Background: Palmitic acid is a saturated fatty acid known to cause lipotoxicity in cells.
Results: Palmitic acid induces autophagy, which is independent of mTOR regulation.
Conclusion: Palmitic acid-mediated autophagy is regulated via PKC-α and acts as a cell survival mechanism.
Significance: Our data reveal a novel mechanism underlying free fatty acid-mediated autophagy and suggest the importance of autophagy in bioactivity of fatty acids.
Keywords: Apoptosis, Autophagy, Diacylglycerol, mTOR, Protein Kinase C (PKC), Palmitic Acid
Abstract
Lipotoxicity refers to the cytotoxic effects of excess fat accumulation in cells and has been implicated as one of the contributing factors to diseases like obesity, diabetes, and non-alcoholic fatty liver. In this study we sought to examine effects of palmitic acid (PA) and oleic acid, two of the common dietary fatty acids on the autophagic process. We found that PA, but not oleic acid, was able to cause an increase in autophagic flux, evidenced by LC3-II accumulation and formation of GFP-LC3 puncta. Notably, PA-induced autophagy was found to be independent of mTOR regulation. Next, in search of the mechanism mediating PA-induced autophagy, we found increased levels of diacylglycerol species and protein kinase C (PKC) activation in PA-treated cells. More importantly, inhibition of classical PKC isoforms (PKC-α) was able to effectively suppress PA-induced autophagy. Finally, we showed that inhibition of autophagy sensitized the cells to PA-induced apoptosis, suggesting the pro-survival function of autophagy induced by PA. Taken together, results from this study reveal a novel mechanism underlying free fatty acid-mediated autophagy. Furthermore, the pro-survival function of autophagy suggests modulation of autophagy as a potential therapeutic strategy in protection of cells against lipotoxicity and lipid-related metabolic diseases.
Introduction
Autophagy is an evolutionarily conserved and highly regulated process whereby intracellular membranes are rearranged to sequester cargo or organelles from the cytoplasm before delivery of the double membrane vesicles called autophagosomes to the lysosomes for degradation (1, 2). This process is essential for maintaining the cellular hemostasis by preventing accumulation of damaged proteins and organelles and helps to sustain metabolism during nutrient deprivation. Recent advances in autophagy studies have also shown that dysregulation of autophagy is implicated in major diseases like cancer and neurodegenerative disorders (3). Autophagy is controlled by a group of autophagy-related genes (ATG genes) that control several sequential steps of the autophagic process, including induction, autophagosome nucleation and elogation, autophagosome docking and fusion with the lysosome, and finally autophagic body breakdown and release of the contents back into the cytosol (4). The serine/threonine-protein kinase mammalian target of rapamycin (mTOR)4 has been identified as a key regulatory protein involved in the induction of autophagy upstream of ATG proteins. In mammalian cells it is known that the initiation process of autophagy depends on a complex consisting of ULK1 (ATG1 homologue), ATG13, and FIP2000 proteins downstream of mTOR. This particular complex is activated with the suppression of mTOR under nutrient-depletion conditions or with mTOR inhibitors (5, 6).
Lipotoxicity refers to excessive accumulation of lipid in non-adipose tissues/cells like hepatocytes, pancreatic cells, and muscle fibers, which leads to loss of cellular functions and finally cell death (7). This phenomenon has been shown in many studies where the excessive levels of serum-free fatty acids (FFA) lead to development of insulin resistance in skeletal muscle (8) and death of mouse myocardiocytes that could lead to heart failure (9). As a result, lipotoxicity has been hypothesized to be the underlying cause of diseases associated with excess lipid accumulation in the body, such as obesity and diabetes (7, 10). High levels of lipid accumulate intracellularly probably due to an imbalance between the levels of FFA import/synthesis and utilization. Excess fatty acids are normally esterified and stored as lipid droplets that can be utilized when broken down by cellular lipases. Notably non-adipose tissues have limited storage capacity for lipid droplets. Therefore, it is believed that excessive uptake of FFA may overwhelm the cell capacity to store and utilize lipid droplets, eventually leading to damage of cellular function and cell death (11).
At present there is accumulating evidence suggesting that autophagy is involved in the physiological and pathological responses of cells to lipid stimulation. For example, autophagy is able to regulate the intracellular levels of lipid droplets in tissues like the liver (12). Other studies have also shown the importance of autophagy in response to non-physiological lipid levels where the autophagic process is required to maintain insulin sensitivity and reduce the levels of endoplasmic reticulum stress during onset of obesity in the hepatocytes (13). Studies in other cell types like pancreatic β-cells have also identified that autophagy plays a role in the maintenance of normal islet morphology and function like insulin secretion especially in mice given a high fat diet (14). Conversely, the loss of autophagy led to development of an insulin-resistant state in the mice and also loss of pancreatic cells function (14). On the other hand, there are still a number of issues regarding autophagy induction by lipid stimulation that remain unresolved. There have been conflicting reports whether FFA stimulation can induce or inhibit autophagy in cells (12, 14–16). Moreover, the signaling pathway linking FFA stimulation to autophagy induction and the physiological role of autophagy in response to FFA also remain to be determined. Therefore, in this study we systematically tested the autophagic responses and the underlying mechanisms induced by the two most common saturated and unsaturated dietary fatty acids, palmitic acid (PA, C16:0) and oleic acid (OA, C18:1) in our experimental system of lipotoxicity. Here, we first found induction of autophagy by PA, but not by OA. Second, we identified a novel signaling pathway involving PKC-α independent of mTOR. Finally, such autophagy was found to serve as a survival mechanism to counter the lipotoxic effects of excessive FFA stimulation. Our study thus suggests that modulation of autophagy can serve as a potential therapeutic strategy in diseases caused by lipotoxic effects of FFA.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
BAPTA-AM, chloroquine diphosphate (CQ), doxycycline, Earle's balanced salt solution (EBSS), 12-O-tetradecanoylphorbol-13-acetate (TPA), rapamycin, myriocin, fumonisin B1, anti-LC3, and anti-tubulin antibodies were purchased from Sigma. PA, OA, and essentially fatty acid-free bovine serum albumin (BSA) were also purchased from Sigma, and both fatty acids were conjugated to the fatty acid-free albumin as described previously (17). Anti-Atg5-Atg12 antibodies were purchased from Nanotools. Anti-PKC-α was from BD Transduction Laboratories. Anti-Atg7 antibody was obtained from ProSci, whereas anti-phosphotyrosine antibody Clone 4G10 and anti-phospho-PKC-α antibodies were obtained from Millipore. All other antibodies were obtained from Cell Signaling. The PKC chemical inhibitors GF109203X, Gö6976, and Rottlerin were purchased from Calbiochem.
Cells and Cell Culture
Mouse embryonic fibroblasts (MEFs) and HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, from Sigma) containing 10% fetal bovine serum (HyClone) and 1% penicillin/streptomycin (Invitrogen) (defined as full medium in this study) in a 5% CO2 atmosphere at 37 °C. The wild-type (WT) and Atg5 knock-out (KO) MEFs and the Tet-off Atg5 MEFs were provided by Dr. N. Misushima (Tokyo Medical and Dental University) (18). In most of the experiments, cells were treated with 0.25 mm PA or OA in high glucose, serum-free DMEM for 4 h unless otherwise stated.
