Background: Caspase-2 deficiency accelerates tumorigenesis in mice, but the underlying mechanisms remain unclear.
Results: Caspase-2 catalytic site Cys-320 and Ser-139 residues sustain G2/M checkpoint and inhibit transformation and NF-κB activation.
Conclusion: Both residues are required for caspase-2 to suppress tumorigenesis in nude mice.
Significance: These findings provide insight into tumor suppression at the cross-roads of apoptosis, cell cycle checkpoint, and NF-κB pathways.
Keywords: Apoptosis, Caspase, Checkpoint Control, NF-kappaB (NF-KB), Tumor Suppressor Gene
Abstract
The multifunctional caspase-2 protein is involved in apoptosis, NF-κB regulation, and tumor suppression in mice. However, the mechanisms of caspase-2 responsible for tumor suppression remain unclear. Here we identified two sites of caspase-2, the catalytic Cys-320 site and the Ser-139 site, to be important for suppression of cellular transformation and tumorigenesis. Using SV40- and K-Ras-transformed caspase-2 KO mouse embryonic fibroblast cells reconstituted with expression of wild-type, catalytic dead (C320A), or Ser-139 (S139A) mutant caspase-2, we demonstrated that similar to caspase-2 deficiency, when Cys-320 and Ser-139 were mutated, caspase-2 lost its ability to inhibit cellular transformation and tumorigenesis. These mutant cells exhibited enhanced cell proliferation, elevated clonogenic activity, accelerated anchorage-independent growth, and transformation and were highly tumorigenic, rapidly producing large tumors in athymic nude mice. Investigation into the underlying mechanism showed that these two residues are needed for caspase-2 to suppress NF-κB activity, promote apoptosis, and sustain the G2/M checkpoint following DNA damage induction. In addition, tumors in nude mice derived from the two mutant cell lines had higher constitutive NF-κB activity and elevated expression of NF-κB targets of antiapoptotic proteins Bcl-xL, XIAP, and cIAP2. A reduction in caspase-2 mRNA was associated with multiple types of cancers in patients. Together, these observations suggest the combined functions of caspase-2 in suppressing NF-κB activation, promoting apoptosis, and sustaining G2/M checkpoint contribute to caspase-2 tumor-suppressing function and that caspase-2 may also impact tumor suppression in humans. These findings provide insight into tumor suppression at the cross-roads of apoptosis, cell cycle checkpoint, and NF-κB pathways.
Introduction
Insufficient apoptosis, aberrant up-regulation of NF-κB activity, and loss of tumor suppressor activity all contribute to malignant cell survival, transformation, and tumor development (1–4). Caspase-2, one of the first mammalian caspases discovered (5–7) and the most evolutionarily conserved of the caspases, has been shown to influence all of the tumor promoting activities listed above (8–12). Caspase-2 promotes apoptosis induced by diverse stimuli, and in DNA damage-induced apoptosis, caspase-2 participates in the mitochondrial (intrinsic) pathway of cell death by cleaving Bid, a premitochondrial step to release cytochrome c and Smac/Diablo from mitochondria, which leads to activation of downstream caspase-3, -6, and -7 and the demise of the cell (13–18). Caspase-2 also promotes the death receptor (extrinsic) apoptotic pathway in response to DNA damage (19, 20). In mitotic catastrophe apoptosis induced by DNA damage, caspase-2 acts at an apical step upstream of cytochrome c release (21–24). In the Chk1-inhibited ATM/ATR-caspase-2 apoptotic pathway, caspase-2 is activated upon Chk1 inactivation and induces apoptosis in coordination with activated ATM and ATR (25). Biochemically, caspase-2 activation for apoptosis can be mediated by the protein complex called PIDDosome, consisting of PIDD,4 RAIDD, and caspase-2. In this PIDDosome, caspase-2 was activated by PIDD, and RAIDD served as a bridging molecule binding to both caspase-2 and PIDD (26, 27).
There are conflicting results on the details of the role of caspase-2 in the NF-κB pathway. One study showed that caspase-2 activates NF-κB in a manner that is dependent on its CARD domain but independent of its catalytic activity and RIP1 cleavage (28), whereas another concluded that caspase-2 functions as an endogenous inhibitor of NF-κΒ-dependent cell survival by proteolytic cleavage of RIP1 (29).
In addition to its influence on apoptosis and NF-κB activity, caspase-2 has a tumor suppressor function in mice. Loss of caspase-2 increases cellular resistance to apoptosis, facilitates cell transformation, and enhances the tumorigenic potential of MEFs transformed by the oncogenes E1A/HRasV12 (30). These transformed caspase-2 KO MEF cells show accelerated tumor development in athymic nude mice, and a deficiency for caspase-2 in mice accelerates lymphomagenesis induced by oncogenic Myc in the Eu-Myc-driven mouse lymphoma model (30).
Although the catalytic activity of caspase-2 is important for many of its functions, it is unclear whether it plays a role in tumor suppression. In addition, it is unclear whether any other site of caspase-2 might be important for its tumor-suppressing function. Here we provide evidence for the requirement of the catalytic site Cys-320 and Ser-139 residue on the tumor-suppressing function of caspase-2.
EXPERIMENTAL PROCEDURES
Cell Culture
All of the cells were cultured in DMEM high glucose with 10% FBS in a 37 °C, 5% CO2 incubator.
Generation of K-Ras-transformed and SV40-immortalized MEF Cells
Retrovirus-mediated expression systems were utilized to create MEF cells that were immortalized by SV40 or with stable expression of oncogene K-Ras in SV40-immortalized MEFs. Briefly, Phoenix packaging cells at 50–70% confluency were transfected with VSVG, Rev, Gag-Pol, and pBABE-Zeo-SV40 Large T or pBABE-puro-K-Ras plasmids at a ratio of 3:3:6:9 (μg), with addition to the cell culture of 500 μl of jetPRIMETM transfection agent (Polyplus-transfection Inc., New York, NY). Thirty-six hours after the transfection, the retroviral supernatants were harvested, and the package cells were removed by filtration (0.45-μm filter). The resulting fresh retroviral solution was used directly to infect the target caspase-2 KO MEFs cells expressing vector (remains Casp2KO), wild-type (Casp2WT), catalytic (Casp2C320A), and Ser-139 mutant (Casp2S139A) caspase-2, at 50% confluency. Forty-eight hours after retroviral infection, MEF cells were selected with 3 μg/ml respective antibiotics, and all of the surviving clones were pooled together for experiments.
MTT Cell Proliferation Assay
The MTT assay was performed as previously described (31) with minor modifications. Briefly, 1 × 104 cells were seeded in 24-well plates (for 24 h), 12-well plates (for 48 h), 60-mm plates (for 72 h), or 100-mm plates (for 96 h) and cultured at 37 °C in a 5% CO2 incubator. At different time points, as indicated above, the medium was removed, and the cells were incubated with 1 mg/ml MTT for 2 h. The solution was then removed, and the converted dye in the solution was resolved with 1 ml of acidic isopropanol. This dye solution was harvested by centrifugation at 13,000 rpm for 5 min. The resulting supernatants were measured for absorbance at 595 nm by using an iMarkTM microplate reader (Bio-Rad).
Trypan Blue Exclusion Assay
1 × 104 cells were seeded and cultured as described for the MTT assay. The cells were harvested at different time points and stained with trypan blue dye. The viable cells (negative for trypan blue) were counted.
Clonogenic Cell Survival Assay
Two hundred cells were seeded in triplicate in 60-mm dishes. Six hours after seeding, the cells were irradiated in an x-ray machine (Faxitron RX-650) with the indicated doses. After 7 days, the cell colonies were stained with crystal violet and manually counted under a microscope (a colony was defined as a cell cluster containing at least 50 cells).
