Background: Reductive carboxylation by isocitrate dehydrogenase (IDH) is required for hypoxic growth, and IDH mutations are associated with cancer.
Results: Reductive carboxylation by IDH is inhibited by NADP+ and isocitrate and inactivated by cancer-associated mutations.
Conclusion: Cancer-associated IDH mutations inactivate reductive carboxylation.
Significance: IDH mutations may reduce the capacity of cells to produce acetyl-CoA via reductive carboxylation.
Keywords: Acetyl Coenzyme A, Cancer, Enzyme Inactivation, Lipid Metabolism, Metabolism, 2-Hydroxyglutarate, α-Ketoglutarate, Isocitrate Dehydrogenase, Reductive Carboxylation
Abstract
Isocitrate dehydrogenase (IDH) is a reversible enzyme that catalyzes the NADP+-dependent oxidative decarboxylation of isocitrate (ICT) to α-ketoglutarate (αKG) and the NADPH/CO2-dependent reductive carboxylation of αKG to ICT. Reductive carboxylation by IDH1 was potently inhibited by NADP+ and, to a lesser extent, by ICT. IDH1 and IDH2 with cancer-associated mutations at the active site arginines were unable to carry out the reductive carboxylation of αKG. These mutants were also defective in ICT decarboxylation and converted αKG to 2-hydroxyglutarate using NADPH. These mutant proteins were thus defective in both of the normal reactions of IDH. Biochemical analysis of heterodimers between wild-type and mutant IDH1 subunits showed that the mutant subunit did not inactivate reductive carboxylation by the wild-type subunit. Cells expressing the mutant IDH are thus deficient in their capacity for reductive carboxylation and may be compromised in their ability to produce acetyl-CoA under hypoxia or when mitochondrial function is otherwise impaired.
Introduction
Cytosolic acetyl-CoA is required to support fatty acid and cholesterol biosynthesis, among other metabolic functions. A major source for acetyl-CoA is glucose-derived pyruvate, which enters the mitochondria and is decarboxylated to acetyl-CoA by pyruvate dehydrogenase and condensed with oxaloacetate by citrate synthase. Citrate exits the mitochondria and is cleaved to acetyl-CoA and oxaloacetate by cytosolic ATP:citrate lyase. However, there is another route to cytosolic citrate and acetyl-CoA known as the reductive carboxylation pathway. In 1948, Ochoa (1) noted the ability of heart extracts to incorporate CO2 into tricarboxylic acids, and subsequent experiments with specifically labeled precursors showed that the reductive carboxylation of αKG2 by IDH was responsible for the fixation of CO2 into tricarboxylic acids (2). IDH1 and IDH2 are closely related homodimers, with the significant difference being that IDH1 is cytosolic and IDH2 is mitochondrial. IDH3 is a mitochondrial heterotrimer that uses NAD+ and has an irreversible function in the tricarboxylic acid cycle (3). Cytosolic IDH1 biochemistry has been studied in some detail. The enzyme has a random order of substrate addition (4, 5) and uses CO2 (not bicarbonate) in the reductive carboxylation reaction (6). Glutamine is thought to make a major contribution to lipogenic acetyl-CoA in brown adipocytes (7, 8). Despite the recognition of the reductive carboxylation pathway in metabolic research, this lipogenic pathway is often overlooked. One reason is that cultured cells grown in 21% O2 and 5% CO2 produce citrate almost exclusively from the condensation of oxaloacetate derived from glutamine and acetyl-CoA from glucose (9). However, a recent series of studies demonstrated that the IDH-dependent reductive carboxylation pathway is essential for citrate synthesis under hypoxic conditions (0.5–1% O2) or when the mitochondrial electron transport chain is damaged (10–12). Thus, reductive carboxylation by IDH contributes to acetyl-CoA synthesis in normal physiological settings and is essential for lipogenesis when mitochondrial function is compromised.