Immunoprecipitation and Immunoblotting
Immunoblotting analysis was performed following standard procedures. The cells were lysed in M2 lysis buffer: 20 mm Tris, pH 7, 0.5% Nonidet P-40, 250 mm NaCl, 3 mm EDTA, 3 mm EGTA, 2 mm dithiothreitol, phosphatase inhibitor mixture (Pierce), and the protease inhibitor mixture (Roche Applied Science). An equal amount of protein for each sample was resolved on SDS-PAGE gel and transferred onto polyvinylidene difluoride membrane (Bio-Rad). After it was blocked with 5% nonfat milk, the membrane was probed with the designated first and second antibodies and subsequently developed with the enhanced chemiluminescence method (Pierce) and visualized by a Kodak Image Station 440CF (Eastman Kodak Co.). The immunoprecipitation assay was performed as described. Briefly, cells after treatment were lysed for 30 min in radioimmunoprecipitation assay buffer: 50 mm Tris HCl, pH 8.0,150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, phosphatase inhibitor mixture (Pierce), and protease inhibitor mixture. Cell lysates containing the same amount of protein were incubated with 1 μg of antibody and mixed overnight at 4 °C. After incubation overnight, 30 μl of Sepharose protein A/G-agarose beads were added to the cell lysates and mixed for 4 h at 4 °C. After incubation, the beads were washed extensively with radioimmunoprecipitation assay buffer five times, and the immunoprecipitated proteins were eluted by boiling for 5 min in sample buffer (Bio-Rad) before being resolved on SDS-PAGE gel and transferred onto polyvinylidene difluoride membrane (Bio-Rad) for Western blotting. Quantification results for the Western blots were obtained using the Kodak 1D 4.5.1 software.
Sample Processing and Imaging for Transmission Electron Microscopy
MEFs were seeded on four-chambered coverglass (NUNC Lab-tek Chambered Coverglass System) at a density of 2 × 104 cells/ml. After the respective treatments for 4 h, cells were fixed with 2.5% glutaraldehyde and washed three times with 1× phosphate buffer saline (PBS). Subsequently, post-fixation with 1% osmium tetroxide was performed followed by dehydration with ascending series of alcohol before being embedded in araldite for 24 h. Ultrathin sections of 99-nm thickness were cut with a glass knife on the Reichert Ultracut E ultramicrotome, mounted onto copper grids, and double-stained with uranyl acetate and lead citrate. Images were acquired using the JEOL JEM1010 transmission electron microscope at voltage level of 100.0 KeV.
Confocal Microscopy
Cells were seeded to a coverglass slide chamber (NUNC Lab-tek Chambered Coverglass System and after the designated treatments, the cells were examined under a confocal microscope (Olympus Fluoview 2000). GFP-LC3 puncta in the cells were quantified by counting the number in cells as described previously (19). Briefly, the GFP-LC3 puncta were manually counted under the confocal microscope. 30 cells were randomly selected from each treatment to calculate the average number of GFP-LC3 puncta per cell. The data shown were from one representative experiment of three independent repeats.
Detection of Cell Death
Flow cytometry was used to determine the cell viability using the live cell propidium iodide (PI) exclusion test, as previously described (20). In brief, cells were trypsinized at the end of the experiments. Cells were washed once with PBS and resuspended in PBS containing 1 mg of PI/ml. The levels of PI incorporation were quantified by flow cytometry using a FACSCalibur flow cytometer. Cell size was evaluated by forward-angle light scattering. PI-negative cells with normal size were considered to be live cells. Morphological changes of treated cells under phase-contrast microscope were also observed to detect cell death after treatment.
Analysis of Lipids Using High Performance Liquid Chromatography/Mass Spectrometry
An Agilent high performance liquid chromatography (HPLC) 1200 system coupled with an Applied Biosystem Triple Quadrupole/Ion Trap mass spectrometer (3200Qtrap) was used to quantitate individual phospholipids. HPLC conditions: Luna 3-μm silica column (inner diameter 150 × 2.0 mm); mobile phase A, chloroform:methanol:ammonium hydroxide, 89.5:10:0.5; mobile phase, chloroform:methanol:ammonium hydroxide:water, 55:39:0.5:5.5; flow rate, 300 μl/min; 5% B for 3 min, then linearly switched to 30% B in 24 min and maintained for 5 min and then linearly changed to 70% B in 5 min and maintained for 7 min. Then the composition of the mobile phase was returned to the original ratio over 5 min and maintained for 6 min before the next sample was analyzed. Multiple reaction monitoring transitions for individual glycerolphospholipid species and sphingolipids were set up at different elution stages for LC-MS analysis (21, 22). Levels of individual lipid levels were quantified using spiked internal standards. Neutral lipids were analyzed using a sensitive HPLC/electrospray ionization/multiple reaction monitoring method modified from a previous method (22). Triacylglycerol (TAGs) were calculated as relative contents to the spiked d5-TAG 48:0 internal standard (CDN isotopes), whereas DAGs were quantified using 4ME 16:0 diether DG (Avanti) as an internal standard.
Transient Small Interfering RNA (siRNA) Transfection
The nonspecific siRNA oligonucleotides and siRNA oligonucleotides targeting mouse and human Atg7 and PKC-α (ON-TARGETplus SMARTpoolTM) were obtained from Dharmacon (Lafayette, CO). The siRNAs was transfected into either MEFs or HepG2 cells using the DharmaFECT 4 transfection reagent according to the manufacturer's protocol.
Statistics
The numeric results are expressed as the mean ± S.D. from at least three independent experiments. The significance level was set at p < 0.05 using Student's t test.
RESULTS
PA, but Not OA, Induces Autophagy
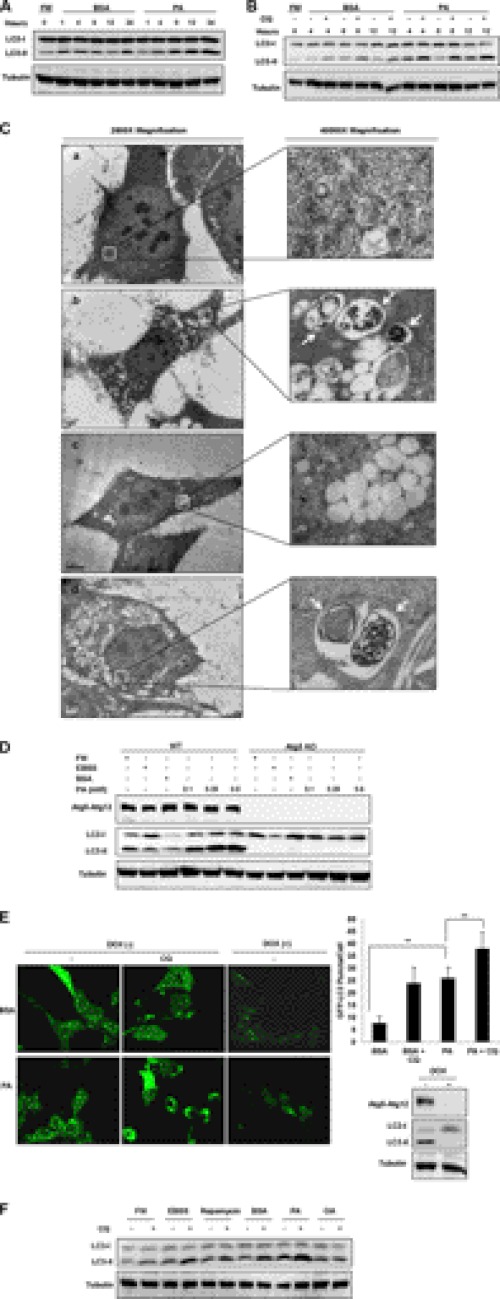
Recent studies have shown that saturated fatty acids such as PA are more cytotoxic compared with unsaturated fatty acids, such as OA (23). However, it is not known whether autophagy is involved in the cytotoxic effect of fatty acids. Here we first tested autophagy level in cells treated with PA. As shown in Fig. 1A, treatment of MEFs with PA resulted in a significant increase in the levels of LC3-II for up to 24 h in comparison to the control cells treated with fatty acid-free BSA. To measure the autophagic flux in cells treated with PA, we used CQ to block lysosomal function and the late degradation stage of autophagy (24). With the addition of CQ, we were able to observe a further increase of LC3-II level in PA-treated cells at the various time points (Fig. 1B), thus clearly suggesting that PA is able to induce autophagy flux in MEFs.
