Assay
See the supplemental material.
Pull-down Assay
A His protein interaction pull-down kit (Thermo Scientific) was used, in which a 4:1 mixture of TBS and lysis buffer provided in the kit was used as the buffer throughout. For immobilizing p21, 100 μg of p21His6 was incubated overnight at 4 °C with 25 μl (50% suspension) of prewashed cobalt chelate resin in 500 μl of the buffer and washed with the buffer four times. Separately, PCNA (500 nm) in 500 μl of the buffer was mixed with the indicated final concentration of T2AA and transferred to the p21-immobilized resin. After the mixture was incubated overnight at 4 °C, the resin was washed with buffer four times, and the bound proteins were eluted with 100 μl of the buffer containing 400 mm imidazole. Each 10 μl of the eluted samples was analyzed by SDS-PAGE with Oriole fluorescent gel stain (Bio-Rad) and a FluorChem imager (Alpha Innotech).
Immunoblotting
The cells treated as indicated were washed twice with ice-cold PBS, collected in a spin tube, and lysed with radioimmune precipitation assay buffer supplemented with Halt protease inhibitor mixture and Halt phosphatase inhibitor mixture (Thermo Scientific) using approximately twice the volume of the cell pellet on ice for 0.5 h. Protein concentration was determined by a BCA assay (Thermo Scientific) according to the manufacturer's recommendation. Normalized amounts of samples were loaded on SDS-PAGE as indicated and electrotransferred to a PVDF membrane. The membrane was blocked with SuperBlock buffer (Thermo Scientific) and incubated with the indicated primary antibodies in SuperBlock at 4 °C overnight, rinsed with TBS, 0.05% Tween 20 three times for 10 min, incubated with the corresponding secondary IgG-HRP conjugate in SuperBlock at room temperature for 1 h, and rinsed with TBS, 0.05% Tween 20 for 15 min. Proteins probed on the membrane were visualized by chemiluminescence using WestPico reagent (Thermo Scientific) and developed on BioMax MR film (Eastman Kodak Co.).
Cell Growth Assay
The cells were treated with drugs as indicated in a black (transparent flat bottom) 384-well or 96-well plate. Cellular viability was determined by using Alamar Blue reagent (Invitrogen) on an EnVision plate reader according to the manufacturer's recommendations and normalized with signal from wells of DMSO treatment = 100% and of no cells = 0%.
Chromatin Immunostaining
Cells grown on a coverslip were incubated with the indicated concentrations of T2AA for 24 h at 37 °C, rinsed once with ice-cold PBS, pre-extracted with ice-cold CSK buffer (100 mm sodium chloride, 3 mm magnesium chloride, 10 mm HEPES, pH 7.4, 300 mm sucrose) containing 0.3% Triton X-100, Halt protease inhibitor mixture and Halt phosphatase inhibitor mixture (Thermo Scientific) for 5 min on ice, and rinsed once with the CSK buffer to remove Triton X-100 from the specimens. The cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature, rinsed three times with PBS for 5 min each, permeabilized with ice-cold methanol for 10 min at −20 °C, and rinsed once with PBS for 5 min. The specimens were blocked with PBS containing 5% FBS and 0.03% Triton X-100 for 60 min, applied anti-PCNA antibody (1:3200) and anti-Polδ3 antibody (2.9 μg/ml) or anti-phospho(Ser33)RPA32 antibody (4.0 μg/ml) in antibody buffer (PBS containing 1% FBS and 0.03% Triton X-100), incubated overnight in a humid chamber at 4 °C, and rinsed with PBS three times for 5 min each at room temperature. The specimens were incubated with Alexa Fluor 555-conjugated goat anti-mouse IgG (Invitrogen) antibody (1:500) in the antibody buffer for 2 h in a humidified chamber at room temperature in the dark, rinsed once with PBS for 5 min, incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen) antibody (1:500) in the antibody buffer for 2 h in a humidified chamber at room temperature in the dark, and rinsed three times with PBS for 5 min. The coverslips were mounted in VectaShield containing DAPI at 1 μg/ml (Vector Laboratories). Immunofluorescence imaging for chromatin was performed on a C1Si microscope (Nikon) with a Cascade 512B photomultiplier (Photometrics). Images were processed using EZC1 (Nikon) and Photoshop (Adobe) software. Phospho-Chk1 and γH2AX staining was performed in the same manner except using anti-phospho(Ser345)Chk1 antibody (1:50) or anti-phospho(Ser139)histone H2AX antibody (1:500) and omitting the pre-extraction and methanol treatment steps.
DNA Replication Assay
DNA replication in U2OS and/or HeLa cells were analyzed by the rates of bromodeoxyuridine (BrdU) incorporation by a method reported previously (13). For immunofluorescence, cells were propagated at a subconfluent density onto a coverslip in a 6-well culture plate the day before the assay. Cells were treated with T2AA for 24 h at the indicated concentrations. During the last 15 min in the incubation period, BrdU was added at a concentration of 10 μm. Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. After permeabilization with 0.2% Triton X-100 in PBS for 15 min, cells were treated with 4 n hydrochloric acid for 10 min and extensively rinsed with PBS. After blocking cells with 3% FBS in PBS for 20 min, anti-BrdU monoclonal antibody (1 μg/ml; clone 44, BD Pharmingen) was added to the cells and incubated at room temperature for 1 h. Cells were rinsed with PBS three times for 5 min each. Anti-mouse IgG conjugated with Alexa Fluor 555 (Invitrogen) was added and incubated at room temperature for 30 min. Cells were rinsed with PBS three times for 5 min each and mounted in VectaShield containing DAPI (1 μg/ml). Fluorescence microscopy was done on an E800 microscope (Nikon) with a DXM1120 digital camera (Nikon). Images were processed using Photoshop (Adobe). For quantitative analyses, BrdU incorporation was measured with the APC BrdU Flow Kit (BD Pharmingen) following the manufacturer's instructions. Cells were pulsed with 20 μm BrdU 15 min before harvesting. The populations of BrdU-positive cells (a) and mean BrdU intensity in S-phase cells (b) were recorded. Total BrdU incorporation was defined as a × b.