Interest in IDH biochemistry increased when mutations in one of the two homodimeric IDHs were associated with several subtypes of cancer, including gliomas and leukemias (13–16). The IDH forward reaction, isocitrate decarboxylation, is inactivated by the common cancer-associated mutations that occur at Arg-132 of IDH1 or the analogous Arg-172 of IDH2 (17–21). Instead, these mutant IDHs produce 2HG from NADPH and αKG. This neomorphic activity leads to the accumulation of 2HG, which acts as an inhibitor of αKG-dependent enzymes, which in turn alters cell physiology by impacting the extent of protein and nucleic acid methylation (22, 23). However, the ability of the mutant IDHs to carry out the NADPH-dependent reductive carboxylation of αKG has not been investigated because the reverse reaction cannot be detected in assays lacking CO2 and pH >7. This study shows that product inhibition, primarily by NADP+ and secondarily by ICT, regulates IDH-dependent reductive carboxylation. The predominant cancer-associated IDH mutations inactivate both the forward and reverse reactions of IDH, leaving only the neomorphic activity producing 2HG from αKG. These properties are consistent with a high cellular reducing environment coupled with low mitochondrial production of citrate as prerequisites activating flux through reductive carboxylation. The expression of mutant IDH enzymes would potentially diminish the capacity of the cell to carry out reductive metabolism and cope with hypoxia or mitochondrial damage.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Mutagenesis
Codon-optimized sequences for high protein expression in Escherichia coli (GeneArt) were obtained for full-length human IDH1 (NCBI reference sequence NM_005896) and a truncated version of human IDH2 (NCBI reference sequence NM_002168) lacking the first 39 amino acids. These IDH1 and IDH2 sequences were cloned between the NdeI and BamHI restriction sites of pET-28b(+), generating constructs pCS59 and pCS68, respectively. pCS59 was mutagenized to obtain IDH1 mutants V71I, G97D, G123R, R132S, and R132H, whereas pCS68 was mutagenized to obtain IDH2 mutants R140Q and R172K. All enzymes expressed from the respective pET-28b-derived constructs contained an N-terminal hexahistidine tag. IDH1(R132H) was subcloned into pET-51b(+) for expression as a Strep-tag® fusion protein (pCS67). The IDH1WT/R132H heterodimer was expressed from BL21(DE3) cells transformed with both pCS59 and pCS67 and grown in double selective medium.
Protein Expression and Purification
Enzymes were expressed in BL21(DE3) cells for 18–20 h at 16 °C following induction with 1 mm isopropyl β-d-thiogalactopyranoside. Lysates were purified using standard nondenaturing nickel-nitrilotriacetic acid column chromatography. To purify the IDH1WT/R132H heterodimer, after elution from nickel-nitrilotriacetic acid, the protein was purified on a Strep-Tactin Superflow Plus column. Purified proteins were dialyzed overnight against 20 mm Tris-HCl (pH 7.5), 200 mm NaCl, 5 mm β-mercaptoethanol, and 10% glycerol. Glycerol was added to 50% to all of the purified proteins for storage at −20 °C. All enzymes were used within 2 weeks of isolation to minimize activity loss and allow batch-to-batch comparisons.
Spectrofluorometric IDH Assay
The forward reaction was measured with 10 μg of enzyme in a total volume of 200 μl containing 20 mm bis-Tris (pH 6.0), 20 mm MgCl2, 400 μm ICT, and 20 μm β-NADP+. The reverse reaction used 10 μg of protein in a volume of 200 μl containing 20 mm bis-Tris (pH 6.0), 20 mm MgCl2, 1 mm αKG, 35 mm NaHCO3 (pH 6.0) and 160 μm β-NADPH. Reaction rates were monitored with a FluoroMax-4 fluorometer. The excitation wavelength was 340 nm (2-nm slit), and the emission was at 460 nm (5-nm slit). Changes in fluorescence were recorded at 25 °C for 400 s starting from the addition of the enzyme. Fluorescence units were converted to picomoles of NADPH using a calibration curve. The slow non-enzymatic rate of NADPH oxidation was subtracted from the rates.
Chromatography-based IDH Assay
Reaction mixtures contained 20 mm bis-Tris (pH 6.0), 20 mm MgCl2, 1 mm [1-14C]αKG (2.5 Ci/mol), 35 mm NaHCO3 (pH 6.0), 500 μm β-NADPH, 0.2 mm DTT, 200 mm Glc-6-P, 0.1 unit of Glc-6-P dehydrogenase, and 1 μg of IDH in a 20-μl volume. Incubation was at 25 °C for 20 min, and the reaction was stopped with 1 μl of formic acid. The reaction mixture (5 μl) was spotted onto a PEI-cellulose plate developed with ethyl ether/formic acid/water (70:20:10, v/v/v). The radioactive products were quantified using a Typhoon PhosphorImager.