FIGURE 1.
PA but not OA induces autophagy. A, MEFs were treated with PA (0.25 mm) conjugated to fatty acid free BSA for the different time points as indicated. Cells treated with BSA acted as the control. FM, full medium (DMEM) with 10% FBS. After treatments, cell lysates were collected and subject to Western blot. B, MEFs were treated with PA (0.25 mm) for the indicated time points with or without the addition of CQ (10 μm). C, MEFs were treated with BSA control (panel a), PA (0.25 mm, panel b), OA (0.25 mm, panel c), or EBSS (panel d) for 4 h before being processed, and cellular images were then taken using an electron microscope at 2,500× and 40,000× magnification. D, WT and Atg5 KO MEFs were treated various concentrations of PA for 4 h. Treatment with EBSS was used as a positive control. E, Atg5 Tet-off MEFs stably expressing GFP-LC3 were treated with PA (0.25 mm) for 4 h, and GFP-LC3 puncta formation was observed using confocal microscopy. Atg5 protein expression was inhibited in the cells cultured with doxycycline (DOX) for 4 days and resulted in the loss of autophagic activity. The number of GFP-LC3 puncta/cell were also counted and presented (**, p < 0.01, student's t test). F, MEFs were treated with either BSA control, PA (0.25 mm) or OA (0.25 mm) for 4 h with or without the addition of CQ. Cells were also treated for the same time with EBSS and the mTOR inhibitor, rapamycin (20 nm), as the positive controls for induction of autophagy.
To further confirm the observation that PA treatment does indeed induce autophagy in the MEFs, transmission electron microscopy studies were performed on MEFs under various treatments. As shown in Fig. 1C, autophagosome-like vacuoles were hardly seen in BSA-treated control cells (panel A). In contrast, we observed an increase in the formation of autophagosome-like structures containing cytosolic contents in the PA-treated MEFs after 4 h (panel C, panel b, white arrows). Similar autophagosome-like vacuoles containing cytosolic contents were also observed in MEFs treated with EBSS as a positive control (Fig. 1C, panel d, white arrows). However, no autophagosome-like vacuoles containing cytosolic contents were observed in MEFs treated with OA for 4 h (Fig. 1C, panel d), suggesting that OA treatment was not able to induce autophagy.
Next, we examined whether PA-induced autophagy is dependent on the essential Atg proteins such as Atg5. As shown in Fig. 1D, no LC3-II was observed in the Atg5 KO MEFs. Similar changes were also found in starved cells treated in EBSS. Therefore, it is believed that PA-induced autophagy is dependent on the canonical autophagy induction machinery. Moreover, we used the inducible Atg5 deletion system with stable expression of GFP-LC3 (18) to further test PA-induced autophagy. As shown in Fig. 1E, there is significant increase of GFP-LC3 puncta in PA-treated cells without the addition of doxycycline. Consistently, the addition of CQ further enhanced both the size and the number of such puncta. Similar to the results in Fig. 1D, deletion of Atg5 by the addition of doxycycline abolished the effect of PA on GFP-LC3 puncta formation.
Finally, we compared the effect of PA with the effect of unsaturated fatty acid, OA on autophagic flux in MEFs. As shown in Fig. 1F, treatment of the MEFs with similar concentrations of OA for 4 h did not cause significant increase in LC3-II conversion even with the presence of CQ, whereas PA together with other known autophagy inducers (EBSS and rapamycin) was able to cause an evident increase in the autophagic flux. Similar observations were also observed in other cell types such as the HepG2 cells, in which treatment with PA significantly enhanced autophagic flux level, whereas OA treatment did not result in any further increase in the autophagic flux when compared with the BSA-treated control cells (supplemental Fig. 1A). Taken together, results from this part of our study confirm that PA, but not OA, is capable of inducing autophagic responses.
PA-induced Autophagy Is Independent of mTOR Signaling Pathway
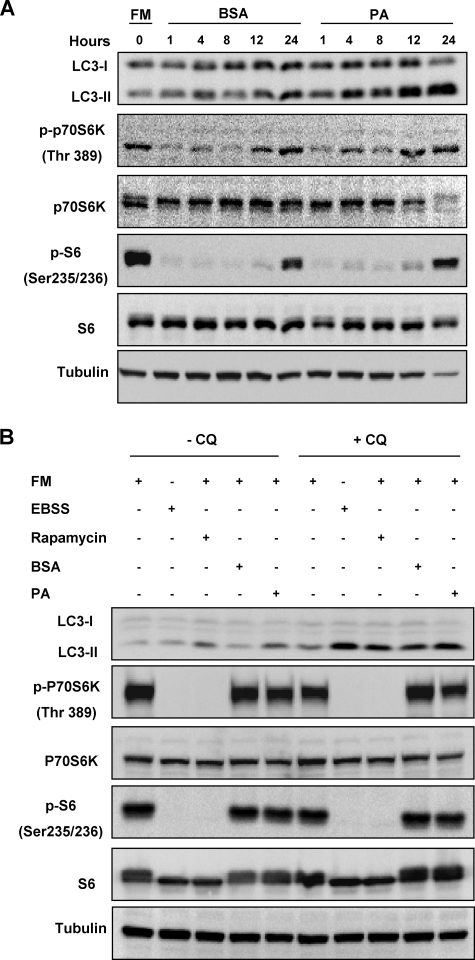
The mTOR signaling pathway has been well established as the key negative regulator of the autophagic process (1, 25). Therefore, we next investigated the role of the mTOR signaling pathway in PA-induced autophagy. The p70 S6 kinase (p70S6K) protein is a direct substrate of mTOR, and its phosphorylation status can be used as an indicator of the activity of the mTOR pathway (26). When MEFs were treated with PA in FBS-free medium, there was a marked reduction of the p-p70S6K level and the addition of BSA or PA failed to affect the level of p-p70S6K especially at the earlier time points (Fig. 2A). Interestingly, PA increased the p-p70S6K level at later time points (12 and 24 h). The same trend was also observed for the direct substrate of p70S6K, the S6 ribosomal protein (Fig. 2A), indicating the possibility that PA may activate mTOR with prolonged treatment.
FIGURE 2.
PA-induced autophagy is independent of the mTOR signaling pathway. A, MEFs were treated with BSA control or PA (0.25 mm) for various time points as indicated. Cell lysates were collected and subject to Western blot. FM, full medium. B, MEFs were treated with BSA control or PA (0.25 mm) for 4 h in full medium. Cells were also treated with EBSS and the mTOR inhibitor, rapamycin (20 nm), for 4 h as positive controls. CQ (10 μm) was added to the treated cells for measuring autophagic flux.
To confirm the above observations, MEFs were treated with PA in full medium, which contains 10% FBS. As shown in Fig. 2B, treatment with PA for 4 h led to an increased level of LC3-II conversion, and the addition of the lysosomal inhibitor CQ further enhanced the LC3-II level, indicating an increase in autophagic flux in PA-treated cells. Consistently, PA failed to reduce the phosphorylation level of p70S6K and S6 ribosomal proteins (Fig. 2B). As expected, common inducers of autophagy like EBSS and rapamycin were able to completely inhibit the mTOR signaling pathway observed by the complete loss of p70S6K and S6 ribosomal protein phosphorylation (Fig. 2B). Consistently, when the HepG2 cells were treated with PA in full medium containing 10% FBS for 4 h, an increase in autophagic flux was also observed with no inhibition of the mTOR signaling pathway (supplemental Fig. 1B). Such results enforced the observation that induction of autophagy by PA is independent of the mTOR signaling pathway.