Flow Cytometry
For cell cycle analysis, cells were cultured at 1 × 106 cells/well of 6-well plates in 2 ml of medium, treated with drugs as indicated, trypsin-lifted, washed once with PBS, suspended in propidium iodide (PI) solution (0.05 mg/ml), and treated with ribonuclease A (2 μg/ml; Calbiochem) at room temperature for 30 min. For apoptosis analysis, cells were cultured at 1 × 105 cells/well of 12-well plates in 1 ml of medium and treated with drugs as indicated. Dead cells detached in the medium were recovered by centrifuge, and live cells attached on the wells were lifted by trypsin. Both dead and live cells were combined and washed once with PBS, and the number of cells in each sample was adjusted to 1–3 × 105. The cells were stained with Annexin-V-FITC reagent (Roche Applied Science) according to the manufacturer's recommendation, followed by staining with PI. For γH2AX staining, cells were cultured at 4 × 105 cells/well of 12-well plates in 1 ml of medium, treated with drugs as indicated, trypsin-lifted and stained by using the FlowCellect H2A.X DNA Damage kit (FCCH025142, Millipore) according to the manufacturer's recommendations. All samples were filtered through 40-μm nylon mesh prior to running flow cytometry. The flow data were acquired using an LSRII flow cytometer equipped with 405-, 488-, 561-, and 640-nm lasers (BD Biosciences).
TLS Assay in Mammalian Cultured Cells
The cisplatin-modified plasmid was constructed, and the TLS assay was performed as previously described (14) with some modifications. Briefly, two oligonucleotides, 5′-GGGAGATCTGGAAGGATCTG-3′ and 5′-AATTCAGATCCTTCCAGATCTCCC-3′, were annealed and inserted between the sites of EcoRV and EcoRI in pUCSV40H. The resultant plasmid was designated as pUCSV40H+BglII. A 13-mer oligonucleotide modified with a 1,3-intrastrand d(GpTpG) platinum cross-link (Pt-GTG) was prepared as described previously (15). This oligonucleotide has the cross-link at the underlined site of 5′-CCTTCGTGCTCCC-3′. An unmodified 13-mer oligonucleotide was used for the construction of control plasmid. These oligonucleotides were purified with HPLC and PAGE. To form a gapped duplex plasmid, pUCSV40H and pUCSV40H+BglII were digested with EcoRV and ScaI, respectively, mixed, and incubated as described previously (14). The gapped duplex plasmid was incubated with a 2-fold molar excess of the purified 13-mer oligonucleotide, which is complementary to the gap sequence except for a bulge in the middle in the presence of ATP (1 mm) and T4 DNA ligase (New England Biolabs, Ipswich, MA) at 16 °C for 6 min. The covalently closed circle plasmid was purified with equilibrium centrifugation on a CsCl gradient. The plasmids with and without the cisplatin adduct are hereafter referred to as Pt-GTG and Mock-GTG, respectively.
For cellular TLS assay, nucleotide excision repair (NER)-deficient XP2OS(SV40) cells (16) were used to avoid elimination of the cross-link by NER. The cells were transfected with Pt-GTG or Mock-GTG plasmid (150 ng) using a Qiagen Effectene Reagent kit (Qiagen GmbH, Hilden, Germany) as described previously (14). Twenty-four hours after the transfection, the cells were exposed to 5 μm T2AA or 0.25% DMSO (as solvent control) for 48 h. The plasmids were extracted from the cells by using the QIAprep spin miniprep kit (Qiagen), digested with the restriction endonuclease DpnI (New England Biolabs) to remove the unreplicated plasmids, and introduced into Escherichia coli strain DH5α. The E. coli was plated onto LB agar plates containing ampicillin, X-gal, and isopropyl-β-d-thiogalactopyranoside. In this system, if the cisplatin-modified strand was replicated as a template (i.e. TLS), the reading frame of the lacZ gene of progeny plasmids would be functional; hence, E. coli colonies became blue. If the opposite strand was replicated (i.e. damage-induced strand loss), progeny plasmids would have a dysfunctional reading frame of the lacZ gene; hence, E. coli colonies showed a white color. Therefore, TLS frequency (%) was defined as (number of blue colonies/number of total colonies) × 100.
RESULTS
T3 Binds to PCNA Cavity That Interacts with PIP-box Sequences
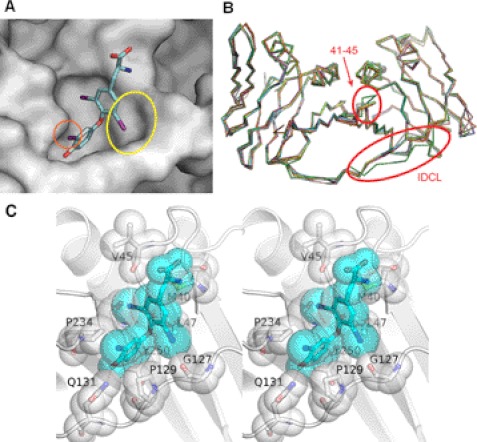
We have established a high-throughput screening protocol to find agents that inhibit biochemical PCNA/PIP-box interactions. This method uses conventional FP for screening chemical libraries by competing the binding of recombinant PCNA protein and fluorescently tagged PL-peptide that possesses a PIP-box sequence, which was previously optimized for high PCNA affinity (IC50 of ∼100 nm in self-competition; data not shown) (11). We have screened ∼38,000 compounds, including FDA-approved drugs and those with known biological activity, and then carefully verified initial hits. After eliminating nonspecific compounds by establishing dose-response curves for the hit compounds with at least two independent experiments, we have ensured that T3 (Fig. 1A) is a true inhibitor of PCNA/PL-peptide binding at IC50 of ∼3 μm (Fig. 1C). To further determine if T3 is binding to the bona fide PIP-box interaction site of PCNA or an allosteric site to trigger conformational change that leads to abolishing PCNA/PL-peptide interaction, we have solved the co-crystal structure of the PCNA-T3 complex (Fig. 2A; the detailed interactions of T3 with PCNA amino acids are depicted in Fig. 2C). T3 binds to the same PCNA cavity that PIP-box protein sequences tightly bind, by inducing an additional binding cavity with allosteric movement of the interdomain connecting loop (IDCL) of PCNA. This IDCL movement around the cavity is unique to the PCNA-T3 complex and is not observed in other PCNA complex structures reported to date (Fig. 2B). This suggests that IDCL has a role in accommodating ligands with additional interactions. Indeed, 3,3′-diiodothyronine, a T3 derivative, showed remarkably lower affinity than that of T3 (data not shown). This is presumably because the 5-iodine atom is missing, and thus it disables the induction of the IDCL movement.