RESULTS
Biochemical Regulation of IDH1 Reductive Carboxylation
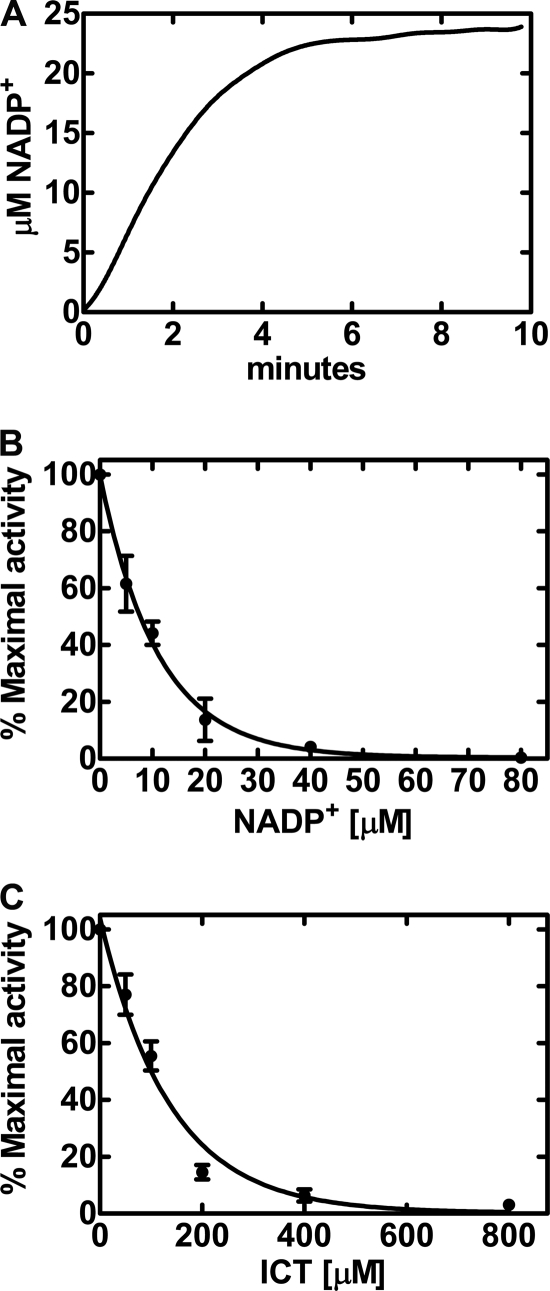
The reductive carboxylation (“reverse reaction”) of IDH1 was studied by following the oxidation of NADPH spectrofluorometrically, which was dependent on the presence of both αKG and CO2. The CO2 source was provided by dissolving 35 mm sodium bicarbonate in pH 6.0 buffer. The apparent Km for bicarbonate was 18 ± 1.2 mm (data not shown). The CO2 Km in these experiments was calculated based on the established pH-dependent equilibrium between bicarbonate and CO2 (6). This equation computes the Km for CO2 as 12.6 ± 1.7 mm at pH 6.0. Reductive carboxylation assays cannot be performed at pH 7.5 because the concentration of CO2 is low regardless of the bicarbonate concentration. This explains why others have overlooked the reverse reaction when analyzing IDH and its mutant derivatives (17–20). IDH1 catalyzed a linear rate of reductive carboxylation that did not continue past ∼3 min (Fig. 1A). The apparent Km for αKG was 1.4 ± 0.2 mm, and that for NADPH was 35 ± 10 μm. The reaction rate slowed and eventually reached equilibrium under these experimental conditions when the amount of NADP+ (and ICT) product reached ∼20 μm. These data suggested that the products were potentially negative regulators of the reverse reaction. The titration of NADP+ into the assay potently inhibited reductive carboxylation, with an apparent Ki of 7.6 ± 1.6 μm (Fig. 1B). ICT also inhibited αKG carboxylation, but was not as potent as NADP+ (Fig. 1C). The apparent Ki for ICT was 110 ± 28 μm. The forward reaction of IDH1 was linear for up to 30 min and proceeded until >90% of the ICT was consumed (data not shown). The apparent Km values for the forward reaction were 9.8 ± 2.4 μm for NADP+ and 43 ± 17 μm for ICT. The kinetic parameters were on the same order as the metabolite concentrations in tissues: αKG, 0.07–3.5 mm; NADPH, 0.12–1.5 mm; NADP+, 20–200 μm; ICT, ∼30 μm; and citrate 70–400 μm (24, 25). These data illustrated that NADP+ and, to a lesser extent, ICT were product inhibitors of IDH1 reductive carboxylation; however, neither NADPH nor αKG was a potent product inhibitor of ICT decarboxylation. This is due to the fact that IDH1 has a higher affinity for NADP+ and ICT than for NADPH and αKG. However, the ability of NADP+ to convert IDH1 into an inactive conformation that can be released only by ICT (26, 27) also contributes to the potent regulation of IDH1 by NADP+. These data suggest that reverse flow through IDH1 is promoted by an intracellular reducing environment (high NAD(P)H/NAD(P)+ ratio) and low production of citrate by the mitochondria.
FIGURE 1.
Regulation of IDH1 reductive carboxylation by NADP+. Reaction rates were determined by following the decrease in NADPH fluorescence as a function of time as described under “Experimental Procedures.” A, the time course for the carboxylation of αKG by IDH1 was linear for only the first few minutes of the assay. Initial rates were calculated from the data in the first 3 min of the reaction. B, inhibition of IDH1 reductive carboxylation of αKG by NADP+ using the spectrofluorometric assay. C, inhibition of IDH1 reductive carboxylation by ICT using the spectrofluorometric assay. The IDH1 specific activity (100% activity) in these experiments was 142 pmol/min/mg, and data points were obtained in triplicate.