Soft Agar Assay
A soft agar assay was performed as previously described with some modifications (30). Specifically, 2 × 104 cells were seeded in 60-mm dishes with 0.35% agar on top of 6% agar (prepared in DMEM with 10% FBS). After 21 days, the cell colonies were stained with crystal violet and manually counted under a microscope.
Tumor Growth in Athymic Nude Mice
Tumor growth in athymic nude mice was performed as previously described with some modifications (30). Specifically, 1 × 106 Casp2WT/SV40/Ras, Casp2KO/SV40/Ras, Casp2C320A/SV40/Ras, and Casp2S139A/SV40/Ras MEF cells were suspended in 100 μl of PBS and injected subcutaneously in the flanks of mice. To avoid variation in immune responses among individual mice, two cell lines expressing different variants (WT versus KO; WT versus C320A; WT versus S139A; KO versus C320A; KO versus S139A; and C320A versus S139A) were injected to the left and right flank of 8-week-old male nude mice (Harlan Laboratories), respectively. These injections were repeated in at least two additional mice, such that each cell line was injected at least nine times into mice. In one set of the experiments, 100 μl of PBS was injected into the flanks of the nude mice as negative controls. All of the animal work conformed to institutional guidelines of the University of Cincinnati Institutional Animal Care and Use Committee.
Preparation of Cell-free Protein Extracts
The cells in culture were harvested, washed with 1× ice-cold PBS, and resuspended in lysis buffer (0.02 m HEPES, pH 7.4, 0.15 m NaCl, 0.001 m EDTA, and 1% Nonidet P-40 (Igepal-CA-630) with a protease inhibitor mixture tablet (one tablet in 10 ml of lysis buffer; Roche Applied Science) and kept on ice for 30 min. After centrifugation at 14,000 × g at 4 °C, the supernatant was collected, and protein concentration was measured by the Bradford method (Bio-Rad). To make tumor tissue lysates, nude mice were euthanized at the termination of experiments (day 14). The tumors were retrieved and washed with 1× ice-cold PBS. A portion of each tumor (∼5 mm × 5 mm) was immediately excised and immersed in the same lysis buffer and sonicated on ice for 25 s. Supernatant after 14,000 × g centrifugation for 10 min were collected, and protein concentrations were measured as described above.
Western Blotting
Aliquots of extracts containing a total of 50 μg of protein were separated on SDS-PAGE, transferred to a PVDF membrane (Bio-Rad). Following blocking with 5% milk/PBST, membranes were incubated with primary antibodies overnight at 4 °C. After a wash with PBST, membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature, developed with ECL (PerkinElmer Life Sciences), and exposed to x-ray films (Phenix Research Products).
Caspase-3 Activity Assay
The cells were first harvested by centrifugation at 700 × g, and then the pellets were washed with PBS twice and lysed in cell lysis buffer containing 10 mm Tris-HCl (pH 7.5), 10 mm NaH2PO4/NaHPO4 (pH 7.5), 130 mm NaCl, 1% Triton X-100, and 10 mm Na4P2O7 on ice for 20 min. After centrifugation at 10,000 × g for 10 min, the supernatants were collected, and protein concentrations were measured by the Bradford method (Bio-Rad). 50 μg of protein from each sample (adjusted to a volume of 25 μl using the lysis buffer) were mixed with 30 mm of substrate DEVD-AFC (BioVision, Mountain View, CA) in 25 μl of 2× assay buffer (40 mm HEPES, pH 7.5, 20% glycerol). Fluorescence was measured at 400-nm excitation and 505-nm emission following 2 h of incubation. Caspase-3 activity was measured by quantifying the cleavage of the fluorogenic peptide, and the results are expressed as the percentages of activity over the controls.
Immunofluorescent Detection of Mitotic Cells
Flow cytometry analysis of the mitotic marker phosphorylated histone H3 was performed on a FACSCalibur (BD Biosciences) using CellQuest software. The cells were harvested by trypsinization, washed with ice-cold PBS once, and fixed in 70% ETOH at −20 °C overnight. Approximately 1 × 106 cells were then washed once with ice-cold PBS and stained with 10 μg/ml fluorescein isothiocyanate-conjugated antibody against phosphorylated histone H3 Ser-10 (catalogue number 16-222; Millipore, Temecula, CA) for 1 h at room temperature in the dark, followed by propidium iodide (20 μg/ml in PBS with 0.1% Triton X-100) and treated with 100 μg/ml RNase on ice for 20 min.
Immunohistochemistry
Paraffin-embedded tissue sections (5 μm thick) from harvested tumor samples were subjected to immunostaining. The tissue sections were deparaffinized in xylene substitute (Shandon Thermo Scientific) and rehydrated in progressively decreasing concentrations of 100, 95, and 70% of ethanol. The slides were washed with 1× PBS once and boiled in antigen retrieval buffer (10 mm citrate) for 5 min in the microwave oven. The slides were left cool for 15 min on the counter and then incubated in 3% H2O2 in 100% methanol for 15 min. The sections were washed and incubated with 5% normal goat serum (Vector Laboratories) in PBST (0.1% Triton X-100) for 1 h at room temperature. The sections were incubated with primary antibodies or normal IgG as a negative control at 4 °C overnight. The slides were rinsed with PBS and incubated with biotinylated secondary antibodies for 1 h at room temperature. Tissue sections were washed in PBS and incubated with ABC reagent (VECTASTAIN® Elite ABC kit; Vector Laboratories) for 30 min at room temperature. Each section was subjected to diaminobenzidine (DAB) substrate mixture (Sigma-Aldrich) and was then counterstained with hematoxylin. The sections were washed with tap water, dehydrated in progressively increasing concentrations of 70, 95, and 100% ethanol and mounted with xylene-based mounting medium. The slides were visualized using the Axioplan 2 imaging microscope (Carl Zeiss Lichtmicroskopie, Göttingen, Germany).
Statistical Analysis of Caspase-2 Gene Expression Data in Human Cancers
Publicly available data sets were retrieved from the NCBI GEO database. The search for appropriate data sets was facilitated by using the academic version of Oncomine. Statistical analysis of gene expression was conducted using the R package. Significance of differential expression was set to p value < 0.001 derived using a t test.
Statistical Analysis
Except caspase-2 gene expression in human cancers, data were expressed as the means ± S.E. and analyzed using two-way analysis of variance, one-way analysis of variance, and post hoc tests as appropriate by GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). The results were considered statistically significant at p < 0.05.
Antibodies
For Western blotting, anticleaved caspase-3 (Cell Signaling catalogue number 9661), p-IκBα (Cell Signaling catalogue number 2859), XIAP (BD Transduction Laboratories catalogue number 610762), cIAP2 (Cell Signaling catalogue number 4952), Bcl-xL (gift from Dr. John C. Reed at the Sanford Burnham Institute), and actin (Cell Signaling catalogue number 3700) were used. For mitotic cell staining, antiphospho-histone H3 (Ser-10), clone 3H10, and FITC conjugate (Millipore catalogue number 16-222) were used. For immunohistochemistry, NF-κB p65 (Cell Signaling catalogue number 3034), Bcl-xL (Santa Cruz Biotechnology catalogue number sc-8392), and cIAP2 (Santa Cruz catalogue number sc-7944) were used.
