Background: Class I myosins contribute to membrane-associated events.
Results: A short segment of basic/hydrophobic amino acids in the tail which binds acidic phospholipids and the actin binding site in the head is required for relocalization of Dictyostelium myosin IB.
Conclusion: Dynamic relocalization results from competition between membrane acidic phospholipids and cytoplasmic F-actin.
Significance: The molecular basis of myosin I relocation is fundamental to understanding cell motility.
Keywords: Cell Motility, Lipids, Lipid-binding Protein, Myosin, Plasma Membrane, BH, Myosin I, PIP2, PIP3, Relocalization
Abstract
Class I myosins have a single heavy chain comprising an N-terminal motor domain with actin-activated ATPase activity and a C-terminal globular tail with a basic region that binds to acidic phospholipids. These myosins contribute to the formation of actin-rich protrusions such as pseudopodia, but regulation of the dynamic localization to these structures is not understood. Previously, we found that Acanthamoeba myosin IC binds to acidic phospholipids in vitro through a short sequence of basic and hydrophobic amino acids, BH site, based on the charge density of the phospholipids. The tail of Dictyostelium myosin IB (DMIB) also contains a BH site. We now report that the BH site is essential for DMIB binding to the plasma membrane and describe the molecular basis of the dynamic relocalization of DMIB in live cells. Endogenous DMIB is localized uniformly on the plasma membrane of resting cells, at active protrusions and cell-cell contacts of randomly moving cells, and at the front of motile polarized cells. The BH site is required for association of DMIB with the plasma membrane at all stages where it colocalizes with phosphoinositide bisphosphate/phosphoinositide trisphosphate (PIP2/PIP3). The charge-based specificity of the BH site allows for in vivo specificity of DMIB for PIP2/PIP3 similar to the PH domain-based specificity of other class I myosins. However, DMIB-head is required for relocalization of DMIB to the front of migrating cells. Motor activity is not essential, but the actin binding site in the head is important. Thus, dynamic relocalization of DMIB is determined principally by the local PIP2/PIP3 concentration in the plasma membrane and cytoplasmic F-actin.
Introduction
All class I myosins have a single heavy chain consisting of an N-terminal globular motor domain that binds actin and has actin-activated ATPase activity, an IQ domain that binds one or more light chains, and a C-terminal nonhelical tail with a basic region adjacent to the motor domain. In addition, long-tail Acanthamoeba and Dictyostelium class I myosins have a glycine/proline/alanine-rich (GPA, Acanthamoeba) or glycine/proline/glutamine-rich (GPQ, Dictyostelium) region and a Src homology 3 (SH3)2 region following the basic region (1–4). Mammalian myosin IC (Myo1C) (5) and Myo1G (6, 7) bind to acidic phospholipids in vitro and in vivo through a putative pleckstrin homology (PH) domain within the basic region that may bind specifically to PIP2. Although Acanthamoeba myosin IC contains a putative PH domain within the basic region (8), AMIC shows no specificity for binding to PIP2 in vitro. AMIC binds to phospholipid vesicles containing either PS, PIP2, or both in proportion to their net negative charge irrespective of their phospholipid composition (9). Moreover, presumably because of the high negative charge of PIP2 and PIP3, endogenous AMIC colocalizes with PIP2/PIP3 in the Acanthamoeba plasma membrane (9).
The basis of the affinity of AMIC for acidic phospholipid in vitro is a short sequence (13 residues) enriched with basic and hydrophobic amino acids (the BH site) that lies within the putative PH domain (9). In vitro studies with synthetic peptides and sequence analysis by a novel computer program (10) identified BH sites in many class I myosins, including Dictyostelium myosin IB, and also nonmyosin proteins, suggesting that plasma membrane-association of proteins through nonspecific BH sites may be widespread. Recently, lipid/membrane binding of mammalian Myo1E was shown to be more similar to the binding of AMIC than the binding of mammalian Myo1C (11).
The colocalization of endogenous AMIC and PIP2/PIP3 in the plasma membrane of Acanthamoeba is consistent with, but does not prove, an important role for the BH site. To determine the importance of the BH site and whether other factors might also be involved in membrane localization in live cells, one needs to be able to express and analyze labeled wild-type and mutant constructs. Therefore, we chose to work with Dictyostelium for which all of the necessary tools are available. When placed in nonnutrient medium, Dictyostelium amoebae chemotax toward aggregation centers initiated by cells secreting cAMP. Chemotaxing cells elongate and polarize, with some proteins moving to the front and others to the rear, and secrete cAMP which attracts neighboring cells thus forming streams of chemotaxing amoebae (12–14). DMIB has been shown to play a role in regulating pseudopod formation and is necessary for persistent chemotactic motility (15, 16). DMIB concentrates at the plasma membrane in axenic cells (17), in the cytoplasm at the front of motile amoebae (17, 18), and at cell-cell contacts (19). We asked whether the BH site is required for the association of DMIB with the plasma membrane, if DMIB shows preference for PIP2/PIP3-enriched regions of the plasma membrane, and what factors, in addition to the BH site, might be required for the dynamic relocalization of DMIB in motile, chemotaxing amoebae.
EXPERIMENTAL PROCEDURES
DNA Constructs
All DMIB expression plasmids were generated using PCR and PCR-based mutagenesis. Regions of the myoB gene were amplified using a full-length clone of the myoB gene (pDTb2) (20) as a template. The 5′ and 3′ oligonucleotides included restriction enzyme sites to enable subsequent cloning to generate GFP fusion proteins (supplemental Table S1). All PCR products were TA-cloned using the Strataclone system (Stratagene), and the full sequence for every clone was verified (BioMedical Genomics Center). The full-length or altered myoB genes were then cloned into a low copy number extrachromosomal plasmid, pTX-GFP (21) except for wild-type GFP-MyoB (DMIB) which was cloned into the related low copy number expression plasmid pLittle (22).
Constructs encoding PH domains of CRAC and PLCδ with C-terminal enhanced GFP were a gift from Dr. C. A. Parent (23). They were transfected into in AX2 myoB− cells (20) and then grown in the same medium as AX2 myoB− cells expressing DMIB.
Cell Culture
AX2 and AX3 cells were grown in HL5 medium (24). DMIB-null cells (myoB− cells) (20) and PI3K1−,5−PTEN− cells (25) were grown in HL5 medium with a final concentration of 7 μg/ml blasticidin S HCl (Invitrogen). myoB− cells and PI3K1–5−,PTEN− cells expressing wild-type or mutant DMIB with N-terminal GFP were grown in HL5 medium with 7 μg/ml blasticidin S HCl and 12 μg/ml G418 sulfate (Mediatech). AX3 cells coexpressing DMIB and ABD-120 (26), a gift from Dr. Goeh Jung, were grown in HL5 medium with 10 μg/ml blasticidin S HCl and 10 μg/ml G418 sulfate.
Dictyostelium amoebae were grown on 10-cm Petri dishes in HL5 medium with appropriate additions (see above), harvested in 10 ml of medium and placed on ice in 15-ml tubes for 20–30 min. Cells were then plated on chambered coverglass (Nalge Nunc International, 155383) in the desired density resulting in about 80% confluence for cells meant to go through starvation cycle and about 50% confluence for cells meant to be observed immediately. Cells were allowed to attach and washed three times for 5 min with starvation buffer (10 mm phosphate buffer, pH 6.2, 2 mm MgSO4, 0.2 mm CaCl2) and left overlaid with starvation buffer for varied periods of time. Freshly plated cells were observed immediately live or were fixed directly in chambered coverglass (10-min fixation with 1% formaldehyde, 0.1% glutaraldehyde, and 0.01% Triton X-100 in 20 mm phosphate buffer, pH 6.2) followed by three washes with 20 mm phosphate buffer, pH 6.2. In other experiments cells were kept at 20 °C in starvation buffer in the dark, to initiate polarization, and were observed at the desired times. Alternatively, plated cells were kept in starvation buffer at 4 °C in the dark overnight and moved to 20 °C the following morning. These cells usually formed streams within 3 h after moving them to 20 °C.
Imaging Live Cells
The absence of DMIB delays development, and overexpression of DMIB significantly slows growth, motility, and development (16, 20, 27–29). Therefore, the developmental time course depends not only on the properties of the expressed mutants but also on the level of their expression and culturing time. For this reason, we did not attempt to phenotype the effects of expressed mutants on properties such as delay in forming streams. Rather, we compared cells that were in a similar stage of development and showed similar morphology, i.e. cells that were freshly plated, randomly moving, elongated, moving directionally or streaming, and had similar levels of overall GFP fluorescence intensity.
The intensity of GFP fluorescence varied depending on the level of protein expression, the protein expressed, the state of cells (vegetative versus starved) and culturing time. For these reasons, as a control, we always monitored expression of DMIB or a previously characterized mutant that was transfected at the same time and treated the same way. This allowed us to compare localization of two or more DMIB mutants in the cells with similar levels of protein expression and make sure that differences between expressed mutants were not caused by differences in their intracellular concentrations.
Each of the developmental stages was monitored for each mutant in at least two independent transfections and during at least two independent starvation cycles, always accompanied by monitoring in parallel at least one control mutant. For freshly plated cells we observed at least 100 cells showing localization at the plasma membrane or its lack for each mutant studied. At this stage fluorescence was stable in time and space, and we quantified it in more detail (see below). In starved cells, localization of DMIB and some of its mutants was transient; it occurred only in cells showing a particular morphology, and even then it happened in a few minutes-long cycles (for example sharp localization at the plasma membrane of the engulfing mouth of streaming cells or diffused localization at the front of elongated cells migrating individually). In a typical experiment, we followed starved cells for 4–6 h starting when cells began to elongate and ending when they formed streams and/or mounds. In these cases we report particular localization for a mutant after registering it for at least 20 cells (and in most cases closer to 100 cells).