Cancer-associated IDH1/IDH2 Mutant Proteins Cannot Catalyze Reductive Carboxylation
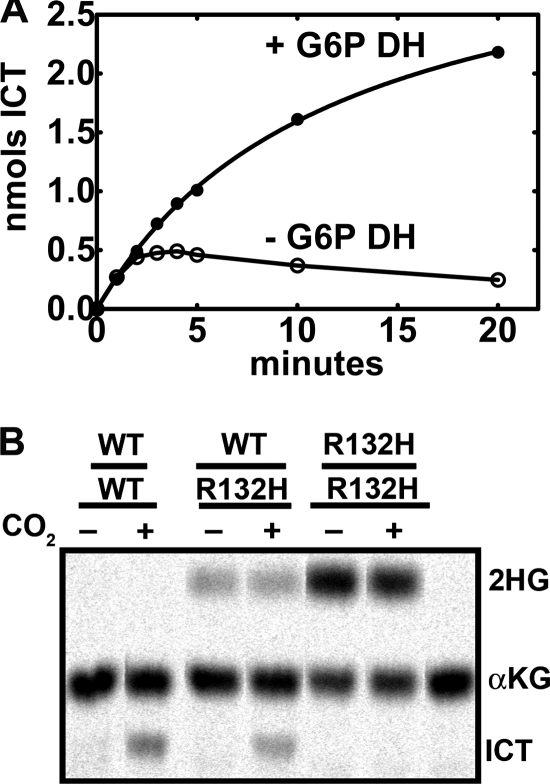
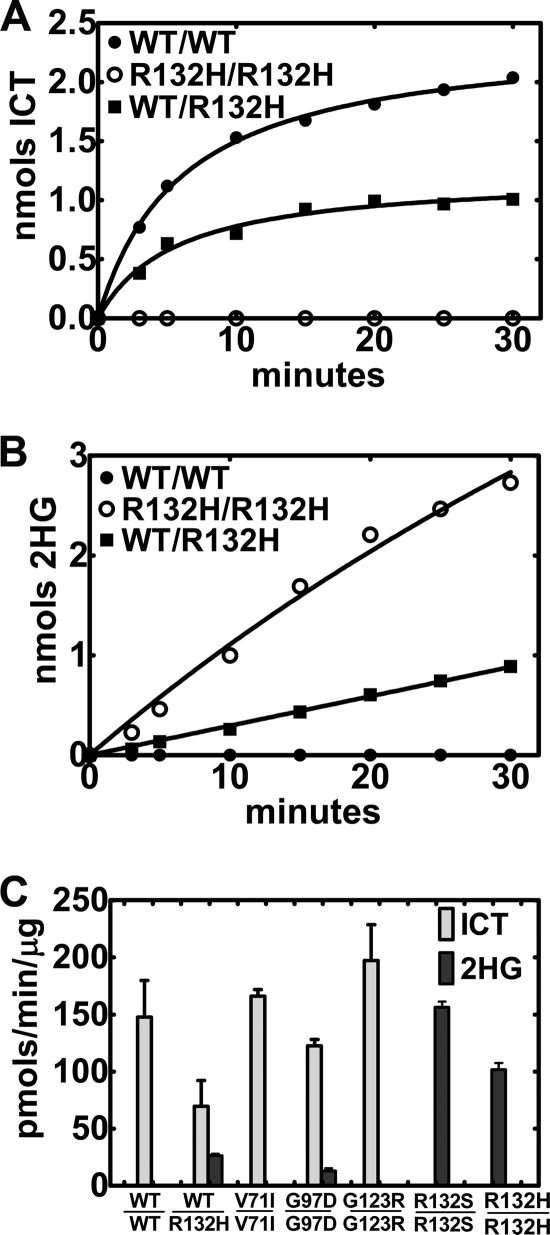
We used the IDH1(R132H) mutant as a model because mutations at Arg-132 are the most common IDH1 alterations associated with cancers (13). IDH1 and IDH2 mutations associated with cancers are known to produce 2HG from αKG and NADPH (17, 18). Measuring the reductive carboxylation rate spectrofluorometrically by following NADPH consumption cannot distinguish between ICT or 2HG formation. A radiochemical/chromatographic assay using [14C]αKG was developed to simultaneously measure these two products. The radiochemical assay was less sensitive than the spectrofluorometric assay, and more product formation was required to obtain a signal above background due to the low specific activity of 1 mm [14C]αKG. This practical issue resulted in rates of product formation that were at the lower level of [14C]αKG detection before the reaction ceased due to NADP+ accumulation (Fig. 1A). After an initial burst of [14C]ICT formation, the amount of the product in the assay actually decreased with time (Fig. 2A). This problem with the reductive carboxylation reaction was eliminated by employing Glc-6-P dehydrogenase as a NADPH-regenerating system to prevent the stringent inhibition of reductive carboxylation by NADP+. The radiochemical assay with the NADPH-regenerating system increased the linearity and extent of IDH1 reductive carboxylation (Fig. 2A). The level of Glc-6-P dehydrogenase required to sustain reductive carboxylation was determined experimentally because the commercial enzyme preparation could convert αKG to 2HG in the absence of IDH1 (data not shown). The extent of the reductive carboxylation reaction was limited only by the accumulation of ICT in the radiochemical assay, resulting in an increase in the amount of product formation (Fig. 2A). This assay was used to compare the reductive carboxylation activities of IDH1, IDH1(R132H) homodimers, and IDH1WT/R132H heterodimers (Fig. 2B). IDH1 produced only ICT from αKG, whereas the mutant IDH1(R132H) homodimers produced only 2HG. These data showed that mutation at Arg-132 inactivated the reverse reaction of IDH1. These single point assays were corroborated by comparing the rates of ICT and 2HG formation by IDH1 and IDH1(R132H) (Fig. 3). IDH1 made only ICT and exhibited an average specific activity of 148 ± 32 pmol/min/μg, whereas IDH1(R132H) homodimers made only 2HG and exhibited an average specific activity of 102 ± 6 pmol/min/μg. We confirmed that IDH1(R132H) was also defective in the forward NADP+-dependent decarboxylation of ICT (data not shown) as reported previously (17–20). Thus, the mutation at the active site Arg-132 inactivates both the forward and reverse reactions of IDH1 and promotes the neomorphic activity of 2HG production.