RESULTS
Enhanced Cell Proliferation in SV40- and SV40/K-Ras-transformed MEF Cells Expressing C320A and S139A Mutant Caspase-2
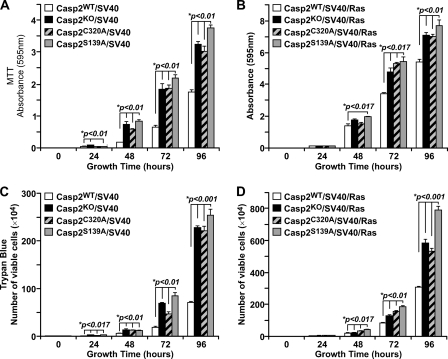
Given that caspase-2 inhibits transformation by the oncogenic E1A/HRasV12 (30), we made stable cell lines by immortalization of caspase-2 (Casp2) KO MEFs with SV40 alone (Casp2/SV40), or followed by transformation with a retroviral construct expressing K-Ras (Casp2/SV40/Ras). The cell line in each case was reconstituted with stable expression of wild-type or Cys-320 mutant caspase-2. These resulting cell lines are isogenic, and expression levels of exogenous caspase-2 protein are similar to endogenous level so that the experimental results would be comparable. We next analyzed the growth properties of these MEFs by the MTT proliferation assay. Consistent with the enhanced proliferation of Casp2 KO MEFs transformed with E1A/HRasV12 (30), our Casp2KO/SV40 and Casp2KO/SV40/Ras cells had significantly enhanced proliferation rates compared with wild-type control MEFs (Fig. 1, A and B). Interestingly, caspase-2 catalytic dead Casp2C320A MEFs, transformed singly by SV40 or doubly by SV40/Ras, also displayed an enhanced proliferation rate, as compared with the wild-type cells (Fig. 1, A and B). These results indicate that caspase-2 suppresses overproliferation, and this function requires the catalytic activity of caspase-2.
FIGURE 1.
A and B, MTT cell proliferation assay with Casp2WT, Casp2KO, Casp2C320A, and Casp2S139A MEFs immortalized with SV40 (A) and with SV40/K-Ras (B). C and D, trypan blue assay of SV40-immortalized cells (C) and SV40/Ras-transformed cells (D). The data are presented as the means ± S.E. of results from experiments performed in triplicate. *p denotes the significant difference compared with wild-type cells.
It is known that caspase-2 is subject to regulation by phosphorylation and that phosphorylation at several serine residues results in suppression of caspase-2 activity and inhibition of apoptosis (20, 22, 32, 33). We here identified a new serine residue, Ser-139, in the prodomain of caspase-2 that regulates various functions of caspase-2 as presented below. When S139A mutant caspase-2 (Casp2S139A) was stably expressed in Casp2KO/SV40 and Casp2KO/SV40/Ras MEFs, it made the cells gain significantly enhanced growth rate displayed by MTT assay (Fig. 1, A and B), which was confirmed by trypan blue exclusion assay (Fig. 1, C and D). These results demonstrate that similar to the catalytic site, Ser-139 is also required for caspase-2 to suppress undesirable overproliferation.
Enhanced Clonogenic Activity in Casp2C320A/SV40 and Casp2S139A/SV40 MEF Cells
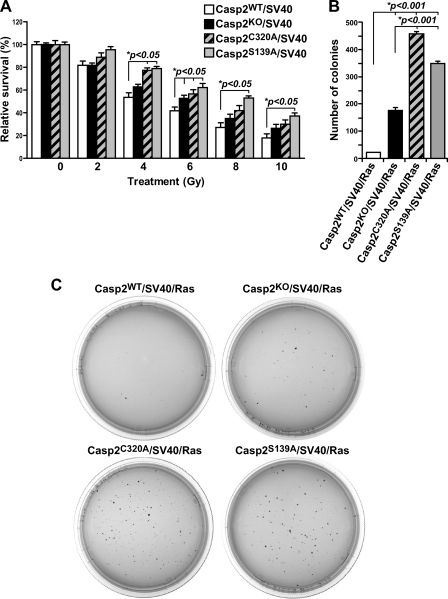
The loss of proliferation inhibition in caspase-2 mutant MEFs prompted us to evaluate their ability to divide by clonogenic assay. DNA-damaging agents activate caspase-2, and this requires the protein complex PIDDosome (8, 10, 27). Therefore, the four Casp2/SV40 cell lines described above were irradiated with IR at various doses (2–10 Gy), and their clonogenic ability over a period of 7 days was examined. We found that, similar to Casp2 KO cells, which is known to have higher clonogenic property (30), cells expressing C320A or S139A mutant caspase-2 formed significantly more colonies than their wild-type counterparts at the same dose of IR starting at 4 Gy (Fig. 2A). This increase in colony formation was also observed in different clones of each cell line (not shown). This result indicates that catalytic site Cys-320 and site Ser-139 are needed for caspase-2 to suppress undesirable increase in cell division and keep the rate of cell division in check in response to DNA damage.
FIGURE 2.
A, clonogenic assay with SV40-immortalized Casp2WT, Casp2KO, Casp2C320A, and Casp2S139A MEFs with colonies counted 7 days after seeding with data presented as the means ± S.E. from three independent experiments with each performed in triplicate. B, quantification of colonies formed in soft agar (in triplicate) by SV40/Ras-transformed Casp2WT, Casp2KO, Casp2C320A, and Casp2S139A cells on day 21. *p denotes significant difference. C, representative images of colonies on soft agar.
Increased Anchorage-independent Growth in Casp2C320A/SV40/Ras and Casp2S139A/SV40/Ras MEF Cells
Given the observed loss of inhibition in proliferation and clonogenic capabilities in caspase-2 mutant MEF cells, we further investigated the impact of these two sites on cellular transformation by determining the capacity of the corresponding cell lines in anchorage-independent growth in soft agar. We found that, similar to Casp2 KO cells, which are known to have enhanced anchorage-independent growth on soft agar (30), MEF cells expressing C320A or S139A mutant caspase-2 formed significantly more colonies in soft agar than the wild-type MEF cells and even more than KO cells (Fig. 2, B and C). This result demonstrates that the catalytic site and Ser-139 are indispensible for caspase-2 to suppress anchorage-independent growth and cellular transformation. It also indicates that both mutations appear to have a dominant negative effect in anchorage-independent growth.
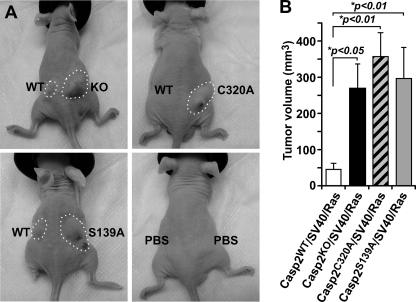
Casp2C320A/SV40/Ras and Casp2S139A/SV40/Ras MEF Cells Are Highly Tumorigenic in Athymic Nude Mice
To further investigate the consequences of the loss of inhibition in proliferation and transformation resulting from mutation in caspase-2 catalytic site and the site Ser-139, we examined the tumorigenic property of these caspase-2 mutant cells in athymic immune-deficient mice. We subcutaneously injected 106 cells (KO, WT, C320A, or S139A) into the flanks of athymic nude mice. Approximately 5 days after the injections, we started to observe growth of cell masses in mice injected with KO MEF cells, but no cell masses were noticeable for WT cells at the same time. Fourteen days after injection, we observed only a mild tumorigenic capability coming from the injected Casp2WT/SV40/Ras cells, as noted by the growth of small cell mass, whereas Casp2KO/SV40/Ras cells were significantly highly tumorigenic and formed larger cell masses (Fig. 3, A and B), which was in agreement with the previous report (30). Interestingly, in addition to KO cells, Casp2C320A/SV40/Ras and Casp2S139A/SV40/Ras MEF cells also started to form cell masses on day 5 after injection and formed large cell masses with the size similar or larger than those grown from KO cells on day 14 (Fig. 3, A and B). It is of note that the experimental results remained the same when bilateral or unilateral tumors were involved (not shown). These results demonstrate that the two residues are required for caspase-2 to suppress tumor development in nude mice.
