Background: RCAN1s are stress-inducible proteins that have been shown to bind to the adenine nucleotide translocator (ANT).
Results: RCAN1-1L can open the mitochondrial permeability transition pore, shift metabolism from oxidative phosphorylation to glycolysis, and induce mitophagy.
Conclusion: RCAN1-1L is a regulator of mitochondrial autophagy (mitophagy).
Significance: This helps to understand how RCAN1s can contribute to cell death/survival and human disease.
Keywords: Autophagy, Metabolism, Mitochondria, Neurodegeneration, Stress, RCAN1
Abstract
Expression of the RCAN1 gene can be induced by multiple stresses. RCAN1 proteins (RCAN1s) have both protective and harmful effects and are implicated in common human pathologies. The mechanisms by which RCAN1s function, however, remain poorly understood. We identify RCAN1s as regulators of mitochondrial autophagy (mitophagy) and demonstrate that induction of RCAN1-1L can cause dramatic degradation of mitochondria. The mechanisms of such degradation involve the adenine nucleotide translocator and mitochondrial permeability transition pore opening. We also demonstrate that RCAN1-1L induction can shift cellular bioenergetics from aerobic respiration to glycolysis, yet RCAN1-1L has very little effect on cell division, whereas it has a cumulative negative effect on cell survival. These results shed the light on mechanisms by which RCAN1s can protect or harm cells and by which they may operate in human pathologies. They also suggest that RCAN1s are important players in autophagy and such elusive phenomena as the mitochondrial permeability transition pore.
Introduction
The RCAN1 gene was discovered to be induced during cellular adaptation to oxidative stress (1). It is localized on chromosome 21 (2), trisomy of which causes Down syndrome. RCAN1 can also be induced by other stresses, including biomechanical stress (3) and psychological stress (4). It has actually been shown that very transient RCAN1 induction, lasting only 2–6 h, can be protective against acute stress (5, 6), but we have hypothesized that chronic induction can have harmful effects (7, 8). Mechanisms by which RCAN1s can protect or harm cells, however, are poorly understood. Many human diseases are associated with chronic stress, and chronic RCAN1 overexpression is associated with Down syndrome and Alzheimer disease (2, 7, 9–11), whereas its deficiency may contribute to cancer (12), cardiac hypertrophy (13), and Huntington disease (14).
The RCAN1 gene can be alternatively spliced/translated to produce a number of isoforms, at least three of which (RCAN1-1L, RCAN1-1S, and RCAN1-4) are produced in humans (6, 15, 16). The first function of RCAN1 proteins demonstrated was to bind and regulate the activity of calcineurin (2, 17–20). However, it soon became clear that like many proteins, RCAN1s could have other functions. The Drosophila homolog of RCAN1, called nebula, can bind to the mitochondrial adenine nucleotide translocator (ANT)2 and regulate its ADP/ATP transporting activity (21). However, besides facilitation of ADP/ATP exchange through the mitochondrial membrane, ANT can carry out another important function; it can form the mitochondrial permeability transition pore (mtPTP), which is a key component of certain forms of apoptotic and necrotic cell death (22, 23). In fact, ANT may currently be the only component of mtPTP on which there is general agreement. Thus, we hypothesized that RCAN1s may regulate mtPTP.
Of all RCAN1 isoforms, RCAN1-1L is the predominant protein isoform found in humans (15, 16). Because the brain exhibits one of the highest levels of RCAN1s, and RCAN1-1L has been implicated in several brain diseases, we used the neuronal ST14A cell line and neural stem cells as models in this study.
We found that RCAN1-1L can indeed regulate mtPTP. Because mtPTP opening has been suggested to initiate autophagy and mitochondrial degradation (24, 25), we tested this possibility and discovered that RCAN1-1L overexpression leads to induction of autophagy and significant mitochondrial loss.
EXPERIMENTAL PROCEDURES
Adenoviral Constructs
A construct carrying RCAN1-1L was created as described previously (14). Null adenoviral vector was acquired from Vector Biolabs Inc. (Philadelphia, PA). To minimize toxic effects, both constructs were purified by centrifugation using two sequential cesium chloride gradients.
Cell Cultures
ST14A cells were grown and differentiated as described previously (14). ENStem cells were acquired from Millipore Corp. (Billerica, MA). Stem cells were grown and differentiated using materials and supplies from the manufacturer and following its protocols. All cultures were maintained at 5% CO2 in medium containing antibiotics (100 units/ml penicillin and 0.1 mg/ml streptomycin).
Fluorescence-activated Cell Sorting (FACS) Analysis
Analysis of mitochondrial mass was performed using a FACScalibur analyzer, whereas analysis of mtPTP was done using a BD LSRII analyzer, both from BD Biosciences. In all experiments, cells were resuspended at a concentration of ∼1 mln/ml in PBS.
Transmission Electron Microscopy (TEM)
Cells were collected, fixed using Karnovsky's solution, dissected, and analyzed using transmission electron microscopy at the USC Center for Electron Microscopy and Microanalysis. All photographs were taken at ×5,000 magnification.
Western Analysis and Antibodies
The analysis was performed following standard protocols, using an ECL detection system from Amersham Biosciences. The signals were detected using a BioChemi HR Camera and quantified using a BioSpectrum AC Imaging System from UVP, LLC, (Upland, CA). In all experiments, arbitrary units were set as follows. The value of light emission of the first sample in each group was set as 1.0. All values were then adjusted according to the loading levels, as measured by β-tubulin emission signal. The “common RCAN1 antibody” was developed against exon 7 (which is common to all RCAN1 isoforms) and characterized in our laboratory as described previously (6, 15, 26). The Lon antibody was also from our laboratory, and it has been described previously (27). β-Tubulin antibody was from Millipore Corp. (catalogue no. 05-661). LC3 antibody was from VWR Scientific (Radnor, PA) (catalogue no. 60941-266). Phospho-p70S6 kinase (Thr-389) antibody was from Cell Signaling Technology Inc. (Danvers, MA) (catalogue no. 9205). PGC-1a antibody was from Calbiochem, an affiliate of Merck (catalogue no. ST1202).
Analysis of Cellular Bioenergetics
Cellular bioenergetics were studied using a Seahorse XF-24 Analyzer from SeahorseBioscience (North Billerica, MA). Embryonic striatal conditionally immortalized cells were cultured on Seahorse XF-24 plates at a density of 50,000 cells/well. On the day of metabolic flux analysis, cells were changed to unbuffered DMEM (DMEM base medium supplemented with 25 mm glucose, 1 mm sodium pyruvate, 31 mm NaCl, 2 mm GlutaMax, pH 7.4) and incubated at 37 °C in a non-CO2 incubator for 1 h. All media and injection reagents were adjusted to pH 7.4 on the day of assay. Four base-line measurements of oxygen consumption rate (OCR; measured by oxygen concentration change) and extracellular acidification rate (ECAR; measured by pH change) were taken before sequential injection of mitochondrial inhibitors. Three readings were taken after each addition of mitochondrial inhibitor. The mitochondrial inhibitors used were oligomycin (ATP synthase inhibitor; 4 μm), carbonylcyanide-4-trifluoromethoxyphenylhydrazone (FCCP) (mitochondrial respiration uncoupler; 1 μm), and rotenone (complex I inhibitor; 1 μm). After the assays, plates were saved, and protein readings were measured for each well to confirm equal cell numbers per well.
