Background: l-Galactono-1,4-lactone dehydrogenase (GLDH) catalyzes the final step of the l-ascorbate biosynthesis pathway and at the same time is essential for complex I accumulation.
Results: The active GLDH is localized within three different subcomplexes of complex I.
Conclusion: Evidence is increasing that GLDH represents a complex I assembly factor.
Significance: New insights into mitochondrial complex I assembly in Arabidopsis thaliana.
Keywords: Arabidopsis, Ascorbic Acid, Mitochondria, Protein Assembly, Respiratory Chain, Complex I, GLDH
Abstract
l-Galactono-1,4-lactone dehydrogenase (GLDH) catalyzes the terminal step of the Smirnoff-Wheeler pathway for vitamin C (l-ascorbate) biosynthesis in plants. A GLDH in gel activity assay was developed to biochemically investigate GLDH localization in plant mitochondria. It previously has been shown that GLDH forms part of an 850-kDa complex that represents a minor form of the respiratory NADH dehydrogenase complex (complex I). Because accumulation of complex I is disturbed in the absence of GLDH, a role of this enzyme in complex I assembly has been proposed. Here we report that GLDH is associated with two further protein complexes. Using native gel electrophoresis procedures in combination with the in gel GLDH activity assay and immunoblotting, two mitochondrial complexes of 470 and 420 kDa were identified. Both complexes are of very low abundance. Protein identifications by mass spectrometry revealed that they include subunits of complex I. Finally, the 850-kDa complex was further investigated and shown to include the complete “peripheral arm” of complex I. GLDH is attached to a membrane domain, which represents a major fragment of the “membrane arm” of complex I. Taken together, our data further support a role of GLDH during complex I formation, which is based on its binding to specific assembly intermediates.
Introduction
Ascorbate (vitamin C) is of central importance for several biological processes. In plants, it was shown to be essential for growth (1), programmed cell death (2), pathogen response (3), signal transduction (4), and the stress response with respect to ozone (5), UV radiation (6), high temperature (7), and high light (8). Ascorbate is the cofactor of several enzymes and one of the major components adjusting the redox state of cells. In plant tissue, it can reach millimolar concentrations and form up to 10% of the soluble carbohydrate content. Biosynthesis of ascorbate in plants mainly takes place via the “l-galactose” also known as “Smirnoff-Wheeler” pathway (9). The terminal step of this pathway, the conversion of l-galactono-1,4-lactone (GL)2 into ascorbate, is catalyzed by l-galactono-1,4-lactone dehydrogenase (GLDH). GLDH is localized in mitochondria. During ascorbate formation, GLDH needs oxidized cytochrome c as the electron acceptor (10–12). Indeed, GL represents a respiratory substrate for oxidative phosphorylation in plants (12, 13).
GLDH has been purified and characterized for several plant species (11, 14, 15). The primary GLDH translation product has a molecular mass of about 68 kDa but is processed to a mature protein of 56–58 kDa. Processing is based on removal of an N-terminal peptide of about 100 amino acids and probably takes place during transport of GLDH into mitochondria (15, 16). GLDH is most active with l-galactono-1,4-lactone but also has some low l-gulono-1,4-lactone activity (14, 16). The enzyme needs noncovalently bound FAD as a co-factor. GLDH so far has not been crystallized but amino acid positions essential for regulation and activity were identified by the investigation of recombinant forms of the enzyme (16–19).
GLDH is localized in the inner mitochondrial membrane (12, 20, 21). Because the mature protein lacks membrane spanning segments (16) it most likely is peripherally attached to the inner mitochondrial membrane. If overexpressed in Escherichia coli, GLDH forms part of the soluble fraction of this bacterium (16). About a decade ago, it surprisingly was discovered that GLDH is attached to the mitochondrial NADH dehydrogenase complex (complex I) of the respiratory chain (22). Complex I has a molecular mass of 1000 kDa and in plants includes at least 48 different subunits, several of which represent proteins specific for this enzyme complex in plants (23, 24). Some of these extra subunits integrate side activities into this respiratory complex, e.g. carboanhydrase (CA) subunits, which were proposed to support CO2 transfer from mitochondria to chloroplasts in plant cells (25). However, GLDH was not found to be attached to the 1000-kDa holoenzyme, but only to a slightly smaller version of complex I, which is of comparatively low abundance (13, 22, 26). This complex has a molecular mass of about 850 kDa and obviously lacks some of the subunits present in the main form of complex I. The identity of these subunits is not known so far. GLDH activity is inhibited in the presence of rotenone, an inhibitor of electron transfer within complex I, if pyruvate and malate are used as respiratory substrates (13). It therefore was speculated that a subpopulation of complex I particles are important for GLDH regulation by monitoring the rate of NADH-driven electron flow through complex I (13).
Silencing of the gene encoding GLDH in tomato does not much affect ascorbate concentration, indicating that GLDH activity is not rate-limiting for ascorbate formation (27). However, silenced plants have a clearly retarded growth and produce smaller fruits. At the same time, the central metabolism of plant mitochondria is significantly changed. It is concluded that GLDH is important for other processes besides ascorbate formation. Recently, characterization of an Arabidopsis knock-out mutant lacking the gene encoding GLDH was found to have drastically reduced amounts of complex I (26). In contrast, the amounts of the other protein complexes of the respiratory chain were not changed. It therefore is speculated that GLDH, besides its role in ascorbate formation, represents an assembly factor for complex I.