Antibodies and Imaging Fixed Cells
For visualization of endogenous DMIB in fixed Dictyostelium cells we used polyclonal rabbit anti-DMIB antibody (18, 19) in 1:100 dilution. For visualization of PIP2 and PIP3 we used commercially available anti-PI(4,5)P2 IgG antibody from Abcam (ab2335) in 1:100 dilution. F-actin was visualized by phalloidin Alexa Fluor 633 (Molecular Probes) in 1:250 dilution or by rhodamine-phalloidin (Sigma) in 1:500 dilution.
For images of fixed cells, Dictyostelium amoebae were grown on coverslips in 6-well chambers or on a chambered coverglass in regular media or in starvation buffer. Cells were washed once with starvation buffer, then fixed with 1% formaldehyde, 0.1% glutaraldehyde, and 0.01% Triton X-100 in 20 mm phosphate buffer, pH 6.2, for 10 min. Cells were than washed and incubated with antibodies as described (30).
Testing Specificity of Anti-PI(4,5)P2 Antibody with Lipid Strips
The specificity of PIP2 antibody was tested with lipid strips from Echelon (P-6002 and P-6001; supplemental Fig. S1). Lipid strips were incubated for 1 h at room temperature in TBS (25 mm Tris, pH 7.5, 150 mm NaCl containing 2% BSA) and then for another hour with TBS buffer containing 2% BSA, 0.1% Tween 20, and PIP2 antibody (1:2,500 dilution). The strips were washed 3 times for 5 min each with TBS containing 0.1%Tween-20. The strips were then incubated for 1 h with secondary antibody (IRDye 700) diluted 10,000 times in TBS with 2% BSA and 0.1% Tween 20, washed three times for 5 min with TBS with 0.1% Tween 20, and rinsed with TBS and stored in TBS. Secondary antibody detection was done using the Odyssey infrared imager (Li-Cor).
We found that PI(4,5)P2 antibody was not highly specific for PI(4,5)P2. The antibody bound to PI(3,4)P2 about 3 times stronger and to PI(3,4,5)P3 only approximately 50% weaker than to PI(4,5)P2. Antibody reaction was negligible with phosphatidylinositol, phosphatidylinositol 3-phosphate, phosphatidylinositol 4-phosphate, phosphatidylinositol 5-phosphate, PS, phosphatidylethanolamine, phosphatidylcholine, sphingosine 1-phosphate, PI(3,5)P2, phosphatidic acid, diacylglycerol, phosphatidylglycerol, cardiolipin, cholesterol, sphingomyelin, and 3-sulfogalactosylceramide.
Other Reagents and Procedures
Dictyostelium cells were transfected with 15 μg of plasmid DNA by electroporation (2 times, 0.9 kV) as described (31). After electroporation PI3K1–5−,PTEN− cells were grown in the presence of heat-killed bacteria until colonies developed. Single colonies were picked and monitored for the presence of GFP fluorescence. Alternatively, and with similar final results, entire transformation plates with multiple fluorescent colonies were cultured. Cells were viewed with a Zeiss LSM 5 LIVE confocal microscope with a 63× lens or with a Zeiss LSM 510 confocal microscope using a 63× lens. The slice thickness was 1 μm, unless stated otherwise. For final illustrations, images were processed in an LSM image browser and Photoshop. Profile scanning of original cell microscopic images was done using Metamorph software. A line scan of each cell was made across two separate regions, and the average maximum fluorescence intensities at the plasma membrane were divided by the average fluorescence intensities in the cytoplasm.
RESULTS
Colocalization of Endogenous DMIB and PIP2/PIP3
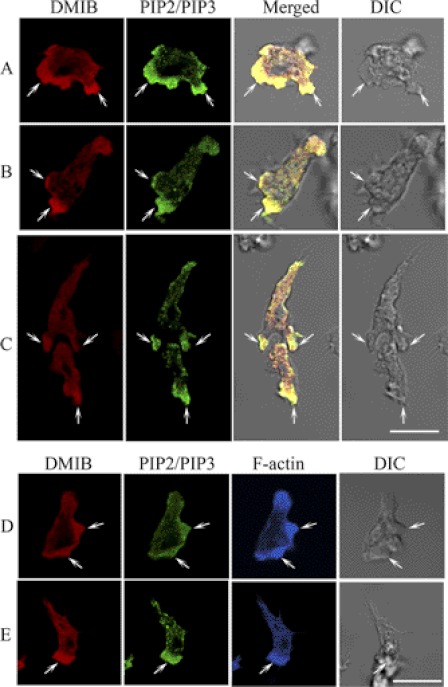
Recent studies have suggested that interactions of some myosins I with phosphoinositides are a major determinant of their in vivo localization (5–7, 9). We visualized the localization of DMIB and PIP2/PIP3 in fixed cells with a polyclonal DMIB-specific antibody and a commercial PIP2 antibody (see “Experimental Procedures”), which we found recognized PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3, but no other lipids tested (supplemental Fig. S1). In nonpolarized, randomly moving cells, DMIB and PIP2/PIP3 colocalized in cell protrusions (Fig. 1A). DMIB and PIP2/PIP3 also colocalized at the front of polarized, chemotaxing cells (Fig. 1B) and, in cell streams, at the front of the leading cell and the engulfing mouth of the following cell (Fig. 1C). Both DMIB and PIP2/PIP3 were also occasionally present at the back of the cells but in concentrations much lower than at the front (Fig. 1, B and C). In most cases, F-actin also localized with DMIB and PIP2/PIP3 (Fig. 1, D and E). The localization of DMIB at the front of motile cells was expected, but the dominant localization of PIP2/PIP3 at the front, and only occasional weak staining at the rear (Fig. 1C), seems to be inconsistent with the known localization of PI(4,5)P2 at the rear and PI(3,4,5)P3 at the front of chemotaxing cells (13, 14, 32). This inconsistency might be explained by the possible presence of PI(3,4)P2, which the antibody recognizes strongly (supplemental Fig. S1) and which is a product of PI(3,4,5)P3 dephosphorylation, at the cell front.
FIGURE 1.
Colocalization of endogenous DMIB and PIP2/PIP3 in fixed AX3 cells. AX3 cells were fixed and endogenous DMIB, and PIP2/PIP3 were visualized with antibodies against DMIB (red) and PIP2/PIP3 (green). F-actin was stained with phalloidin Alexa Fluor 633 (blue). In nonpolarized cells, DMIB colocalizes with PIP2/PIP3 in random cell protrusions (A). In polarized cells, DMIB and PIP2/PIP3 colocalize at the cell front (B). In chemotaxing cells, DMIB and PIP2/PIP3 colocalize at the front of the leading cell and in the engulfing mouth of the following cell (C). Actin colocalizes with DMIB and PIP2/PIP3 at random cell protrusions (D) and at the front of polarized cells (E). Arrows mark the sites of colocalization of DMIB with PIP2/PIP3 (A–C) and with F-actin (D and E). Scale bars, 10 μm. DIC, differential interference control microscopy.
Localization of Expressed DMIB
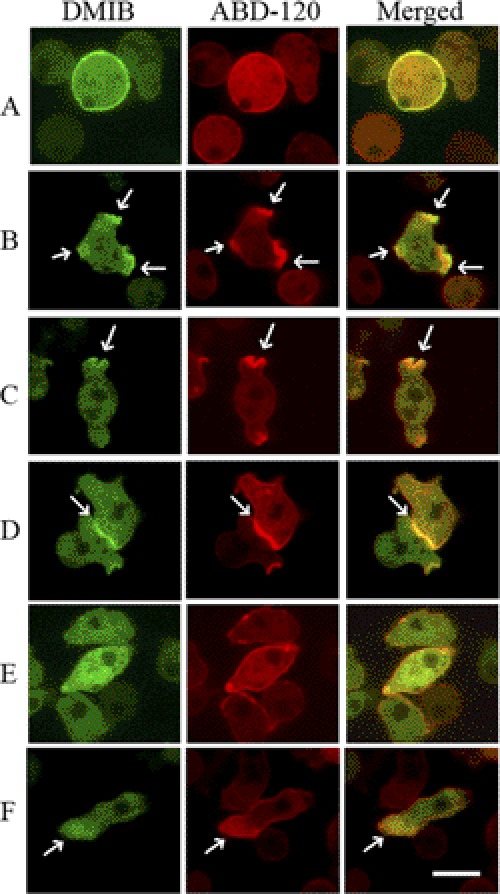
DMIB and ABD-120 (to monitor F-actin localization) (26) fused to GFP and mRFP, respectively, were coexpressed in wild-type AX3 cells and their localizations followed during the morphological changes induced by starvation. Cells freshly plated in nonnutrient medium (see “Experimental Procedures”) have low motility, and DMIB initially localized uniformly to the plasma membrane in these cells (Fig. 2A). After about 0.5–2 h, when random cell movement increased, DMIB became enriched in pseudopods (Fig. 2B), cups (Fig. 2C), and random cell-cell contacts (Fig. 2D and supplemental Movie S1). The protruding myosin-enriched regions were usually also enriched in F-actin (Fig. 2, B–D). Upon longer starvation (4–5 h), when the cell cortex was not very active and cells form fewer pseudopods and cups, DMIB became diffused predominantly in the cytoplasm (Fig. 2E) whereas F-actin remained mostly cortical (Fig. 2E). Finally, after 6–8 h starvation, when cells became highly polarized and elongated (Fig. 2F), DMIB was localized at the cell front. The localization of expressed DMIB on the plasma membrane, in cell protrusions, at cell-cell contacts, and at the front of elongated polarized live cells agrees with the localization of endogenous DMIB in fixed cells, as shown in Fig. 1 and reported by others (17–19).
FIGURE 2.