FIGURE 3.
Caspase-2 inhibits tumor formation in athymic nude mice. A, representative mice with tumors circled in dashed outlines were derived from 1 × 106 SV40/K-Ras-transformed Casp2WT, Casp2KO, Casp2C320A, and Casp2S139A MEFs injected (subcutaneously) in hind flanks of the mice and PBS as negative control. B, tumors were measured by caliper on day 14 after cell injection, and tumor volume was calculated by the equation V = (L × W2) × 0.5, where L is the length, and W is the width of the tumor. The data represent the average tumor volumes from nine mice for each cell line and are expressed as the means ± S.E. *p denotes significant difference compared with cell mass generated by wild-type cells.
Reduction in mRNA Levels of Caspase-2 Is Associated with Lymphomas and Other Tumors in Humans
Because the loss of caspase-2 accelerates lymphoma development in Eμ-Myc-driven lymphoma in mice (28), we examined whether the caspase-2 mRNA level is reduced in cancer patients. Using the publicly available data from the NCBI Gene Expression Omnibus (GEO) repository, we searched for cancer microarray experiments showing a significant change in the mRNA levels of caspase-2 in different types of human tumors compared with normal tissues. By doing this, we gathered a body of evidence on a significant reduction in the expression of caspase-2 in various tumor and cancer states. Specifically, we found that caspase-2 is underexpressed in human Burkitt's, centroblastic, and diffuse large B-cell lymphomas when compared with normal lymphoid tissues (Table 1) (34). Furthermore, we found a significant down-regulation of caspase-2 mRNA in many other types of cancers (Table 1) (35–43). This table suggests that not only does caspase-2 have a tumor-suppressing function in lymphomagenesis in mice as previously reported in Ref. 28, but it may also contribute to the suppression of various types of malignancies in humans.
TABLE 1.
Summary of cancer types in human patients where caspase-2 was found down-regulated in comparison with normal tissue
Analysis of differential gene expression is based on publicly available data from the NCBI GEO database (see “Experimental Procedures” for details).
| Type of cancer | p value | Sample size (cancer/normal) | GEO data set identification number | Reference |
|---|---|---|---|---|
| Burkitt's lymphoma | 1.44 × 10−5 | 17/25 | GSE2350 | Basso et al. (34) |
| Centroblastic lymphoma | 2.90 × 10−10 | 28/25 | GSE2350 | Basso et al. (34) |
| Diffuse large B-cell lymphoma | 1.47 × 10−4 | 32/25 | GSE2350 | Basso et al. (34) |
| Chronic lymphocytic leukemia | 7.37 × 10−13 | 34/25 | GSE2350 | Basso et al. (34) |
| Chronic lymphocytic leukemia | 2.10 × 10−5 | 12/31 | GSE60 | Alizadeh et al. (36) |
| Chronic lymphocytic leukemia | 6.23 × 10−38 | 448/74 | GSE13159 | Haferlach et al. (35) |
| Hairy cell leukemia | 3.38 × 10−8 | 16/25 | GSE2350 | Basso et al. (34) |
| Hepatocellular carcinoma | 2.02 × 10−5 | 22/21 | GSE14520 | Roessler et al. (39) |
| Cirrhosis | 1.99 × 10−5 | 58/19 | GSE14323 | Mas et al. (40) |
| Invasive breast carcinoma | 2.99 × 10−7 | 53/6 | GSE9014 | Finak et al. (43) |
| Infiltrating bladder urothelial carcinoma | 2.43 × 10−6 | 13/14 | GSE3167 | Dyrskjot et al. (37) |
| Superficial bladder cancer | 1.47 × 10−6 | 28/14 | GSE3167 | Dyrskjot et al. (37) |
| Ovarian mucinous adenocarcinoma | 9.33 × 10−6 | 13/4 | GSE6008 | Hendrix et al. (38) |
| Ovarian endometrioid adenocarcinoma | 3.03 × 10−5 | 37/4 | GSE6008 | Hendrix et al. (38) |
| Glioblastoma | 2.28 × 10−7 | 22/3 | GSE4536 | Lee et al. (41) |
| Barrett's esophagus | 4.01 × 10−5 | 15/28 | GSE13898 | Kim et al. (42) |
| Esophageal adenocarcinoma | 6.29 × 10−4 | 75/28 | GSE13898 | Kim et al. (42) |
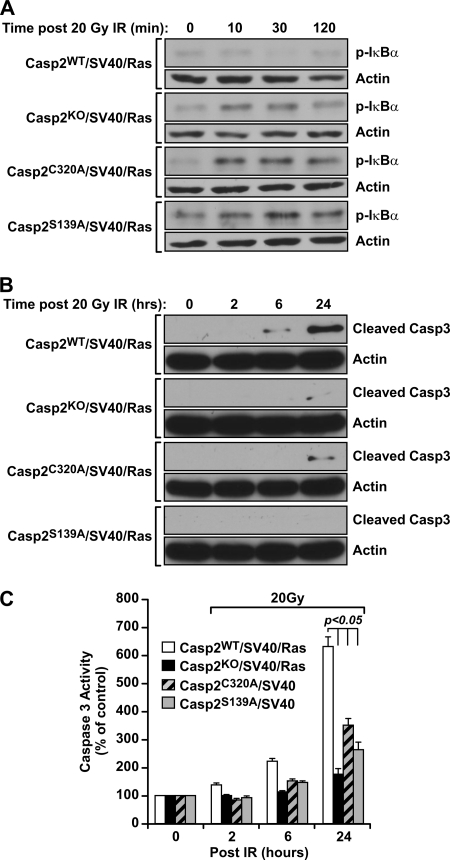
NF-κB Activity Induced by DNA Damage Is Elevated in Casp2C320A/SV40/Ras and Casp2S139A/SV40/Ras MEF Cells over Wild-type Casp2 Cells
NF-κB is a transcription factor important for many cellular processes, among which constitutive NF-κB activity is associated with cell tumorigenic activity (2). It has been reported that caspase-2 is an endogenous inhibitor of NF-κB (29). Thus, we examined the effect of caspase-2 on IR-induced activation of NF-κB. The four cell lines of Casp2/SV40/Ras were irradiated with x-ray (20 Gy) and harvested at various time points up to 2 h. Activation of NF-κB was assessed by Western blot analysis using an antibody specific for IκBα phosphorylation at serine 32 (p-IκBα), a signal that induces IκBα degradation and subsequent NF-κB nuclear translocation and activation (2). Consistent with caspase-2 being an endogenous inhibitor of NF-κB reported in Ref. 29, we observed a significant increase in IκBα phosphorylation in irradiated KO MEF cells, as compared with nonirradiated KO cells (Fig. 4A). This was also in contrast to the WT cells, where no significant increase in IκBα phosphorylation following irradiation was observed (Fig. 4A). Interestingly, irradiated C320A and S139A cells also displayed an increase in IR-induced IκBα phosphorylation, and this increase was sustained for up to 2 h following IR treatment (Fig. 4A). In addition, an increase of p65 nuclear translocation was observed in caspase-2 KO and C320A and S139A mutant cell lines compared with WT cells (not shown). These results demonstrate that the catalytic site and Ser-139 residue are responsible for caspase-2 to suppress DNA damage-induced NF-κB activation.