Here we present a biochemical investigation on GLDH localization within plant mitochondria. Protein complexes of the inner mitochondrial membrane were carefully solubilized by the use of nonionic detergents and resolved protein complexes were separated by blue native PAGE. Using a newly developed in gel GLDH activity assay and immunoblotting, three distinct GLDH containing protein complexes of 850, 470, and 420 kDa were discovered. The 850-kDa complex represents the known smaller version of mitochondrial complex I. GLDH is shown to be attached to the membrane arm of the 850-kDa complex I. Subunits of the novel 470 and 420 kDa complexes were identified by mass spectrometry. Like the 850-kDa complex, they also include complex I subunits. We propose that GLDH has a more extended function in complex I assembly by specifically binding to several of its assembly intermediates.
EXPERIMENTAL PROCEDURES
Arabidopsis thaliana Cultivation and Isolation of Mitochondria
A. thaliana cells (var. Columbia-0) were cultivated as previously described (28). Isolation of mitochondria was performed according to Werhahn et al. (29).
Gel Electrophoresis Procedures and Immunoblotting
One-dimensional BN-PAGE and two-dimensional BN/SDS-PAGE was performed as previously described (30). Two-dimensional BN/BN-PAGE was carried out as outlined in Sunderhaus et al. (31). For the experiments of the current investigation, first dimension BN-PAGE was carried out in the presence of digitonin, second dimension BN-PAGE in the presence of Triton X-100. Proteins were visualized by Coomassie colloidal staining (32, 33). After separation on polyacrylamide gels proteins were blotted onto a nitrocellulose membrane using the Trans Blot Cell from Bio-Rad. The transfer of proteins was performed as described in Kruft et al. (34). Immunostainings were carried out using the VectaStain ABC Kit (Vector Laboratories, Burlingame, CA). The carbonic anhydrase antibody was provided by Eduardo Zabaleta (Mar del Plata University, Argentina). The GLDH antibody was purchased from Agrisera Antibodies (Vännäs, Sweden).
In Gel Activity Stainings
In gel staining for NADH-ubiquinone-oxidoreductase was carried out as previously described (35).
In gel activity staining of GLDH was performed as follows. After half-completion of the electrophoretic run of a BN-PAGE the Coomassie-containing cathode buffer was replaced by a cathode buffer without Coomassie for dye reduction within the gel. The gel was incubated in 100 ml of GLDH staining solution (40 mm Tris(hydroxymethyl)aminomethane, 2 mm l-galactono-1,4-lactone, 1 mg/ml of nitro blue tetrazolium chloride, 200 μm phenazine methosulfate) in the dark. The pH of the solution was adjusted to 8.8 (HCl). GLDH activity becomes visible as purple bands or spots after 15–30 min. The activity staining was stopped by rinsing the gel with water. To improve visualization and destain the background the gel was transferred into destaining solution (40% methanol and 10% acetic acid) overnight. The resulting gels were finally scanned on a transmission scanner (PowerLook III, UMAX).
Mass Spectrometry (MS)
Tryptic digestion of proteins and MS were performed as described previously (23). Protein identifications were based on the MASCOT search algorithm using the A. thaliana protein data base, release TAIR10 (www.arabidopsis.org).
RESULTS
Identification of GLDH Containing Protein Complexes
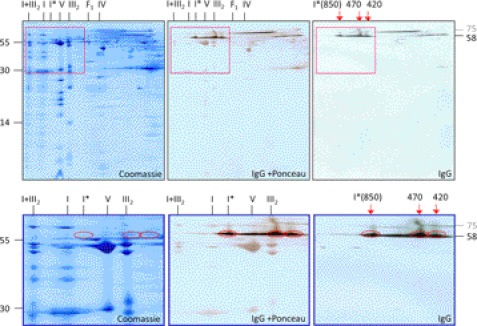
Immunoblotting experiments were carried out to first get information on GLDH localization in plant mitochondria. For this approach, mitochondria were isolated from a suspension cell culture of A. thaliana. Isolated organelles were solubilized by 5% digitonin and protein complexes were subsequently separated by two-dimensional blue native (BN)/SDS-PAGE (Fig. 1). Upon Coomassie staining, subunits of the mitochondrial protein complexes are visible as reported before (36). The main form of complex I runs at about 1000 kDa. In addition, a slightly smaller version of complex I is visible in accordance with previous investigations (13, 22, 26). On our gels, it runs at 850 kDa on the first gel dimension and is designated complex I*. It also includes an additional 58-kDa subunit not present in the main form of complex I. This protein represents GLDH as shown by a parallel immunoblotting experiment. Furthermore, the 58-kDa immune signal is detectable at two further regions on the two-dimensional gels, which correspond to 470 and 420 kDa on the native gel dimension. Finally, the 58-kDa GLDH signal is visible in the <100 kDa region of the native gel dimension. This signal represents the monomeric form of GLDH, which was reported previously (36); see the two-dimensional BN/SDS-PAGE GelMap of Arabidopsis mitochondria (www.gelmap.de/47, spot 116). We conclude that GLDH not only forms part of the 850-kDa complex I*, but additionally of two unknown protein complexes of 470 and 420 kDa.