ATP Level Measurements
Cell lysates were centrifuged at 12,000 × g, and the supernatants were collected. ATP levels were quantitatively measured by a bioluminescence assay that uses recombinant firefly luciferase and d-luciferin (assay kit from Invitrogen).
RESULTS
Cell Models and RCAN1-1L Overexpression
To overexpress RCAN1-1L, we have created an adenoviral construct that encodes the full-size RCAN1-1L (14). We used two cell models: ST14A cells that were developed from embryonic rat striatal neurons and neural progenitor ENStem cells that were developed from human embryonic stem cells. Cells were transfected using different concentrations of our construct and analyzed at several time points after the transfection. Cells infected with this construct produce an RCAN1-1L protein that has an expected size of about 38 kDa (supplemental Fig. 1). In ENStem cells, we used 15 MOI of the construct, and this viral amount did not produce any significant toxic effects, as judged by analysis of cell growth (supplemental Fig. 1A). ST14A cells are significantly larger than ENStem cells, and we therefore used higher amounts of the construct (30 MOI). At this concentration, the adenoviral vector did not have any toxic effects on ST14A cells (supplemental Fig. 1B). Using 30 MOI of the construct produced the highest levels of RCAN1-1L, and increasing amounts of the construct did not lead to higher RCAN1-1L overexpression. RCAN1-1L was overexpressed at all tested time points (24, 48, and 72 h (Supplemental Fig. 1B)) up to 2 weeks, beyond which we no longer analyzed its expression.
RCAN1-1 Overexpression Leads to Reduction in Mitochondrial Mass in Dividing and Differentiating ST14A Cells
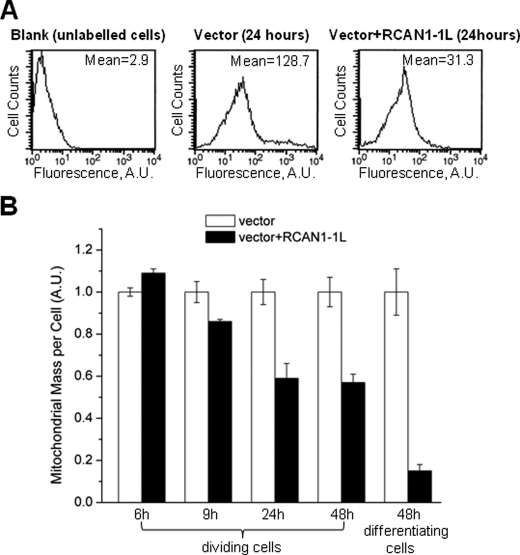
We first evaluated mitochondrial mass using FACS analysis. In this analysis, live cells were labeled with MitoTracker Green (Molecular Probes, Inc.), which binds to mitochondria, and total signals were measured per cell (Fig. 1). ST14A cells were transfected using RCAN1-1L adenoviral construct and analyzed at several time points after the transfection. We found that overexpression of RCAN1-1L for 9 h or longer caused a significant decrease in mitochondrial mass. In dividing cells, RCAN1-1L overexpression produced the greatest mitochondrial loss at 24 h, after which no further change was seen (Fig. 1B). RCAN1-1L overexpression also had a similar effect on differentiating cells, except the decrease in mitochondrial mass was even more pronounced (Fig. 1B). FACS analysis permits the separation of live and apoptotic cells; however, the results were similar for both groups of cells.
FIGURE 1.
Mitochondrial mass evaluation in ST14A cells using FACS analysis. Cells were transfected with vector carrying RCAN1-1L fragment and with null vector (control). A, example of FACS analysis. Mitochondria were labeled with MitoTracker Green (Molecular Probes, Inc.), and signals were measured using the FACS analyzer (as described under “Experimental Procedures”). Mitochondrial mass was evaluated by comparison of the mean fluorescence signals between cells transfected with null vector and vector carrying RCAN1-1L. B, summary of mitochondrial mass FACS analysis. Dividing and differentiating cells were analyzed at the indicated time periods after transfection. Mean values were converted to arbitrary units (A.U.) to make the results from different experiments directly comparable with each other. Fluorescence values in samples transfected with null construct (control) were set as 1.0. Results (means ± S.E. (error bars)) represent measurements from three independent experiments. At the 6 h time point, mitochondrial mass was increased, whereas at all other time points it was significantly decreased, in cells overexpressing RCAN1-1L as compared with controls, as evaluated by Student's t test (p < 0.05).
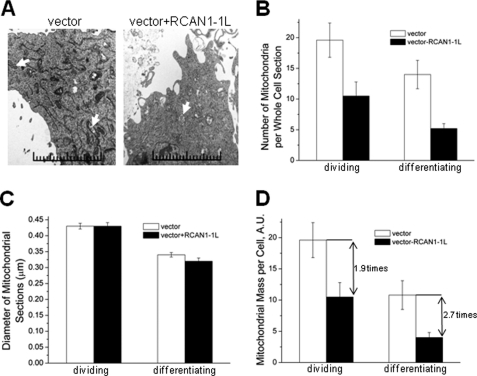
Such significant reduction in mitochondrial mass after RCAN1-1L overexpression was surprising, and to confirm these results, we performed an independent mitochondrial analysis using TEM, which is a more direct method. In this analysis of mitochondrial mass, we measured two parameters: number of mitochondria per cell and mitochondrial size (Fig. 2). Dividing cells were collected at 24 h after transfection, whereas differentiating cells were collected at 48 h, a little longer time to allow for cell differentiation. Loss of mitochondria was readily apparent from TEM photographs (Fig. 2A). TEM analysis revealed a significant drop in mitochondrial number after RCAN1-1L overexpression in both dividing and differentiating cells, confirming and extending our findings from FACS analysis. In dividing cells, the amount of mitochondria per cell was reduced by almost 50%, whereas, in differentiating cells, the mitochondrial loss was even greater (Fig. 2B). On the other hand, mitochondrial size was exactly the same in both dividing and differentiating control cells and cells overexpressing RCAN1-1L, supporting the conclusion that changes in mitochondrial number were not due, for the most part, to altered fission/fusion dynamics (Fig. 2C). To make a final estimate of mitochondrial mass per cell, we combined our data for mitochondrial number and mitochondrial size (Fig. 2D). Our TEM analysis reveals that overexpression of RCAN1-1L in dividing ST14A cells for 24 h led to a mitochondrial mass reduction of about 50%, and overexpression of RCAN1-1L in differentiating ST14A cells for 48 h led to a mitochondrial mass decrease of almost 70%.
FIGURE 2.
Electron microscopy analysis of mitochondria in ST14A cells. Cells were incubated to semiconfluence, and RCAN1-1L was overexpressed using our adenoviral construct. Dividing cells were collected in 24 h, whereas differentiating cells were collected 48 h after transfection. Cells were fixed and analyzed using transmission electron microscopy. A, representative photographs of cell sections (×5,000 magnification). Examples of mitochondria are marked by arrows. B, mitochondrial number. Whole cell sections were reconstructed, and, to ensure comparison only of cells cut close to the middle, only sections that spread into about three photomicrographs were analyzed. Twenty reconstructed cell sections were analyzed for each sample. Mitochondria were counted manually. In both cases, mitochondrial number was significantly decreased in cells overexpressing RCAN1-1L as compared with control cases, as evaluated by Student's t test (p < 0.05). C, mitochondrial size. Whole cell sections were reconstructed as in B, and the diameter of each mitochondrion was measured using a digital ruler (when mitochondria had an oval shape, diameter was calculated as (shortest diagonal + longest diagonal/2)). In differentiating cells, mitochondrial size was slightly (∼6%) but statistically significantly decreased in cells overexpressing RCAN1-1L as compared with control cases, as evaluated by Student's t test (p < 0.05). D, mitochondrial mass, in which the data from B and C were combined. Mitochondrial mass is ∼1.9 times reduced in dividing cells and ∼2.7 times reduced in differentiating cells after RCAN1-1L overexpression. This down-regulation is statistically significant at the p < 0.05 level, as evaluated by Student's t test. A.U., arbitrary units.