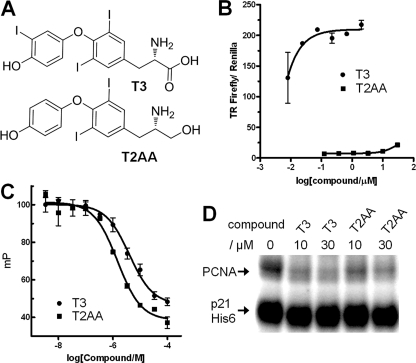
FIGURE 1.
Inhibition of PCNA protein-protein interaction by T3 and T2AA. A, chemical structure of T3 and T2AA. T2AA lacks 3′-iodine and carboxylic acid of T3 that are essential for the thyroid hormone activity of T3. B, thyroid hormone activity of T3 and T2AA. HepG2 cells were transiently transfected with an expression vector, CMV-TRβ, and a firefly luciferase reporter vector that contains thyroid hormone-responsive element, and were stimulated by titration of T3 (circle) or T2AA (square). T3 fully activated TRβ-TRE reporter at EC50 < 10 nm, and T2AA showed almost no activation. Error bars, S.E. C, inhibition of PCNA/PIP-box peptide interaction by T3 and T2AA. Fluorescent polarization value (in millipolarization units (mP)) was measured for titration of T3 (circle) or T2AA (square) in triplicate in a mixture of PCNA protein (100 nm) and fluorescein-tagged-SAVLQKKITDYFHPKK peptide (10 nm) that contains a PIP-box motif (underlined). Error bars, S.E. The inhibition curve was fitted by Prism (GraphPad) to determine IC50 as 3 μm (T3) and 1 μm (T2AA). D, inhibition of PCNA/full-length p21 interaction by T3 and T2AA. p21-His6 protein was immobilized on cobalt beads and incubated overnight with PCNA (500 nm) containing the indicated concentration of T3 or T2AA. PCNA bound on the p21-immobilized beads was analyzed by SDS-PAGE/Oriole Orange staining.
FIGURE 2.
Co-crystal structure of PCNA-T3 complex. A, close-up view of PCNA-T3 interaction site (Protein Data Bank code 3VKX). PCNA is shown as a gray surfaced model. T3 (Fig. 2A) is shown as a stick model, in which carbon is blue, oxygen is red, nitrogen is indigo, iodine is violet, and hydrogen is omitted. Water molecules packed in the crystal were omitted. The 5-iodine interacts with the IDCL loop, inducing an extra cavity (yellow circle). The 3′-iodine (orange circle) juts out from the PCNA interaction interface. B, superimposition of PCNA structures bound to T3 (green), p21 peptide (cyan, magenta, and yellow) (Protein Data Bank code 1AXC), and DNA Polη peptide (pink, white, and orange) (Protein Data Bank code 2ZVK). The averaged root mean square deviation value is 0.84 Å for corresponding Cα atoms. All PCNA structures except that of the PCNA-T3 complex are structurally very similar, whereas the PCNA-T3 complex possesses significant perturbation of IDCL and the region of Asp41–Val45. C, stereo diagram of detailed interaction between T3 (cyan) and PCNA (white). T3 and residues of PCNA involved in the interaction with T3 are shown by sticks and transparent spheres. A hydrogen bond is shown by orange dots.
T2AA, T3 Derivative Lacking TR Activity, Inhibits PCNA Protein-Protein Interaction
T3 is a very potent thyroid hormone, and this property precludes its use as a PCNA inhibitor. The carboxylic acid and 3′-iodine of T3 is essential for its thyroid hormone activity (17). In the PCNA-T3 crystal structure, the 3′-iodine atom of T3 is positioned out from the cavity (Fig. 2A), suggesting that it is dispensable for binding. Therefore, we have created a T3 derivative compound in which the carboxylic acid of T3 is replaced by an alcohol and the 3′-iodine is removed. The new compound T2AA (Fig. 1A) showed almost no thyroid hormone activity in a TR reporter assay (Fig. 1B) but inhibited the PCNA/PL-peptide binding at an IC50 value of ∼1 μm, a slightly better potency than that of T3 (Fig. 1C).
We next investigated if T2AA can inhibit PCNA interaction with a full-length protein. p21 is a protein containing a PIP-box with the highest affinity to PCNA known to date (5). Thus, if T2AA can inhibit PCNA-p21 interaction, theoretically T2AA can perturb any PCNA/PIP-box protein interactions in cells. A pull-down assay was carried out by using recombinant p21 protein immobilized on cobalt beads. PCNA protein was incubated with it in the presence of T3 or T2AA. They diminished the intensity of the PCNA band on the immobilized p21 dose-dependently (Fig. 1D). This result suggests that T2AA theoretically can inhibit any PCNA protein-protein interactions known to date once it reaches PCNA in the cells. Given the fact that PCNA-p21 interaction is mediated also at the IDCL region in addition to the PIP-box (18), the ability of T2AA to eliminate this interaction is noteworthy and demonstrates that the PIP-box interaction is the primary determinant for the PCNA-protein interaction.
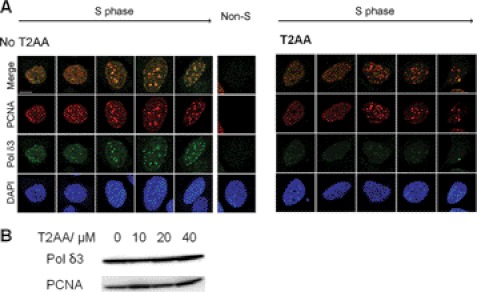
T2AA Inhibits PCNA-DNA Polymerase δ Interaction on Replication Foci in Cells but Does Not Remove Chromatin-bound PCNA
Upon observing that T2AA disrupts PCNA-p21 interaction in vitro, we investigated if T2AA inhibits PCNA interactions in cells, in the same way that p21 does to control the cell cycle (19). DNA polymerase δ is recruited to the DNA replication fork by PCNA (20) to synthesize the lagging strand, in which the subunit 3 (Polδ3) directly binds to PCNA with its PIP-box (21). Affinity of the Polδ3 PIP-box to PCNA is much smaller than that of p21, which possibly allows dynamic replication regulation, such as polymerase switching and replication inhibition by p21 (22). Therefore, theoretically T2AA should disrupt PCNA-Polδ3 interaction if enough concentration is achieved on the replication fork. To verify this, we have imaged PCNA and Polδ3 in chromatin of S-phase cells upon treatment of T2AA. Cells were treated by T2AA, pre-extracted to remove proteins unbound to chromatin, and immunostained. PCNA and Polδ3 are clearly colocalized in the absence of T2AA. However, when cells were treated with T2AA, Polδ3 was exclusively washed out from the chromatin, but PCNA was not (Fig. 3A). Therefore, T2AA dissociated Polδ3 from PCNA but did not dissociate PCNA from the replication fork. On the other hand, expressions of Polδ3 protein in whole cell lysate were not reduced (Fig. 3B), showing that the elimination of chromatin Polδ3 by T2AA is not due to its down-regulation.