FIGURE 1.
Immunological detection of l-galactono-1,4-lactone dehydrogenase in a mitochondrial protein fraction of A. thaliana. Proteins were separated by BN/SDS-PAGE and either stained by Coomassie Blue (left) or blotted onto nitrocellulose membranes for immunological GLDH detection (center and right). The blot in the center additionally was stained with Ponceau for background visualization. Enlargements of the boxed regions are given in the lower line of the figure. Red circles indicate GLDH. A molecular mass standard is given to the left. Nomenclature of protein complexes: I, complex I; I*, 850-kDa subcomplex of complex I; V, complex V; III2, dimeric complex III; F1, F1 part of complex V; IV, complex IV; I+III2, supercomplex composed of complex I + dimeric complex III; 850, 470, and 420 kDa, GLDH containing protein complexes (the 850-kDa complex corresponds to I*). GLDH is detected on the second gel dimension at 58 kDa. The 58-kDa signal on the right side of the two-dimensional gel represents monomeric GLDH. Another signal at about 75 kDa represents a cross-reaction with an unknown protein (see supplemental Table S1 and supplemental Fig. 1).
An in gel activity assay was developed to investigate if the 850-, 470-, and 420-kDa complexes have GLDH activity. The assay includes 2 mm l-galactono-1,4-lactone to avoid substrate inhibition, which was reported to take place at higher concentrations (16). Oxidized cytochrome c was substituted by the electron acceptor phenazine methosulfate, which increases sensitivity of the assay. The pH of the assay solution was adjusted to 8.8 in accordance with the pH optimum of GLDH reported previously (16). Finally, nitro blue tetrazolium was used for GLDH activity visualization. This redox dye is yellow in its oxidized form and purple upon reduction into formazan (37). Reduction takes place in the presence of ascorbate. Additionally, reduced phenazine methosulfate can directly reduce nitro blue tetrazolium, which may enhance the ascorbate-mediated color reaction. Nitro blue tetrazolium itself cannot be reduced directly by the GLDH. Using this assay, GLDH activity becomes visible on native protein gels as purple bands or spots. The principle of the assay is summarized in Fig. 2A and the details are given under “Experimental Procedures.”
FIGURE 2.
l-Galactono-1,4-lactone dehydrogenase in gel activity assay. A, reaction scheme of the in gel activity assay. B, in gel activity assay for GLDH. Mitochondrial membrane proteins were solubilized by 5% digitonin and subsequently separated by one-dimensional blue native PAGE. Co, Coomassie stain. GL−/GL+, in gel GLDH activity stain in the presence (GL+) or absence (GL−) of the substrate l-galactono-1,4-lactone. IgG, immunoblot for GLDH detection. The identities of protein complexes are given on the right, the GLDH-containing protein complexes are indicated in the center (for nomenclature see Fig. 1). The faint signals between the 470- and 850-kDa complexes are caused by local overloading of the gel in the regions of complexes III and V.
For performing the GLDH in gel activity assay, mitochondria were solubilized by digitonin and protein complexes were subsequently separated by one-dimensional BN PAGE (Fig. 2B). The 850-, 470-, and 420-kDa complexes exhibit strong GLDH activity. No activity is detectable in the absence of GL. The activity-stained bands exactly correspond to the signals obtained by immunoblotting (Fig. 2B). The bands at 850, 470, and 420 kDa are not visible on a parallel Coomassie-stained gel indicating that the in gel GLDH activity assay has very high sensitivity.
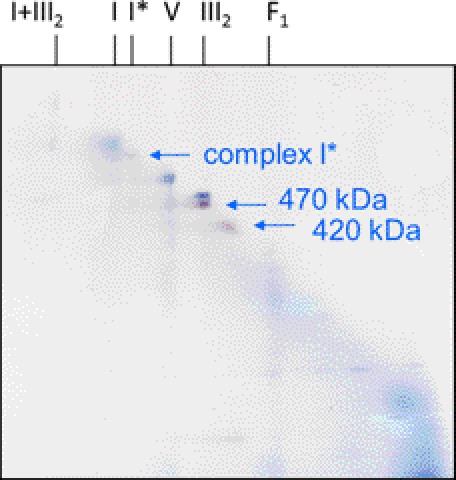
For further investigation of the 470- and 420-kDa complexes, a mitochondrial protein fraction was resolved by two-dimensional BN/BN PAGE (Fig. 3). First dimension BN PAGE was carried out in the presence of digitonin, second dimension BN PAGE in the presence of Triton X-100. On the resulting two-dimensional gels, most protein complexes are positioned on a diagonal line, but resolution is increased in comparison to one-dimensional BN PAGE due to differential effects of the two detergents on the individual protein complexes. Visualization of 850-, 470-, and 420-kDa complexes was carried out by in gel GLDH activity staining (Fig. 3). Both, the 470- and 420-kDa complexes again were not visible on a parallel Coomassie-stained BN/BN gel due to their low concentration (supplemental Fig. S3). The 470-kDa complex runs close to dimeric complex III (500 kDa).
FIGURE 3.