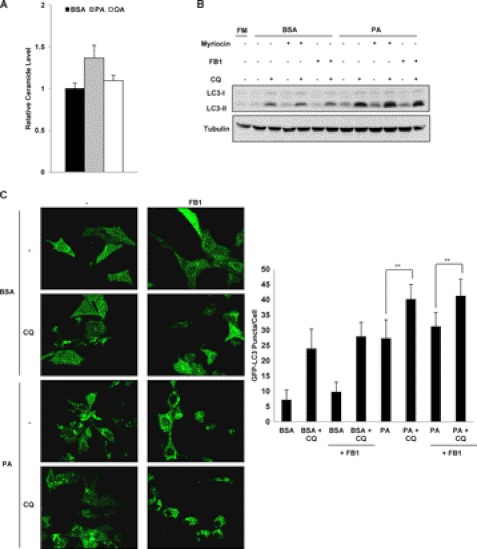
Accumulation of Intracellular Ceramide Is Not Related to PA-induced Autophagy
Ceramides belong to the class of lipids known as sphingolipids, and the accumulation of intracellular ceramides have been linked to the progression of diseases like obesity and other metabolic diseases (27). Furthermore, recent studies have also shown that short chain ceramides were able to induce autophagy in a variety of cancer cell lines (28, 29). Therefore, we next investigated whether ceramides are involved in PA-induced autophagy. Here we first measured the levels of intracellular ceramides in cells treated with different types of FFA. Treatment with PA for 4 h resulted in an increase of ceramides by about 1.4-fold compared with the cells treated with BSA control, whereas a marginable increase was found in OA-treated cells (Fig. 3A). To test the effect of intracellular ceramides on PA-induced autophagy, cells were treated with the chemical inhibitor of serine palmitoyltransferase, myriocin, or the inhibitor of sphingosine N-acyltransferase fumonisin B1 (FB1) as both of them are known to inhibit ceramide production in cells (28, 29). The addition of myriocin or FB1 to PA-treated cells did not change the autophagic flux level observed with LC3-II conversion (Fig. 3B). Furthermore, the addition of FB1 also failed to reduce the formation of GFP-LC3 puncta formed in PA-treated cells with or without the presence of CQ (Fig. 3C). These results suggest that accumulation of endogenous ceramides is not related to PA-induced autophagy.
FIGURE 3.
Autophagy induction by PA is not caused by the accumulation of intracellular ceramide levels. A, MEFs were treated with BSA control, PA (0.25 mm) or OA (0.25 mm) for 4 h, and the total ceramide levels were then quantified using LC-MS as described under “Experimental Procedures.” The relative ceramide levels of cells from the various treatments were calculated by normalizing to the ceramide levels of the cells treated with BSA. Data were presented as the means ± S.D. of three independent experiments. B, MEFs were treated with BSA control or PA (0.25 mm) for 4 h with or without the presence of either myriocin (1 μm) or FB1 (10 μm) to inhibit ceramide production in the cells. CQ (10 μm) was also added to the treated cells to observe the changes in the autophagic flux. FM, full medium. C, Atg5 Tet-off MEFs stably expressing GFP-LC3 were treated with either BSA control or PA (0.25 mm) for 4 h with or without the presence of FB1 (10 μm), and GFP-LC3 puncta formation was observed using confocal microscopy. The number of GFP-LC3 puncta/cell were also counted and presented. **, p < 0.01, Student's t test.
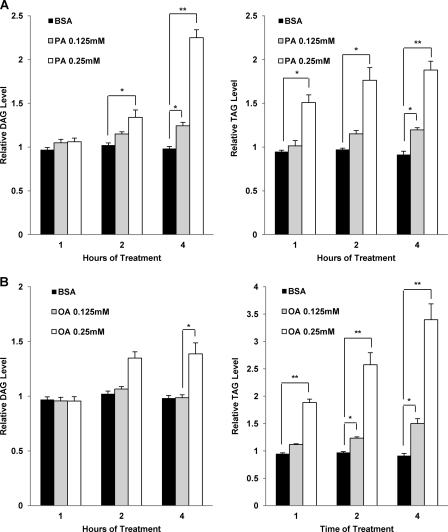
PA, but Not OA, Induces Accumulation of Intracellular Diacylglycerol
After excluding the involvement of ceramides, we next measured the levels of other lipid metabolites upon PA and OA stimulation, including DAG and TAG. Both DAG and TAG are glycerolipids, with different number of fatty acid molecules linked to the glycerol molecule. When taken by cells, excess FFAs can be converted into their respective acyl-CoA derivatives (30). These acyl-CoA derivatives can then be incorporated and stored in the cells as neutral lipids like DAG and TAG (23, 31). Thus, we would expect that the levels of DAG and TAG will increase substantially during PA or OA treatment. MEFs were, therefore, treated with either BSA as control or 0.125 and 0.25 mm of either PA (Fig. 4A) or OA (Fig. 4B) for 1, 2, and 4 h. We then measured the relative levels of DAG and TAG accumulation in the treated cells. Indeed, we observed that treatment with either PA or OA resulted in a dose- and time- dependent increase in the relative levels of DAG and TAG compared with BSA-treated control cells (Fig. 4A, left panel). Furthermore, although TAG levels in the PA-treated cells also increased significantly at treatment times up to 4 h, we observed that the rate of increase in the TAG level was not proportional to that of DAG with 0.25 mm of PA treatment (Fig. 4A, right panel).
FIGURE 4.
PA and OA treatment induce differential accumulation of intracellular DAG and TAG that is both time- and dose-dependent. A, MEFs were treated with BSA control and PA (0.125 and 0.25 mm) for 1, 2, and 4 h. The total DAG (left panel) and TAG (right panel) levels were then quantified using LC-MS as described under “Experimental Procedures.” The relative DAG and TAG levels were calculated by normalizing their respective levels in each treatment at different time points to the levels present in the untreated cells. B, cells were treated with BSA control and OA (0.125 and 0.25 mm) for 1, 2, and 4 h. The total DAG (left panel) and TAG (right panel) levels were then quantified using LC-MS. The relative DAG and TAG levels were calculated as described in A. Data were presented as the means ± S.D. of three independent experiments, and Student's t tests were calculated between the BSA treated cells and either PA- or OA-treated cells at each respective time point. *, p < 0.05; **, p < 0.01, Student's t test.
On the other hand, in cells treated with different concentrations of OA, there was no significant accumulation of DAG (Fig. 4B, left panel). Instead, we observed a significant time and dose-dependent increase in TAG accumulation in the cells treated with OA (Fig. 4B, right panel). Similar results were also observed in HepG2 cells treated with PA and OA. Only PA treated HepG2 cells had significantly increased DAG accumulation by up to 4 h of treatment, but this was not seen in OA-treated HepG2 cells (supplemental Fig. 2). PA treatment also resulted in a significant increase in TAG levels by 4 h (supplemental Fig. 2A, right panel), although the magnitude of increase was smaller than OA-treated HepG2 cells. (supplemental Fig. 2B, right panel). Previous studies have suggested that the enzyme responsible for the conversion of DAG to TAG, diacylglycerol acyltransferase (DGAT), has a higher specificity for fatty acyl-CoAs that contain the 18C:1 chain derived from OA compared with other fatty acyl-CoAs that have the 16C:0 chain coming from PA (31, 32). Therefore, such a substrate specificity by DGAT could well explain our data that in the OA-treated cells, the TAG level increased more evidently than that of DAG (Fig. 4B), whereas in cells treated with PA the accumulation of DAG was more significant than that of TAG (Fig. 4A).