FIGURE 3.
Inhibition of PCNA-DNA polymerase interaction on chromatin by T2AA. A, U2OS cells were untreated or treated with T2AA (20 μm) for 24 h. Proteins unbound to chromatin were extracted away by CSK buffer/Triton X-100, and each indicated protein was immunostained. DNA Polδ3 is colocalized well with PCNA in untreated S-phase cells but was removed by the extraction, suggesting that the PCNA-Polδ3 interaction on chromatin was disrupted by the T2AA treatment, but PCNA loading on replication forks was not. A non-S-phase cell is shown as a negative control for PCNA staining. DAPI served as nuclear stain. B, immunoblotting for Polδ3 in whole lysates of U2OS cells treated with T2AA showed no down-regulation. PCNA served as a loading control.
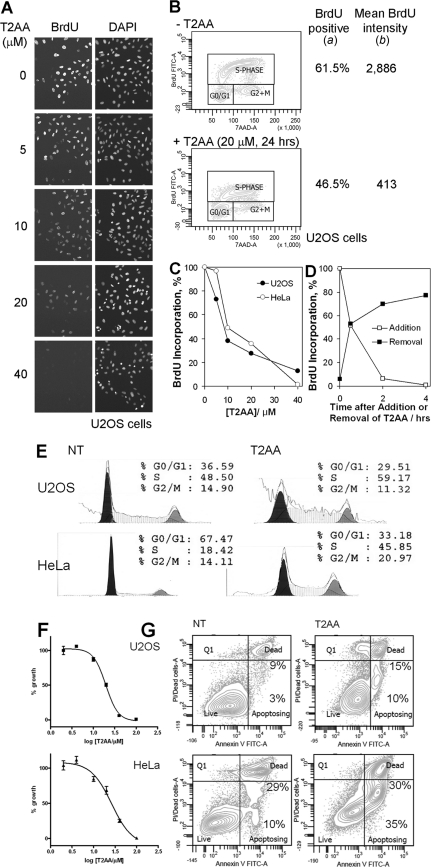
T2AA Inhibits DNA Replication in Cancer Cells and Their Proliferation
Because T2AA disrupted PCNA-Polδ3 interaction on chromatin, next we investigated if T2AA could achieve relevant functional effects in cells. DNA polymerase δ synthesizes de novo DNA strands, which is measurable by nucleotide incorporation. When cells were treated with BrdU under titration of T2AA, T2AA inhibited the BrdU incorporation significantly in a dose-dependent manner (Figs. 4, A–C), which is consistent with the elimination of Polδ3 from chromatin. By performing the assays in a time-dependent manner after the addition or removal of T2AA, pharmacological action of T2AA appeared fairly rapid and was reversible. The BrdU incorporations were >90% inhibited within 2 h of T2AA treatment and >70% recovered after 2 h of T2AA release (Fig. 4D), which was ∼100% recovered after 18 h (data not shown). Parallel to the inhibition of DNA synthesis, T2AA arrested cells at S-phase (Fig. 4E), similarly to other agents that inhibit DNA replication/synthesis, such as aphidicolin and hydroxyurea. Further, when cells were cultured with T2AA, growth of both U2OS (p53 WT) and HeLa cells (p53 destroyed by E6) was inhibited with a similar efficacy (Fig. 4F), suggesting that growth inhibition by T2AA is p53-independent. The growth inhibition was accompanied by increased cellular populations in early apoptosis but not necrosis (Fig. 4G), suggesting that T2AA induced the growth inhibition not by nonspecific toxicity.
FIGURE 4.
Functional response of cells upon T2AA treatment. A, T2AA inhibits DNA replication in cells. DNA replication was analyzed by BrdU incorporation after a 24-h treatment of U2OS cells with T2AA. Cells were treated with the drug at the indicated concentrations and labeled with 10 μm BrdU for 15 min. Incorporated BrdU was detected by immunostaining using anti-BrdU antibody. B and C, the BrdU incorporation and dose dependence of T2AA treatment were quantified by flow cytometry. The populations of cells positive for BrdU incorporation (S-phase) (a) and mean intensity of BrdU signal in the cells within the S-phase fraction (b) were determined. Total BrdU incorporation was defined as a × b. D, time dependence of the inhibition of DNA replication by T2AA was examined by flow cytometry. U2OS cells were treated with 20 μm T2AA in an indicated period or released from a treatment with T2AA at 20 μm for 4 h. Cells were labeled with 20 μm BrdU for 15 min and analyzed by flow cytometry as in B and C. t½ of the response was read as ∼0.5 h in both the T2AA addition and release. E, T2AA arrested cells at S-phase. Cells were treated with T2AA (20 μm) for 24 h, stained by PI, and sorted by DNA contents. F, T2AA suppressed growth of cells. Cells were treated with the indicated titration of T2AA triplicate in 384-well plate for 3 days. Signals of each well were measured and normalized with that of well containing no cells (0%) and cells with no treatment (100%). Growth inhibition curve was fitted by Prism (GraphPad) to determine IC50 as 20 μm (U2OS) and 26 μm (HeLa). Error bars, S.E. G, T2AA induced early apoptosis. Cells were treated with T2AA (20 μm) for 5 days, stained, and sorted by Annexin-V and PI. The population percentages of early apoptosis (Apoptosing) and late apoptosis (Dead) cells are indicated. NT, no treatment.