RCAN1-1L Stimulates Autophagy
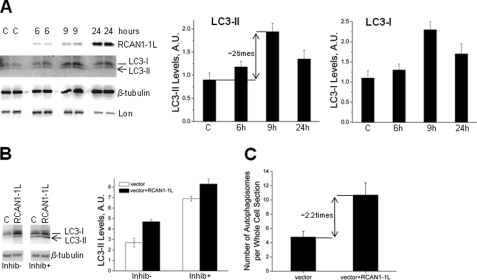
Mitochondrial mass can change either due to mitochondrial amplification (fission) or due to mitochondrial degradation. Because the decrease in mitochondrial mass after RCAN1-1L overexpression was quite rapid, it is more probable that this was due to increased mitochondrial degradation rather than to insufficient fission. The most common mechanism of mitochondrial degradation involves autophagy, which is also called mitophagy. This process involves sequestration of mitochondria inside double membrane vesicles (autophagosomes) that then fuse with lysosomes to form autophagolysosomes, where their contents are degraded via acidic lysosomal hydrolases. A widely accepted method for monitoring autophagosomes in mammalian cells is to measure the levels of a modified form of the LC3 protein (homolog of yeast Atg8 protein). LC3 is normally present in the cytoplasm, and it needs to be proteolytically cleaved (LC3-I) and lipidated (LC3-II) to be recruited to forming autophagosomal membranes. LC3-II is a validated marker of autophagic compartments in mammalian cells (28, 29). We assessed LC3-II levels in dividing ST14A cells at several time points after RCAN1-1L overexpression, using Western analysis (Fig. 3A). Our results demonstrate that LC3-II levels are slightly increased as early as 6 h and maximally (∼2-fold) at about 9 h after RCAN1-1L overexpression. This is consistent with our findings that mitochondrial mass begins to decrease around 9 h after RCAN1-1L overexpression. At 24 h after RCAN1-1L overexpression, LC3-II levels remained increased, but they were lower than at the 9 h time point. This is also consistent with our observation that mitochondrial mass did not further decrease between 24 and 48 h in dividing ST14A cells (Fig. 1). Clearly, to remain viable, cells still need some mitochondria. It is interesting that levels of LC3-I, from which LC3-II is produced, were also consistently elevated after the RCAN1-1L induction. This suggests that expression of LC3 may be up-regulated to sustain the enhanced autophagic activity induced by RCAN1-1L.
FIGURE 3.
RCAN1-1L-induced mitochondrial degradation involves activation of autophagy. A, levels of LC3 (I and II) are increased after RCAN1-1L overexpression. ST14A cells were transfected with vector carrying RCAN1-1L fragment or with null vector (C) for the indicated time periods. Control (C), in this particular case, represents cells transfected with null construct for 9 h. Signals were detected and quantified as described under “Experimental Procedures.” Probing with β-tubulin was used to control protein loadings. Results represent data from two separate experiments, in which two separate cell samples were used for each time point. Values shown are means ± S.E. (error bars) At 9 h, levels of LC3-II and LC3-I were markedly (∼2 times) increased in cells overexpressing RCAN1-1L as compared with controls as evaluated by Student's t test (p < 0.05). B, autophagic flux after RCAN1-1L overexpression. Cells were transfected, and at the 6 h time point after transfection, they were incubated for another 3 h (total 9 h after transfection) either with (Inhib+) or without (Inhib−) inhibitors of lysosomal proteolysis (20 mm NH4Cl + 100 μm leupeptin) to prevent autophagosome clearance. Levels of LC3-II were measured as in A. Three separate cell samples were used for each time point. Values shown are means ± S.E. C, the number of autophagosomes is increased after RCAN1-1L overexpression. Dividing ST14A cells were collected at 24 h after RCAN1-1L overexpression and analyzed using TEM as shown in supplemental Fig. 2. Whole cell sections were reconstructed, and, to ensure comparison only of cells cut close to the middle, only sections that spread into at least three photomicrographs were analyzed. Ten reconstructed cell sections were analyzed for each sample. Autophagosomes were counted manually. Autophagosome number was significantly (∼2.2-fold) increased in cells overexpressing RCAN1-1L in comparison with control cells, as evaluated by Student's t test (p < 0.05). A.U., arbitrary units.
Elevated levels of LC3-II can be caused by increased new production of autophagosomes or by inhibition of clearance of existing autophagosomes (autophagic flux). To distinguish between these possibilities, we designed experiments in which autophagosome clearance was prevented using inhibitors of lysosomal proteolysis (20 mm NH4Cl + 100 μm leupeptin). As previously, LC3-II levels were increased ∼2-fold at 9 h after transfection (Fig. 3B, inhib−). Blockage of lysosomal proteolysis resulted in a marked increase in levels of LC3-II in both control and RCAN1-1L-transfected cells. These results support the idea that the increase in LC3-II levels observed upon RCAN1-1L expression was not a result of blockage of autophagosome clearance but rather was a manifestation of enhanced autophagic activity in these cells. These conclusions are also confirmed by observation of TEM photographs (Fig. 3C), in which autophagosomes are seen at higher (∼2.2 times) numbers in cells overexpressing RCAN1-1L. The fact that most of the autophagic vesicles in RCAN1-1L-expressing cells show lower density than the surrounding cytosol (supplemental Fig. 2) supports the presence of proteases already in this compartment and provides further evidence that autophagosomes fuse normally with lysosomes in these cells.
Besides autophagic degradation, mitochondrial proteases have been also suggested to play a role in mitochondrial turnover (30). We have tested whether the Lon protease may be involved and found that, instead, its level is decreased after RCAN1-1L overexpression (Fig. 3A). This indicates that mitochondrial proteases are unlikely culprits for mitochondrial loss, and it probably reflects the reduction in mitochondrial mass, confirming our above findings that RCAN1-1L overexpression can cause mitochondrial loss.
RCAN1-1L Inhibits mTOR Signaling and Regulates PGC-1α
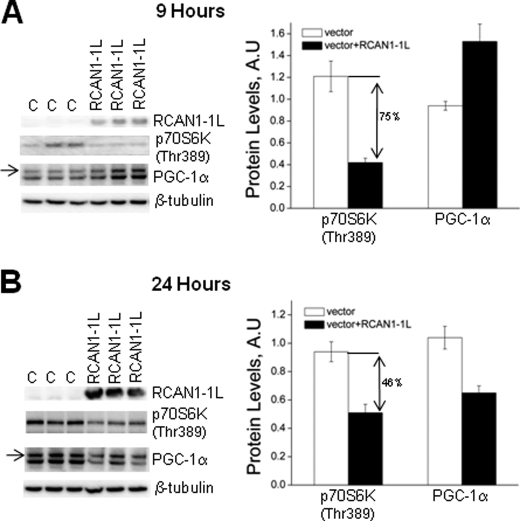
The kinase mTOR is one of the main regulators of autophagy, and the induction of autophagy by inhibition of TOR is conserved from yeast to mammals (31, 32). Because RCAN1-1L can induce autophagy, we tested whether it may also inhibit mTOR signaling. This was done using a commonly accepted method that measures the levels of p70S6 kinase (p70S6K) phosphorylated at Thr-389, which is an epitope directly phosphorylated by mTOR. We found that the levels of phosphorylated p70S6K were, indeed, inhibited following RCAN1-1L overexpression. Following 9 h of RCAN1-1L overexpression, levels of p70S6K(Thr-389) were significantly (∼75%) decreased in cells overexpressing RCAN1-1L as compared with controls (Fig. 4A). The levels were less (∼46%) reduced at 24 h following RCAN1-1L overexpression (Fig. 4B), which is in agreement with our results showing that levels of LC3-II are also less reduced at 24 h as compared with 9 h following RCAN1-1L overexpression (Fig. 3A). Overall, these results indicate that RCAN1-1L may induce autophagy through inhibition of mTOR signaling.