FIGURE 4.
Caspase-2 suppresses NF-κB activation and promotes apoptosis following 20 Gy of IR. Casp2WT, Casp2KO, Casp2C320A, and Casp2S139A MEFs were irradiated (20 Gy), and total cell lysates were prepared at indicated time points following IR and subjected to immunoblotting using an anti-phospho-IκBα (S32) antibody. A, actin was served as a protein-loading control. B and C, resistance to high dose (20 Gy) of IR-induced apoptosis in MEFs deficient in caspase-2 or reconstituted with catalytic dead or S139A mutant caspase-2 determined by caspase-3 cleavage using Western blotting (B) and caspase-3 activity using fluorogenic substrate assays (C).
Casp2C320A/SV40/Ras and Casp2S139A/SV40/Ras MEF Cells Are More Resistant to Apoptosis Induced by DNA Damage than Wild-type Caspase-2 Cells
The ability of cancer cells to escape apoptosis and continue to proliferate is one of the fundamental mechanisms for cancer development. In addition, one of the mechanisms for NF-κB to promote cell survival is to suppress apoptosis. We evaluated sensitivity of caspase-2 MEF cell lines to apoptosis induced by irradiation (20 Gy). Wild-type cells displayed significant cleavage of caspase-3, an indicator of apoptosis, at 6 and 24 h following IR (Fig. 4B). In contrast, there was less amount of caspase-3 cleavage in KO, C320A, and S139A cells at 6 and 24 h post-IR (Fig. 4B). This reduced caspase-3 activation was further confirmed by the measurement of caspase-3 activity using cleavage of fluorogenic caspase-3 substrates (Fig. 4C). These results indicate that wild-type caspase-2, in response to a relatively high dose of ionizing radiation at 20 Gy, sensitizes cells to apoptosis, which requires the catalytic Cys-320 and Ser-139 sites.
Caspase-2 Impacts G2/M DNA Damage Checkpoint in Response to Low Dose γ-Irradiation
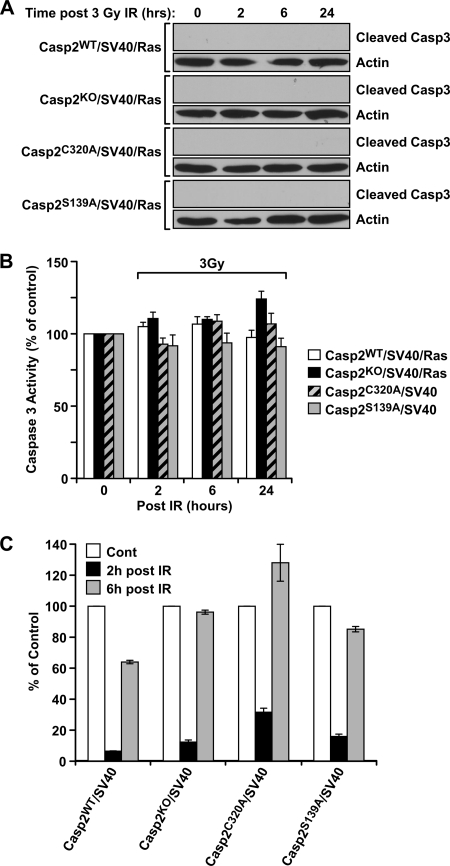
In contrast to apoptotic sensitivity of wild-type caspase-2 cells to a high dose of IR (20 Gy), when a lower dose of 3 Gy was used to irradiate the cells, no significant difference in apoptotic sensitivity was detected by caspase-3 cleavage and fluorescent caspase-3 substrate assays in all four cell lines up to 24 h following IR (Fig. 5, A and B). This result indicates that following a low dose of IR, caspase-2 does not sensitize cells to apoptosis or at least not yet up to 24 h. Intrigued by this result and the rather important role of Cys-320 and Ser-139 in all of the above cellular functions and tumorigenesis in nude mice, we investigated whether Cys-320 or Ser-139 could play a role in other cellular processes following DNA damage induction.
FIGURE 5.
Caspase-2 regulates G2/M checkpoint following low dose of IR. A and B, SV40-immortalized Casp2WT, Casp2KO, Casp2C320A, and Casp2S139A MEFs were irradiated (3 Gy), and total cell lysates were prepared at the indicated time following IR and subjected to immunoblotting with an anticleaved caspase-3 antibody (A) and caspase-3 fluoregenic substrate assay (B). C, analysis of G2/M checkpoint in MEFs showing flow cytometric profiles of mitotic cells before (0 h) and after low dose (4 Gy) of IR at the indicated time points. The cells were fixed, stained for DNA content (PI) and phospho-H3 (FITC), and assessed by flow cytometry. The data were expressed as the average of two experiments and normalized to untreated controls (Cont) of each cell line. PI, Propidium iodide.
Although insufficient to trigger apoptosis, a low dose of IR can activate DNA damage cell cycle checkpoints, and such checkpoints are antitumor mechanisms that halt the progression of cells from one phase of the cell cycle to the next to allow the time for DNA repair and maintenance of genome stability. As a result, impaired DNA damage checkpoint is intimately related to cancer development (3). Because caspase-2-deficient MEF cells were abnormal in cell cycle progression in responses to IR (30), we examined the integrity of the G2/M DNA damage checkpoint, which prevents mitotic entry by arresting cells at the G2 phase following induction of DNA damage. At various time points following irradiation at 4 Gy, the aforementioned four cell lines were labeled with an antibody recognizing phosphorylated histone H3 at serine 10, a marker of mitotic cells (44). As expected, there was an IR-induced arrest of the cells at G2 phase of cell cycle in all four cell lines 2 h following irradiation (Fig. 5C), indicating that the caspase-2 does not affect the activation of the G2/M checkpoint. However, at 6 h post-IR, a higher percentage of mitotic cells (relative to nonirradiated cells of the same cell line) was observed in caspase-2-deficient cells (96%) than in wild-type cells (64%) (Fig. 5C), demonstrating that caspase-2-deficient cells failed to sustain G2/M checkpoint arrest for longer periods of time. This result revealed that wild-type caspase-2 has a role in sustaining the G2/M checkpoint by delaying cells with DNA damage from entering mitosis, therefore allowing repair. Similar to caspase-2 deficiency, mutation in Cys-320 or Ser-139 of caspase-2 also rendered cells unable to sustain the G2/M checkpoint at 6 h post-IR, with a mitotic percentage of 128 and 85%, respectively (Fig. 5C). Collectively, these data revealed a new function of caspase-2 in sustaining the G2/M DNA damage checkpoint, and this function requires its catalytic site Cys-320 and Ser-139 residue.