Detection of GLDH-containing protein complexes on a two-dimensional BN/BN gel by activity staining. Isolated mitochondria from A. thaliana were treated with digitonin for protein solubilization and protein complexes were subsequently separated by two-dimensional BN-digitonin/BN-Triton X-100 PAGE. The gel was stained for GLDH activity (purple spots). Background complexes are visible due to Coomassie Blue present during the electrophoresis run (the gel was not stained with Coomassie after completion of the electrophoresis run). Identities of protein complexes are given on top of the gels (for nomenclature, see Fig. 1). The arrows indicate GLDH-containing complexes identified by the in gel activity assay. Another GLDH signal at the lower border of the two-dimensional gel below the 420-kDa complex most likely represents GLDH, which became detached from the 420-kDa complex during the second native gel dimension. Two more extensively GLDH activity stained BN/BN gels are presented in supplemental Fig. S2. A comparison of a two-dimensional BN/BN gel before and after GLDH activity staining is presented in supplemental Fig. S3.
Although present at extremely low concentrations, gel spots representing the 470- and 420-kDa complexes were further analyzed by mass spectrometry. As expected, both protein complexes include GLDH (Table 1). Furthermore, and as expected, MS analysis of the 470-kDa complex revealed identification of complex III subunits (supplemental Table S2). This complex is of very high abundance and migrates in very close proximity to the 470-kDa complex on the BN/BN gel. However, the GLDH activity stain clearly is not at the position of complex III (Fig. 3). Strikingly, the 470-kDa complex additionally includes the CA2, CAL2, and Grim-19 subunits, which form part of the membrane arm of complex I in plants. Similarly, MS analysis of the 420-kDa complex revealed, besides GLDH, several subunits of the membrane arm of complex I: CA2, CA3, CAL2, and NAD2 (Table 1). Further complex I subunits were not identified, probably due to the low abundance of the 470- and 420-kDa complexes and due to the fact that the membrane arm of complex I mainly includes very hydrophobic subunits, which are difficult to detect by MS. Besides, some proteins of the HSP60 and malic enzyme complexes and the F1-part of ATP synthase were identified in the spot representing the 420-kDa complex (supplemental Table S2). However, as in the case of the 470-kDa complex, identification of these subunits rather reflects spot overlappings on our BN/BN gel than physical association of these proteins with GLDH, because these complexes run in very close proximity to the 420-kDa complex and are of very high abundance.
TABLE 1.
Proteins of the two-dimensional BN/BN gel (Fig. 3) identified by MS
Only complex I subunits are shown. For complete list of the identified proteins see supplemental Table S2.
| Samplea | Accession No.b | Proteinc | Massd | Mascot scoree | No. peptidesf | S.C.g | Complex |
|---|---|---|---|---|---|---|---|
| kDa | % | ||||||
| 470 kDa | At3g47930 | GLDH | 68.5 | 201 | 4 | 11.8 | Complex I |
| 470 kDa | At1g47260 | CA2 | 30.0 | 178 | 4 | 25.5 | Complex I |
| 470 kDa | At3g48680 | CAL2 | 27.9 | 154 | 3 | 17.6 | Complex I |
| 470 kDa | At1g04630 | GRIM19 | 16.1 | 65 | 1 | 7.0 | Complex I |
| 420 kDa | At1g47260 | CA2 | 30.0 | 204 | 4 | 24.5 | Complex I |
| 420 kDa | At3g48680 | CAL2 | 27.9 | 177 | 3 | 13.7 | Complex I |
| 420 kDa | At3g47930 | GLDH | 68.5 | 170 | 4 | 8.5 | Complex I |
| 420 kDa | At5g66510 | CA3 | 27.8 | 101 | 1 | 15.5 | Complex I |
| 420 kDa | AtMg00285 | NAD 2A | 54.8 | 84 | 2 | 4.2 | Complex I |
a Analyzed protein complex (Fig. 3).
b Accession numbers of identified proteins as given by TAIR.
c Names of identified proteins.
d Calculated molecular mass of the identified proteins as deduced from the corresponding gene.
e Probability score for the protein identifications based on MS analysis and MASCOT search.
f Number of unique peptides.
g Sequence coverage of the proteins by identified peptides.
MS analyses of the 850-kDa complex revealed, as expected, several complex I subunits, which form part of the membrane and the peripheral arm (supplemental Table S2). Also in this data set, some subunits of other protein complexes were identified, which represent components of highly abundant protein complexes running in close proximity to the 850-kDa complex on the BN/BN gel.
Localization of GLDH within Complex I
GLDH forms part of the 850-kDa complex I* and possibly of further complex I subcomplexes. However, precise localization of GLDH within complex I so far is not clear. Complex I consist of two longish domains, which together form an L-like structure. One domain is embedded in the inner mitochondrial membrane (“membrane arm”), whereas the other protrudes into the mitochondrial matrix (“peripheral arm“). NADH oxidation takes place at the peripheral arm, whereas proton translocation is mediated by the membrane arm. Both processes are probably coupled through conformational changes (38–40).
At which position does GLDH bind to mitochondrial complex I in plants? To address this question, further experiments were carried out using the two-dimensional BN/BN PAGE system in combination with in gel activity staining and immunoblotting. First dimension BN PAGE was carried out in the presence of digitonin, second dimension BN PAGE in the presence of Triton X-100. Because Triton X-100 has a slightly reduced mildness during membrane solubilization compared with digitonin, some protein complexes partly get dissected into subcomplexes during the second gel dimension. On the resulting two-dimensional gels, these subcomplexes migrate below the diagonal line. For example, the 1500-kDa I + III2 supercomplex, which is composed of complexes I and III2, is dissected into its two components on the second native gel dimension (28). Furthermore, complex I is partially dissected on the second native gel dimension into two subcomplexes representing the membrane arm (550 kDa) and the peripheral arm (370 kDa) (23, 28).