FIGURE 4.
mTOR signaling and expression of PGC-1α after RCAN1-1L overexpression. ST14A cells were transfected with vector carrying RCAN1-1L fragment and with null vector (C). Signals were detected and quantified as described under “Experimental Procedures.” p70S6K(Thr-389) was detected as a ∼68-kDa size protein. The expected ∼113-kDa size PGC-1α protein is marked by an arrow. Probing with β-tubulin was used to control protein loading. Results represent data from two separate experiments, in which three separate cell samples were used for each time point. Values shown are means ± S.E. (error bars). A, 9 h after RCAN1-1L overexpression. Levels of p70S6K(Thr-389) were significantly (∼75% or ∼3-fold) decreased in cells overexpressing RCAN1-1L as compared with controls, as evaluated by Student's t test (p < 0.05). Levels of PGC-1α were significantly (62%) increased in cells overexpressing RCAN1-1L as compared with controls, as evaluated by Student's t test (p < 0.05). B, 24 h after RCAN1-1L overexpression. Levels of both p70S6K(Thr-389) and PGC-1α were significantly (46 and 38%, respectively) decreased in cells overexpressing RCAN1-1L compared with controls, as evaluated by Student's t test (p < 0.05). A.U., arbitrary units.
Because PGC-1α is the main coordinator of mitochondrial biogenesis and content (33, 34), we also tested whether RCAN1-1L-induced degradation of mitochondria may be coordinated with decreased mitochondrial biogenesis. Interestingly, levels of PGC-1α first increase (∼62%) following 9 h of RCAN1-1L overexpression but then decrease (∼38%) following 24 h of RCAN1-1L overexpression (Fig. 4C). Our blots revealed two products: the expected ∼113-kDa PGC-1α protein (shown by an arrow) and a smaller protein, which may be a spliced PGC-1α form. However, the smaller protein was regulated in exactly the same manner as the 113-kDa protein, so, regardless of quantification of only one or both forms, the results still remain the same. These results suggest that mitochondrial biogenesis may be stimulated following short, transient, RCAN1-1L induction but inhibited if RCAN1-1L is induced for long periods of time. Eventual inhibition of mitochondrial biogenesis also seems logical, because it would be wasteful to eliminate mitochondria, by mitophagy, while simultaneously continuing mitochondrial synthesis. Overall, our data indicate that RCAN1-1L-induced mitophagy may be also accompanied by decreased mitochondrial synthesis. However, more detailed studies, analyzing other mitochondrial biogenesis factors, will need to be performed in order to test this idea.
RCAN1-1L Regulates mtPTP
How mitochondrial autophagy is initiated still remains unknown. Some data suggest that mtPTP plays a role in initiating this process (24, 25). Here we have tested whether RCAN1-1L may regulate mtPTP function.
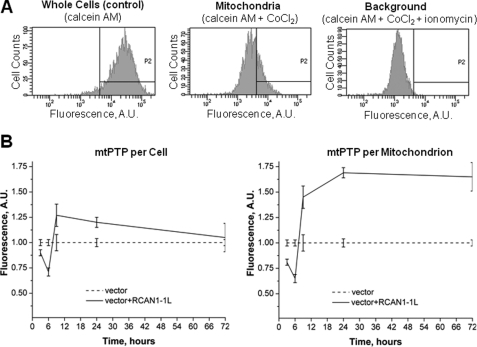
There are many ways to measure mtPTP. Unfortunately, none of them is perfect. Some (older) techniques measure mtPTP per unit of mitochondria, whereas others measure mtPTP per cell. The advantage of the techniques that evaluate mtPTP per cell is that they permit the measurements of mtPTP in intact cells, whereas techniques that measure mtPTP per unit of mitochondria require isolated mitochondria. Because, in the end, we would like to determine potential effects of RCAN1-1L on cell viability, we felt that making measurements in isolated mitochondria would be less appropriate for our purposes. In addition, during the isolation procedure, mitochondria in some samples may be more fragile than in others and may even be lost. Therefore, we made our mtPTP measurements in intact cells. This was done using the fluorescent dye calcein, as first described by Petronilli et al. (35), using the mtPTP assay kit from Molecular Probes, Inc. Cells were collected and incubated with calcein dye, which penetrates into the cytoplasm and mitochondria. Next, the fluorescence from cytosolic dye was quenched by the addition of CoCl2. Then half of the cells were left as is, and the other half were treated with ionomycin to trigger mtPTP opening and subsequent loss of mitochondrial calcein fluorescence. The samples were studied with the FACS analyzer, and mitochondrial signals were calculated as follows: fluorescence intensity in the cells treated with CoCl2 alone minus the fluorescence intensity in the cells treated with CoCl2 plus ionomycin (Fig. 5A). Samples in which pores are open the most have the highest values.
FIGURE 5.
mtPTP dynamics in dividing ST14A cells after RCAN1-1L overexpression. Cells were transfected with vector carrying RCAN1-1L fragment and with null vector (control). A, example of FACS analysis. Assays were performed in intact cells using the calcein fluorescence dye as described under “Results.” B, summary of PTP analysis. Cells were analyzed at the indicated time periods after transfection. Mean values were converted to arbitrary units (A.U.) to make the results from different experiments directly comparable with each other. Fluorescence values in samples transfected with null construct (control) were set as 1.0. Results (means ± S.E. (error bars)) represent measurements from three independent experiments. Obtained numbers represent mtPTP values per cell, which also were expressed per mitochondrion, taking into account data shown in Figs. 2 and 3. At the 3 and 6 h time points after RCAN1-1L overexpression, mtPTP values were significantly decreased either per cell or per mitochondrion (Student's t test, p < 0.05). At the 12 and 24 h time points, mtPTP values were significantly increased either per cell or per mitochondrion (Student's t test, p < 0.05). At the 72 h time point after RCAN1-1L overexpression, mtPTP values become equal as expressed per cell; however, they remained elevated when expressed per mitochondrion (Student's t test, p < 0.05).
Our results reveal that initial (3–6 h) RCAN1-1L overexpression leads to a brief and transient mtPTP closing, whereas longer (9–72 h) overexpression causes a reversal of this effect, with more sustained mtPTP opening, as measured per cell (Fig. 5B). Because we have already estimated mitochondrial mass per cell (Figs. 2 and 3), our mtPTP data can also be recalculated per unit of mitochondria. When mtPTP data are expressed per unit of mitochondria, the results are similar, with one noticeable exception; although mtPTP returns to normal by 72 h after RCAN1-1L overexpression as expressed per cell, it seems to remain strongly open as expressed per mitochondrion. Overall, our results clearly identify RCAN1-1L as a regulator of mammalian mtPTP.