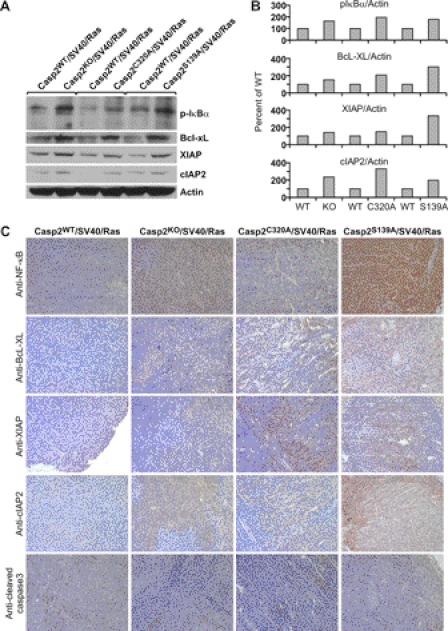
Tumors Derived from Casp2KO/SV40/Ras, Casp2C320A/SV40/Ras, or Casp2S139A/SV40/Ras MEFs Have Elevated Constitutive Activation of NF-κB and Increased Levels of Candidate NF-κB-targeted Antiapoptotic Proteins
Many types of tumors rely on constitutive NF-κB activity for survival, and constitutive NF-κB activity is present in a variety of human tumors, including lymphomas (37). To understand whether NF-κB activation and resistance to apoptosis are associated with the tumor development observed in our nude mouse model, we evaluated the phosphorylation of IκBα in cell-free protein extracts prepared from the tumor mass derived in nude mice by Western blot analyses. We found that IκBα was constitutively phosphorylated and also with higher levels of expression in the tumor extracts derived from Casp2KO/SV40/Ras, Casp2C320A/SV40/Ras, and Casp2S139A/SV40/Ras MEF cells, as compared with those derived from Casp2WT/SV40/Ras cells (Fig. 6, A and B). Candidate protein targets of NF-κB known to confer apoptotic resistance such as Bcl-xL, XIAP, and cIAP2 were also up-regulated in the cell masses derived from caspase-2 KO, Cys-320, and Ser-139 mutant MEF cell lines compared with WT controls (Fig. 6, A and B). These results were complemented by up-regulation of p65 subunit of NF-κB and the three antiapoptotic proteins in KO, Ser-139, and Cys-320 samples by immunohistochemical analysis of paraffin-embedded tumor samples (Fig. 6C). Consistent with the up-regulation of these antiapoptotic proteins, expression of active pro-apoptotic caspase-3 (cleaved caspase-3) was not detectable in tumor samples derived from caspase-2 KO and two point mutant cell lines, whereas it was readily detectable in wild-type samples (Fig. 6C).
FIGURE 6.
Caspase-2 suppresses NF-κB activation in tumors. Western blotting analysis of protein expression of p-IκBα and NF-κB targeted antiapoptotic proteins as indicated in tumors derived from MEFs deficient in caspase-2 or expressing catalytic dead or S139A mutant caspase-2. A and B, total lysates from day 14 tumor mass derived from each cell lines as indicated were subjected to Western blotting and probed with antibodies recognizing the indicated proteins (A), and the results were quantified using actin as control and normalized to wild-type control by using ImageJ 1.37c (National Institutes of Health) (B). C, immunohistochemical analysis of protein expression of the p65 subunit of NF-κB and NF-κB-targeted antiapoptotic proteins as indicated in formalin-fixed, paraffin-embedded tumor sections derived from each MEF cell lines as indicated.
DISCUSSION
This study provides new insight into how caspase-2 suppresses cell proliferation, transformation, and tumorigenesis at the cross-roads of apoptosis, cell cycle checkpoint, and NF-κB pathways. Based on the experimental data presented in this study, we propose the following working model. As a multifunctional protein, the catalytic site Cys-320 and the Ser-139 residue are essential for caspase-2 to suppress cellular hyperproliferation and aberrant activation of NF-κB, prolong DNA damage-induced G2/M checkpoint, and promote pro-apoptotic cell death. These functions together contribute to the role of caspase-2 in the suppression of tumor development induced by oncogenic K-Ras in nude mice.
This model is supported by the findings summarized below. The aberrant NF-κB activity and the resultant up-regulation of antiapoptotic NF-κB target proteins could contribute to the apoptotic resistance in caspase-2 KO, C320A, and S139A mutant cells. In the same cell populations, the failure to sustain G2/M checkpoint for a longer period of time could accelerate cell transformation and tumorigenesis by allowing cells with damaged DNA and genome instability to escape cell cycle arrest and go through cell division. Resistance to apoptosis accompanied by uncontrollable cell division could promote growth of tumor mass in the nude mice. This working model is also supported by the knowledge that a G2/M checkpoint defect promotes tumorigenesis (3) and that there is close association of up-regulation of three such antiapoptotic proteins, Bcl-xL, XIAP, and cIAP2, with constitutive activation of NF-κB and resistance to apoptosis in a variety of human malignancies (45–50).
Although the role of caspase-2 catalytic activity is well established in cell death, whether it is also involved in antitumor function of caspase-2 was not determined (30). This study provides the first indication for the requirement of catalytic site Cys-320 for antitumor function of caspase-2. Furthermore, this study identified a new site Ser-139 of caspase-2 that is also involved in antitumor function of caspase-2. Caspase-2 with a single mutation at Cys-320 or Ser-139 rendered cells unable to sustain G2/M checkpoint for a longer period of time and more resistant to apoptosis. Meanwhile, they exhibited enhanced NF-κB activity and were highly tumorigenic in nude mice. Similar to Cys-320 mutant caspase-2, Ser-139 mutant caspase-2 loses its apoptotic function despite the fact that Cys-320 is still wild type. Based on this, we speculated that Ser-139 site promotes the catalytic activity of caspase-2. Because this residue is a serine, it could be a putative site of phosphorylation and subject to post-translational modification following DNA damage similar to other reported serine residues on caspase-2 (20, 22, 32, 33). Although we do not know at this time whether S139A MEF cells actually have genomic instability, the reported increase in genomic instability in caspase-2 KO MEFs compared with wild-type cells (30) warrants further examination of genomic instability in S139A MEF cells. Therefore, this study lays a foundation for future investigation into the molecular mechanism underlying the two sites of caspae-2 in tumor suppression.
It is also noted in this study that mutations at the catalytic site and Ser-139 of caspase-2 confer cells with enhanced cellular activities over caspase-2 KO cells in many of the functions that we analyzed in this study. This indicates traits of gain of function mutations in these two sites. How these mutations give rise to such gain of function phenotypes is yet to be investigated. We speculate that these mutations may change the conformation of caspase-2, leading to changes of the dynamics of caspase-2 to interact with other proteins and gain a new ability to interact with certain proteins that wild-type caspase-2 would not interact with.
This study opens new avenues for further studies of the tumor-suppressing function of caspase-2. The primary cleavage target of caspase-2 in promoting DNA damage-induced apoptosis is Bid, which mediates activation of the mitochondrial pathway of apoptotic cell death. It is important to examine whether the resistance to apoptosis in cells expressing the two mutant forms of caspase-2 proteins is due to a lack of or attenuated Bid cleavage. Furthermore, cleavage target(s) of caspase-2 that mediates NF-κB pathway needs to be investigated. Because the catalytic activity is required for NF-κB suppression, it is reasonable to postulate that the target(s) of caspase-2 in the regulation of NF-κB is a protein(s) that can be catalytically cleaved by caspase-2. Along this line, it was reported that caspase-2 cleavage of RIP1 resulted in NF-κB suppression, whereas down-regulation of caspase-2 resulted in NF-κB activation (29). Our findings that wild-type caspase-2 keeps NF-κB activity in check and at a low level is in agreement with the above report. However, another report showed that caspase-2, in forming a complex with TRAF2 and RIP1, activates NF-κB, which requires its caspase recruitment domain and is independent of its enzymatic activity and RIP1 cleavage (28). This discrepancy could be due to different experimental conditions and cell types. Moreover, this study warrants identification of protein targets of caspase-2 that are responsible for mediating the G2/M DNA damage checkpoint function of caspase-2. Identification of these proteins involved in caspase-2 tumor-suppressing function will provide insight toward understanding the mechanism of tumor suppression mediated by caspase-2 and the potential for developing agents capable of modulating the level and activity of caspase-2 for the suppression of malignant transformation and tumorigenesis.