T2AA Induces DNA Replication Stress in Cells by Stalling Single-stranded DNA
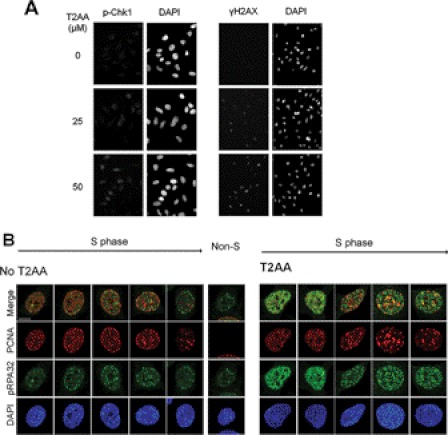
Because T2AA inhibits DNA replication, next we investigated if T2AA actually induces DNA replication stress. When DNA replication is arrested in S-phase, cells contain unreplicated single-stranded DNA (ssDNA) in chromatin and subsequently activate the ATR-Chk1 pathway, leading to histone H2AX phosphorylation (23). Chk1 and H2AX were phosphorylated upon T2AA treatment (Fig. 5A). ATR phosphorylated RPA32 that was accumulated on ssDNA, and such RPA32 phosphorylation was observed in cells treated with aphidicolin or hydroxyurea (24). To verify this for T2AA, we have investigated the RPA32 phosphorylation in chromatin by immunostaining. After 24 h of treatment, T2AA increased the immunofluorescence of phospho-RPA32 in nuclei (Fig. 5B). The phospho-RPA32 foci were not colocalized well with PCNA foci, which is consistent with a previous observation that phosphorylation of RPA32 prevents RPA association to replication forks (25) where PCNA should exist. These observations suggest that T2AA-mediated DNA replication arrest activates the ATR-Chk1 pathway and accumulates stalled ssDNA.
FIGURE 5.
Induction of DNA replication stress by T2AA. A, HeLa cells were treated with indicated concentrations of T2AA for 4 h. Immunostaining was performed for Ser345-phosphorylated Chk1 or Ser139-phosphorylated histone H2AX. B, U2OS cells were untreated or treated with T2AA (40 μm) for 24 h. Proteins unbound to chromatin were extracted away by CSK buffer/Triton X-100, and each indicated protein was immunostained. RPA32 on the chromatins of S-phase cells was Ser33-phosphorylated upon T2AA treatment. A non-S-phase cell is shown as a negative control for PCNA staining. DAPI served as nuclear staining agent.
T2AA Inhibits TLS in Cells
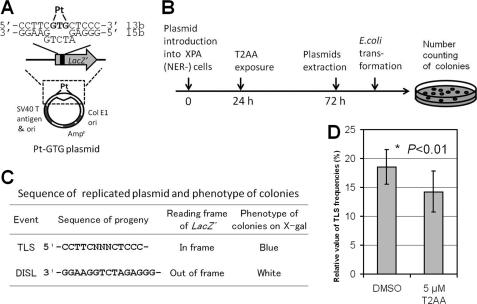
One of the important roles of PCNA is polymerase switching from regular replication polymerases (such as DNA Polδ) to translesion synthesis polymerases (such as DNA Polη) when the replication fork meets a damaged site on the DNA template (such as a cisplatin intrastrand cross-link, the major reaction product of DNA and cisplatin) (26). Because T2AA inhibited DNA synthesis in cells (Fig. 4A), we investigated if T2AA also can inhibit TLS that is PCNA-dependent (7), at least in part. To verify this in cells, we used a cellular TLS assay that uses replications of a plasmid DNA containing an intrastrand cisplatin cross-link (Pt-GTG) in a coding region of the lacZ gene (Fig. 6A), in which another plasmid lacking the cross-link serves as a control for non-TLS plasmid replication (14). To avoid removal of the cross-link by NER, we used XP2OS(SV40), an NER-deficient XPA cell line (16). The cells were transfected with the plasmid in the presence of T2AA. The replicated plasmids in the cells were recovered and analyzed for the TLS event by E. coli transformation/colony selection on X-gal plates (Fig. 6, B and C). T2AA significantly reduced the occurrence of TLS compared with that of DMSO control (14.2 versus 18.5% (i.e. 23% reduction); Fig. 6D).
FIGURE 6.
Inhibition of TLS across the cisplatin Pt-GTG cross-link in mammalian cultured cells by T2AA. Shown are the structure of the cisplatin-cross-linked Pt-GTG plasmid (A) and experimental protocol used in this study (B). C, predicted sequences of replicated plasmid and phenotypes of colonies on the X-gal LB plates; DISL, damage induced strand loss. D, T2AA inhibited TLS across the cisplatin-cross-linked Pt-GTG adduct. Relative values of the TLS frequency (percentages) across Pt-GTG adduct in XPA cells treated with DMSO (18.5 ± 3.0%) or T2AA (14.2 ± 3.6%) are shown, which is the ratio of the TLS frequency of the modified plasmid to that of the non-modified plasmid (i.e. (TLS frequency of Pt-GTG plasmid/TLS frequency of Mock-GTG plasmid) × 100). Data are means with S.D. values (error bars) from at least three independent experiments. The value significantly decreased (*, p < 0.01) from that of the DMSO control. Statistical comparison was carried out using Student's t test for one-tailed comparison. Actual numbers of the colony counting are given in supplemental Table S1.
T2AA Increases Cellular DNA Damage Induced by Cisplatin
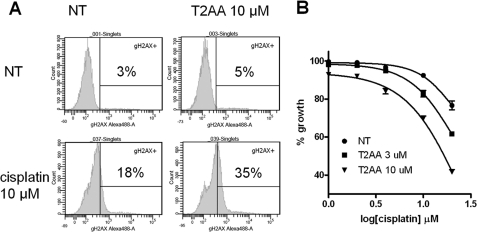
Because T2AA inhibited the TLS in cells, next we investigated if T2AA could actually increase cellular DNA damage upon treatment with cisplatin. U2OS cells were treated with T2AA, cisplatin, or both, and then the DNA damage response was measured by γH2AX staining. As expected, cisplatin significantly induced γH2AX, whereas T2AA induced γH2AX only a little. In contrast, when both T2AA and cisplatin were added together, the γH2AX level was further increased much higher than the level combined from each single treatment (Fig. 7A). To observe the influence of the cisplatin/T2AA-induced DNA damage on cell growth, the drugs were removed to release the cells from S-phase arrest. The cells were cultured under drug-free conditions, and their viability was measured. U2OS cells are relatively resistant to cisplatin, and their growth is not significantly affected by the presence of up to 10 μm cisplatin, and it was reduced only less than 10% with 10 μm T2AA. In contrast, when cells received both compounds, their growth was significantly reduced (Fig. 7B).