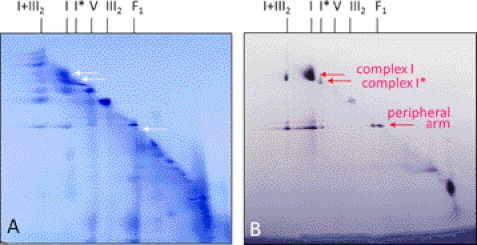
In the first experiment, a mitochondrial fraction of Arabidopsis was separated by two-dimensional BN/BN PAGE and gels were activity stained for NADH oxidation (Fig. 4). As expected, the main form of complex I (1000 kDa) becomes visible as well as its peripheral arm (370 kDa). Both complexes are also visible as dissection products of the I + III2 supercomplex. The peripheral arm of complex I additionally is visible on the diagonal line on the two-dimensional gel system, indicating that a small proportion of complex I was dissected into its arms within our mitochondrial fraction. The GLDH containing 850-kDa complex I* has NADH oxidation activity. Furthermore, the 850-kDa complex is also partially dissected into its arms on the second gel dimension. The peripheral arm of the 850-kDa complex has the same size as the peripheral arm of the 1000-kDa main form of complex I. We conclude that the peripheral arm of the 850-kDa complex I* includes a complete set of subunits. Therefore, the unknown subunits of complex I, which are absent in complex I*, must form part of the membrane arm.
FIGURE 4.
Detection of NADH dehydrogenase complexes on a two-dimensional BN/BN gel by activity staining. Isolated mitochondria from A. thaliana were treated with digitonin for protein solubilization and protein complexes were subsequently separated by two-dimensional BN-digitonin/BN-Triton X-100 PAGE. A, Coomassie colloidal stained gel. B, NADH dehydrogenase activity stained gel. Identities of protein complexes are given on top of the gels (for nomenclature see Fig. 1). The arrows indicate NADH dehydrogenase complexes identified by the in gel activity assay. The signals in the low molecular mass region of the two-dimensional gel (bottom, right) represent alternative NADH-dehydrogenases.
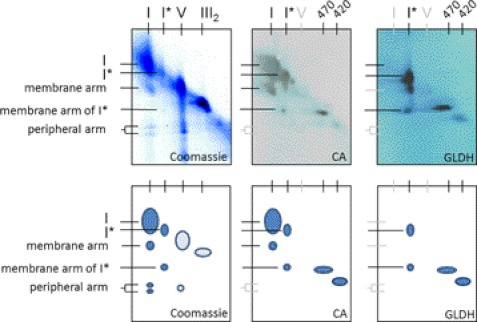
In the second experiment, a mitochondrial fraction of Arabidopsis was separated by two-dimensional BN/BN PAGE and the resulting gels were used for immunoblotting experiments using antibodies directed against GLDH and the carbonic anhydrase subunit CA2 of complex I (Fig. 5). The latter subunit is known to form part of the membrane arm of complex I (23, 28). As expected, the CA antibody recognizes complex I (1000 kDa) and its membrane arm (570 kDa) (Fig. 5, center). Furthermore, complex I* (850 kDa) is recognized as well as its membrane arm (470 kDa), which is of slightly reduced size compared with the membrane arm of the holoenzyme (550 kDa). Finally, the 470- and 420-kDa complexes are recognized on the diagonal line. This verifies the MS data indicating that the 470- and 420-kDa complexes represent parts of the membrane arm of complex I (including the CA2 subunit). The GLDH antibody does not recognize complex I (1000 kDa), which was expected because it was never reported to form part of the main form of the NADH dehydrogenase complex. In contrast, the 850-kDa complex I* is recognized as well as its membrane arm at 470 kDa. As expected, the 470- and 420-kDa complexes are additionally recognized on the diagonal line of protein complexes visible on the two-dimensional BN/BN gels. We conclude that GLDH is attached to the membrane arm of complex I* (470 kDa), which represents a large but incomplete fragment of the membrane arm of complex I.
FIGURE 5.
Detection of CA- and GLDH-containing protein complexes on a two-dimensional BN/BN gel using antibodies. Isolated mitochondria from Arabidopsis were treated with digitonin for protein solubilization and protein complexes were subsequently separated by two-dimensional BN-digitonin/BN-Triton X-100 PAGE. The resulting two-dimensional gels were either stained by Coomassie Blue (left) or blotted onto nitrocellulose membranes for immunological detection of CA (center) or GLDH (right). Identities of protein complexes are given on the top and left of the gels (for nomenclature see Fig. 1; black letters, visible spots; gray letters, positions of protein complexes not visible on one or two of the subfigures). The schemes below further illustrate the identity of the resolved protein complexes (dark blue circles, complex I and derived protein complexes; light blue circles, other protein complexes).