RCAN1-1L Reduces OCR but Increases Glycolysis (ECAR)
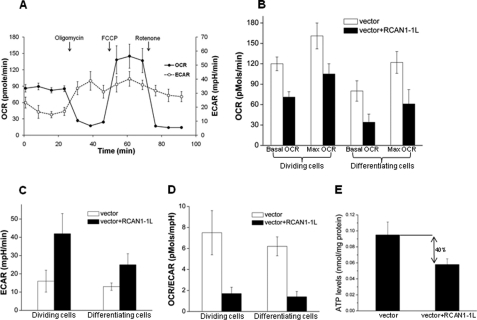
Because RCAN1-1L caused a dramatic reduction in mitochondrial mass, we wondered how this affected cellular bioenergetics. Analysis of bioenergetic pathways in ST14A cells was achieved with the SeahorseTM XF-ExtraFlux analyzer, which, simultaneously and non-invasively, quantifies cellular mitochondrial respiration as OCR and glycolytic flux (lactate generation) as ECAR. The sequential addition of oligomycin, FCCP, and rotenone denote ATP turnover, maximal respiration, and non-mitochondrial respiration, respectively. Oligomycin inhibits mitochondrial ATP synthesis, so that mitochondrial respiration measured in the absence/presence of oligomycin provides some measure of respiratory control. In the presence of FCCP, ATP/ADP levels, membrane potential, and pH gradient no longer limit respiration, and, hence, OCR represents the maximal capacity of substrate oxidation by the mitochondria. The residual rate in the presence of rotenone is taken to measure largely non-mitochondrial oxygen consumption. As shown in Fig. 6A, our ST14A cells responded appropriately to all conditions.
FIGURE 6.
RCAN1-1L reduces OCR but increases ECAR. RCAN1-1L was overexpressed using our adenoviral construct. Dividing cells were analyzed 24 h after transfection, and differentiating cells were analyzed 48 h after transfection. A, example of real-time analysis of bioenergetic pathways in ST14A cells. OCR and ECAR were monitored over time using the Seahorse metabolic analyzer. Cells were grown in 24-well plates to semiconfluence. To control cell amount, total protein content was determined in each well after the measurements, and data were normalized relative to protein content. Cells were incubated with oligomycin, FCCP, and rotenone. Results are means ± S.E. (error bars) of four independent samples. B, OCR. Values before the addition of oligomycin were used to calculate basal mitochondrial respiration. Values in the presence of FCCP were used to calculate maximal mitochondrial respiration. Results represent measurements from three experiments ± S.E. Both basal and maximal OCR were significantly reduced after RCAN1-1L overexpression in dividing as well as differentiating cells, as evaluated by Student's t test (p < 0.05). C, ECAR. Results represent measurements from three experiments ± S.E. ECAR were significantly increased after RCAN1-1L overexpression in both dividing and differentiating cells, as evaluated by Student's t test (p < 0.05). D, basal OCR/ECAR ratio. Results represent measurements from three experiments ± S.E. The ratios were significantly decreased after RCAN1-1L overexpression in both dividing and differentiating cells evaluated by Student's t test (p < 0.05). E, ATP levels are decreased after RCAN1-1L overexpression. Dividing ST14A cells were collected at 24 h after RCAN1-1L overexpression and analyzed as described under “Experimental Procedures.” ATP levels were significantly (∼40%) decreased in cells overexpressing RCAN1-1L in comparison with control cells, as evaluated by Student's t test (p < 0.05).
We found that both basal and maximal OCR were significantly decreased in dividing ST14A cells after RCAN1-1L overexpression (Fig. 6B). This decrease was even more pronounced in differentiated cells. At the same time, ECAR values were increased (Fig. 6C), suggesting a compensatory mechanism; therefore, OCR/ECAR ratios in both dividing and differentiating cells after RCAN1-1L overexpression (Fig. 6D) were dramatically reduced. As could be expected, this was accompanied by decreased production of ATP. Overexpression of RCAN1-1L for 24 h lead to significantly (∼40%) lowered ATP levels (Fig. 6E). These findings are in logical agreement with our data demonstrating that RCAN1-1L leads to mitochondrial degradation (Figs. 2 and 3), and they suggest that RCAN1-1L induction can cause a dramatic shift from aerobic respiration to glycolysis.
RCAN1-1L Overexpression Reduces Cell Survival
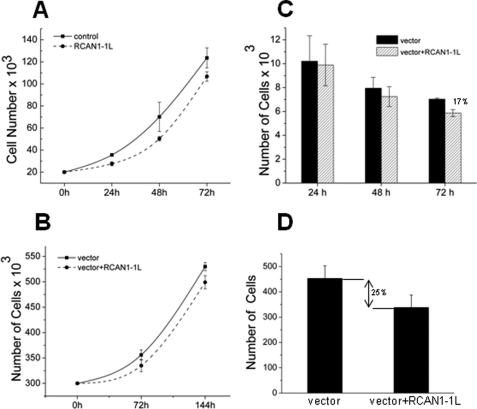
Because RCAN1-1L caused reduction of mitochondrial mass and a shift from aerobic respiration to glycolysis, we wondered how this might affect cell physiology, cell division, and survival. We first tested the effect of RCAN1-1L on dividing cells. We analyzed ST14A cells and ENStem cells and found that RCAN1-1L overexpression had only a slightly negative effect on cell number in both cell types (Fig. 7, A and B). This effect occurred as soon as in 24 h and did not significantly increase with time. If RCAN1-1L affected cell division, then one would expect a progressive decrease in cell number in samples that overexpress RCAN1-1L compared with controls. Because this is not the case, we conclude that RCAN1-1L does not significantly affect cell division.
FIGURE 7.
Effect of RCAN1-1L overexpression on cell division and survival. A, dividing ST14A Cells. Cell number was measured using a Coulter Counter (Coulter Corp.). The results represent mean values of three independent experiments ± S.E. (error bars). After 72 h, the number of cells in the samples overexpressing RCAN1-1L was significantly (∼17%) lower than in controls as evaluated by Student's t test (p < 0.05). B, dividing ENStem cells. Cell number was measured using a Coulter Counter. The results represent mean values of three independent experiments ± S.E. After 72 h, the number of cells in the samples overexpressing RCAN1-1L was significantly (∼10%) lower than in controls, as evaluated by Student's t test (p < 0.05). C, differentiating ST14A cells. Cells were analyzed microscopically and counted manually. Only live differentiating cells were counted. The results represent the means of three independent experiments ± S.E. After 72 h, the number of cells in the samples overexpressing RCAN1-1L was significantly (∼17%) lower than in controls, as evaluated by Student's t test (p < 0.05). D, differentiating ENStem cells. Cells were grown and differentiated for 12 days (288 h). They were then analyzed microscopically and counted manually. Only live differentiating cells were counted. The results represent the means of three independent experiments ± S.E. The number of cells in the samples overexpressing RCAN1-1L was significantly (∼25%) lower than in controls, as evaluated by Student's t test (p < 0.05).