Inhibition of Protein Kinase C Inhibits Autophagy Induction Caused by PA Treatment
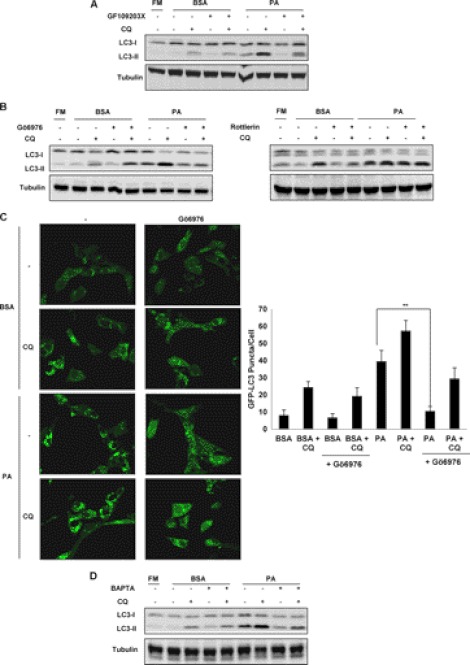
Because significant accumulation of DAG was observed in PA, but not in OA-treated cells, we then investigated the signaling pathways associated with DAG accumulation in cells. One of the well known signaling pathways activated by DAG is the PKC family, where DAG serves as a natural agonist to recruit PKC proteins to membrane for activation (33). There are essentially 3 classes of PKC family comprising a total of 10 PKC isozymes in the mammalian system: classical, novel, and atypical PKCs (34). Here we made use of different chemical inhibitors that target all PKCs or specific classes of PKC to investigate the involvement of PKC in PA-induced autophagy. First, GF109203X, a general PKC inhibitor, was able to markedly reduce LC3-II level in cells treated with PA with or without the presence of CQ (Fig. 5A), suggesting that inhibition of PKC signaling pathway impairs PA-induced autophagy. Next, we tested the effects of Gö6976 and Rottlerin, which inhibit the classical and novel classes of PKC, respectively. As shown in Fig. 5B, left panel), the classical PKC inhibitor Gö6976 was found to reduce the LC3-II level in PA-treated cells, similar to the effect of GF109203X. In contrast, the addition of the novel PKC inhibitor Rottlerin failed to suppress the autophagic flux induced by PA (Fig. 5B, right panel). We further confirmed the effect of Gö6976 by examining the changes of GFP-LC3 puncta formation in the MEFs with stable expression of GFP-LC3 (Fig. 5C). Similar results were obtained with HepG2 cells when they were treated with PA in the presence of Gö6976 as well (supplemental Fig. 1C). All these findings indicate a possibility that the classical PKC is implicated in PA-induced autophagy. Activation of the classical class of PKC members requires both calcium ions (Ca2+) and DAG (34); therefore, we tried to investigate whether PA-induced autophagy is sensitive to the loss of free intracellular Ca2+ ions in the cells. To this end, we made use of BAPTA-AM, an intracellular Ca2+ ion chelator (35). As expected, the addition of BAPTA-AM resulted in an overall decrease in autophagic flux observed in the PA-treated cells (Fig. 5D), suggesting that the free intracellular Ca2+ ions are involved in PA-induced autophagy. Overall, the results from this part of our study support the notion that the classical group of PKC is implicated in PA-induced autophagy via enhanced intracellular level of DAG and calcium ions.
FIGURE 5.
PA-induced autophagy requires the activation of PKC family. A, MEFs were treated with BSA control or PA (0.25 mm) for 4 h with or without the presence of the general PKC inhibitor GF109203X (1 μm). CQ (10 μm) was added to the treated cells to determine the levels of autophagic flux. FM, full medium. B, MEFs were treated with BSA control or PA (0.25 mm) for 4 h with or without the presence of either the classical PKC inhibitor Gö6976 (1 μm) (left panel) or the novel PKC inhibitor, rottlerin (10 μm) (right panel). CQ (10 μm) was added to the treated cells to determine the levels of autophagic flux. C, Atg5 Tet-off MEFs stably expressing GFP-LC3 were treated with either BSA control or PA (0.25 mm) for 4 h with or without the presence of classical PKC inhibitor Gö6976 (1 μm), and GFP-LC3 puncta formation was observed using confocal microscopy. The number of GFP-LC3 puncta/cell were also counted and presented (**, p < 0.01, Student's t test). D, MEFs were treated with BSA control or PA (0.25 mm) for 4 h with or without the presence of BAPTA-AM (10 μm).
PKC-α Is Involved in Induction of Autophagy in PA-treated Cells
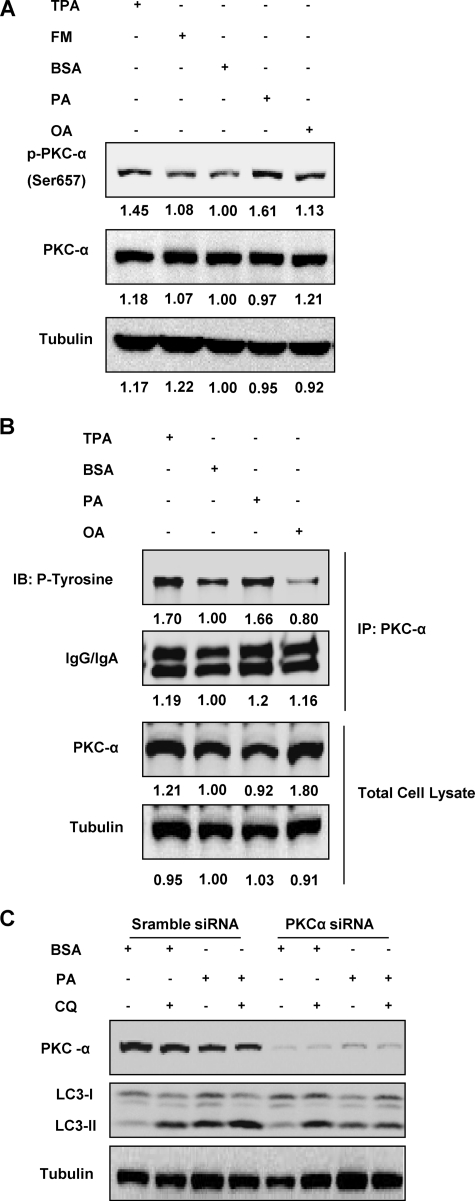
After establishing the role of classical PKCs in PA-induced autophagy, we then tried to identify the member of PKC within the classical group responsible for autophagy induction in PA-treated cells. As shown in Fig. 6A, there was enhanced phosphorylated PKC-α in PA-treated cells compared to the BSA-treated control group, whereas no evident changes were found in OA-treated cells. As expected, a similar increase was also found in cells treated with TPA, the positive control that is known to activate members of the PKC family including PKC-α (36). To support the results obtained above, we further examined PKC-α activation by measuring the phosphorylation levels of tyrosine residues from immune-precipitated PKC-α proteins. As shown in Fig. 6B, we observed that PKC-α tyrosine phosphorylation level was significantly increased after PA stimulation compared with the BSA control cells, similar to the TPA-treated cells. Consistently, PA has a much stronger effect than OA in enhancing the PKC-α tyrosine phosphorylation level (Fig. 6B). Finally, siRNA knockdown of PKC-α was carried out to confirm the involvement of PKC-α in PA-induced autophagy. When cells with PKC-α knocked down were treated with PA for 4 h, there was a decrease in the levels of LC3-II and autophagic flux compared with the PA-treated cells that were given scrambled siRNA as control (Fig. 6C). Thus, data presented in this part of our study clearly demonstrate that PKC-α activation is involved as the upstream signal for the induction of autophagy in cells stimulated with PA.
FIGURE 6.
Activation of PKC-α is required for PA-induced autophagy. A, MEFs were treated with BSA control, PA (0.25 mm), or OA (0.25 mm) for 4 h, and activation status of PKC-α was observed using immunoblotting. Cells treated with TPA (100 nm) for 20 min were used as a positive control. The respective lane intensity was quantified using Kodak Imaging Software as a -fold change to the BSA control treatment. B, MEFs were treated with BSA control, PA (0.25 mm), or OA (0.25 mm) for 4 h, and PKC-α was then immunoprecipitated (IP) and blotted for levels of phosphorylated tyrosine as another indicator of PKC-α activation. TPA (100 nm) was added to the cells for 20 min to act as a positive control. The respective lane intensity was quantified using Kodak Imaging Software as a -fold change to the BSA control treatment. C, PKC-α in MEFs was knocked down with PKC-α siRNA, whereas the control cells were knocked down with scramble siRNA as described. The cells were then treated with either BSA control or PA (0.25 mm) for 4 h with or without the presence of CQ (10 μm).