FIGURE 2.
Reductive formation of ICT and 2HG by wild-type and mutant IDH1 proteins. A, comparison of the radiochemical ([14C]αKG conversion to ICT and 2HG) assay for IDH1 in the presence and absence of a Glc-6-P dehydrogenase (G6P DH) NADPH-regenerating system. B, analysis of the products formed by IDH1 homodimers (WT/WT), IDH1WT/R132H heterodimers (WT/R132H), and IDH1(R132H) homodimers (R132H/R132H) using the radiochemical assay with the NADPH-regenerating system to simultaneously detect [14C]ICT and [14C]2HG by thin-layer chromatography as described under “Experimental Procedures.”
FIGURE 3.
Production of ICT and 2HG by wild-type and mutant IDH1 proteins. A, representative time course for the formation of ICT by IDH1 homodimers (WT/WT; ●), IDH1(R132H) homodimers (R132H/R132H; ○), and IDH1WT/R132H heterodimers (WT/R132H; ■). Reaction rates slowed with time due to product inhibition by ICT. B, representative time course for the formation of 2HG by the three types of IDH1 proteins. C, the IDH1 specific activities for individual IDH1 proteins was determined in at least two different lots of individually purified proteins using the radiochemical assay to detect both [14C]ICT and [14C]2HG formation.
Reductive Carboxylation in Wild-type/Mutant Heterodimers
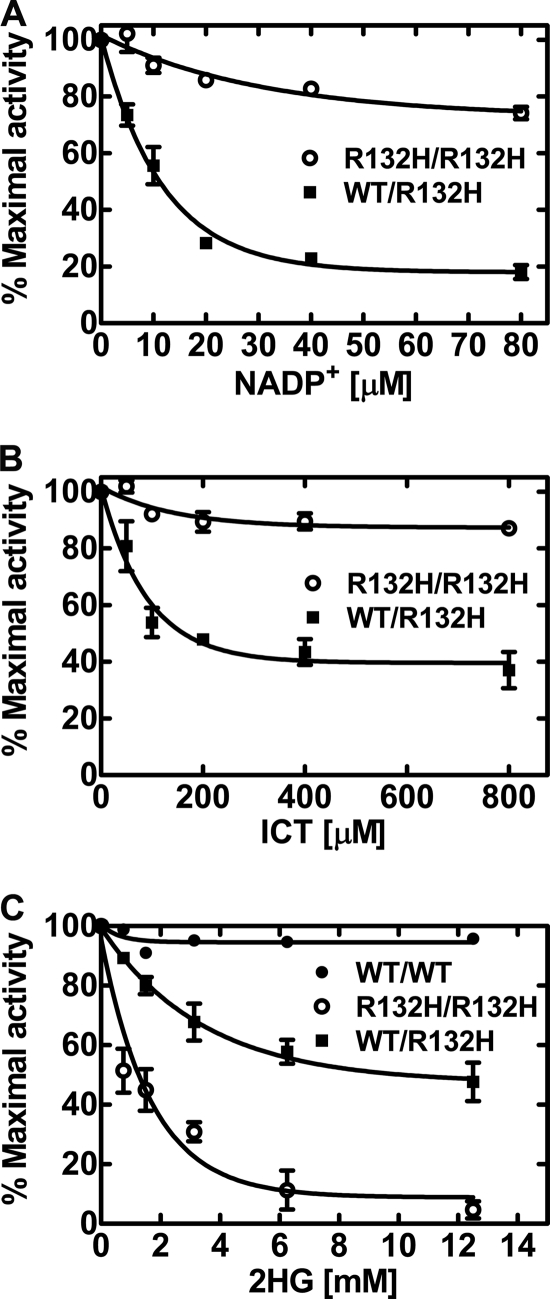
In all cases, cancer-associated IDH mutations occur in only one allele (13). Therefore, heterodimers between wild-type and mutant subunits are predicted to compose 50% of the IDH enzyme in these tumors. The initial characterization of wild-type/mutant heterodimers concluded that the mutant subunit had a dominant-negative effect on the wild-type subunit (21). Subsequent work concluded that each half of the heterodimer was functionally independent in the forward reaction (19). The IDH1WT/R132H heterodimer produced both ICT and 2HG (Fig. 2B). IDH1WT/R132H heterodimers produced ICT at an average specific activity of 70 ± 22 pmol/min/μg and 2HG at an average specific activity of 27 ± 2 pmol/min/μg (Fig. 3, A and B). ICT formation by IDH1WT/R132H heterodimers exhibited biphasic inhibition by NADP+ using the spectrofluorometric assay (Fig. 4A). A portion of NADPH utilization was blocked by low concentrations of NADP+ as stringently as in the wild-type enzyme (Fig. 1B); however, there remained a proportion of the NADPH oxidation that was refractory to NADP+ regulation that corresponded to the production of 2HG by the mutant monomer. This conclusion was corroborated by the observation that IDH1(R132H) homodimers were insensitive to NADP+ inhibition (Fig. 4A). Likewise, ICT also inhibited IDH1WT/R132H NADPH oxidation, but there also remained a significant rate of oxidation that was refractory to ICT inhibition (Fig. 4B). The IDH1(R132H) homodimers were insensitive to the presence of ICT. The fact that the maximal inhibition of NADPH consumption by NADP+ in the IDH1WT/R132H heterodimers was greater than the maximal ICT inhibition was consistent with NADP+ binding converting IDH1 to an open inactive conformation that is overcome only by ICT (26, 27). ICT was initially absent in the NADP+ titration in Fig. 4A, but ICT was