DISCUSSION
Using native gel electrophoresis procedures, which are combined with in gel activity stains and immunoblotting, novel insights into GLDH localization in plant mitochondria were achieved: (i) GLDH forms part of three mitochondrial protein complexes of 850, 470, and 420 kDa; (ii) all three complexes exhibit GLDH activity upon analysis by a newly developed in gel GLDH activity assay; (iii) the 850-kDa complex represents a known smaller version of complex I (13, 22, 26), here termed complex I*, but also the 470- and 420-kDa complexes representing subcomplexes of complex I; (iv) 850-kDa complex I* has a complete peripheral arm; and (v) The GLDH containing 470 kDa complex represents the membrane arm of complex I*, which represents a large fragment of the membrane arm of the holoenzyme.
Very recently, the x-ray structure of bacterial complex I, which has a much simpler subunit composition, was resolved by x-ray crystallography (38, 40). Also, the overall structure of mitochondrial complex I is known by single particle electron microscopy and meanwhile also by partial x-ray crystallography (39, 41–43). All complex I particles analyzed so far have the characteristic L-like shape. Mitochondrial complex I from plants is very special, because the L-motif is markedly modified (28, 44–46). Most notably, it has a second matrix exposed domain, which is attached to the membrane arm at a central position. It was shown to include carbonic anhydrase subunits (25, 28), which belong to the set of subunits special to complex I in plants (23, 47). GLDH is another protein of this set of plant-specific complex I subunits. However, it is absent in the 1000-kDa holo complex but present only in a slightly smaller 850-kDa complex I*, which is of comparatively low abundance. Because the amount of complex I is very much reduced in an Arabidopsis knock-out line lacking the gene for GLDH, its involvement in complex I assembly was suggested (26).
Assembly of complex I was extensively studied in fungi and mammals (reviewed in Refs. 48 and 49–51) but not much is known about this process in plants. Due to its unique shape, the assembly pathway of plant complex I most likely differs substantially from the pathways taking place in other groups of organisms. Using low-SDS treatment of isolated complex I from Arabidopsis, subcomplexes were systematically generated and analyzed for subunit composition (23, 47). This experimental approach gave insights into subunit arrangement within plant complex I. However, disassembly of complex I does not necessarily reflect its assembly pathways. Other insights into complex I assembly in plants came from analyses of mutants defective in complex I accumulation (26, 52). Most recently, Arabidopsis knock-out lines defective in 7 different complex I subunits were systematically analyzed by native gel electrophoresis procedures (53). Assembly of the membrane arm of complex I was shown to involve intermediates of 200, 400, 450, and 650 kDa. Because abundances of these assembly intermediates are extremely low, they were only detectable by immunoblotting. Their subunit compositions therefore are largely unknown. However, based on information about which intermediates accumulate in which knock-out line, some first results became clear. Most notably, the plant-specific carbonic anhydrase subunit CA2 forms part of all assembly intermediates and therefore is involved in the initial events leading to the formation of the membrane arm.
We present evidence that the newly described GLDH containing protein complexes of 470 and 420 kDa represent assembly intermediates of complex I. Due to the fact that the GLDH only is attached to the subcomplexes and not to the holo-complex I, it is unlikely that they represent degradation fragments of intact complex I. Because the assembly intermediates are of very low abundance, they are only detectable by immunoblotting or by the GLDH in gel activity assay. Their subunit composition therefore is not known. However, analyses by mass spectrometry and immunoblotting allowed identifying subunits of the membrane arm of complex I, most notably CA2, CAL2, CA3, NAD2, and Grim-19 (Fig. 3, Table 1). All other identified proteins form part of protein complexes of very high abundance, which are localized on the BN/BN gels in close proximity to the 470- and 420-kDa complexes and that most likely were detected due to spot overlappings. We presume that GLDH containing 470- and 420-kDa complexes represent the 450- and 400-kDa assembly intermediates described by Meyer et al. (53), which were both shown to include CA2 (the size difference of the complexes as described by the two studies can be explained by a slight variation in molecular mass calibration of the native gels used for electrophoresis). The 850-kDa complex I* is partially dissected into a 470-kDa membrane fragment, which includes GLDH and CA and which exactly co-migrates on our gels with the 470-kDa complex identified by GLDH in gel activity staining (Fig. 5). The 470-kDa dissection product of the 850-kDa complex also exhibits GLDH activity (supplemental Fig. S2).
In summary, our data point to a more extensive role of GLDH in complex I assembly. GLDH binds to the 420- and 470-kDa complex I assembly intermediates, which at a later stage form the 850-kDa intermediate. Formation of the 1000-kDa complex I holoenzyme is preceded by detachment of GLDH. Because we do not see smaller complex I subcomplexes on our GLDH activity stained two-dimensional BN/BN gels, we believe that the 420-kDa complex is the smallest complex I subcomplex that includes GLDH. We so far cannot answer if GLDH binding to the 420- and 470-kDa complexes is a prerequisite for formation of the 850-kDa complex.
GLDH integrated into the 850-, 470-, and 420-kDa complexes is active in ascorbate formation as revealed by our in gel activity assay. It currently cannot be decided whether GLDH activity (conversion of GL into ascorbate) is required for its assembly function. The biological reason of the bifunctionality of this protein so far remains a mystery and should be further investigated.
Supplementary Material
Acknowledgment
We thank Dagmar Lewejohann for expert technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grant Br 1829/10-1.

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- GL
- l-galactono-1,4-lactone
- GLDH
- l-galactono-1,4-lactone dehydrogenase
- CA
- carbonic anhydrase
- BN
- blue native.