Autophagy Is Important Cell Survival Mechanism for Cells against Lipotoxicity Caused by PA
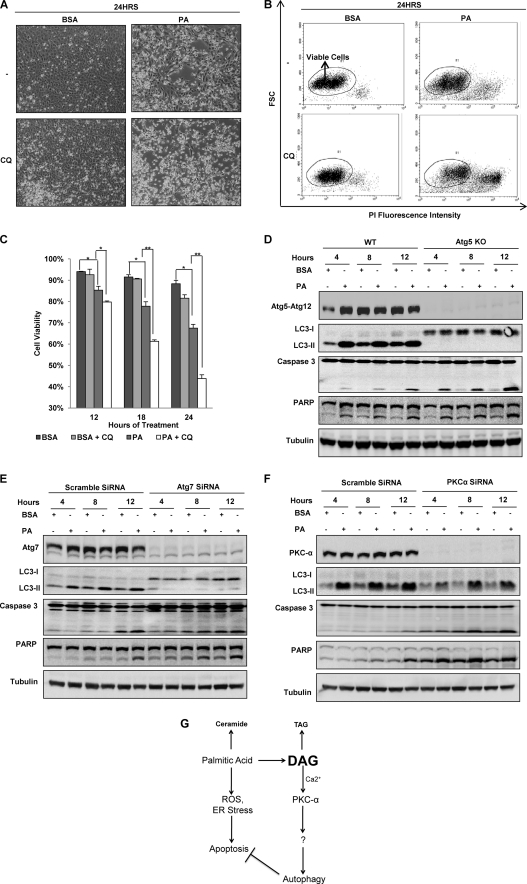
In this part of our study we aimed to investigate the physiological relevance of autophagy induced by PA. Recent studies have shown that PA possesses cytotoxic properties and many non-adipose cells are killed by long term treatment of PA (23, 37). Using phase-contrast microscopy, we observed that treatment with PA up to 24 h resulted in a significant increase in the number of dead cells as compared with the BSA-treated control cells (Fig. 7A). This observation was further validated using the PI live cell exclusion assay coupled with flow cytometry as shown in Fig. 7, B and C. Notably, the addition of CQ markedly enhanced cell death induced by PA in both assays (Fig. 7, A–C). The addition of CQ to the BSA control cells for up to 24 h did not induce any significant decrease in cell viability, suggesting that CQ alone is not cytotoxic to the cells (Fig. 7C). Similar results were also observed when HepG2 cells were subjected to the same assays (supplemental Fig. 3, A and B). Because CQ is known to block autophagy by suppressing the lysosomal function, such findings thus indicate that PA-induced autophagy may serve as a pro-survival function to protect against PA-mediated lipotoxicity. To further confirm that autophagy plays a cell survival role in response to PA, we compared PA-induced cell death between WT and Atg5 KO MEFs. As shown in Fig. 7D, there were higher levels of caspase 3 and poly(ADP-ribose) polymerase cleavage in Atg5 KO MEFs compared to the WT MEFs, suggesting the possibility that autophagy is important for cell survival in times of lipotoxic stress. We further confirmed this observation by knocking down Atg7 using Atg7 siRNA in MEFs. As shown in Fig. 7E, Atg7 knockdown led to (i) complete suppression of LC3-II conversion induced by PA and (ii) significant increase of apoptotic cell death, shown by an increase of both caspase 3 and poly(ADP-ribose) polymerase cleavage, which are well established markers of apoptosis. Similar results were also observed in HepG2 cells when Atg7 was knocked down (supplemental Fig. 3C), further supporting our conclusion that autophagy is a pro-cell survival mechanism in during lipotoxic stress.
FIGURE 7.
Autophagy acts as a cell survival mechanism against lipotoxicity caused by PA. A, cell morphology was observed under a phase contrast microscopy (×200) of MEFs treated with either BSA control or PA (0.25 mm) for 24 h with or without the presence of CQ (10 μm). B, shown are representative dot-plots of flow cytometry data of the PI exclusion test. MEFs were treated as described in panel A. C, quantification of the cell viability data from panel B is shown. Data were presented as the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01, Student's t test). D, WT and Atg5 KO MEFs were treated with either BSA control or PA (0.25 mm) for the indicated time points. PARP, poly(ADP-ribose) polymerase. E, Atg7 was knocked down with Atg7 siRNA, and the cells were treated with either BSA control or PA (0.25 mm) for the indicated time points. F, PKC-α was knocked down with PKC-α siRNA, and the cells were treated with either BSA control or PA (0.25 mm) for the indicated time points. G, shown is a summary of the proposed signaling pathways involved in PA-mediated autophagy and its pro-survival role in PA-induced apoptosis and lipotoxicity. ROS, reactive oxygen species.
Because our earlier data establish the role of PKC-α in PA-induced autophagy, we next investigated whether PKC-α knockdown would affect the lipotoxicity of PA. It was found that PKC-α knockdown first reduced the LC3-II protein level, consistent with the earlier findings that the general and classical PKC inhibitors are capable of inhibiting PA-induced autophagy (Fig. 5, A--C). More importantly, PKC-α knockdown also markedly enhanced both caspase 3 and poly(ADP-ribose) polymerase cleavage, thus further supporting the pro-survival function of autophagy induced by PA (Fig. 7F). Interestingly, we also observed that reduction of the LC3-II level in PKC-α knockdown was not as effective as Atg7 knockdown in suppression of autophagy (LC3-II conversion), whereas the sensitization on PA-induced apoptosis (caspase 3 and poly(ADP-ribose) polymerase cleavage) was similar or even stronger (Fig. 7, E and F). Such a discrepancy suggests the possibility that PKC-α may also mediate other pro-survival mechanisms in addition to autophagy to protect against PA-induced cytotoxicity.
DISCUSSION
Lipotoxicity has been thought to be the main contributor to the progression of various diseases associated with excess lipid accumulation in the body, such as obesity and diabetes (7, 10). On the other hand, the autophagic process has been well documented as a cell survival mechanism and implicated in several disease models such as cancer and neurodegenerative diseases (3, 38). At present, autophagy has been shown to be able to regulate lipid metabolism (12), but little is known about whether lipids like FFA can regulate autophagic activity. In this study we provide evidence that autophagy can be induced by the saturated fatty acid, PA, but not by the unsaturated fatty acid, OA. Furthermore, we showed that autophagy induction by PA is independent of mTOR activity. Instead PA-induced autophagy is mediated by a novel signaling pathway involving PKC-α, most probably via enhanced intracellular level of DAG produced from PA. We also presented evidence suggesting that autophagy plays a pro-survival function to protect against PA-mediated lipotoxicity. Our findings are generally consistent with earlier reports that FFA such as PA is capable of inducing autophagy in pancreatic β-cells (14, 15). On the other hand, it has been reported that both PA and OA treatment prevented fusion of autophagosome and lysosome and thus inhibited autophagy (16). Another recently published paper also reported that only OA but not PA was capable of inducing autophagy in hepatocytes (39). It is believed that the conflicting results could be attributed to factors such as the cell types used, whether the model used is in vivo or in vitro, the concentration and duration of FFA treatment, and the ratio of conjugated BSA to FFA used.