continuously present in the titration in Fig. 4B. We next explored the regulation of IDH1 by the neomorphic product 2HG. 2HG had a minimal effect on reductive carboxylation by IDH1 (Fig. 4C). In contrast, 2HG blocked NADPH utilization by IDH1(R132H) homodimers, with an apparent Ki of 1.2 ± 0.3 mm. The addition of 2HG to the IDH1WT/R132H reaction blocked 50% of the NADPH oxidation. In the radiochemical assay of IDH1WT/R132H, 2HG blocked the formation of radiolabeled 2HG but did not inhibit the formation of ICT (data not shown). These data were consistent with the model in which 2HG inhibited the mutant monomer and did not inhibit the wild-type subunit. Thus, the two IDH1 subunits in the IDH1WT/R132H heterodimer were both operational.
FIGURE 4.
NADP+, ICT, and 2HG regulation of IDH1 and IDH1(R132H). A, NADP+ inhibition of NADPH oxidation by IDH1(R132H) homodimers (R132H/R132H; ○) and IDH1WT/R132H heterodimers (WT/R132H; ■). B, ICT regulation of NADPH oxidation by IDH1(R132H) homodimers (○) and IDH1WT/R132H heterodimers (■). C, 2HG inhibition of NADPH oxidation by IDH1 homodimers (WT/WT; ●), IDH1(R132H) homodimers (○), and IDH1WT/R132H heterodimers (■). The spectrofluorometric assay following αKG-dependent NADPH oxidation was used in these experiments. Means ± S.E. of triplicate measurements are plotted.
The most prevalent mutations occur at Arg-132 in IDH1 and the equivalent Arg-172 in IDH2, and the substitution of these amino acids inactivates the forward reaction and leads to the formation of 2HG in the presence of NADPH (17, 18). Both the IDH1(R132S) and IDH1(R132H) mutants were defective in reductive carboxylation and produced copious amounts of 2HG (Fig. 3C). The properties of the mutant enzymes correlated with the absence of an active site arginine rather than the specific type of amino acid substituted for the arginine. Large-scale sequencing efforts have identified other IDH1 mutations that occur infrequently in gliomas and leukemias compared with mutations at Arg-132 (13). IDH1(V71I) and IDH1(G123R) were as active as IDH1 in reductive carboxylation and did not produce 2HG (Fig. 3C). The IDH1(G97D) mutant was deficient in reductive carboxylation and produced a small amount of 2HG compared with the IDH1(R132H) mutant. IDH2 proteins expressed poorly and were obtained in much lower yields than IDH1. IDH2 also carried out robust reductive carboxylation under the same assay conditions used for IDH1, with a specific activity that averaged 136 ± 15 pmol/min/μg. Cancer-associated IDH2 mutations at the active site arginines (IDH2(R172K) and IDH2(R140Q)) were also defective in reductive carboxylation in addition to producing 2HG (data not shown), as others have reported (16, 18). These results were consistent with the high degree of structural similarity between IDH1 and IDH2 (26, 28). Thus, the frequently encountered mutations at the active site arginines inactivated both the forward and reverse IDH reactions and transformed the monomer subunit into an αKG reductase that generated only 2HG. Infrequent mutations at other sites did not fit this paradigm, and their catalytic properties cannot be predicted.