FIGURE 7.
Increasing cisplatin-induced DNA damage by T2AA. A, T2AA increased cisplatin-induced DNA damage response. U2OS cells were treated with the indicated combination of T2AA and cisplatin for 18 h, stained for γH2AX, and analyzed by flow cytometry. B, growth of cells treated with cisplatin and T2AA. U2OS cells were treated with the indicated titration of cisplatin with T2AA for 18 h, replated in fresh medium to remove the drugs, and cultured for 3 days. Viability of the cells was measured by Alamar Blue reagent, and signals were normalized with those of no cell well (0%) and cells with no drugs (100%). NT, no treatment. Error bars, S.D.
DISCUSSION
Importance of Ubiquitous Mediators of Cell Maintenance in Cancer
Based on the success of pathway target therapeutics for cancers, such as Herceptin, numerous efforts have been made for finding and characterizing further novel oncogenic targets. Although these approaches have been successful, fundamental limitations appeared; they are effective only when cancers depend on the pathway (“cancer addiction”), and resistance often occurs rapidly by mutating the target. Therefore, the importance of ubiquitous molecules in supporting cancers began to be the focus of investigation (4). Principally, therapeutics targeting non-oncogenic molecules may not be cancer-selective. However, these therapeutics are more likely to be applicable for diverse cancers with low occurrence of resistance; thus, such therapeutics can be robust and reliable, like traditional chemotherapies. Given the fact that chemotherapy and/or radiation therapy is indispensable in the majority of cancer treatments, adjuvant therapeutics that can be utilized with them are very useful in the clinical setting.
Role of PCNA for Cell Growth and DNA Repair
Since being cloned in 1987 (27), PCNA has been extensively characterized for its numerous functions. Because PCNA is indispensable for DNA replication and damage repair, it has been anticipated as a promising non-oncogenic therapeutic target (9), but no small molecule inhibitor of PCNA is known to date. In this study, we demonstrate that an inhibitor of PCNA functions can be created by focusing on protein-protein interaction aspects of PCNA. The inhibitor actually inhibited DNA replication and enzymatic TLS that is a mechanism of DNA repair in cancer cells. It also increased cisplatin-induced DNA damage.
Affinity of PCNA Ligands for Pathway Switching and Possibility of Controlling Them with Small Molecules
PCNA interacts with numerous proteins, of which more than half are PIP-box proteins (5). Assuming that they interact with the same PCNA cavity, their interaction must be strictly controlled by timing and orders to organize the PCNA complex on the replication fork working smoothly, which include not only DNA polymerization in both leading and lagging strands but also ligation of Okazaki fragment and winding DNA on histones to reconstruct nucleosomes. Given that mammalian cells have only one PCNA homolog that clamps DNA, it is quite surprising that such dynamic control is successfully accomplished on a simple symmetric PCNA homotrimer. A possible mechanism is switching PCNA binding molecules based on their PCNA affinities. As such, p21 is the tightest PCNA ligand and can compete with any PIP-box molecules on PCNA to stop a process ongoing on it. DNA Polδ and FEN-1 are PCNA ligands that are essential for DNA replication, but their PCNA affinities are 3 orders of magnitude weaker than p21 (21). Thus, they can be competed away by p21 to stop the DNA replication, which would be the mechanism of T2AA in inhibiting the DNA replication (Figs. 3 and 4, A–D). In this model, pharmacology of a PCNA inhibitor is defined by which PCNA interacting proteins are eliminated and therefore could be defined by the balance of PCNA affinities of the proteins and the inhibitor. This hypothesis could be validated by a competitive proteomics study in which proteome samples are isolated by PCNA affinity (28) in the presence of PCNA inhibitors with divergent PCNA affinity and by interpreting the relationship between their pharmacology and proteins that are competed away from the PCNA.
How Does T2AA Act on PCNA Machinery?
To effectively inhibit PCNA functions on the replication fork, do all three PCNAs on the homotrimer need to be inhibited, or is inhibiting one sufficient? PCNA machineries are loaded to the replication fork by RFC, a complex of PIP-box proteins (29). In this model, if a compound dissociates interactions of all three PCNAs, it could liberate PCNA from the DNA replication fork. One of our observations suggested that DNA Polδ3 was eliminated from chromatin, but PCNA was not (Fig. 3). Therefore, T2AA may have disrupted PCNA-Polδ3 but not PCNA-RCF, which may be due to the difference of affinities of these two PCNA interactions or because holding of PCNA on the replication fork is not entirely PIP-box interaction-dependent.
How Does T2AA Induce Replication Fork Stress?
In eukaryotic cells, leading strand and lagging strand are replicated by different polymerases: DNA Polδ for lagging strand and DNA Polϵ for leading strand. Both of them are regulated by PCNA but in a different manner. PCNA directly binds and catalyzes Polδ very effectively. In contrast, PCNA binds and catalyzes Polϵ much less effectively but facilitates loading of Polϵ on DNA (30). Furthermore, on lagging strands, PCNA facilitates Okazaki fragment ligation by interacting with FEN-1 (31) and DNA ligase I (32). Therefore, theoretically, PCNA inhibition could stall replication of only lagging strands. Lagging strands have much more ssDNA (between Okazaki fragments) than leading strands have; therefore, T2AA could have induced high numbers of the unreplicated ssDNA that were detected by RPA32 phosphorylation (Fig. 5B).
Replication Fork Stress and Lethality of T2AA Treatments
PCNA is incomparably multifunctional, and its inhibition could thus cause promiscuous effects in regard to cell growth. However, the actual effect of T2AA treatment was not entirely cytotoxic (Fig. 4H). When cells are faced with DNA replication stress, they attempt to remove the stalled site by transiently inducing double strand breaks, in which BLM helicase, Mus81 endonuclease, and ATR kinase are needed (33). PCNA is dissociated from chromatin during this process (34), suggesting that replication forks are disassembled and therefore T2AA would no longer be effective. Replication stress induced by T2AA could become lethal when this process is inhibited, such as when co-administrating it with an ATR inhibitor.