In this study we attempted to identify the molecular mechanisms underlying PA-induced autophagy, with several important findings. First, we exclude the involvement of ceramides in PA-induced autophagy, as the inhibition of ceramide synthesis by myriocin or FB1 is unable to block autophagy (Fig. 3, B and C). Previous reports have shown that treatment with C2 and C6 ceramides are able to induce autophagy in human cancer cells via inhibition of Akt and mTOR activity (28, 29). One possible explanation for this discrepancy is the fact that the type of ceramides used in their study is different from those accumulated in PA-treated cells. In our study the type of ceramide metabolites found in PA-treated cells consisted of mainly the Cer(d18:1/16:0) (N-(palmitoyl)-ceramide) species (data not shown). Our finding is indeed consistent with an earlier report that ceramide accumulation only plays a nominal role in affecting cellular homeostasis observed in cells treated with PA (37).
The second important finding in our study is that PA-induced autophagy is mTOR-independent, as PA fails to inhibit mTOR activity in cells cultured under full medium (Fig. 2B and supplemental Fig. 1B), and it may even activate mTOR in cells under starvation (Fig. 2A). It is known that high levels of FFA lead to constant activation of mTOR activity, a process related to the development of diseases such as diabetes and obesity (40). Furthermore, it has also been reported that PA has the ability to induce activation of mTOR via another member of the PKC family, PKC-δ (41). Taken together, it is obvious that PA-induced autophagy is not mediated via suppression of mTOR activity.
Then how does PA induce autophagy when mTOR activity is not impaired? Members of the PKC family such as PKC-δ and PKC-θ have been reported to be involved in autophagy induction under different conditions (35, 42). In this study we have identified a member of the classical PKC family, PKC-α, as an important mediator in PA-induced autophagy based on the following observations: (i) general and typical PKC inhibitors are able to block PA-induced autophagy (Fig. 5 and supplemental Fig. 1C), (ii) PA, but not OA, induces PKC-α activation (Fig. 6), and (iii) siRNA knockdown of PKC-α significantly reduced PA-induced autophagy (Fig. 7F). Our results indicate the importance of intracellular DAG as the main stimulus for induction of autophagy via the activation of PKC-α, based on the common understanding that DAG is required for activation of classical PKC (33) and also our observation of an increase of DAG levels in cells treated by PA, not by OA (Fig. 4A). Previous studies have demonstrated that DGAT, the enzyme responsible for conversion of DAG to TAG, has a higher specificity for the 18C:1-CoA substrate derived from OA compared with the 16C:0-CoA substrate derived from PA (31, 32), and this could be the reason why OA treatment resulted in a more significant increase in total TAG levels but much lesser accumulation of DAG compared with PA (Fig. 4 and supplemental Fig. 2).
However, it should also be noted that there have been 2 isoforms of DGAT enzymes identified in mammalian cells, namely DGAT1 and DGAT2 (30). Although both enzymes contribute to TAG synthesis, there may still be functional difference between the two as Dgat1−/− mice are resistant to diet-induced obesity (43), whereas Dgat2−/− mice die shortly after birth (44). At present, we have some preliminary results showing that DGAT-1 knocked-down was able to promote autophagy in OA-treated cells (data not shown). This indicates that accumulation of intracellular DAG does play a role in activating the autophagic process. More studies will need to be carried out in the future to explore the role of the two respective DGAT enzymes in regulating both DAG and TAG levels in response to exogenous FFA challenge.
Results from this study also raise an interesting possibility that DAG accumulation in the cells has undesirable effects on the cellular homeostasis and the induction of autophagy via PKC activation may serve the purpose of alleviating such adverse effects. The activation of various members of the PKC family by accumulation of intracellular DAG induced by PA treatment has been well studied (45–47). Moreover, there is evidence suggesting that DAG-mediated PKC-δ function plays a critical role in mammalian antibacterial autophagy (48). These may also help us to understand the fact that lack of DAG accumulation in OA-treated cells did not lead to PKC activation and thus no autophagy induction. Such a hypothesis is also supported by an earlier study that increased TAG accumulation and decreased levels of DAG in cells by overexpression of DGAT1 helped to prevent cell death caused by PA treatment (23). At present, how PKC mediates autophagy independent of mTOR is still largely unknown. Therefore, studying the molecular mechanism underlying PKC-regulated autophagy and identification of the downstream molecular targets of PKC would be very important topics for future studies.
After establishing the role of DAG-PKC in PA-mediated autophagy, we next examined the role of autophagy in PA-mediated lipotoxicity. The cytotoxicity of common dietary long chain saturated FFAs like PA has been well established (23, 37). Several factors like increased ROS production (49) and induction of endoplasmic reticulum stress (50) have been implicated in lipotoxicity. On the other hand, it has been well established that autophagy generally serves as an important cell survival mechanism under various stress conditions, such as starvation, oxidative stress, and DNA damage (3). Data from this study also reveal that PA-induced autophagy plays a pro-survival function, and suppression of autophagy by either CQ (Fig. 7, A–C, and supplemental Fig. 3, A–C) or knockdown of Atg7 (Fig. 7E and supplemental Fig. 3C) markedly enhanced PA-induced apoptosis. At present, it is still not clear how autophagy protects against PA-mediated cell death. One possibility remaining to be tested is that autophagy is involved in degrading and clearing the accumulated DAG from the cells via lipases that are present in the lysosomes. An earlier study has shown that autophagy plays an important role in lipid metabolism (12) in cells by affecting the intracellular levels of TAG in the cells, and thus it is possible that autophagy can also play a role in regulation of intracellular levels of DAG to promote cell survival in times of exogenous FFA stress.
Taken together, as summarized in Fig. 7G, here we reveal a novel mechanism in regulating PA-induced autophagy; PA promotes the accumulation of intracellular DAG, which in turn activates PKC-α as an upstream signaling mechanism for autophagy. Moreover, such inducible autophagy plays an important pro-survival role in mitigating PA-induced apoptosis and lipotoxicity.
Supplementary Material
Acknowledgment
We thank Dr. N. Mizushima (Tokyo Medical and Dental University) for providing the Atg5−/− MEFs and the Tet-off Atg5 MEFs with stable expression of GFP-LC3.
This work was supported in part by research grants from the Singapore National Medical Research Council (NMRC/1260/2010) and the Singapore Biomedical Research Council (BMRC/08/1/21/19/554) (to H.-M. S.).

This article contains supplemental Figs. 1–3.
- mTOR
- mammalian target of rapamycin
- FFA
- serum-free fatty acid
- PA
- palmitic acid
- OA
- oleic acid
- CQ
- chloroquine diphosphate
- EBSS
- Earle's balanced salt solution
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- MEF
- mouse embryonic fibroblast
- PI
- propidium iodide
- TAG
- triacylglycerol
- p70S6K
- p70 S6 kinase
- FB1
- fumonisin B1
- DAG
- diacylglycerol
- DGAT
- diacylglycerol acyltransferase.