REFERENCES
- 1. Pignocchi C., Foyer C. H. (2003) Apoplastic ascorbate metabolism and its role in the regulation of cell signaling. Curr. Opin. Plant Biol. 6, 379–389 [DOI] [PubMed] [Google Scholar]
- 2. de Pinto M. C., Paradiso A., Leonetti P., De Gara L. (2006) Hydrogen peroxide, nitric oxide, and cytosolic ascorbate peroxidase at the cross-road between defense and cell death. Plant J. 48, 784–795 [DOI] [PubMed] [Google Scholar]
- 3. Barth C., Moeder W., Klessig D. F., Conklin P. L. (2004) The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin C-1. Plant Physiol. 134, 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barth C., De Tullio M., Conklin P. L. (2006) The role of ascorbic acid in the control of flowering time and the onset of senescence. J. Exp. Bot. 57, 1657–1665 [DOI] [PubMed] [Google Scholar]
- 5. Conklin P. L., Barth C. (2004) Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senesence. Plant Cell Environ. 27, 959–970 [Google Scholar]
- 6. Gao Q., Zhang L. (2008) Ultraviolet-B-induced oxidative stress and antioxidant defense system responses in ascorbate-deficient vtc1 mutants of Arabidopsis thaliana. J. Plant Physiol. 165, 138–148 [DOI] [PubMed] [Google Scholar]
- 7. Larkindale J., Hall J. D., Knight M. R., Vierling E. (2005) Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 138, 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Müller-Moulé P., Golan T., Niyogi K. K. (2004) Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol. 134, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wheeler G. L., Jones M. A., Smirnoff N. (1998) The biosynthetic pathway of vitamin C in higher plants. Nature. 393, 365–369 [DOI] [PubMed] [Google Scholar]
- 10. Mapson L. W., Isherwood F. A., Chen Y. T. (1954) Biological synthesis of l-ascorbic acid. The conversion of l-galactono-γ-lactone into l-ascorbic acid by plant mitochondria. Biochem. J. 56, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mapson L. W., Breslow E. (1958) Biological synthesis of ascorbic acid. l-Galactono-γ-lactone dehydrogenase. Biochem. J. 68, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartoli C. G., Pastori G. M., Foyer C. H. (2000) Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 123, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Millar A. H., Mittova V., Kiddle G., Heazlewood J. L., Bartoli C. G., Theodoulou F. L., Foyer C. H. (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol. 133, 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oba K., Ishikawa S., Nishikawa M., Mizuno H., Yamamoto T. (1995) Purification and properties of l-galactono-γ-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J. Biochem. 117, 120–124 [DOI] [PubMed] [Google Scholar]
- 15. Ostergaard J., Persiau G., Davey M. W., Bauw G., Van Montagu M. (1997) Isolation of a cDNA coding for l-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. Purification, characterization, cDNA cloning, and expression in yeast. J. Biol. Chem. 272, 30009–30016 [DOI] [PubMed] [Google Scholar]
- 16. Leferink N. G., van den Berg W. A., van Berkel W. J. (2008) l-Galactono-γ-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. FEBS J. 275, 713–726 [DOI] [PubMed] [Google Scholar]
- 17. Leferink N. G., Jose M. D., van den Berg W. A., van Berkel W. J. (2009) Functional assignment of Glu-386 and Arg-388 in the active site of l-galactono-γ-lactone dehydrogenase. FEBS Lett. 583, 3199–3203 [DOI] [PubMed] [Google Scholar]
- 18. Leferink N. G., van Duijn E., Barendregt A., Heck A. J., van Berkel W. J.(2009) Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol. 150, 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leferink N. G., Fraaije M. W., Joosten H. J., Schaap P. J., Mattevi A., van Berkel W. J. (2009) Identification of a gatekeeper residue that prevents dehydrogenases from acting as oxidases. J. Biol. Chem. 284, 4392–4397 [DOI] [PubMed] [Google Scholar]
- 20. Imai T., Karita S., Shiratori G., Hattori M., Nunome T., Oba K., Hirai M. (1998) l-Galactono-γ-lactone dehydrogenase from sweet potato. Purification and cDNA sequence analysis. Plant Cell Physiol. 39, 1350–1358 [DOI] [PubMed] [Google Scholar]
- 21. Siendones E., Gonzalez-Reyes J. A., Santos-Ocana C., Navas P., Cordoba F. (1999) Biosynthesis of ascorbic acid in kidney bean. l-Galactono-γ-lactone dehydrogenase is an intrinsic protein located at the mitochondrial inner membrane Plant Physiol. 120, 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heazlewood J. L., Howell K. A., Millar A. H. (2003) Mitochondrial complex I from Arabidopsis and rice. Orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta 1604, 159–169 [DOI] [PubMed] [Google Scholar]
- 23. Klodmann J., Sunderhaus S., Nimtz M., Jänsch L., Braun H. P. (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell. 22, 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klodmann J., Braun H. P. (2011) Proteomic approach to characterize mitochondrial complex I from plants. Phytochemistry 72, 1071–1080 [DOI] [PubMed] [Google Scholar]
- 25. Braun H. P., Zabaleta E. (2007) Carbonic anhydrase subunits of the mitochondrial NADH dehydrogenase complex (complex I) in plants. Physiol. Plant. 129, 114–122 [Google Scholar]