In addition to having a tumor-suppressing function in mice, caspase-2 may also have an impact on tumor suppression in humans, as suggested by the significant reduction in caspase-2 gene expression in multiple types of human malignancies. This possibility is also supported by the following studies. For instance, the human caspase-2 gene resides on chromosome 7q, which is frequently deleted in leukemia (51). In addition, caspase-2 can be activated by cyclin D3 (52), can be up-regulated by the candidate tumor suppressor RBM5 (53), and is linked to Mdm2 and the tumor suppressor p53 (30, 54–57). Furthermore, a portion of caspase-2 in human tumor cell lines is localized to the promyelocytic leukemia nuclear bodies (58), a subnuclear structure associated with apoptosis and the maintenance of genome stability, and also to the centrosome (59), the organelle that ensures the fidelity of cell division. Future work will need to address whether caspase-2 has a tumor suppressor function in humans and whether there are somatic mutations at Cys-320 and Ser-139 and other sites of caspase-2 that are linked with human cancers.
Acknowledgments
We thank Drs. David R. Plas for flow cytometry support and Bcl-XL antibody (for immunohistochemistry), Siva Vallabhapurapu for discussion of NF-κB, Maria T. Diaz-Meco and Jorge Moscat for SV40 and K-Ras retroviral constructs, Jinsong Zhang for GAPDH primers for RT-PCR, Jerry B Lingrel and Sohaib Khan for helpful advice and critical reading of the manuscript, Glenn Doerman for graphics preparation, Maryellen Daston and Jason B. Garrison for editing this manuscript, and Du lab members for helpful discussion of this project.
This work was supported by the startup fund of the Department of Cancer and Cell Biology, University of Cincinnati College of Medicine.
- PIDD
- p53-induced protein with a death domain
- MEF
- mouse embryonic fibroblast
- MTT
- methylthiazolyldiphenyl-tetrazolium bromide
- Casp
- caspase.
REFERENCES
- 1. Grivennikov S. I., Greten F. R., Karin M. (2010) Immunity, inflammation, and cancer. Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karin M. (2006) Nuclear factor-κB in cancer development and progression. Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 3. Kastan M. B., Bartek J. (2004) Cell-cycle checkpoints and cancer. Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 4. Evan G. I., Vousden K. H. (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348 [DOI] [PubMed] [Google Scholar]
- 5. Kumar S., Kinoshita M., Noda M., Copeland N. G., Jenkins N. A. (1994) Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 β-converting enzyme. Genes Dev. 8, 1613–1626 [DOI] [PubMed] [Google Scholar]
- 6. Kumar S., Tomooka Y., Noda M. (1992) Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 185, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 7. Wang L., Miura M., Bergeron L., Zhu H., Yuan J. (1994) Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 78, 739–750 [DOI] [PubMed] [Google Scholar]
- 8. Kumar S. (2009) Caspase 2 in apoptosis, the DNA damage response and tumour suppression. Enigma no more? Nat. Rev. 9, 897–903 [DOI] [PubMed] [Google Scholar]
- 9. Kitevska T., Spencer D. M., Hawkins C. J. (2009) Caspase-2. Controversial killer or checkpoint controller? Apoptosis 14, 829–848 [DOI] [PubMed] [Google Scholar]
- 10. Zhivotovsky B., Orrenius S. (2005) Caspase-2 function in response to DNA damage. Biochem. Biophys. Res. Commun. 331, 859–867 [DOI] [PubMed] [Google Scholar]
- 11. Krumschnabel G., Manzl C., Villunger A. (2009) Caspase-2. Killer, savior and safeguard. Emerging versatile roles for an ill-defined caspase. Oncogene 28, 3093–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouchier-Hayes L., Green D. R. (2012) Caspase-2. The orphan caspase. Cell Death Differ. 19, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du C., Fang M., Li Y., Li L., Wang X. (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42 [DOI] [PubMed] [Google Scholar]
- 14. Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Induction of apoptotic program in cell-free extracts. Requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 15. Guo Y., Srinivasula S. M., Druilhe A., Fernandes-Alnemri T., Alnemri E. S. (2002) Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J. Biol. Chem. 277, 13430–13437 [DOI] [PubMed] [Google Scholar]
- 16. Robertson J. D., Enoksson M., Suomela M., Zhivotovsky B., Orrenius S. (2002) Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 277, 29803–29809 [DOI] [PubMed] [Google Scholar]
- 17. Gao Z., Shao Y., Jiang X. (2005) Essential roles of the Bcl-2 family of proteins in caspase-2-induced apoptosis. J. Biol. Chem. 280, 38271–38275 [DOI] [PubMed] [Google Scholar]
- 18. Wagner K. W., Engels I. H., Deveraux Q. L. (2004) Caspase-2 can function upstream of bid cleavage in the TRAIL apoptosis pathway. J. Biol. Chem. 279, 35047–35052 [DOI] [PubMed] [Google Scholar]
- 19. Lavrik I. N., Golks A., Baumann S., Krammer P. H. (2006) Caspase-2 is activated at the CD95 death-inducing signaling complex in the course of CD95-induced apoptosis. Blood 108, 559–565 [DOI] [PubMed] [Google Scholar]
- 20. Shin S., Lee Y., Kim W., Ko H., Choi H., Kim K. (2005) Caspase-2 primes cancer cells for TRAIL-mediated apoptosis by processing procaspase-8. EMBO J. 24, 3532–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castedo M., Perfettini J. L., Roumier T., Valent A., Raslova H., Yakushijin K., Horne D., Feunteun J., Lenoir G., Medema R., Vainchenker W., Kroemer G. (2004) Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene 23, 4362–4370 [DOI] [PubMed] [Google Scholar]
- 22. Andersen J. L., Johnson C. E., Freel C. D., Parrish A. B., Day J. L., Buchakjian M. R., Nutt L. K., Thompson J. W., Moseley M. A., Kornbluth S. (2009) Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J. 28, 3216–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castedo M., Perfettini J. L., Roumier T., Andreau K., Medema R., Kroemer G. (2004) Cell death by mitotic catastrophe. A molecular definition. Oncogene 23, 2825–2837 [DOI] [PubMed] [Google Scholar]
- 24. Vakifahmetoglu H., Olsson M., Zhivotovsky B. (2008) Death through a tragedy. Mitotic catastrophe. Cell Death Differ. 15, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 25. Sidi S., Sanda T., Kennedy R. D., Hagen A. T., Jette C. A., Hoffmans R., Pascual J., Imamura S., Kishi S., Amatruda J. F., Kanki J. P., Green D. R., D'Andrea A. A., Look A. T. (2008) Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 133, 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tinel A., Eckert M. J., Logette E., Lippens S., Janssens S., Jaccard B., Quadroni M., Tschopp J. (2011) Regulation of PIDD auto-proteolysis and activity by the molecular chaperone Hsp90. Cell Death Differ. 18, 506–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tinel A., Tschopp J. (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304, 843–846 [DOI] [PubMed] [Google Scholar]
- 28. Lamkanfi M., D'hondt K., Vande Walle L., van Gurp M., Denecker G., Demeulemeester J., Kalai M., Declercq W., Saelens X., Vandenabeele P. (2005) A novel caspase-2 complex containing TRAF2 and RIP1. J. Biol. Chem. 280, 6923–6932 [DOI] [PubMed] [Google Scholar]