DISCUSSION
A key finding of this work is that the commonly found mutations at the active site Arg-132, recently associated with several subsets of cancers, inactivate the IDH1 reverse reaction (NADPH/CO2-dependent αKG carboxylation), in addition to blocking the forward reaction (NADP+-dependent ICT decarboxylation) (17). IDH2 is also active in reductive carboxylation, and mutations at either of the two active site arginines inactive the reverse reaction and transform the enzyme into a producer of 2HG. The inactivation of reductive carboxylation has important implications for understanding the contribution of mutant IDH to metabolism. The experiments with the IDH1WT/R132H heterodimers showed that the mutant subunit does not inactivate the normal reactions carried out by the wild-type subunit; thus, the net effect of expressing one mutant IDH allele is to reduce the overall capacity of the cell to carry out reductive carboxylation by about half. In addition, the αKG carbon destined for citrate is instead converted to 2HG, which reduces αKG levels (29). This may have little effect on lipogenesis in cells grown with abundant glutamine in high O2 concentrations, but the ability to produce acetyl-CoA when mitochondrial function is compromised or under hypoxic conditions may be diminished by the inactivation of 50% of the cell's capacity to carry out reductive carboxylation. These metabolic issues may contribute to the reduced aggressiveness of glioma cell lines expressing IDH1(R132H) mutations (30). However, our biochemical studies, coupled with knockdown studies in cultured cells (12), show that both IDH1 and IDH2 function interchangeably supports reductive carboxylation. This conclusion is not surprising considering the high degree of sequence and structural similarity between IDH1 (26) and IDH2 (28). Therefore, a general conclusion about the effect of mutations in IDH1 or IDH2 on reductive metabolism is difficult because the effect depends on both the remaining wild-type enzyme activity and the expression level of the other isoform in a particular cell line. The inactivation of both the forward and reverse reactions of IDH by the recurrent cancer-associated mutations in the active site arginines is consistent with the idea that 2HG formation by mutant IDH is an important event that alters cell physiology (22, 23).
This work was supported, in whole or in part, by National Institutes of Health Grants GM062896 (to S. J.) and GM034496 (to C. O. R.) and Cancer Center (CORE) Support Grant CA21765. This work was also supported by the American Syrian Associated Charities.
- αKG
- α-ketoglutarate
- IDH
- isocitrate dehydrogenase
- 2HG
- R(−)2-hydroxyglutarate
- ICT
- isocitrate
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Ochoa S. (1948) Biosynthesis of tricarboxylic acids by carbon dioxide fixation. III. Enzymatic mechanisms. J. Biol. Chem. 174, 133–157 [PubMed] [Google Scholar]
- 2. D'Adamo A. F., Jr., Haft D. E. (1965) An alternate pathway of α-ketoglutarate catabolism in the isolated, perfused rat liver. I. Studies with dl-glutamate-2- and -5-14C. J. Biol. Chem. 240, 613–617 [PubMed] [Google Scholar]
- 3. Ramachandran N., Colman R. F. (1980) Chemical characterization of distinct subunits of pig heart DPN-specific isocitrate dehydrogenase. J. Biol. Chem. 255, 8859–8864 [PubMed] [Google Scholar]
- 4. Northrop D. B., Cleland W. W. (1974) The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. II. Dead-end and multiple inhibition studies. J. Biol. Chem. 249, 2928–2931 [PubMed] [Google Scholar]
- 5. Uhr M. L., Thompson V. W., Cleland W. W. (1974) The kinetics of pig heart triphosphopyridine nucleotide-isocitrate dehydrogenase. I. Initial velocity, substrate and product inhibition, and isotope exchange studies. J. Biol. Chem. 249, 2920–2927 [PubMed] [Google Scholar]
- 6. Dalziel K., Londesborough J. C. (1968) The mechanisms of reductive carboxylation reactions. Carbon dioxide or bicarbonate as substrate of nicotinamide-adenine dinucleotide phosphate-linked isocitrate dehydrogenase and malic enzyme. Biochem. J. 110, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoo H., Stephanopoulos G., Kelleher J. K. (2004) Quantifying carbon sources for de novo lipogenesis in wild-type and IRS-1 knockout brown adipocytes. J. Lipid Res. 45, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 8. Yoo H., Antoniewicz M. R., Stephanopoulos G., Kelleher J. K. (2008) Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J. Biol. Chem. 283, 20621–20627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., Thompson C. B. (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., Thompson C. B. (2011) Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., Stephanopoulos G. (2012) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., DeBerardinis R. J. (2012) Reductive carboxylation supports growth in tumor cells with defective mitochondria. Nature 481, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dang L., Jin S., Su S. M. (2010) IDH mutations in glioma and acute myeloid leukemia. Trends Mol. Med. 16, 387–397 [DOI] [PubMed] [Google Scholar]