To address the effect of RCAN1-1L on cell survival, we analyzed non-dividing cells. Differentiated ST14A cells do not survive very long in cell culture, so we performed such analyses only at 24, 48, and 72 h after RCAN1-1L overexpression with these cells. Opposite to our observations with dividing cells, we found that RCAN1-1L had no significantly negative effect during the initial hours of its overexpression, but we observed lower cell survival after longer time periods of RCAN1-1L overexpression (Fig. 7C). We then tested if such effects may accumulate in differentiated cells even more with time by employing another cell model, ENStem cells, which can survive in cell culture up to 3 weeks after differentiation. RCAN1-1L was overexpressed in these cells as previously, and cells were differentiated for 12 days (288 h) and analyzed (Fig. 7D). We found that RCAN1-1L overexpression led to an almost 25% reduction in cell number. This indicates that the negative effects of RCAN1-1L, indeed, accumulate in differentiated cells. Thus, our data suggest that RCAN1-1L has no effect on cell division, but it has a cumulative negative effect on cell survival. It has been recently demonstrated that RCAN1 can induce apoptosis (36), so we tested whether cell loss in our studies is also mediated by apoptosis. For these experiments, we labeled cells with one of the earliest markers of apoptosis, Annexin V (supplemental Fig. 4). We found that, after prolonged time periods (e.g. 24 h), RCAN1-1L did indeed increase apoptosis, in agreement with a recent report (36). Overall, these data indicate that prolonged RCAN1-1L induction may lead to initiation of apoptosis and cell death.
DISCUSSION
In the present study, we analyzed the cellular and metabolic effects of RCAN1-1L overexpression. RCAN1-1L can be induced by multiple stresses as well as various human diseases (4). Depending on the condition, it can be induced severalfold (by some stressors) or just 2–3-fold (as in Down syndrome and Alzheimer disease). It is possible that RCAN1-1L effects differ depending on the levels of its overexpression. Also, in contrast to Down syndrome and Alzheimer disease, we have found that RCAN1 deficiency can contribute to Huntington disease (14). Thus, the effects of RCAN1-1L can be expression time-, expression level-, and cell type-dependent, and more detailed studies will clearly be needed to address these questions. However, it is now clear that RCAN1-1L, which is the predominantly expressed RCAN1 protein isoform in the human brain (15), can be a regulator of mitochondrial degradation. This finding is in agreement with the previous report that human fetal brains with chromosome 21 trisomy show reduced mitochondrial DNA content (21). The RCAN1 gene is localized on chromosome 21, and it is overexpressed in chromosome 21 trisomies (2, 7), such as Down syndrome. Thus, it is possible that reduced mitochondrial DNA content in Down syndrome may be due to the overexpression of RCAN1-1L.
Our results demonstrate that RCAN1-1L overexpression can lead to mitochondrial degradation in both dividing and non-dividing cells. RCAN1-1L overexpression is followed by induction of autophagosome synthesis, and we show that activation of autophagy and mitochondrial degradation after RCAN1-1L overexpression coincides with opening of mtPTP, which confirms the theory that mtPTP opening can be a signal for mitochondrial degradation (24, 25). We were particularly interested in measures of basal and maximal respiration, in order to correlate changes in mtPTP formation and ADP/ATP translocation activity with diminished respiratory control. Our results showing mtPTP opening after RCAN1-1L overexpression are also in good agreement with the observation that cellular respiration (oxygen consumption) was diminished at the same time.
Remarkably, even drastic (more than 50%) degradation of the total cellular mitochondrial mass did not affect cell division and had no dramatic effect on cell survival. However, cell survival was also lowered with time, and the negative effect on this cell function was cumulative. Our results indicate that this may be due to the fact that, as mitochondria are lost, cells shift their bioenergetics from aerobic respiration to glycolysis, which enables them to survive, grow, and divide. RCAN1-1L can be induced by multiple stresses, and it is possible that although its induction might slightly reduce cell survival, it can also significantly increase resistance to stress factors. Thus, RCAN1-1L induction could have overall beneficial effects during short term stress. Overall, our findings have several important implications, as discussed below.
mtPTP
Although the mtPTPs were described some 3 decades ago, many have recently questioned all aspects of their structure and function (for reviews, see Refs. 23 and 24). Several proteins have been suggested to form mtPTP; however, they have all been debated and questioned recently. The ANT protein is the most commonly accepted player in mtPTP, and it is believed that this protein forms the core of the pore. Because RCAN1s can bind to ANT, we have tested their role in mtPTP opening and have found that RCAN1s indeed regulate mtPTP. Interestingly, RCAN1-1L initially leads to a brief mtPTP closing and then a much more sustained opening (Fig. 5). This observation argues against an alternative hypothesis of mtPTP, suggesting that no particular proteins play a role in mtPTP, but rather mtPTPs are formed from aggregates of any misfolded membrane proteins (37). If this was true and RCAN1-1L simply caused mitochondrial membrane proteins to misfold, then RCAN1-1L overexpression would have only one effect, either causing opening or closing. Of note, our model, in which RCAN1-1L can bind to ANT, also does not completely explain why mtPTP initially opens and then closes. These findings rather suggest that other players must be involved in order to switch mtPTP from the closed to the open state.
Autophagy
RCAN1-1L overexpression is followed by increased LC3 protein levels, particularly of LC3-II, which is a well accepted marker of increased autophagy. Increased levels of autophagosomes can be caused by their accelerated synthesis, decreased degradation (autophagic flux), or both. We demonstrate that autophagic flux does not change after RCAN1-1L overexpression, suggesting that RCAN1-1L may induce de novo synthesis of autophagosomes.
Our data suggest that the molecular mechanisms by which RCAN1-1L might induce formation of autophagosomes involve inhibition of mTOR signaling, which can be mediated, at least partially, through changes in intracellular ATP availability (Fig. 6E). mTOR can be regulated through several pathways: (a) amino acids, (b) insulin, or (c) glucose and ATP levels (38). We found that RCAN1-1L induction can lower cellular levels of ATP. Its overexpression for 24 h led to significantly (∼40%) lowered ATP levels (Fig. 6E), suggesting that RCAN1-1L can inhibit mTOR signaling and activate autophagy in an ATP-dependent manner. Because autophagy is an ATP-dependent process, it is possible that the reduction of ATP becomes self-limiting at some point, providing an explanation for the decrease in autophagy observed at 24 h (Fig. 3A).
It has been suggested that mtPTP as well as reduction of mitochondrial membrane potential may play a role in initiating autophagy. Our results suggest that mtPTP is an unlikely player in initiation of this process, because mtPTP pore opening begins after 9 h of RCAN1-1L overexpression (Fig. 5), whereas maximal initiation of autophagy is observed before/at 9 h of RCAN1-1L overexpression (Fig. 3). We also tested whether the collapse in mitochondrial membrane potential could be involved (supplemental Fig. 3). We found that indeed mitochondrial membrane potential was significantly reduced at 9 h of RCAN1-1L overexpression and returned to normal at 24 h. These data suggest that initiation of autophagy by RCAN1-1L may be at least partially driven by reduction of mitochondrial membrane potential. Thus, our data indicate that RCAN1-1L-induced autophagy may be initiated through inhibition of mTOR signaling regulated by ATP. For example, inhibition of ATP production may lead to increased AMP/ATP ratio and activation of AMPK, which then leads to inhibition of mTOR and activation of autophagy. RCAN1-1L-induced autophagy may also be initiated through reduction of mitochondrial membrane potential or even through a combination of mTOR and mitochondrial membrane potential pathways. However, the exact contributions of each of the RCAN1-1L induced pathways remain to be analyzed in more detail.