Localization of expressed GFP-DMIB in live cells. AX3 cells were cotransfected with DMIB and ABD-120 fused to GFP (green) and RFP (red), respectively. Live cells images are shown. DMIB is localized uniformly on the plasma membrane of freshly plated cells (A), in pseudopods (B), cups (C), and at cell-cell contacts (D) of randomly moving cells. DMIB is mostly diffuse in cells starved for about 4 h (E) and is localized to the cell front in elongated polarized cells (F). In all cases except E, DMIB colocalizes with F-actin. Arrows mark sites of DMIB and F-actin colocalization. Scale bar, 10 μm. See also supplemental Movie S1.
DMIB Mutants
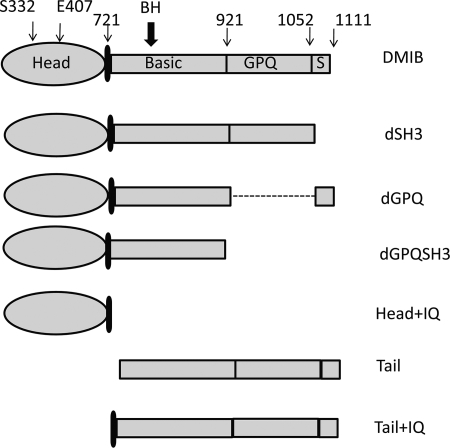
The roles of the major regions of DMIB (Fig. 3) in determining the localization of DMIB at different stages of cell motility were investigated. Previous studies have established that the SH3 region (S in Fig. 3) binds CARMIL, a scaffolding protein that also binds G-actin, actin-capping protein, and Arp2/3 (33). The GPQ region contains an ATP-insensitive actin binding site (34); the basic region contains the BH site (801KKKVLVHTLIRR812), which binds acidic phospholipids in vitro (10); the IQ region provides the light chain binding site and, like the heads of all myosins, Head (the motor domain in Fig. 3) has an ATP-sensitive actin binding site and actin-activated ATPase and motor activities (4).
FIGURE 3.
Schematic representation of DMIB and its mutants expressed in Dictyostelium cells. The major regions that were deleted are labeled (S = SH3). The positions of residues mutated within the head and the position of BH site are indicated. The point mutations in the head in full-length DMIB were: S332A (DMIB-S332A), which results in motor-dead myosin, and E407K (DMIB-E407K), which results in severely reduced binding to F-actin. Mutations of the BH sites in full-length DMIB, dGPQSH3, and the tail were: deletion of entire BH site (dBH), substitution of 5 basic residues within the BH site with Ala (BH-Ala), and point mutation I810D. See supplemental Fig. S2 for more detailed description of DMIB-I810D.
The following DMIB large deletion mutants were studied: mutant dSH3 had the C-terminal SH3 domain deleted, dGPQ had the GPQ region deleted, dGPQSH3 lacked both GPQ and SH3 retaining only the basic region of the tail, mutant Head+IQ had the entire tail deleted, Tail had the head and IQ domains deleted, Tail+IQ lacked the head domain, and Tail lacked the head domain and IQ region (Fig. 3).
The role of the recently described BH site in DMIB localization in vivo was investigated by expressing mutants in which the BH site was deleted (dBH) or its five basic residues were replaced by Ala (BH-Ala) in DMIB, dGPQSH3, and Tail. From our earlier data on the binding of peptides to phospholipid vesicles (9, 10) and from BH-plots (supplemental Fig. S2A), all of these mutations in the BH site would be expected to reduce binding to acidic lipids drastically. In addition, and to test further the relevance of BH-search and properties of the BH site, the hydrophobic residue Ile-810 within the BH site of DMIB was replaced with acidic Asp. This mutation was not tested previously with synthetic peptides, but, according to the prediction of the BH-plot (supplemental Fig. S2A), it should greatly reduce lipid binding by DMIB.
Also, three point mutations were made in the DMIB head: one to inactivate its actin-activated ATPase activity (S322A) and two others to perturb its binding to F-actin (E407K and N154A). Actin-activated ATPase activity of ameboid myosins requires phosphorylation of a serine or threonine residue in the middle part of the head domain, the TEDS site (35–37); the S332A mutation of DMIB results in essentially complete loss of its actin-activated ATPase activity (38).
The E407K mutation of DMIB is homologous to the E476K mutation of Dictyostelium myosin II, which was shown to bind but not hydrolyze ATP, thus substantially reducing binding of Dictyostelium myosin II to F-actin in the presence of ATP (39, 40). The homologous mutations of mammalian myosin X (E456K) (41), Aspergillus myosin I (E444K) (42), and mammalian MIB (E409K) (43) have similar effects.
The DMIB N154A mutation is homologous to the (N233A) mutation of Dictyostelium myosin II, which makes the myosin unable to bind nucleotides, and, therefore, the myosin forms a stable, rigor complex with F-actin (44). The homologous mutation of mammalian MIB (N160A) has a similar effect on its interaction with F-actin (43).
Based on the homology of the ATP binding site of all myosins, and the results with other myosin Is, we expect the E407K mutation to severely weaken and the N154A mutation to greatly strengthen binding of DMIB to F-actin. All of the constructs, with GFP added to their N termini, were expressed in Dictyostelium AX2 myoB− cells (20) to avoid competition with endogenous DMIB.
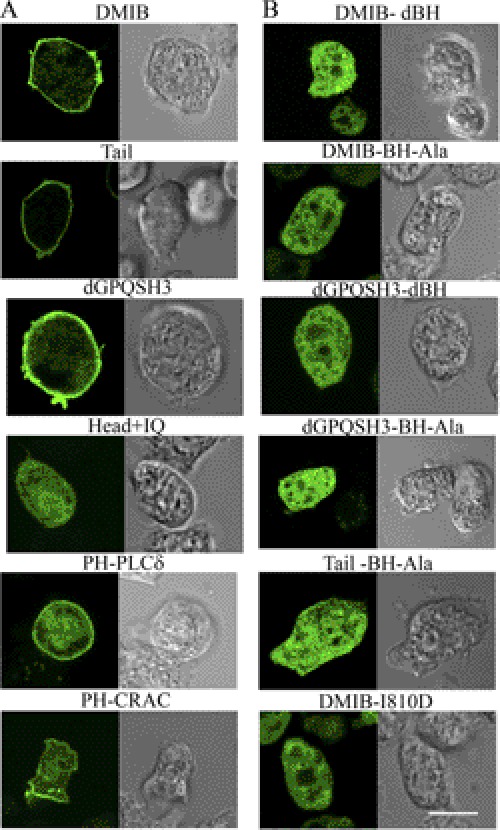
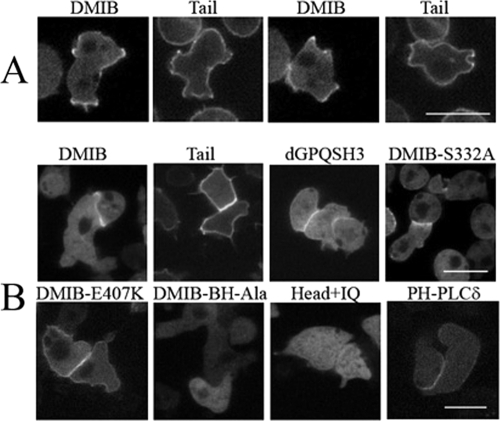
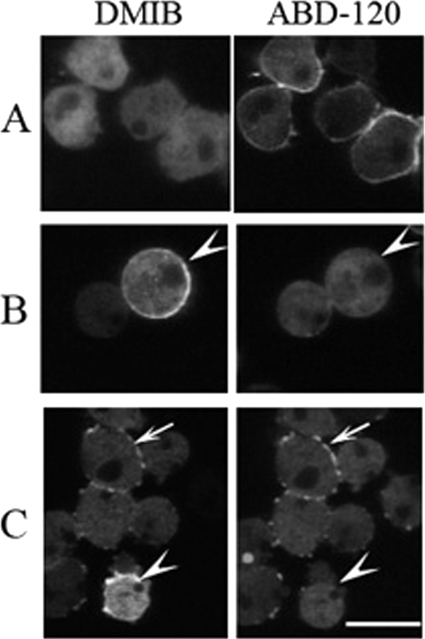
Tail BH Site Is Necessary for Uniform Localization of DMIB to Plasma Membrane
We had shown earlier that the BH site of DMIB is sufficient for binding to vesicles containing acidic phospholipids in vitro (10). Here, we investigate the importance of the BH site in vivo by expressing the DMIB BH mutants described above. In cells freshly plated in nonnutrient buffer, DMIB, Tail, and dGPQSH3 localized uniformly on the plasma membrane whereas the Head+IQ, lacking the C-terminal tail, was exclusively cytoplasmic (Fig. 4A), demonstrating that the basic region is required and sufficient for plasma membrane localization at this stage. Consistent with these results, expressed DMIB-S332A, DMIB-E407K, Tail+IQ, dGPQ, and dSH3 also localized to the plasma membrane (data not shown). In addition to sharp peripheral fluorescence, there was some diffuse cytoplasmic fluorescence, more in cells expressing DMIB than in those expressing Tail (Fig. 4A), consistent with earlier fractionation experiments showing that a significant amount of DMIB is cytoplasmic (45). The PH domains of PLCδ and CRAC were also uniformly enriched in the plasma membrane of freshly plated cells (Fig. 4A), indicative of uniform distributions of PI(4,5)P2 and PI(3,4,5)P3 (46), respectively, in the plasma membrane. Deletion of the BH site (dBH) or replacement of its five hydrophobic amino acids with Ala (BH-Ala) resulted in the loss of plasma membrane localization and diffuse cytoplasmic staining of DMIB, Tail, and dGPQSH3 (Fig. 4B). A point mutation of DMIB within the BH site, DMIB-I810D (Fig. 4B), also resulted in cytoplasmic localization as predicted by the BH-plot (supplemental Fig. S2B).
FIGURE 4.