REFERENCES
- 1. Klionsky D. J., Emr S. D. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mizushima N. (2007) Autophagy. Process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 3. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He C., Klionsky D. J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizushima N. (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 [DOI] [PubMed] [Google Scholar]
- 7. Brookheart R. T., Michel C. I., Schaffer J. E. (2009) As a matter of fat. Cell Metab. 10, 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J. K., Michael M. D., Previs S. F., Peroni O. D., Mauvais-Jarvis F., Neschen S., Kahn B. B., Kahn C. R., Shulman G. I. (2000) Redistribution of substrates to adipose tissue promotes obesity in mice with selective insulin resistance in muscle. J. Clin. Invest. 105, 1791–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu H. C., Kovacs A., Ford D. A., Hsu F. F., Garcia R., Herrero P., Saffitz J. E., Schaffer J. E. (2001) A novel mouse model of lipotoxic cardiomyopathy. J. Clin. Invest. 107, 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chavez J. A., Summers S. A. (2010) Lipid oversupply, selective insulin resistance, and lipotoxicity molecular mechanisms. Biochim. Biophys. Acta 1801, 252–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaffer J. E. (2003) Lipotoxicity. When tissues overeat. Curr. Opin. Lipidol. 14, 281–287 [DOI] [PubMed] [Google Scholar]
- 12. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang L., Li P., Fu S., Calay E. S., Hotamisligil G. S. (2010) Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., Azuma K., Hirose T., Tanaka K., Kominami E., Kawamori R., Fujitani Y., Watada H. (2008) Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high fat diet. Cell Metab. 8, 325–332 [DOI] [PubMed] [Google Scholar]
- 15. Choi S. E., Lee S. M., Lee Y. J., Li L. J., Lee S. J., Lee J. H., Kim Y., Jun H. S., Lee K. W., Kang Y. (2009) Protective role of autophagy in palmitate-induced INS-1 beta cell death. Endocrinology 150, 126–134 [DOI] [PubMed] [Google Scholar]
- 16. Koga H., Kaushik S., Cuervo A. M. (2010) Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 24, 3052–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Listenberger L. L., Brown D. A. (2007) Fluorescent detection of lipid droplets and associated proteins. Curr. Protoc. Cell Biol., Chapter 24, Unit 24.2 [DOI] [PubMed] [Google Scholar]
- 18. Hosokawa N., Hara Y., Mizushima N. (2007) Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 581, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 19. Wu Y. T., Tan H. L., Shui G., Bauvy C., Huang Q., Wenk M. R., Ong C. N., Codogno P., Shen H. M. (2010) Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 285, 10850–10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu Y. T., Tan H. L., Huang Q., Kim Y. S., Pan N., Ong W. Y., Liu Z. G., Ong C. N., Shen H. M. (2008) Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy 4, 457–466 [DOI] [PubMed] [Google Scholar]
- 21. Shui G., Stebbins J. W., Lam B. D., Cheong W. F., Lam S. M., Gregoire F., Kusonoki J., Wenk M. R. (2011) Comparative plasma lipidome between human and cynomolgus monkey. Are plasma polar lipids good biomarkers for diabetic monkeys? PLoS One 6, e19731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shui G., Guan X. L., Low C. P., Chua G. H., Goh J. S., Yang H., Wenk M. R. (2010) Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry. Application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosyst. 6, 1008–1017 [DOI] [PubMed] [Google Scholar]
- 23. Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S, Schaffer J. E. (2003) Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Codogno P., Meijer A. J. (2005) Autophagy and signaling. Their role in cell survival and cell death. Cell Death Differ. 12, 1509–1518 [DOI] [PubMed] [Google Scholar]
- 26. Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 27. Holland W. L., Summers S. A. (2008) Sphingolipids, insulin resistance, and metabolic disease. New insights from in vivo manipulation of sphingolipid metabolism. Endocr. Rev. 29, 381–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scarlatti F., Bauvy C., Ventruti A., Sala G., Cluzeaud F., Vandewalle A., Ghidoni R., Codogno P. (2004) Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J. Biol. Chem. 279, 18384–18391 [DOI] [PubMed] [Google Scholar]
- 29. Pattingre S., Bauvy C., Carpentier S., Levade T., Levine B., Codogno P. (2009) Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J. Biol. Chem. 284, 2719–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coleman R. A., Mashek D. G. (2011) Mammalian triacylglycerol metabolism. Synthesis, lipolysis, and signaling. Chem. Rev. 111, 6359–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li L. O., Klett E. L., Coleman R. A. (2010) Acyl-CoA synthesis, lipid metabolism, and lipotoxicity. Biochim. Biophys. Acta. 1801, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coleman R., Bell R. M. (1976) Triacylglycerol synthesis in isolated fat cells. Studies on the microsomal diacylglycerol acyltransferase activity using ethanol-dispersed diacylglycerols. J. Biol. Chem. 251, 4537–4543 [PubMed] [Google Scholar]
- 33. Newton A. C. (2001) Protein kinase C. Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 101, 2353–2364 [DOI] [PubMed] [Google Scholar]
- 34. Schmitz-Peiffer C., Biden T. J. (2008) Protein kinase C function in muscle, liver, and beta cells and its therapeutic implications for type 2 diabetes. Diabetes 57, 1774–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakaki K., Wu J., Kaufman R. J. (2008) Protein kinase Cθ is required for autophagy in response to stress in the endoplasmic reticulum. J. Biol. Chem. 283, 15370–15380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gschwendt M., Kittstein W., Marks F. (1991) Protein kinase C activation by phorbol esters. Do cysteine-rich regions and pseudosubstrate motifs play a role? Trends Biochem. Sci. 16, 167–169 [DOI] [PubMed] [Google Scholar]
- 37. Listenberger L. L., Ory D. S., Schaffer J. E. (2001) Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J. Biol. Chem. 276, 14890–14895 [DOI] [PubMed] [Google Scholar]
- 38. Levine B., Klionsky D. J. (2004) Development by self-digestion. Molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 39. Mei S., Ni H. M., Manley S., Bockus A., Kassel K. M., Luyendyk J. P., Copple B. L., Ding W. X. (2011) Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J. Pharmacol. Exp. Ther. 339, 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muoio D. M., Newgard C. B. (2008) Mechanisms of disease. Molecular and metabolic mechanisms of insulin resistance and beta cell failure in type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9, 193–205 [DOI] [PubMed] [Google Scholar]
- 41. Wang X., Yu W., Nawaz A., Guan F., Sun S., Wang C. (2010) Palmitate induced insulin resistance by PKCθ-dependent activation of mTOR/S6K pathway in C2C12 myotubes. Exp. Clin. Endocrinol. Diabetes 118, 657–661 [DOI] [PubMed] [Google Scholar]
- 42. Chen J. L., Lin H. H., Kim K. J., Lin A., Ou J. H., Ann D. K. (2009) PKCδ Signaling. A dual role in regulating hypoxic stress-induced autophagy and apoptosis. Autophagy 5, 244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr. (2000) Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 25, 87–90 [DOI] [PubMed] [Google Scholar]
- 44. Stone S. J., Myers H. M., Watkins S. M., Brown B. E., Feingold K. R., Elias P. M., Farese R. V., Jr. (2004) Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J. Biol. Chem. 279, 11767–11776 [DOI] [PubMed] [Google Scholar]
- 45. Yu C., Chen Y., Cline G. W., Zhang D., Zong H., Wang Y., Bergeron R., Kim J. K., Cushman S. W., Cooney G. J., Atcheson B., White M. F., Kraegen E. W., Shulman G. I. (2002) Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 277, 50230–50236 [DOI] [PubMed] [Google Scholar]
- 46. Benoit S. C., Kemp C. J., Elias C. F., Abplanalp W., Herman J. P., Migrenne S., Lefevre A. L., Cruciani-Guglielmacci C., Magnan C., Yu F., Niswender K., Irani B. G., Holland W. L., Clegg D. J. (2009) Palmitic acid mediates hypothalamic insulin resistance by altering PKCθ subcellular localization in rodents. J. Clin. Invest. 119, 2577–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eitel K., Staiger H., Rieger J., Mischak H., Brandhorst H., Brendel M. D., Bretzel R. G., Häring H. U., Kellerer M. (2003) Protein kinase Cδ activation and translocation to the nucleus are required for fatty acid-induced apoptosis of insulin-secreting cells. Diabetes 52, 991–997 [DOI] [PubMed] [Google Scholar]
- 48. Shahnazari S., Yen W. L., Birmingham C. L., Shiu J., Namolovan A., Zheng Y. T., Nakayama K., Klionsky D. J., Brumell J. H. (2010) A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 8, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Piro S., Anello M., Di Pietro C., Lizzio M. N., Patanè G., Rabuazzo A. M., Vigneri R., Purrello M., Purrello F. (2002) Chronic exposure to free fatty acids or high glucose induces apoptosis in rat pancreatic islets. Possible role of oxidative stress. Metabolism 51, 1340–1347 [DOI] [PubMed] [Google Scholar]
- 50. Borradaile N. M., Han X., Harp J. D., Gale S. E., Ory D. S., Schaffer J. E. (2006) Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 47, 2726–2737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.