Cell Damage and Protection
The RCAN1 gene is transiently induced during acute cell adaptation to oxidative/calcium stress (1). Therefore, it was hypothesized that its induction may protect against such stress, which appears to be correct (5, 6). Chronic RCAN1 induction, however, is associated with Down syndrome and Alzheimer disease (2, 7), which suggests that the gene can also be harmful. To reconcile these findings, it was hypothesized that RCAN1 may exert protective effects if induced for only a short time but harmful effects if induced chronically (8). However, the mechanisms by which RCAN1s may protect or harm are incompletely understood.
Our data provide some new clues on possible mechanisms of RCAN1 function. RCAN1-1L overexpression eventually leads to mtPTP opening and mitochondrial degradation. Both mtPTP opening and mitochondrial degradation are generally accepted as features of lowered cell viability. Thus, we propose that the mechanism(s) by which RCAN1s may transiently protect cells during short term acute stress may involve mtPTP closing, as we demonstrate in Fig. 5B. On the other hand, if RCAN1 overexpression persists chronically, it may cause persistent mtPTP opening and mitochondrial degradation, which eventually decreases cell survival. In addition, short (9-h) RCAN1-1L induction may also stimulate mitochondrial biogenesis, whereas longer RCAN1-1L induction may cause the opposite effect (Fig. 4A).
These data are in agreement with the fact that acute stress induces RCAN1 gene only for several h, with maximal expression around 5 h after stress, with a return to normal levels by 24 h (1). Also, considering that many publications employ the MTT assay as a measure of cell survival, it is important to keep in mind that RCAN1 induction can lead mitochondrial degradation. This assay measures MTT reductase activity, and if mitochondrial mass decreases, then the assay would suggest that cell survival is decreased even if cells are growing normally. Thus, this method may produce misleading results, and it may not be appropriate in experiments manipulating RCAN1 expression. For example, it was reported that elevated levels of RCAN1s increase neuronal susceptibility to oxidative stress, as measured by an MTT assay (39). However, because elevated levels of RCAN1s can cause lower mitochondrial mass, MTT reductase activity could appear lower even if cells survive better.
Human Disease and Aging
Mitochondrial activity and autophagy have been suggested to play major roles in several common human diseases. For example, it has been recently recognized that mitochondrial dysfunction is an important factor in induction of Alzheimer disease (40, 41). We have previously proposed that overexpression of RCAN1s may lead to Alzheimer disease by inducing Tau hyperphosphorylation (7, 8, 15, 42). Now our results suggest that it may also contribute to this disease by diminishing mitochondrial functions. On the other hand, it is also possible that decreased mitochondrial mass and increased glycolysis may decrease production of reactive oxygen species, and such decreases may be beneficial for cell survival, at least in the short run. However, RCAN1s can also inhibit calcineurin and induce GSK-3β (15) both of which can lead to accumulation of the hyperphosphorylated Tau protein and neurodegeneration. Thus, it is possible that both protective and damaging effects of RCAN1s take place simultaneously and that the damaging effect overwhelms the protective one in the long run in cases such as Down syndrome and Alzheimer disease.
The results presented here also provide new clues about possible RCAN1-1L involvement in Huntington disease. We have recently shown that RCAN1-1L is deficient in this disease, and its overexpression protects against mutant huntingtin protein toxicity (14). It has been suggested that this protection involves huntingtin phosphorylation. However, it has been recently found that insufficient autophagy may also play a role in Huntington disease (43). Thus, our results indicate that RCAN1-1L may also protect against this disease by inducing autophagy.
Our findings that RCAN1-1L can switch cellular bioenergetics from aerobic respiration to glycolysis without affecting cell division could also be of interest for understanding cancers because this may help to explain how cancers can tolerate hypoxia. Finally, because mitochondrial activity is dramatically reduced in type II diabetes and aging (44), it appears possible that RCAN1s may also play an important role in these cases.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-ES003598 (and American Recovery and Reinvestment Act supplement 3RO1-ES 003598-22S2) (to K. J. A. D.) and R01AG016718 (to E. C.).

This article contains supplemental Figs. 1–4.
- ANT
- adenine nucleotide translocator
- mtPTP
- mitochondrial permeability transition pore
- OCR
- oxygen consumption rate
- ECAR
- extracellular acidification rate
- FCCP
- carbonylcyanide-4-trifluoromethoxyphenylhydrazone
- MOI
- multiplicities of infection
- p70S6K
- p70S6 kinase
- mTOR
- mammalian target of rapomycin
- TEM
- transmission electron microscopy.
REFERENCES
- 1. Crawford D. R., Leahy K. P., Abramova N., Lan L., Wang Y., Davies K. J. (1997) Hamster adapt78 mRNA is a Down syndrome critical region homologue that is inducible by oxidative stress. Arch. Biochem. Biophys. 342, 6–12 [DOI] [PubMed] [Google Scholar]
- 2. Fuentes J. J., Genescà L., Kingsbury T. J., Cunningham K. W., Pérez-Riba M., Estivill X., de la Luna S. (2000) DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum. Mol. Genet. 9, 1681–1690 [DOI] [PubMed] [Google Scholar]
- 3. Wang Y., De Keulenaer G. W., Weinberg E. O., Muangman S., Gualberto A., Landschulz K. T., Turi T. G., Thompson J. F., Lee R. T. (2002) Direct biomechanical induction of endogenous calcineurin inhibitor Down syndrome critical region-1 in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 283, H533–H539 [DOI] [PubMed] [Google Scholar]
- 4. Ermak G., Pritchard M. A., Dronjak S., Niu B., Davies K. J. (2011) Do RCAN1 proteins link chronic stress with neurodegeneration? FASEB J. 25, 3306–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leahy K. P., Crawford D. R. (2000) adapt78 protects cells against stress damage and suppresses cell growth. Arch. Biochem. Biophys. 379, 221–228 [DOI] [PubMed] [Google Scholar]
- 6. Ermak G., Harris C. D., Davies K. J. (2002) The DSCR1 (Adapt78) isoform 1 protein calcipressin 1 inhibits calcineurin and protects against acute calcium-mediated stress damage, including transient oxidative stress. FASEB J. 16, 814–824 [DOI] [PubMed] [Google Scholar]
- 7. Ermak G., Morgan T. E., Davies K. J. (2001) Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer's disease. J. Biol. Chem. 276, 38787–38794 [DOI] [PubMed] [Google Scholar]
- 8. Ermak G., Davies K. J. (2003) DSCR1(Adapt78). A Janus gene providing stress protection but causing Alzheimer's disease? IUBMB Life 55, 29–31 [DOI] [PubMed] [Google Scholar]
- 9. Cook C. N., Hejna M. J., Magnuson D. J., Lee J. M. (2005) Expression of calcipressin1, an inhibitor of the phosphatase calcineurin, is altered with aging and Alzheimer's disease. J. Alzheimers Dis. 8, 63–73 [DOI] [PubMed] [Google Scholar]
- 10. Harris C. D., Ermak G., Davies K. J. (2005) Multiple roles of the DSCR1 (Adapt78 or RCAN1) gene and its protein product calcipressin 1 (or RCAN1) in disease. Cell Mol. Life Sci. 62, 2477–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baek K. H., Zaslavsky A., Lynch R. C., Britt C., Okada Y., Siarey R. J., Lensch M. W., Park I. H., Yoon S. S., Minami T., Korenberg J. R., Folkman J., Daley G. Q., Aird W. C., Galdzicki Z., Ryeom S. (2009) Down's syndrome suppression of tumor growth and the role of the calcineurin inhibitor DSCR1. Nature 459, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minami T., Horiuchi K., Miura M., Abid M. R., Takabe W., Noguchi N., Kohro T., Ge X., Aburatani H., Hamakubo T., Kodama T., Aird W. C. (2004) Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J. Biol. Chem. 279, 50537–50554 [DOI] [PubMed] [Google Scholar]