BH site is necessary for localization of DMIB to the plasma membrane. Constructs are identified as described in Fig. 3 and “Results.” A, localization of GFP-labeled DMIB, DMIB deletion constructs, PH-PLCδ, and PH-CRAC in AX2 myoB− cells newly plated in nonnutrient buffer. B, localization of mutants with BH site deleted (dBH) or mutated (BH-Ala, I810D). Left panels in A and B show GFP images, and right panels show differential interference contrast microscopy images. Scale bar, 10 μm.
These data show that binding of DMIB to the plasma membrane of vegetative cells newly plated in nonnutrient buffer requires the BH site within the basic region of the tail. Neither the head nor IQ domains, nor the GPQ and SH3 domains in the tail are significantly involved in binding of DMIB to the plasma membrane. These conclusions are consistent with earlier in vitro data for DMIB and AMIC (9, 10) and with the uniform distribution of PIP2 and PIP3 in the plasma membrane of cells at this stage.
Head Is Required for Relocalization of DMIB in Motile Cells
As shown in Fig. 2 and supplemental Movie S1, in randomly moving cells (starved about 2 h) expressed DMIB relocalized to active cell protrusions and sites of random cell-cell contacts. In the first stage of this process, the cell began to lose its round shape, and the uniform plasma membrane distribution of DMIB was lost. In cells undergoing similar shape changes, DMIB located to protrusions and was not present in other regions of the plasma membrane whereas Tail remained uniformly distributed on the entire plasma membrane (Fig. 5A and supplemental Movies S2 and S3). Similarly, in cells forming random cell-cell contacts, DMIB was enriched at the contact sites, and Tail was uniformly distributed on the plasma membrane with only moderate enrichment at contact sites (Fig. 5B). DMIB-BH-Ala and Head+IQ remained exclusively in the cytoplasm (Fig. 5B). Contact sites were enriched in PI(4,5)P2 as monitored by PH-PLCδ (Fig. 5B).
FIGURE 5.
DMIB head is required for dissociation of DMIB from the plasma membrane, and BH site is required for DMIB localization to cell-cell contacts in randomly moving cells. A, within about 2 h, DMIB (supplemental Movie S2) relocates to cell protrusions whereas Tail remains uniformly concentrated on the plasma membrane (supplemental Movie S3). B, DMIB, dGPQSH3 (supplemental Movie S4), DMIB-S332A, and DMIB-E407K are enriched at random cell-cell contacts, where PIP2 (PH-PLCδ) also concentrates, and are absent from other regions of the plasma membrane. Tail remains relatively uniformly associated with the plasma membrane and DMIB-BH-Ala, and Head+IQ remains diffused in the cytoplasm. Scale bars, 10 μm.
Thus, DMIB head is required for release of DMIB from its uniform distribution on the plasma membrane, and the BH site is required for relocalization of DMIB to protrusions and cell-cell contacts, probably because of the high local charge density from PIP2 at those sites. Neither motor activity, actin binding through the head, nor the GPQ and SH3 domains are required for localization of DMIB to cell-cell contacts because DMIB-S322A, DMIB-E407K, and dGPQSH3 located to cell-cell contacts similarly to DMIB (Fig. 5B and supplemental Movie S4).
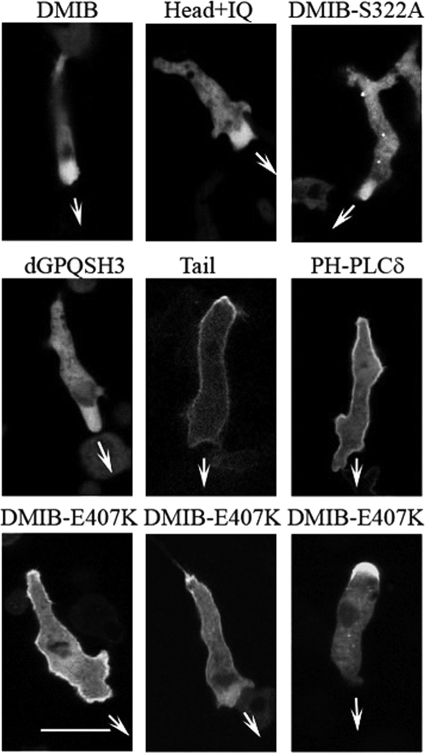
Upon prolonged starvation (about 8 h), Dictyostelium amoebae elongate, polarize, and start moving directionally, eventually forming head-to-tail cell streams. In elongated, directionally moving cells, expressed DMIB was transiently and diffusely localized predominantly at the cell front (Fig. 6), as was endogenous DMIB (Fig. 1), with the occasional presence of DMIB at the rear (Fig. 6). This diffuse localization at the front of individually migrating cells required only the motor domain, Head+IQ exhibited this diffuse localization (Fig. 6 and supplemental Movie S5), but motor activity was not required as motor-dead DMIB-S332A also localized diffusely at the front (Fig. 6). In agreement with this conclusion, dGPQSH3 (Fig. 6) as well as a mutants with compromised BH sites (dGPQSH3-dBH and DMIB-BH-Ala) also localized diffusely at the cell front (data not shown). On the other hand, Tail remained tightly associated with the entire plasma membrane of elongated cells with some transient enrichment at the rear and no detectable diffuse enrichment at the front (Fig. 6 and supplemental Movie S6). PI(4,5)P2 was located similarly to Tail along the entire plasma membrane and transiently enriched at the rear, as determined by the localization of PH-PLCδ (Fig. 6 and supplemental Movie S7).
FIGURE 6.
DMIB head is required for relocation of DMIB to front of polarized cells. Images of live starved elongated cells expressing proteins as marked at the top of the panels are shown. DMIB and Head+IQ show the same diffuse localization at the front as does motor-dead DMIB mutant (DMIB-S332A) and DMIB minus GPQ and SH3 domains (dGPQSH3). Tail and PIP2 (as monitored with PH-PLCδ) localize mostly uniformly on the plasma membrane with enrichment at the rear. DMIB point mutant with weakened actin binding (DMIB-E407K) localizes on plasma membrane with strong enrichment at the rear but shows stronger cytoplasmic presence than does Tail. Arrows mark the direction of cell movement. Scale bar, 10 μm. See also supplemental Movies S5–S7.
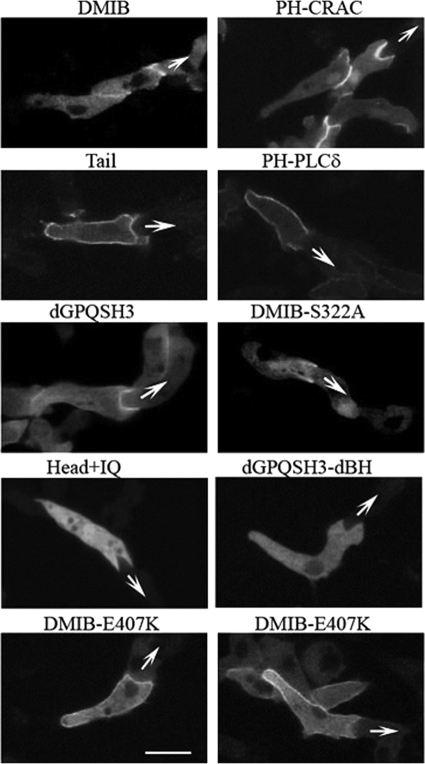
In streaming cells, whose fronts came into contact with the rears of other cells, DMIB localized much more sharply, but transiently, at the engulfing mouth (Fig. 7) as did also PI(3,4,5)P3, as monitored by PH-CRAC (Fig. 7). The transient localization of PIP3 (47) was probably responsible for the transient localization of DMIB. Tail, however, remained mostly uniformly localized on the plasma membrane with some enrichment at the rear but no significant enrichment at the front (Fig. 7 and supplemental Movie S8), as did PIP2 (PH-PLCδ, Fig. 7 and supplemental Movie S9). Thus, proper localization to the front of the cell at this stage requires the presence of the myosin head. Myosin lacking the GPQ and SH3 domains (dGPQSH3, supplemental Movie S10) and myosin with greatly reduced actin-activated Mg-ATPase activity (DMIB-S332A) also localized sharply to the engulfing mouth (although DMIB-S332A more weakly), indicating that the GPQ and SH3 domains and motor activity are not essential for this localization (Fig. 7). However, the BH site is essential because neither Head+IQ nor dGPQSH3dBH localized sharply to the engulfing mouth (Fig. 7 and supplemental Movie S11).
FIGURE 7.
Localization of DMIB in streaming cells. Images of live streaming cells expressing proteins as marked at the top of the panels are shown. Expressed DMIB colocalizes with PIP3 (PH-CRAC) at the engulfing mouth of cells in chemotaxing streams as does motor-dead DMIB-S332A and DMIB missing the GPQ and SH3 domains (dGPQSH3). Head+IQ and mutant missing BH site (dGPQSHdBH) are fully cytoplasmic. Expressed Tail and PIP2 (PH-PLCδ) remain mostly uniformly distributed on the plasma membrane. DMIB point mutant with weakened actin binding (DMIB-E470K) also localizes primarily to the plasma membrane but has a higher cytoplasmic component than does Tail. See also supplemental Movies S8–S11.
Binding of Head to Cytoplasmic F-actin Is Important for Localization of DMIB
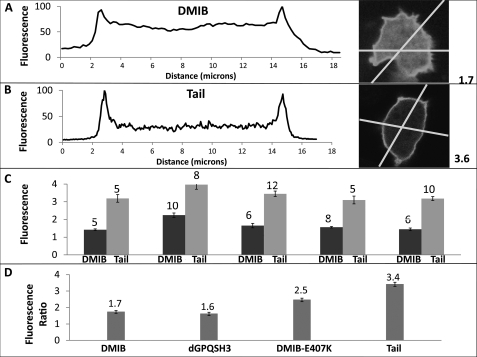
Although both expressed DMIB and expressed Tail were mostly uniformly distributed on the plasma membrane of freshly plated, nonmotile cells (Fig. 4A), there seemed to be more DMIB than Tail in the cytoplasm. We quantified this difference by line-scanning fluorescent images of cells transfected with either DMIB or Tail and processed in parallel (Fig. 8, A and B). The ratio of the peak fluorescence intensity on the plasma membrane to the average fluorescence intensity in the cytoplasm was consistently lower for DMIB than for Tail (Fig. 8C) with an overall average of 1.7 for DMIB and 3.4 for Tail (Fig. 8D). The ratio value for DMIB-E407K (weak actin binding mutant) was in between the values for DMIB and Tail and was 2.5. The ratio for dGPQSH3 was essentially the same as for DMIB and equal to 1.6 (Fig. 8D).