- 26. Pineau B., Layoune O., Danon A., De Paepe R. (2008) l-Galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 283, 32500–32505 [DOI] [PubMed] [Google Scholar]
- 27. Alhagdow M., Mounet F., Gilbert L., Nunes-Nesi A., Garcia V., Just D., Petit J., Beauvoit B., Fernie A. R., Rothan C., Baldet P. (2007) Silencing of the mitochondrial ascorbate synthesizing enzyme l-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol. 145, 1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sunderhaus S., Dudkina N. V., Jänsch L., Klodmann J., Heinemeyer J., Perales M., Zabaleta E., Boekema E. J., Braun H. P. (2006) Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J. Biol. Chem. 281, 6482–6488 [DOI] [PubMed] [Google Scholar]
- 29. Werhahn W., Niemeyer A., Jänsch L., Kruft V., Schmitz U. K., Braun H. (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol. 125, 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heinemeyer J., Lewejohann D., Braun H. P. (2007) Blue native gel electrophoresis for the characterization of protein complexes in plants. Methods Mol. Biol. 355, 343–352 [DOI] [PubMed] [Google Scholar]
- 31. Sunderhaus S., Eubel H., Braun H. P. (2007) Two-dimensional blue native/blue native polyacrylamide gel electrophoresis for the characterization of mitochondrial protein complexes and supercomplexes. Methods Mol. Biol. 372, 315–324 [DOI] [PubMed] [Google Scholar]
- 32. Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 33. Neuhoff V., Stamm R., Pardowitz I., Arold N., Ehrhardt W., Taube D. (1990) Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11, 101–117 [DOI] [PubMed] [Google Scholar]
- 34. Kruft V., Eubel H., Jänsch L., Werhahn W., Braun H. P. (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- 35. Zerbetto E., Vergani L., Dabbeni-Sala F. (1997) Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18, 2059–2064 [DOI] [PubMed] [Google Scholar]
- 36. Klodmann J., Senkler M., Rode C., Braun H. P. (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol. 157, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eadie M. J., Tyrer J. H., Kukums J. R., Hooper W. D. (1970) Aspects of tetrazolium salt reduction relevant to quantitative histochemistry. Histochemie 21, 170–180 [DOI] [PubMed] [Google Scholar]
- 38. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 39. Hunte C., Zickermann V., Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 40. Efremov R. G., Sazanov L. A. (2011) Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 [DOI] [PubMed] [Google Scholar]
- 41. Grigorieff N. (1998) Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J. Mol. Biol. 277, 1033–1046 [DOI] [PubMed] [Google Scholar]
- 42. Radermacher M., Ruiz T., Clason T., Benjamin S., Brandt U., Zickermann V. (2006) The three-dimensional structure of complex I from Yarrowia lipolytica. A highly dynamic enzyme. J. Struct. Biol. 154, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clason T., Ruiz T., Schägger H., Peng G., Zickermann V., Brandt U., Michel H., Radermacher M. (2010) The structure of eukaryotic and prokaryotic complex I. J. Struct. Biol. 169, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dudkina N. V., Heinemeyer J., Keegstra W., Boekema E. J., Braun H. P. (2005) Structure of dimeric ATP synthase from mitochondria. An angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett. 579, 5769–5772 [DOI] [PubMed] [Google Scholar]
- 45. Peters K., Dudkina N. V., Jänsch L., Braun H. P., Boekema E. J. (2008) A structural investigation of complex I and I+III2 supercomplex from Zea mays at 11–13-Å resolution. Assignment of the carbonic anhydrase domain and evidence for structural heterogeneity within complex I. Biochim. Biophys. Acta 1777, 84–93 [DOI] [PubMed] [Google Scholar]
- 46. Bultema J. B., Braun H. P., Boekema E. J., Kouril R. (2009) Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim. Biophys. Acta 1787, 60–67 [DOI] [PubMed] [Google Scholar]
- 47. Klodmann J., Lewejohann D., Braun H. P. (2011) Low-SDS blue native PAGE. Proteomics 11, 1834–1839 [DOI] [PubMed] [Google Scholar]
- 48. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 49. Vogel R. O., Smeitink J. A., Nijtmans L. G. (2007) Human mitochondrial complex I assembly. A dynamic and versatile process. Biochim. Biophys. Acta 1767, 1215–1227 [DOI] [PubMed] [Google Scholar]
- 50. Remacle C., Barbieri M. R., Cardol P., Hamel P. P. (2008) Eukaryotic complex I. Functional diversity and experimental systems to unravel the assembly process. Mol. Genet. Genomics 280, 93–110 [DOI] [PubMed] [Google Scholar]
- 51. Lazarou M., Thorburn D. R., Ryan M. T., McKenzie M. (2009) Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793, 78–88 [DOI] [PubMed] [Google Scholar]
- 52. Perales M., Eubel H., Heinemeyer J., Colaneri A., Zabaleta E., Braun H. P. (2005) Disruption of a nuclear gene encoding a mitochondrial γ-carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 350, 263–277 [DOI] [PubMed] [Google Scholar]
- 53. Meyer E. H., Solheim C., Tanz S. K., Bonnard G., Millar A. H. (2011) Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 286, 26081–26092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.