Mechanistically, PCNA inhibitors can be selective to replicating cells and valuable as long as they are cooperative with cytotoxic therapies. Induction of DNA stress in actively replicating cells generates many stalled replication forks where DNA is disassembled from nucleosomes. The formed linker DNA portion is unprotected from the histone core and could thus be liable against DNA damage inducers, such as cisplatin. In fact, cisplatin and other alkylation agents react with DNA preferentially in the linker portion rather than in the nucleosome region in vitro (35). T2AA could thus have sensitized cells for inducing cisplatin-mediated DNA damage (Fig. 7A). Another possible mechanism is that T2AA could also inhibit TLS that makes cells exempt from DNA damage, which was verified in the TLS inhibition (Fig. 6D). The majority of DNA damage induced by cisplatin is intrastrand cross-links that are not highly toxic to cells if they are allowed to be repaired (36). The γH2AX induction observed in cells co-treated by cisplatin and T2AA (Fig. 7A) could represent such cross-links. It was shown that cells are not expected to die unless they retain γH2AX foci 24 h after cisplatin treatment (37). Thus, investigating if T2AA also prevents cisplatin-induced DNA damage repair that is facilitated by PCNA (such as NER) and if T2AA stabilizes the γH2AX foci long enough could be valuable for validating lethality of cisplatin/T2AA co-treatment. This is very likely because T2AA inhibits interaction of PCNA and Polδ3 in chromatin (Fig. 3A), and both of them are absolutely essential in the late step of NER (gap-filling synthesis) (38). Actually, perturbation of this step reportedly induced γH2AX (39), which could be the mechanism of the γH2AX up-regulation by cisplatin/T2AA co-treatment.
Future Investigation
The PCNA-T3 co-crystal structure is useful to rationally design new leads with improved PCNA affinity. The observation that IDCL and the Asp41–Val45 region of PCNA are specifically perturbed upon T2AA association (Fig. 2B) indicates that they are key interaction sites to dynamically adopt a small molecule. Lead optimization efforts under this hypothesis are ongoing to produce new compounds with higher PCNA affinity. Such compounds will be evaluated in animal tumor models for chemotherapy sensitization, which thus will validate a new strategy for sensitizing cancer cells to cisplatin treatment by targeting PCNA. T2AA could functionally inhibit other processes that PCNA coordinates, such as origin refiring (40), chromatin assembly (41), epigenetic inheritance (42) and sister chromatid cohesion (43, 44). Functional characterization of T2AA in these processes will elucidate their functional significance from the chemotherapeutic viewpoint.
Supplementary Material
Acknowledgments
We thank David Smithson, Armand Guigemde, Heather Ross, and Enas Ahmed for high-throughput screening; Andy Lemoff, Cynthia Jeffries, and Bing Yan for analytical chemistry; Marcelo Actis and Anand Mayasundari for synthetic chemistry support; Robert Cassell for peptide synthesis; Scott Perry, Dana Lucas, Ann-Marie Hamilton-Easton, and Richard Ashmun for flow cytometry; Jennifer Peters and Victoria Frohlich for light microscopy; the beamline staff of SPring-8 and Photon Factory Japan for the crystallography data collection; and Youngsoo Lee and Peter McKinnon for scientific advice. Toshiki Tsurimoto, Kip Guy, and Richard Kriwacki kindly provided plasmids for this study (see supplemental material).
This work was supported by the American Lebanese Syrian Associated Charities, American Cancer Society Research Scholar Grant RSG CDD-120969 (to A. I. and N. F.), Grant-in-aid for Scientific Research for Young Scientists B 19710058 from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to M. K.), and KAKENHI and the Targeted Proteins Research Program from MEXT, Japan (to A. H., H. H., and M. S.).

This article contains supplemental Materials and Methods, Table S1, and References.
The atomic coordinates and structure factors (code 3VKX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PCNA
- proliferating cell nuclear antigen
- CSK
- cytoskeletal
- γH2AX
- phospho(Ser139)histone H2AX
- NER
- nucleotide excision repair
- PIP-box
- PCNA-interacting protein box
- T2AA
- T2 amino alcohol ((S)-4-(4-(2-amino-3-hydroxypropyl)-2,6-diiodophenoxy)phenol)
- T3
- 3,3′,5-triiodothyronine
- TLS
- translesion DNA synthesis
- TR
- thyroid hormone receptor
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- FP
- fluorescence polarization
- IDCL
- interdomain connecting loop
- Polδ
- Polϵ, and Polη, polymerase δ, ϵ, and η, respectively.