- 14. Mardis E. R., Ding L., Dooling D. J., Larson D. E., McLellan M. D., Chen K., Koboldt D. C., Fulton R. S., Delehaunty K. D., McGrath S. D., Fulton L. A., Locke D. P., Magrini V. J., Abbott R. M., Vickery T. L., Reed J. S., Robinson J. S., Wylie T., Smith S. M., Carmichael L., Eldred J. M., Harris C. C., Walker J., Peck J. B., Du F., Dukes A. F., Sanderson G. E., Brummett A. M., Clark E., McMichael J. F., Meyer R. J., Schindler J. K., Pohl C. S., Wallis J. W., Shi X., Lin L., Schmidt H., Tang Y., Haipek C., Wiechert M. E., Ivy J. V., Kalicki J., Elliott G., Ries R. E., Payton J. E., Westervelt P., Tomasson M. H., Watson M. A., Baty J., Heath S., Shannon W. D. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G. J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K. W., Velculescu V. E., Vogelstein B., Bigner D. D. (2009) IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andersson A. K., Miller D. W., Lynch J. A., Lemoff A. S., Cai Z., Pounds S. B., Radtke I., Yan B., Schuetz J. D., Rubnitz J. E., Ribeiro R. C., Raimondi S. C., Zhang J., Mullighan C. G., Shurtleff S. A., Schulman B. A., Downing J. R. (2011) IDH1 and IDH2 mutations in pediatric acute leukemia. Leukemia 25, 1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Vander Heiden M. G., Su S. M. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., Cross J. R., Fantin V. R., Hedvat C. V., Perl A. E., Rabinowitz J. D., Carroll M., Su S. M., Sharp K. A., Levine R. L., Thompson C. B. (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pietrak B., Zhao H., Qi H., Quinn C., Gao E., Boyer J. G., Concha N., Brown K., Duraiswami C., Wooster R., Sweitzer S., Schwartz B. (2011) A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of αHG. Biochemistry 50, 4804–4812 [DOI] [PubMed] [Google Scholar]
- 20. Jin G., Reitman Z. J., Spasojevic I., Batinic-Haberle I., Yang J., Schmidt-Kittler O., Bigner D. D., Yan H. (2011) 2-Hydroxyglutarate production, but not dominant-negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS ONE 6, e16812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y., Ding J., Lei Q., Guan K. L., Xiong Y. (2009) Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S. H., Ito S., Yang C., Wang P., Xiao M. T., Liu L. X., Jiang W. Q., Liu J., Zhang J. Y., Wang B., Frye S., Zhang Y., Xu Y. H., Lei Q. Y., Guan K. L., Zhao S. M., Xiong Y. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chowdhury R., Yeoh K. K., Tian Y. M., Hillringhaus L., Bagg E. A., Rose N. R., Leung I. K., Li X. S., Woon E. C., Yang M., McDonough M. A., King O. N., Clifton I. J., Klose R. J., Claridge T. D., Ratcliffe P. J., Schofield C. J., Kawamura A. (2011) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williamson J. R., Corkey B. E. (1979) Assay of citric acid cycle intermediates and related compounds: update with tissue metabolite levels and intracellular distribution. Methods Enzymol. 55, 200–222 [DOI] [PubMed] [Google Scholar]
- 25. Albe K. R., Butler M. H., Wright B. E. (1990) Cellular concentrations of enzymes and their substrates. J. Theor. Biol. 143, 163–195 [DOI] [PubMed] [Google Scholar]
- 26. Xu X., Zhao J., Xu Z., Peng B., Huang Q., Arnold E., Ding J. (2004) Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 279, 33946–33957 [DOI] [PubMed] [Google Scholar]
- 27. Yang B., Zhong C., Peng Y., Lai Z., Ding J. (2010) Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 20, 1188–1200 [DOI] [PubMed] [Google Scholar]
- 28. Ceccarelli C., Grodsky N. B., Ariyaratne N., Colman R. F., Bahnson B. J. (2002) Crystal structure of porcine mitochondrial NADP+-dependent isocitrate dehydrogenase complexed with Mn2+ and isocitrate. Insights into the enzyme mechanism. J. Biol. Chem. 277, 43454–43462 [DOI] [PubMed] [Google Scholar]
- 29. Reitman Z. J., Jin G., Karoly E. D., Spasojevic I., Yang J., Kinzler K. W., He Y., Bigner D. D., Vogelstein B., Yan H. (2011) Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. U.S.A. 108, 3270–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bralten L. B., Kloosterhof N. K., Balvers R., Sacchetti A., Lapre L., Lamfers M., Leenstra S., de Jonge H., Kros J. M., Jansen E. E., Struys E. A., Jakobs C., Salomons G. S., Diks S. H., Peppelenbosch M., Kremer A., Hoogenraad C. C., Smitt P. A., French P. J. (2011) IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann. Neurol. 69, 455–463 [DOI] [PubMed] [Google Scholar]