FIGURE 8.
Comparison of cytoplasmic and membrane-associated fractions of DMIB, Tail, DMIB-N154A, and dGPQSH3. A and B, fluorescence images of freshly plated Dictyostelium cells expressing DMIB or Tail were scanned. Examples of individual linear cross-scans for DMIB (A) and Tail (B) are shown. Each cell was scanned twice across two different lines. Scans were normalized for each cell taking the average maximum fluorescence intensity of membrane peaks as 100%. The ratios of the maximum fluorescence intensity on the plasma membrane to the average fluorescence intensity in the cytoplasm for single cells shown in A and B were 1.7 and 3.6, respectively, as indicated at the sides of the panels. C, ratio of the maximum fluorescence intensity on the plasma membrane to the average fluorescence intensity in the cytoplasm in five experiments. Each pair of bars represents a separate experiment, and the number of cells scanned in each experiment is indicated on the top of bars. D, average of fluorescence ratios for DMIB, dGPQSH3, DMIB-E407K, and Tail. The average fluorescence ratios are indicated at the tops of the bars. The number of scanned cells and independent experiments were as follow: 35 cells from 5 experiments for DMIB, 22 cells from 3 experiments for dGPQSH3, 30 cells from 3 experiments for DMIB-E407K, and 40 cells from 5 experiments for Tail. Error bars, S.E.
These results suggest that the head reduces DMIB association with the plasma membrane, presumably by binding to some cytoplasmic protein, and the fluorescence ratio for DMIB-E407K suggests that this protein may be actin. Actin is the most abundant cytoplasmic protein known to bind to the myosin head. In addition to highly concentrated cortical actin, which is easily visualized by fluorescence microscopy of cells stained with rhodamine phalloidin or other probes, Dictyostelium has a pool of cytoplasmic F-actin (48) that is only diffusely stained (individual actin filaments are below the resolution of fluorescence microscopy, and Dictyostelium does not have stress fibers).
The localization of DMIB with reduced affinity for F-actin, DMIB-E407K, was intermediate between the localization of DMIB and Tail. In freshly plated cells, DMIB-E407K localized uniformly to the plasma membrane from which it mostly dissociated in time (data not shown), which was more like DMIB than like Tail. In randomly moving cells, DMIB-E407K localized to cell-cell contacts (Fig. 5B), again more like DMIB than like Tail. However, in elongated cells, a substantial fraction of DMIB-E407K was associated with the plasma membrane, like Tail, with only weak, transient diffuse presence in the cytoplasm at the front of the cell (Fig. 6) where DMIB strongly localized with F-actin (Figs. 2 and 6). Some of the cells that did not show uniform localization of DMIB-E407K on the plasma membrane showed strong accumulation in the cytoplasm at the rear of the cell (Fig. 6). In streaming cells, DMIB-E407K localized mostly uniformly on the plasma membrane with some enrichment at the rear and front (Fig. 7). Thus, DMIB-E407K behaved more like Tail than like DMIB, remaining mostly associated with the plasma membrane, but with a higher fraction than Tail in the cytoplasm. These results are consistent with binding of DMIB to cytoplasmic F-actin via its motor domain being an important, but not the only, factor responsible for the cytoplasmic localization of DMIB.
The localization of DMIB-N154A (the strong actin-binding mutant) was also different from the localization of DMIB. However, cells expressing DMIB-N154A could not be compared directly with cells expressing other DMIB constructs because expression of DMIB-N154A changed the cell morphology, even in freshly plated cells (Fig. 9). The most striking feature of cells expressing DMIB-N154A was the strong presence of wave-like structures at the cell periphery and within the cell body. These waves often had a patched appearance and moved through the cell (Fig. 9, Ab and B and supplemental Movies S12 and S13) in a manner reminiscent of actin waves described previously under different circumstances (49–51). The waves contained DMIB-N154A, which was present only at the bottom of the wave, and F-actin, which was also present in higher focal planes. The region inside the wave was depleted of F-actin (supplemental Fig. S3). These features are also characteristics of actin waves described by others (49, 52). Note that actin waves sequester a large portion of cytoplasmic F-actin.
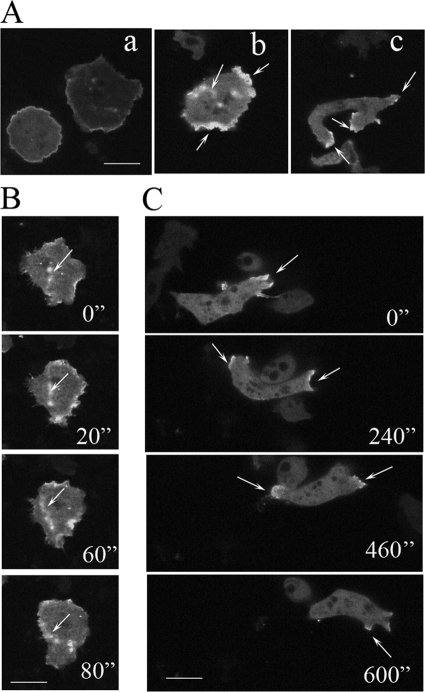
FIGURE 9.
Localization of DMIB-N154A. Aa, freshly plated cells. Ab, cells starved for 2 h. Ac, cells starved for 8 h. B, more detail of DMIB N154A distribution in cells represented by cell in Ab. C, more detail of DMIB N154A distribution in cells represented by cell in Ac. Arrows point to the sites of DMIB N154A location. Scale bars, 10 μm.
Cells with waves were present in freshly plated cells and increased after 4–6 h of starvation (the time when DMIB was mostly diffuse). The waves were transient and not uniform even in freshly plated cells, most likely representing a mixture of prewave, wave, and postwave stages. In cells with strong waves, DMIB-N154A was associated with the waves and not with the plasma membrane. The cortical staining of the cell shown in Fig. 9Ab is diffuse staining associated with an actin wave that reached the cell periphery (supplemental Movie S13) and not plasma membrane-associated staining.
In freshly plated cells that did not show waves or had only weak waves, DMIB-N154A was located on the plasma membrane (Fig. 9Aa), as would be expected for a DMIB mutant with an intact BH site region, and the borders of these cells often contained multiple small protrusions. In elongated, migrating cells, DMIB-N154A was present at both the front and back, moving between these two locations (Fig. 9C). DMIB-N154A cells differed from DMIB-transfected cells. The former tended to have long, sideways protrusions (Fig. 9Ac) and a forked leading edge (Fig. 9C, 0′' and 600”) with DMIB-N154A in the protrusions and the forked extensions.
Thus, both the E407K and N154A mutations change the localization of DMIB but in different ways, as expected because the E407K mutation should greatly reduce and the N154K mutation greatly increase binding of DMIB to F-actin. These results are consistent with the interaction of the DMIB head with F-actin being crucial for the proper localization of DMIB.
Competition between Binding of DMIB to Plasma Membrane and Cytoplasmic F-actin
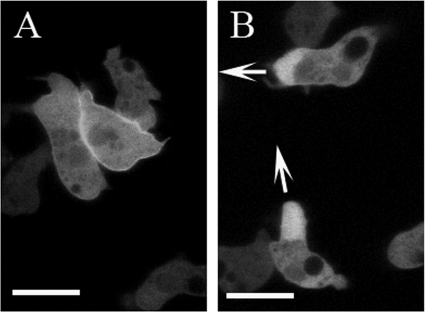
We investigated further the probability of DMIB localization being affected by its binding to cytoplasmic F-actin by treating cells with latrunculin A (LatA). To monitor F-actin and DMIB simultaneously we used AX3 cells cotransfected with ABD-120 and DMIB. We chose cells at the stage when plasma membrane association of DMIB was low, i.e. cells that were starved for 4 h but had not yet elongated. At this stage, cortical actin was easily detectable but DMIB is mostly diffuse in the cytoplasm (Figs. 2E and 10A).
FIGURE 10.
Relocation of DMIB to plasma membrane in cells treated with LatA. AX3 cells cotransfected with DMIB and F-actin probe ABD-120 were starved for 4 h and treated with 7.5 μm LatA. A, in cells before treatment DMIB is mostly diffused and cortical actin is present. B and C, after 20-min exposure to LatA, cortical F-actin is absent, and DMIB reappears on the plasma membrane, alone (arrowheads) or accompanied by F-actin patches (arrows). Scale bar, 10 μm.
LatA treatment causes rapid disappearance of cortical F-actin; by 5 min all cortical F-actin was essentially gone (data not shown and Ref. 53). At that time, DMIB began to appear slowly at the plasma membrane, peaking at about 20 min (Fig. 10, B and C). Images of randomly chosen nontreated, control cells (324 cells) and cells treated with LatA (440 cells) were visually scored for the presence of DMIB at the plasma membrane. The percentage of cells with DMIB on the plasma membrane increased from 7% for nontreated cells to 54% for LatA-treated cells.
However, by 20 min F-actin reappeared on the plasma membrane in the form of patches. The appearance of actin patches, despite the presence of LatA, is not well understood but has been routinely observed by us and others (e.g. Fig. 4 in Ref. 53). Approximately 33% of the LatA-treated cells that had DMIB localized on the plasma membrane did not have any actin patches (Fig. 9B) and, in the 66% of cells with F-actin patches, DMIB localized with the F-actin (Fig. 10C).