- 13. Rothermel B. A., McKinsey T. A., Vega R. B., Nicol R. L., Mammen P., Yang J., Antos C. L., Shelton J. M., Bassel-Duby R., Olson E. N., Williams R. S. (2001) Myocyte-enriched calcineurin-interacting protein, MCIP1, inhibits cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. U.S.A. 98, 3328–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ermak G., Hench K. J., Chang K. T., Sachdev S., Davies K. J. (2009) Regulator of calcineurin (RCAN1-1L) is deficient in Huntington disease and protective against mutant huntingtin toxicity in vitro. J. Biol. Chem. 284, 11845–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ermak G., Harris C. D., Battocchio D., Davies K. J. (2006) RCAN1 (DSCR1 or Adapt78) stimulates expression of GSK-3β. FEBS J. 273, 2100–2109 [DOI] [PubMed] [Google Scholar]
- 16. Davies K. J., Ermak G., Rothermel B. A., Pritchard M., Heitman J., Ahnn J., Henrique-Silva F., Crawford D., Canaider S., Strippoli P., Carinci P., Min K. T., Fox D. S., Cunningham K. W., Bassel-Duby R., Olson E. N., Zhang Z., Williams R. S., Gerber H. P., Pérez-Riba M., Seo H., Cao X., Klee C. B., Redondo J. M., Maltais L. J., Bruford E. A., Povey S., Molkentin J. D., McKeon F. D., Duh E. J., Crabtree G. R., Cyert M. S., de la Luna S., Estivill X. (2007) Renaming the DSCR1/Adapt78 gene family as RCAN. Regulators of calcineurin. FASEB J. 21, 3023–3028 [DOI] [PubMed] [Google Scholar]
- 17. Görlach J., Fox D. S., Cutler N. S., Cox G. M., Perfect J. R., Heitman J. (2000) Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 19, 3618–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kingsbury T. J., Cunningham K. W. (2000) A conserved family of calcineurin regulators. Genes Dev. 14, 1595–1604 [PMC free article] [PubMed] [Google Scholar]
- 19. Rothermel B., Vega R. B., Yang J., Wu H., Bassel-Duby R., Williams R. S. (2000) A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 275, 8719–8725 [DOI] [PubMed] [Google Scholar]
- 20. Cao X., Kambe F., Miyazaki T., Sarkar D., Ohmori S., Seo H. (2002) Novel human ZAKI-4 isoforms. Hormonal and tissue-specific regulation and function as calcineurin inhibitors. Biochem. J. 367, 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang K. T., Min K. T. (2005) Drosophila melanogaster homolog of Down syndrome critical region 1 is critical for mitochondrial function. Nat. Neurosci. 8, 1577–1585 [DOI] [PubMed] [Google Scholar]
- 22. Javadov S., Karmazyn M. (2007) Mitochondrial permeability transition pore opening as an end point to initiate cell death and as a putative target for cardioprotection. Cell Physiol. Biochem. 20, 1–22 [DOI] [PubMed] [Google Scholar]
- 23. Zorov D. B., Juhaszova M., Yaniv Y., Nuss H. B., Wang S., Sollott S. J. (2009) Regulation and pharmacology of the mitochondrial permeability transition pore. Cardiovasc. Res. 83, 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodriguez-Enriquez S., He L., Lemasters J. J. (2004) Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int. J. Biochem. Cell Biol. 36, 2463–2472 [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez-Enriquez S., Kai Y., Maldonado E., Currin R. T., Lemasters J. J. (2009) Roles of mitophagy and the mitochondrial permeability transition in remodeling of cultured rat hepatocytes. Autophagy 5, 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ermak G., Cheadle C., Becker K. G., Harris C. D., Davies K. J. (2004) DSCR1(Adapt78) modulates expression of SOD1. FASEB J. 18, 62–69 [DOI] [PubMed] [Google Scholar]
- 27. Bota D. A., Davies K. J. (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4, 674–680 [DOI] [PubMed] [Google Scholar]
- 28. Mizushima N. (2004) Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 29. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Droge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fesus L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., Gonzalez-Estevez C., Gorski S., Gottlieb R. A., Haussinger D., He Y. W., Heidenreich K., Hill J. A., Hoyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jaattela M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovacs A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., Lopez-Otin C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Melendez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Munz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nurnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Talloczy Z., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcategui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mijaljica D., Prescott M., Devenish R. J. (2007) Different fates of mitochondria. Alternative ways for degradation? Autophagy 3, 4–9 [DOI] [PubMed] [Google Scholar]
- 31. Blommaart E. F., Luiken J. J., Blommaart P. J., van Woerkom G. M., Meijer A. J. (1995) Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 [DOI] [PubMed] [Google Scholar]
- 32. Noda T., Ohsumi Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 [DOI] [PubMed] [Google Scholar]
- 33. Romanello V., Sandri M. (2010) Mitochondrial biogenesis and fragmentation as regulators of muscle protein degradation. Curr. Hypertens. Rep. 12, 433–439 [DOI] [PubMed] [Google Scholar]
- 34. Scarpulla R. C. (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 35. Petronilli V., Miotto G., Canton M., Brini M., Colonna R., Bernardi P., Di Lisa F. (1999) Transient and long-lasting openings of the mitochondrial permeability transition pore can be monitored directly in intact cells by changes in mitochondrial calcein fluorescence. Biophys. J. 76, 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun X., Wu Y., Chen B., Zhang Z., Zhou W., Tong Y., Yuan J., Xia K., Gronemeyer H., Flavell R. A., Song W. (2011) Regulator of calcineurin 1 (RCAN1) facilitates neuronal apoptosis through caspase-3 activation. J. Biol. Chem. 286, 9049–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He L., Lemasters J. J. (2002) Regulated and unregulated mitochondrial permeability transition pores. A new paradigm of pore structure and function? FEBS Lett. 512, 1–7 [DOI] [PubMed] [Google Scholar]
- 38. Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. (2010) mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Porta S., Martí E., de la Luna S., Arbonés M. L. (2007) Differential expression of members of the RCAN family of calcineurin regulators suggests selective functions for these proteins in the brain. Eur. J. Neurosci. 26, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 40. Querfurth H. W., LaFerla F. M. (2010) Alzheimer's disease. N. Engl. J. Med. 362, 329–344 [DOI] [PubMed] [Google Scholar]
- 41. Yao J., Irwin R. W., Zhao L., Nilsen J., Hamilton R. T., Brinton R. D. (2009) Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 106, 14670–14675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harris C. D., Ermak G., Davies K. J. (2007) RCAN1-1L is overexpressed in neurons of Alzheimer's disease patients. FEBS J. 274, 1715–1724 [DOI] [PubMed] [Google Scholar]
- 43. Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D., Cuervo A. M. (2010) Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 13, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petersen K. F., Befroy D., Dufour S., Dziura J., Ariyan C., Rothman D. L., DiPietro L., Cline G. W., Shulman G. I. (2003) Mitochondrial dysfunction in the elderly. Possible role in insulin resistance. Science 300, 1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.