REFERENCES
- 1. Weinstein I. B., Joe A. K. (2006) Mechanisms of disease. Oncogene addiction. A rationale for molecular targeting in cancer therapy. Nat. Clin. Pract. Oncol. 3, 448–457 [DOI] [PubMed] [Google Scholar]
- 2. Gorre M. E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P. N., Sawyers C. L. (2001) Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 293, 876–880 [DOI] [PubMed] [Google Scholar]
- 3. Yauch R. L., Dijkgraaf G. J., Alicke B., Januario T., Ahn C. P., Holcomb T., Pujara K., Stinson J., Callahan C. A., Tang T., Bazan J. F., Kan Z., Seshagiri S., Hann C. L., Gould S. E., Low J. A., Rudin C. M., de Sauvage F. J. (2009) Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo J., Solimini N. L., Elledge S. J. (2009) Principles of cancer therapy. Oncogene and non-oncogene addiction. Cell 136, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 6. Haracska L., Johnson R. E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. (2001) Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell Biol. 21, 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hishiki A., Hashimoto H., Hanafusa T., Kamei K., Ohashi E., Shimizu T., Ohmori H., Sato M. (2009) Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 284, 10552–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoimenov I., Helleday T. (2009) PCNA on the crossroad of cancer. Biochem. Soc. Trans. 37, 605–613 [DOI] [PubMed] [Google Scholar]
- 10. Acharya N., Yoon J. H., Gali H., Unk I., Haracska L., Johnson R. E., Hurwitz J., Prakash L., Prakash S. (2008) Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 17724–17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kontopidis G., Wu S. Y., Zheleva D. I., Taylor P., McInnes C., Lane D. P., Fischer P. M., Walkinshaw M. D. (2005) Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. U.S.A. 102, 1871–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hishiki A., Shimizu T., Serizawa A., Ohmori H., Sato M., Hashimoto H. (2008) Crystallographic study of G178S mutant of human proliferating cell nuclear antigen. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 819–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taddei A., Roche D., Sibarita J. B., Turner B. M., Almouzni G. (1999) Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147, 1153–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sawai T., Kawanishi M., Takamura-Enya T., Yagi T. (2009) Establishment of a method for analyzing translesion synthesis across a single bulky adduct in human cells. Genes Environ. 31, 24–30 [Google Scholar]
- 15. Shivji M. K., Moggs J. G., Kuraoka I., Wood R. D. (2006) Assaying for the dual incisions of nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 314, 435–456 [DOI] [PubMed] [Google Scholar]
- 16. Kawanishi M., Enya T., Suzuki H., Takebe H., Matsui S., Yagi T. (1998) Mutagenic specificity of a derivative of 3-nitrobenzanthrone in the supF shuttle vector plasmids. Chem. Res. Toxicol. 11, 1468–1473 [DOI] [PubMed] [Google Scholar]
- 17. Leeson P. D., Ellis D., Emmett J. C., Shah V. P., Showell G. A., Underwood A. H. (1988) Thyroid hormone analogues. Synthesis of 3′-substituted 3,5-diiodo-l-thyronines and quantitative structure-activity studies of in vitro and in vivo thyromimetic activities in rat liver and heart. J. Med. Chem. 31, 37–54 [DOI] [PubMed] [Google Scholar]
- 18. Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 19. Chen J., Jackson P. K., Kirschner M. W., Dutta A. (1995) Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature 374, 386–388 [DOI] [PubMed] [Google Scholar]
- 20. Bravo R., Frank R., Blundell P. A., Macdonald-Bravo H. (1987) Cyclin/PCNA is the auxiliary protein of DNA polymerase-δ. Nature 326, 515–517 [DOI] [PubMed] [Google Scholar]
- 21. Bruning J. B., Shamoo Y. (2004) Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-δ p66 subunit and flap endonuclease-1. Structure. 12, 2209–2219 [DOI] [PubMed] [Google Scholar]
- 22. Li R., Waga S., Hannon G. J., Beach D., Stillman B. (1994) Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature 371, 534–537 [DOI] [PubMed] [Google Scholar]
- 23. Flynn R. L., Zou L. (2011) ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 36, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vassin V. M., Anantha R. W., Sokolova E., Kanner S., Borowiec J. A. (2009) Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA replication stress. J. Cell Sci. 122, 4070–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vassin V. M., Wold M. S., Borowiec J. A. (2004) Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell Biol. 24, 1930–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lehmann A. R. (2006) Translesion synthesis in mammalian cells. Exp. Cell Res. 312, 2673–2676 [DOI] [PubMed] [Google Scholar]
- 27. Matsumoto K., Moriuchi T., Koji T., Nakane P. K. (1987) Molecular cloning of cDNA coding for rat proliferating cell nuclear antigen (PCNA)/cyclin. EMBO J. 6, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohta S., Shiomi Y., Sugimoto K., Obuse C., Tsurimoto T. (2002) A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2–5 complex as a novel PCNA-binding protein. J. Biol. Chem. 277, 40362–40367 [DOI] [PubMed] [Google Scholar]
- 29. Bowman G. D., O'Donnell M., Kuriyan J. (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]
- 30. Chilkova O., Stenlund P., Isoz I., Stith C. M., Grabowski P., Lundström E. B., Burgers P. M., Johansson E. (2007) The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 35, 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu X., Li J., Li X., Hsieh C. L., Burgers P. M., Lieber M. R. (1996) Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 24, 2036–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levin D. S., McKenna A. E., Motycka T. A., Matsumoto Y., Tomkinson A. E. (2000) Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 10, 919–922 [DOI] [PubMed] [Google Scholar]
- 33. Hanada K., Budzowska M., Davies S. L., van Drunen E., Onizawa H., Beverloo H. B., Maas A., Essers J., Hickson I. D., Kanaar R. (2007) The structure-specific endonuclease Mus81 contributes to replication restart by generating double strand DNA breaks. Nat. Struct. Mol. Biol. 14, 1096–1104 [DOI] [PubMed] [Google Scholar]
- 34. Shimura T., Torres M. J., Martin M. M., Rao V. A., Pommier Y., Katsura M., Miyagawa K., Aladjem M. I. (2008) Bloom's syndrome helicase and Mus81 are required to induce transient double strand DNA breaks in response to DNA replication stress. J. Mol. Biol. 375, 1152–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galea A. M., Murray V. (2010) The influence of chromatin structure on DNA damage induced by nitrogen mustard and cisplatin analogues. Chem. Biol. Drug Des. 75, 578–589 [DOI] [PubMed] [Google Scholar]
- 36. Siddik Z. H. (2003) Cisplatin. Mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279 [DOI] [PubMed] [Google Scholar]
- 37. Banáth J. P., Klokov D., MacPhail S. H., Banuelos C. A., Olive P. L. (2010) Residual γH2AX foci as an indication of lethal DNA lesions. BMC Cancer. 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ogi T., Limsirichaikul S., Overmeer R. M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N. G., Mullenders L. H., Yamashita S., Fousteri M. I., Lehmann A. R. (2010) Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37, 714–727 [DOI] [PubMed] [Google Scholar]
- 39. Matsumoto M., Yaginuma K., Igarashi A., Imura M., Hasegawa M., Iwabuchi K., Date T., Mori T., Ishizaki K., Yamashita K., Inobe M., Matsunaga T. (2007) Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 120, 1104–1112 [DOI] [PubMed] [Google Scholar]
- 40. Arias E. E., Walter J. C. (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 41. Shibahara K., Stillman B. (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 [DOI] [PubMed] [Google Scholar]
- 42. Iida T., Suetake I., Tajima S., Morioka H., Ohta S., Obuse C., Tsurimoto T. (2002) PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells 7, 997–1007 [DOI] [PubMed] [Google Scholar]
- 43. Moldovan G. L., Pfander B., Jentsch S. (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23, 723–732 [DOI] [PubMed] [Google Scholar]
- 44. Inoue A., Li T., Roby S. K., Valentine M. B., Inoue M., Boyd K., Kidd V. J., Lahti J. M. (2007) Loss of ChlR1 helicase in mouse causes lethality due to the accumulation of aneuploid cells generated by cohesion defects and placental malformation. Cell Cycle 6, 1646–1654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.