We do not know why DMIB did not associate with cortical F-actin prior to LatA treatment but seemed to associate with F-actin patches after cells were exposed to LatA. Nevertheless, the appearance of DMIB on the plasma membrane after LatA treatment, especially in the absence of actin patches, agrees with the assumption that depolymerization of cytoplasmic F-actin should allow F-actin-associated DMIB to bind to acidic phospholipids, most likely PS, in the plasma membrane.
To explore further the roles that F-actin and PI(3,4,5)P3 play in relocalization of DMIB to the front of migrating cells, we expressed dGPQSH3 in PI3K1–5−,PTEN− cells (25) (Fig. 11). Our previous results were consistent with diffuse localization of DMIB at the front of individually migrating cells being due to association with F-actin whereas sharp localization at the mouth of cells in front-back contact reflects DMIB association with PIP3 (Figs. 6 and 7). PI3K1–5−,PTEN− cells lack the five phosphoinositide 3-kinases responsible for conversion of PIP(4,5)P2 to PIP(3,4,5)P3, and PTEN, which catalyzes the conversion of PIP(3,4,5)P3 to PIP(4,5)P2. These cells are not depleted of PI(4,5)P2 but cannot form a PI(3,4,5)P3 gradient in response to cAMP (25). However PI3K1–5−,PTEN− cells can still elongate and chemotax, and F-actin becomes polarized in response to cAMP (25). If DMIB relocates to the front of these cells it cannot be PIP3-driven. We used dGPQSH3 in this experiment to eliminate any possible contribution of the ATP-independent actin binding site in the GPQ domain and the CARMIL binding site in the SH3 domain to the localization of DMIB.
FIGURE 11.
Localization of dGPQSH3 in PI3K1–5−,PTEN− cells. Freshly plated cells (A) and cells starved for 6 h (B) are shown. Arrows indicate the direction of cell movement. Scale bars, 10 μm.
Under our conditions, PI3K1–5−,PTEN− cells did not form streams but were capable of some elongation and directional movement for short periods of time. dGPQSH3 localized diffusely at the front of migrating cells (Fig. 11B). This observation agrees with the proposal that F-actin, not PI(3,4,5)P3, is primarily responsible for the diffuse localization of DMIB at the front of elongated cells. In freshly plated PI3K1–5−,PTEN− cells, dGPQSH3 localized at the plasma membrane (Fig. 11A), most likely bound to PIP2.
DISCUSSION
Myosins I are required for the proper formation and function of both pseudopodia and endocytic structures in amoeboid cells (15, 16, 27–29). These functions require precise control of the recruitment of myosin I to the plasma membrane (54). We have demonstrated that the dynamic localization of DMIB requires both plasma membrane targeting by its tail and F-actin targeting by its head domain.
Furthermore, we have shown unequivocally that the BH site is required in vivo for association of DMIB with the plasma membrane in resting, randomly motile, and chemotaxing Dictyostelium amoebae. This includes uniform localization at the plasma membrane in freshly plated cells, localization at the sites of cell-cell contacts of randomly moving cells, and at the engulfing mouths of chemotaxing cells. Endogenous and expressed wild type-DMIB localized to these regions, but mutants with a deleted or nonfunctional BH site did not. The regions enriched in DMIB were also enriched in either or both PI(4,5)P2 and PI(3,4,5)P3, as determined by the localization of the PH domains of PLCδ and CRAC, respectively. Both PIP2 and PIP3 were dispersed uniformly on the plasma membrane of freshly plated cells, cell-cell contacts were enriched in PIP2, and PIP3 was present at the engulfing mouths of streaming cells. These observations agree with those reported by others (23, 46, 47, 55–57; for reviews see Refs. 13, 32).
However, although required for plasma membrane association, the BH site alone is not sufficient for proper relocalization of DMIB during random cell movement and starvation-induced cell polarization because localization of tail alone was different from the localization of full-length myosin. In freshly plated cells, both DMIB and Tail localized uniformly to the plasma membrane, but the fraction of DMIB in the cytoplasm was higher than for Tail. In randomly moving cells, both DMIB and Tail were enriched in protrusions and cell-cell contacts, but Tail was also present at the remaining regions of the plasma membrane from which DMIB was absent. The most striking difference between DMIB and Tail was that upon starvation DMIB relocated to the front of elongated cells whereas Tail remained relatively evenly distributed on the entire plasma membrane with some enrichment at the rear.
The differences between Tail and DMIB localization could be explained by the presence of a cytoplasmic factor that interacts with the myosin head, pulling myosin off the membrane into the cytoplasm. Such a factor was proposed earlier based on fractionation studies (45, 58) but never identified. Now we have shown that the cytoplasmic factor(s) that keeps full-length DMIB off the membrane involves F-actin.
We have shown that disturbing DMIB binding to F-actin, by two different mutations, changes the localization of DMIB. The localization of DMIB-E407K, in which binding to F-actin through the head is greatly weakened, was intermediate between the localization of DMIB and the localization of Tail. DMIB-E407K lingered on the plasma membrane and at the rear of migrating cells. In freshly plated cells, the cytoplasmic fraction of DMIB-E407K was lower than for DMIB whereas the cytoplasmic fraction of the dGPQSH3 truncation mutant was very similar to that of DMIB.
The localization of the strong actin-binding mutant DMIB-N154A was different from the localization of DMIB and DMIB-E407K. DMIB-N154A was present at both rear and front of elongated cells, but the morphology of the cells was also changed. The most striking difference was the strong presence of DMIB-N154A in actin waves. DMIB is a known component of actin waves (49), but it associates with wave by a treadmilling mechanism, and under our conditions we did not detect expressed DMIB or any other DMIB mutants in actin waves. The strong presence of DMIB-N154A in actin waves most likely reflected its high affinity for F-actin. It is also likely that DMIB-N154A enhances formation of actin waves.
In summary, localization of all mutants with either a deleted or mutated BH site in the tail or a deleted or mutated actin binding site in the head was different from the localization of DMIB. Therefore, at least in all situations that we have studied, the two main factors that determine the localization of DMIB are binding to the plasma membrane through the BH site in the tail and binding to cytoplasmic F-actin through the ATP-sensitive actin binding site in the head. Competition between these two major interactions can explain the dynamic localization of DMIB.
In freshly plated cells, expressed DMIB and Tail both localize uniformly on the plasma membrane bound by the BH site in the tail to acidic phospholipids PI(4,5)P2, PI(3,4,5)P3, and PS in the plasma membrane. Brief starvation causes PIP2 and PIP3 to disappear from the plasma membrane, thus weakening binding of the BH site, and binding of the DMIB head to F-actin relocates DMIB from the membrane to the cytoplasm. Then, in randomly moving cells, DMIB moves from the cytoplasm to new PIP2/PIP3-enriched sites in protrusions and cell-cell contacts. Absent the Head actin binding sites, expressed Tail remains on the plasma membrane in the absence of PIP2 and PIP3 bound to PS and subsequently is only moderately enriched in regions containing PI(4,5)P2 at cell-cell contacts.
But why would DMIB bind to PIP3 at the front of the cell preferentially to PIP2 at the rear? Perhaps DMIB is present at high concentration in the cytoplasm at the front of the cell in association with highly concentrated F-actin, as shown by the intense diffuse fluorescence at the front of chemotaxing cells observed with DMIB and Head but not with Tail (Fig. 6). The diffuse presence of dGPQSH3 at the front of PI3K1–5−,PTEN−-cells that lack PI(3,4,5)P3 gradients supports this explanation (Fig. 11).
And why would expressed Tail be enriched in the plasma membrane at the rear of chemotaxing cells? In most cells, the concentration of PI(4,5)P2 is higher than that of PI(3,4,5)P3 (59). Possibly the concentration of PI(4,5)P2 at the rear of the cell is higher than the concentration of PI(3,4,5)P3 at the front.
Cells that had been starved for 4 h had well defined cortical F-actin but no associated DMIB (Fig. 2E) whereas chemotaxing cells had well defined cortical F-actin along their sides (Fig. 2F) with no enrichment of DMIB in the cortex. On the other hand, both expressed DMIB and Head colocalized with F-actin at the front of chemotaxing cells (Fig. 2F). We do not know why DMIB associates differently with different F-actin pools, but cytoplasmic F-actin, which is not easily visualized by standard fluorescence microscopy (47), may have higher affinity than cortical F-actin for DMIB. It may be relevant that mammalian Myo1B does not associate with stable actin bundles or stress fibers (60) and mammalian Myo1A does not localize to stress fibers (61). Interestingly, myosins other than class I have been shown to have preferences for different regions of the actin cytoskeleton (62).
Localization of expressed dGPQdSH3 and DMIB-S332A was very similar to the localization of DMIB, indicating that neither the GPQ domain, which is required for ATP-independent binding of the tail to F-actin (34), the SH3 domain, which is required for binding to CARMIL (33), nor motor activity is required for the dynamic relocalization of DMIB. This agrees with earlier work showing that although the SH3 domain and motor activity are essential for DMIB function neither is required for its localization to the plasma membrane of vegetative cells or to the leading edge of chemotaxing cells (17). Interactions of DMIB with other proteins through its tail (33, 63) may, however, play a supportive role in DMIB localization and/or be more important in situations not investigated in our current work. For example, DMIB is present in early endosomes (64) and in eupodias/knobby feet (65) which we did not study.
Because motor activity seems not to be essential for DMIB localization what is responsible for its dynamic relocalization? As we discussed above, plasma membrane-bound DMIB is most likely recruited from an adjacent (and at least partially actin-bound) cytoplasmic pool. Relocalization of DMIB to the front of polarized cells may be coupled to relocation of F-actin. For example, DMIB may be translocated in association with actin waves without need for myosin motor activity. The movement of DMIB-N154A with actin waves clearly illustrates such a possibility. DMIB could then be recruited from highly concentrated wave-associated pools by other myosin receptors that the wave contacts. Relocation of highly concentrated regions of DMIB with actin waves has been observed by others, although under conditions different from ours (49).