- 29. Guha M., Xia F., Raskett C. M., Altieri D. C. (2010) Caspase 2-mediated tumor suppression involves survivin gene silencing. Oncogene 29, 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho L. H., Taylor R., Dorstyn L., Cakouros D., Bouillet P., Kumar S. (2009) A tumor suppressor function for caspase-2. Proc. Natl. Acad. Sci. U.S.A. 106, 5336–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hansen M. B., Nielsen S. E., Berg K. (1989) Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 119, 203–210 [DOI] [PubMed] [Google Scholar]
- 32. Nutt L. K., Margolis S. S., Jensen M., Herman C. E., Dunphy W. G., Rathmell J. C., Kornbluth S. (2005) Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell 123, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nutt L. K., Buchakjian M. R., Gan E., Darbandi R., Yoon S. Y., Wu J. Q., Miyamoto Y. J., Gibbon J. A., Andersen J. L., Freel C. D., Tang W., He C., Kurokawa M., Wang Y., Margolis S. S., Fissore R. A., Kornbluth S. (2009) Metabolic control of oocyte apoptosis mediated by 14–3-3ζ-regulated dephosphorylation of caspase-2. Dev. Cell 16, 856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390 [DOI] [PubMed] [Google Scholar]
- 35. Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G. T., Béné M. C., De Vos J., Hernández J. M., Hofmann W. K., Mills K. I., Gilkes A., Chiaretti S., Shurtleff S. A., Kipps T. J., Rassenti L. Z., Yeoh A. E., Papenhausen P. R., Liu W. M., Williams P. M., Foà R. (2010) Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia. Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 28, 2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson J., Jr., Lu L., Lewis D. B., Tibshirani R., Sherlock G., Chan W. C., Greiner T. C., Weisenburger D. D., Armitage J. O., Warnke R., Levy R., Wilson W., Grever M. R., Byrd J. C., Botstein D., Brown P. O., Staudt L. M. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 [DOI] [PubMed] [Google Scholar]
- 37. Dyrskjøt L., Kruhøffer M., Thykjaer T., Marcussen N., Jensen J. L., Møller K., Ørntoft T. F. (2004) Gene expression in the urinary bladder. A common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 64, 4040–4048 [DOI] [PubMed] [Google Scholar]
- 38. Hendrix N. D., Wu R., Kuick R., Schwartz D. R., Fearon E. R., Cho K. R. (2006) Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 66, 1354–1362 [DOI] [PubMed] [Google Scholar]
- 39. Roessler S., Jia H. L., Budhu A., Forgues M., Ye Q. H., Lee J. S., Thorgeirsson S. S., Sun Z., Tang Z. Y., Qin L. X., Wang X. W. (2010) A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70, 10202–10212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mas V. R., Maluf D. G., Archer K. J., Yanek K., Kong X., Kulik L., Freise C. E., Olthoff K. M., Ghobrial R. M., McIver P., Fisher R. (2009) Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol. Med. 15, 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N. M., Pastorino S., Purow B. W., Christopher N., Zhang W., Park J. K., Fine H. A. (2006) Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 [DOI] [PubMed] [Google Scholar]
- 42. Kim S. M., Park Y. Y., Park E. S., Cho J. Y., Izzo J. G., Zhang D., Kim S. B., Lee J. H., Bhutani M. S., Swisher S. G., Wu X., Coombes K. R., Maru D., Wang K. K., Buttar N. S., Ajani J. A., Lee J. S. (2010) Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS One 5, e15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finak G., Bertos N., Pepin F., Sadekova S., Souleimanova M., Zhao H., Chen H., Omeroglu G., Meterissian S., Omeroglu A., Hallett M., Park M. (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 14, 518–527 [DOI] [PubMed] [Google Scholar]
- 44. Wang B., Matsuoka S., Carpenter P. B., Elledge S. J. (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 45. Akyurek N., Ren Y., Rassidakis G. Z., Schlette E. J., Medeiros L. J. (2006) Expression of inhibitor of apoptosis proteins in B-cell non-Hodgkin and Hodgkin lymphomas. Cancer 107, 1844–1851 [DOI] [PubMed] [Google Scholar]
- 46. Runnebaum I. B., Brüning A. (2005) Glucocorticoids inhibit cell death in ovarian cancer and up-regulate caspase inhibitor cIAP2. Clin. Cancer Res. 11, 6325–6332 [DOI] [PubMed] [Google Scholar]
- 47. Hu S., Du M. Q., Park S. M., Alcivar A., Qu L., Gupta S., Tang J., Baens M., Ye H., Lee T. H., Marynen P., Riley J. L., Yang X. (2006) cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J. Clin. Invest. 116, 174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kashkar H., Seeger J. M., Hombach A., Deggerich A., Yazdanpanah B., Utermöhlen O., Heimlich G., Abken H., Krönke M. (2006) XIAP targeting sensitizes Hodgkin lymphoma cells for cytolytic T-cell attack. Blood 108, 3434–3440 [DOI] [PubMed] [Google Scholar]
- 49. Kashkar H., Haefs C., Shin H., Hamilton-Dutoit S. J., Salvesen G. S., Kronke M., Jurgensmeier J. M. (2003) XIAP-mediated caspase inhibition in Hodgkin's lymphoma-derived B cells. J. Exp. Med. 198, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vega M. I., Jazirehi A. R., Huerta-Yepez S., Bonavida B. (2005) Rituximab-induced inhibition of YY1 and Bcl-xL expression in Ramos non-Hodgkin's lymphoma cell line via inhibition of NF-κB activity. Role of YY1 and Bcl-xL in Fas resistance and chemoresistance, respectively. J. Immunol. 175, 2174–2183 [DOI] [PubMed] [Google Scholar]
- 51. Kumar S., White D. L., Takai S., Turczynowicz S., Juttner C. A., Hughes T. P. (1995) Apoptosis regulatory gene NEDD2 maps to human chromosome segment 7q34–35, a region frequently affected in haematological neoplasms. Hum Genet 95, 641–644 [DOI] [PubMed] [Google Scholar]
- 52. Mendelsohn A. R., Hamer J. D., Wang Z. B., Brent R. (2002) Cyclin D3 activates Caspase 2, connecting cell proliferation with cell death. Proc. Natl. Acad. Sci. U.S.A. 99, 6871–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fushimi K., Ray P., Kar A., Wang L., Sutherland L. C., Wu J. Y. (2008) Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc. Natl. Acad. Sci. U.S.A. 105, 15708–15713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vakifahmetoglu H., Olsson M., Orrenius S., Zhivotovsky B. (2006) Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25, 5683–5692 [DOI] [PubMed] [Google Scholar]
- 55. Baptiste-Okoh N., Barsotti A. M., Prives C. (2008) A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tyagi A., Singh R. P., Agarwal C., Agarwal R. (2006) Silibinin activates p53-caspase 2 pathway and causes caspase-mediated cleavage of Cip1/p21 in apoptosis induction in bladder transitional-cell papilloma RT4 cells. Evidence for a regulatory loop between p53 and caspase 2. Carcinogenesis 27, 2269–2280 [DOI] [PubMed] [Google Scholar]
- 57. Oliver T. G., Meylan E., Chang G. P., Xue W., Burke J. R., Humpton T. J., Hubbard D., Bhutkar A., Jacks T. (2011) Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell 43, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tang J., Xie W., Yang X. (2005) Association of caspase-2 with the promyelocytic leukemia protein nuclear bodies. Cancer Biol. Ther 4, 645–649 [DOI] [PubMed] [Google Scholar]
- 59. Narine K. A., Keuling A. M., Gombos R., Tron V. A., Andrew S. E., Young L. C. (2010) Defining the DNA mismatch repair-dependent apoptotic pathway in primary cells. Evidence for p53-independence and involvement of centrosomal caspase 2. DNA repair 9, 161–168 [DOI] [PubMed] [Google Scholar]