Other groups (5–7, 11, 60, 61, 63, 66–76) have investigated the factors determining the localization of class I myosins I in various cells with only one consensus requirement. At least part of the basic region in the tail is necessary for localization to biological membranes, binding to acidic phospholipids, and many of the multiple processes involving class I myosins require their binding to membranes (2, 54, 77–82).
The known membrane-association sites in the basic region of myosin-I tails are the PH domain and the BH site. Mutations of conserved residues within PH domains affect the in vivo localization of mammalian Myo1C (5) and Myo1B (67). However, the PH domain is not sufficient for proper localization of mammalian Myo1G (7), and mutation of a conserved residue in the putative PH domain of mammalian Myo1E had no significant effect on its in vivo localization suggesting, together with other data, that this myosin I binds to acid phospholipids through less specific electrostatic interactions (11), as does DMIB. Interestingly a BH plot (10) of Myo1E reveals a well defined BH site in its tail.
In summary, we have shown that the BH site is absolutely required for association of DMIB with the plasma membrane in vivo but that relocalization of DMIB during a starvation cycle requires interaction of DMIB with cytoplasmic F-actin. Our findings on the role of the BH site in DMIB should be applicable to other proteins because other class I myosins, myosin VI and several other cytoskeletal proteins (partially listed in Ref. 10) have similar BH sites. For example a dibasic motif in the tail of myosin XIV is an essential determinant of its plasma membrane localization (83), the basic region of Dictyostelium WASP that binds PIP3 and PIP2 with similar affinities is responsible for WASP localization to the leading edge (84), and, similarly, the basic region of WAVE2 is responsible for its localization to PIP3-enriched lamellipodia in platelets (85). Binding of proteins to membrane lipids by basic-hydrophobic, or just basic, regions seems to be a common phenomenon, and our results show that such sites can play a critical role in membrane localization and relocalization of proteins.
Supplementary Material
Acknowledgments
We thank Dr. Carole A Parent (NCI, National Institutes of Health) for the gift of plasmids encoding PH domains of CRAC and PLCδ, Dr. Goeh Jung (NHLBI, National Institutes of Health) for the gift of Dictyostelium cells cotransfected with DMIB and ABD-120, Dr. Oliver Hoeller and Dr. Robert K. Kay (Cambridge, United Kingdom) for very helpful advice on culturing and transfection of PI3K1–5−,PTEN− cells, Dr. Shunji Senda for help in preparation of plasmids encoding DMIB, and Godefroy Chery for help with cell culturing and data processing.
This work was supported, in whole or in part, by National Institutes of Health Grant GM046486 (to M. A. T.). This work was also supported by the NHLBI Intramural Research Program, National Institutes of Health.

This article contains supplemental Figs. S1–S3, Table S1, and Movies S1–S13.
- SH3
- Src homology 3
- ABD-120
- actin binding domain of actin-binding protein 120
- AMIC
- Acanthamoeba myosin IC
- BH
- basic-hydrophobic
- CRAC
- cytosolic regulator of adenylyl cyclase
- DMIB
- Dictyostelium myosin IB/MYOB
- Lat A
- latrunculin A
- PH
- pleckstrin homology
- PIP2/PIP3
- phosphoinositide bisphosphate/phosphoinositide trisphosphate
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PI(3,5)P2
- phosphatidylinositol 3,5-bisiphosphate
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PLCδ
- phospholipase Cδ
- PS
- phosphatidylserine
- TBS
- Tris-buffered saline.
REFERENCES
- 1. Pollard T. D., Doberstein S. K., Zot H. G. (1991) Myosin I. Annu. Rev. Physiol. 53, 653–681 [DOI] [PubMed] [Google Scholar]
- 2. Kim S. V., Flavell R. A. (2008) Myosin I: from yeast to human. Cell. Mol. Life Sci. 65, 2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coluccio L. (2008) in Myosins: A Superfamily of Molecular Motors (Coluccio L. M., ed) pp. 95–124, Springer, Dordrecht, The Netherlands [Google Scholar]
- 4. de la Roche M. A., Côté G. P. (2001) Regulation of Dictyostelium myosin I and II. Biochim. Biophys. Acta 1525, 245–261 [DOI] [PubMed] [Google Scholar]
- 5. Hokanson D. E., Laakso J. M., Lin T., Sept D., Ostap E. M. (2006) Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol. Biol. Cell 17, 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olety B., Wälte M., Honnert U., Schillers H., Bähler M. (2010) Myosin 1G (Myo1G) is a haematopoietic specific myosin that localises to the plasma membrane and regulates cell elasticity. FEBS Lett. 584, 493–499 [DOI] [PubMed] [Google Scholar]
- 7. Patino-Lopez G., Aravind L., Dong X., Kruhlak M. J., Ostap E. M., Shaw S. (2010) Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH). J. Biol. Chem. 285, 8675–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hwang K. J., Mahmoodian F., Ferretti J. A., Korn E. D., Gruschus J. M. (2007) Intramolecular interaction in the tail of Acanthamoeba myosin IC between the SH3 domain and a putative pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 104, 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brzeska H., Hwang K. J., Korn E. D. (2008) Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J. Biol. Chem. 283, 32014–32023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brzeska H., Guag J., Remmert K., Chacko S., Korn E. D. (2010) An experimentally based computer search identifies unstructured membrane binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J. Biol. Chem. 285, 5738–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feeser E. A., Ignacio C. M., Krendel M., Ostap E. M. (2010) Myo1e binds anionic phospholipids with high affinity. Biochemistry 49, 9353–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weijer C. J. (2009) Collective cell migration in development. J. Cell Sci. 122, 3215–3223 [DOI] [PubMed] [Google Scholar]
- 13. Janetopoulos C., Firtel R. A. (2008) Directional sensing during chemotaxis. FEBS Lett. 582, 2075–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iglesias P. A., Devreotes P. N. (2008) Navigating through models of chemotaxis. Curr. Opin. Cell Biol. 20, 35–40 [DOI] [PubMed] [Google Scholar]
- 15. Wessels D., Murray J., Jung G., Hammer J. A., 3rd, Soll D. R. (1991) Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil. Cytoskeleton 20, 301–315 [DOI] [PubMed] [Google Scholar]
- 16. Jung G., Hammer J. A., 3rd (1990) Generation and characterization of Dictyostelium cells deficient in a myosin I heavy chain isoform. J. Cell Biol. 110, 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Novak K. D., Titus M. A. (1998) The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol. Biol. Cell 9, 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukui Y., Lynch T. J., Brzeska H., Korn E. D. (1989) Myosin I is located at the leading edges of locomoting Dictyostelium amoebae. Nature 341, 328–331 [DOI] [PubMed] [Google Scholar]
- 19. Morita Y. S., Jung G., Hammer J. A., 3rd, Fukui Y. (1996) Localization of Dictyostelium myoB and myoD to filopodia and cell-cell contact sites using isoform-specific antibodies. Eur. J. Cell Biol. 71, 371–379 [PubMed] [Google Scholar]
- 20. Novak K. D., Peterson M. D., Reedy M. C., Titus M. A. (1995) Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J. Cell Biol. 131, 1205–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levi S., Polyakov M., Egelhoff T. T. (2000) Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid 44, 231–238 [DOI] [PubMed] [Google Scholar]
- 22. Patterson B., Spudich J. A. (1995) A novel positive selection for identifying cold-sensitive myosin II mutants in Dictyostelium. Genetics 140, 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Comer F. I., Lippincott C. K., Masbad J. J., Parent C. A. (2005) The PI3K-mediated activation of CRAC independently regulates adenylyl cyclase activation and chemotaxis. Curr. Biol. 15, 134–139 [DOI] [PubMed] [Google Scholar]
- 24. Sussman M. (1987) Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28, 9–29 [DOI] [PubMed] [Google Scholar]
- 25. Hoeller O., Kay R. R. (2007) Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 17, 813–817 [DOI] [PubMed] [Google Scholar]
- 26. Pang K. M., Lee E., Knecht D. A. (1998) Use of a fusion protein between GFP and an actin binding domain to visualize transient filamentous-actin structures. Curr. Biol. 8, 405–408 [DOI] [PubMed] [Google Scholar]
- 27. Novak K. D., Titus M. A. (1997) Myosin I overexpression impairs cell migration. J. Cell Biol. 136, 633–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jung G., Wu X., Hammer J. A., 3rd (1996) Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J. Cell Biol. 133, 305–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Falk D. L., Wessels D., Jenkins L., Pham T., Kuhl S., Titus M. A., Soll D. R. (2003) Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB, and MyoF) in motility and chemotaxis in Dictyostelium. J. Cell Sci. 116, 3985–3999 [DOI] [PubMed] [Google Scholar]
- 30. Brzeska H., Szczepanowska J., Matsumura F., Korn E. D. (2004) Rac-induced increase of phosphorylation of myosin regulatory light chain in HeLa cells. Cell Motil. Cytoskeleton 58, 186–199 [DOI] [PubMed] [Google Scholar]
- 31. Gaudet P., Pilcher K. E., Fey P., Chisholm R. L. (2007) Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 32. Bagorda A., Parent C. A. (2008) Eukaryotic chemotaxis at a glance. J. Cell Sci. 121, 2621–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung G., Remmert K., Wu X., Volosky J. M., Hammer J. A., 3rd (2001) The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153, 1479–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosenfeld S. S., Rener B. (1994) The GPQ-rich segment of Dictyostelium myosin IB contains an actin binding site. Biochemistry 33, 2322–2328 [DOI] [PubMed] [Google Scholar]
- 35. Brzeska H., Lynch T. J., Martin B., Korn E. D. (1989) The localization and sequence of the phosphorylation sites of Acanthamoeba myosins I: an improved method for locating the phosphorylated amino acid. J. Biol. Chem. 264, 19340–19348 [PubMed] [Google Scholar]
- 36. Brzeska H., Korn E. D. (1996) Regulation of class I and class II myosins by heavy chain phosphorylation. J. Biol. Chem. 271, 16983–16986 [DOI] [PubMed] [Google Scholar]
- 37. Bement W. M., Mooseker M. S. (1995) TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil. Cytoskeleton 31, 87–92 [DOI] [PubMed] [Google Scholar]
- 38. Tsiavaliaris G., Fujita-Becker S., Dürrwang U., Diensthuber R. P., Geeves M. A., Manstein D. J. (2008) Mechanism, regulation, and functional properties of Dictyostelium myosin-1B. J. Biol. Chem. 283, 4520–4527 [DOI] [PubMed] [Google Scholar]
- 39. Friedman A. L., Geeves M. A., Manstein D. J., Spudich J. A. (1998) Kinetic characterization of myosin head fragments with long-lived myosin ATP states. Biochemistry 37, 9679–9687 [DOI] [PubMed] [Google Scholar]
- 40. Ruppel K. M., Spudich J. A. (1996) Structure-function studies of the myosin motor domain: importance of the 50-kDa cleft. Mol. Biol. Cell 7, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kerber M. L., Jacobs D. T., Campagnola L., Dunn B. D., Yin T., Sousa A. D., Quintero O. A., Cheney R. E. (2009) A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 19, 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu X., Osherov N., Yamashita R., Brzeska H., Korn E. D., May G. S. (2001) Myosin I mutants with only 1% of wild-type actin-activated MgATPase activity retain essential in vivo function(s). Proc. Natl. Acad. Sci. U.S.A. 98, 9122–9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Almeida C. G., Yamada A., Tenza D., Louvard D., Raposo G., Coudrier E. (2011) Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat. Cell Biol. 13, 779–789 [DOI] [PubMed] [Google Scholar]
- 44. Shimada T., Sasaki N., Ohkura R., Sutoh K. (1997) Alanine scanning mutagenesis of the switch I region in the ATPase site of Dictyostelium discoideum myosin II. Biochemistry 36, 14037–14043 [DOI] [PubMed] [Google Scholar]
- 45. Senda S., Lee S. F., Côté G. P., Titus M. A. (2001) Recruitment of a specific amoeboid myosin I isoform to the plasma membrane in chemotactic Dictyostelium cells. J. Biol. Chem. 276, 2898–2904 [DOI] [PubMed] [Google Scholar]
- 46. Sasaki A. T., Janetopoulos C., Lee S., Charest P. G., Takeda K., Sundheimer L. W., Meili R., Devreotes P. N., Firtel R. A. (2007) G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178, 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dormann D., Weijer G., Parent C. A., Devreotes P. N., Weijer C. J. (2002) Visualizing PI3 kinase-mediated cell-cell signaling during Dictyostelium development. Curr. Biol. 12, 1178–1188 [DOI] [PubMed] [Google Scholar]
- 48. Giffard R. G., Spudich J. A., Spudich A. (1983) Ca2+-sensitive isolation of a cortical actin matrix from Dictyostelium amoebae. J. Muscle Res. Cell Motil. 4, 115–131 [DOI] [PubMed] [Google Scholar]
- 49. Bretschneider T., Anderson K., Ecke M., Müller-Taubenberger A., Schroth-Diez B., Ishikawa-Ankerhold H. C., Gerisch G. (2009) The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys. J. 96, 2888–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vicker M. G. (2002) Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp. Cell Res. 275, 54–66 [DOI] [PubMed] [Google Scholar]
- 51. Schroth-Diez B., Gerwig S., Ecke M., Hegerl R., Diez S., Gerisch G. (2009) Propagating waves separate two states of actin organization in living cells. HFSP J. 3, 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gerisch G., Ecke M., Wischnewski D., Schroth-Diez B. (2011) Different modes of state transitions determine pattern in the phosphatidylinositide-actin system. BMC Cell Biol. 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gerisch G., Bretschneider T., Müller-Taubenberger A., Simmeth E., Ecke M., Diez S., Anderson K. (2004) Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys. J. 87, 3493–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hartman M. A., Finan D., Sivaramakrishnan S., Spudich J. A. (2011) Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 27, 133–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dormann D., Weijer G., Dowler S., Weijer C. J. (2004) In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J. Cell Sci. 117, 6497–6509 [DOI] [PubMed] [Google Scholar]
- 56. Jin T., Zhang N., Long Y., Parent C. A., Devreotes P. N. (2000) Localization of the G protein βγ complex in living cells during chemotaxis. Science 287, 1034–1036 [DOI] [PubMed] [Google Scholar]
- 57. Lee S., Comer F. I., Sasaki A., McLeod I. X., Duong Y., Okumura K., Yates J. R., 3rd, Parent C. A., Firtel R. A. (2005) TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16, 4572–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Senda S., Titus M. A. (2000) A potential mechanism for regulating myosin I binding to membranes in vivo. FEBS Lett. 484, 125–128 [DOI] [PubMed] [Google Scholar]
- 59. Lemmon M. A. (2003) Phosphoinositide recognition domains. Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 60. Tang N., Ostap E. M. (2001) Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 11, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 61. Tyska M. J., Mooseker M. S. (2002) MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys. J. 82, 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brawley C. M., Rock R. S. (2009) Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 106, 9685–9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Krendel M., Osterweil E. K., Mooseker M. S. (2007) Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 581, 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neuhaus E. M., Soldati T. (2000) A myosin I is involved in membrane recycling from early endosomes. J. Cell Biol. 150, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fukui Y., Inoué S. (1997) Amoeboid movement anchored by eupodia, new actin-rich knobby feet in Dictyostelium. Cell Motil. Cytoskeleton 36, 339–354 [DOI] [PubMed] [Google Scholar]
- 66. Yengo C. M., Ananthanarayanan S. K., Brosey C. A., Mao S., Tyska M. J. (2008) Human deafness mutation E385D disrupts the mechanochemical coupling and subcellular targeting of myosin-1a. Biophys. J. 94, L5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Komaba S., Coluccio L. M. (2010) Localization of myosin 1b to actin protrusions requires phosphoinositide binding. J. Biol. Chem. 285, 27686–27693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Benesh A. E., Nambiar R., McConnell R. E., Mao S., Tabb D. L., Tyska M. J. (2010) Differential localization and dynamics of class I myosins in the enterocyte microvillus. Mol. Biol. Cell 21, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kong H. H., Pollard T. D. (2002) Intracellular localization and dynamics of myosin-II and myosin-IC in live Acanthamoeba by transient transfection of EGFP fusion proteins. J. Cell Sci. 115, 4993–5002 [DOI] [PubMed] [Google Scholar]
- 70. Lee W. L., Bezanilla M., Pollard T. D. (2000) Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stöffler H. E., Honnert U., Bauer C. A., Höfer D., Schwarz H., Müller R. T., Drenckhahn D., Bähler M. (1998) Targeting of the myosin-I myr 3 to intercellular adherens type junctions induced by dominant active Cdc42 in HeLa cells. J. Cell Sci. 111, 2779–2788 [DOI] [PubMed] [Google Scholar]
- 72. Ruppert C., Godel J., Müller R. T., Kroschewski R., Reinhard J., Bähler M. (1995) Localization of the rat myosin I molecules myr 1 and myr 2 and in vivo targeting of their tail domains. J. Cell Sci. 108, 3775–3786 [DOI] [PubMed] [Google Scholar]
- 73. Sokac A. M., Schietroma C., Gundersen C. B., Bement W. M. (2006) Myosin-1c couples assembling actin to membranes to drive compensatory endocytosis. Dev. Cell 11, 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yamashita R. A., Osherov N., May G. S. (2000) Localization of wild type and mutant class I myosin proteins in Aspergillus nidulans using GFP fusion proteins. Cell Motil. Cytoskeleton 45, 163–172 [DOI] [PubMed] [Google Scholar]
- 75. Doberstein S. K., Pollard T. D. (1992) Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J. Cell Biol. 117, 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maravillas-Montero J. L., Gillespie P. G., Patiño-López G., Shaw S., Santos-Argumedo L. (2011) Myosin 1c participates in B cell cytoskeleton rearrangements, is recruited to the immunologic synapse, and contributes to antigen presentation. J. Immunol. 187, 3053–3063 [DOI] [PubMed] [Google Scholar]
- 77. Ostap E. M., Pollard T. D. (1996) Overlapping functions of myosin-I isoforms? J. Cell Biol. 133, 221–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Soldati T., Schliwa M. (2006) Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 7, 897–908 [DOI] [PubMed] [Google Scholar]
- 79. Tuxworth R. I., Titus M. A. (2000) Unconventional myosins: anchors in the membrane traffic relay. Traffic 1, 11–18 [DOI] [PubMed] [Google Scholar]
- 80. Krendel M., Mooseker M. S. (2005) Myosins: tails (and heads) of functional diversity. Physiology 20, 239–251 [DOI] [PubMed] [Google Scholar]
- 81. Gillespie P. G., Müller U. (2009) Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell 139, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Titus M. A. (2006) Myosin I and actin dynamics: the frogs weigh in. Dev. Cell 11, 594–595 [DOI] [PubMed] [Google Scholar]
- 83. Hettmann C., Herm A., Geiter A., Frank B., Schwarz E., Soldati T., Soldati D. (2000) A dibasic motif in the tail of a class XIV apicomplexan myosin is an essential determinant of plasma membrane localization. Mol. Biol. Cell 11, 1385–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Myers S. A., Han J. W., Lee Y., Firtel R. A., Chung C. Y. (2005) A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell 16, 2191–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oikawa T., Yamaguchi H., Itoh T., Kato M., Ijuin T., Yamazaki D., Suetsugu S., Takenawa T. (2004) PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 6, 420–426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.