Background: Yeast mitochondrial leucyl-tRNA synthetase (ymLeuRS) aids splicing of group I introns.
Results: β-Strand extensions that link editing and aminoacylation domains have adapted for splicing.
Conclusion: The ymLeuRS editing domain functionally diverged to accommodate splicing.
Significance: Housekeeping proteins modulate their original activities to acquire secondary functions.
Keywords: Aminoacyl tRNA Synthetase, Protein Synthesis, Ribozyme, RNA Splicing, Transfer RNA (tRNA)
Abstract
The yeast mitochondrial leucyl-tRNA synthetase (ymLeuRS) performs dual essential roles in group I intron splicing and protein synthesis. A specific LeuRS domain called CP1 is responsible for clearing noncognate amino acids that are misactivated during aminoacylation. The ymLeuRS CP1 domain also plays a critical role in splicing. Herein, the ymLeuRS CP1 domain was isolated from the full-length enzyme and was active in RNA splicing in vitro. Unlike its Escherichia coli LeuRS CP1 domain counterpart, it failed to significantly hydrolyze misaminoacylated tRNALeu. In addition and in stark contrast to the yeast domain, the editing-active E. coli LeuRS CP1 domain failed to recapitulate the splicing activity of the full-length E. coli enzyme. Although LeuRS-dependent splicing activity is rooted in an ancient adaptation for its aminoacylation activity, these results suggest that the ymLeuRS has functionally diverged to confer a robust splicing activity. This adaptation could have come at some expense to the protein's housekeeping role in aminoacylation and editing.
Introduction
The diverse family of aminoacyl tRNA-synthetases has been recruited to host alternate activities in the cell (1, 2) in addition to their housekeeping function in protein synthesis (3). This includes aiding splicing of group I introns in the mitochondria, which is essential to certain lower eukaryotes. Two fungal mitochondrial aminoacyl tRNA-synthetases, leucyl-tRNA synthetase (LeuRS or NAM2p) (4–6) and tyrosyl-tRNA synthetase (TyrRS or CYT-18p) (7, 8), are required protein splicing factors to facilitate splicing of group I introns in Saccharomyces cerevisiae and Neurospora crassa, respectively (2).
Yeast mitochondrial LeuRS (ymLeuRS)2 was originally implicated in group I intron splicing when suppressor mutations in LeuRS rescued RNA processing in the absence of a functional bI4 maturase (9). In addition to the bI4 maturase (10, 11) and also Mss116p (12), LeuRS aids splicing of the related bI4 and aI4α group I introns from the mitochondrial genes encoding cytochrome b (cob) and the α subunit of cytochrome oxidase (cox1α). We have demonstrated that LeuRS and bI4 maturase bind independently to the bI4 intron and stimulate RNA splicing (13, 14). However, the mechanistic details of how LeuRS promotes splicing remain unclear.
The connective polypeptide 1 (CP1) domain (15, 16) of LeuRS has been proposed to play a role in RNA splicing (17, 18). The ∼170-amino acid CP1 insertion splits the ATP binding Rossmann-fold that characterizes the aminoacylation active site of class I aminoacyl tRNA synthetases (16, 19) and folds discretely into a separate domain. The CP1 domain is connected to the rest of the enzyme via two flexible β-strands (Fig. 1A) (20). Although the LeuRS CP1 domain is best known for its role in proofreading or editing misacylated tRNALeu (21, 22), genetic rescue experiments identified splicing sensitive sites within and around the CP1 domain (9, 23). We had also shown that the isolated CP1 domain from ymLeuRS stimulated bI4 intron RNA splicing in vivo (17).
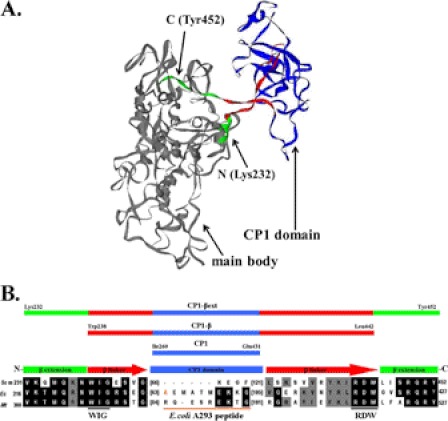
FIGURE 1.
Primary and tertiary structure of the LeuRS CP1 domain. A, shown is a homology model of yeast mitochondrial LeuRS (17). B, multiple sequence alignment of LeuRS CP1 domain and flanking regions containing both the N- and C-terminal β-strand linkers and extensions is shown. The CP1 domain, β-strand linkers, and β-strand extensions are shown in blue, red, and green, respectively. Bracketed numbers indicate the number of amino acids of a peptide insert. The E. coli LeuRS A293 residue (37, 38) is highlighted in orange, and the corresponding peptide indicated by an orange line. It is absent in the yeast mitochondrial LeuRS. Each of the yeast mitochondrial CP1 domain constructs is indicated at the top of the figure as follows: CP1 (Ile-260 to Glu-431), CP1-β (Trp-238 to Leu-442), and CP1-βext (Lys-232 to Tyr-452). The N- and C-terminal ends of the CP1-βext construct are also indicated in the homology model. Scm, S. cerevisiae mitochondrial; Ec, Escherichia coli; Mt, M. tuberculosis.
Herein, we utilized a LeuRS-dependent in vitro splicing assay (14) to isolate and characterize molecular determinants within the ymLeuRS CP1 domain and the connecting β-strands that directly impact splicing. We show how the LeuRS CP1 domain adapted to and functionally diverged to accommodate its dual essential activities in aminoacylation and splicing. In some cases LeuRS adaptations for splicing appear to have evolved at some expense to the enzyme housekeeping function in aminoacylation.
EXPERIMENTAL PROCEDURES
Materials
Oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA). Radiolabeled amino acids and nucleotides were purchased from Amersham Biosciences and PerkinElmer Life Sciences, respectively. Cloned Pfu DNA polymerase and dNTP mixture were obtained from Stratagene (La Jolla, CA). Restriction endonucleases and T4 DNA ligase were procured from New England Biolabs Inc. (Ipswich, MA) or Promega (Madison, WI). E. coli strains DH5α and BL21 (DE3) codon PLUS were obtained from Stratagene (La Jolla, CA). The plasmid pGP1–2 expressing T7 RNA polymerase was generously provided by Dr. Tracy Palmer (University of East Anglia, UK).
Plasmids and Mutagenesis
The plasmid pBETeCP1–2-35 (21) contains the gene fragment encoding the CP1 domain of E. coli LeuRS and was used for in vitro expression of the protein ecCP1-βext LeuRS. The ymLeuRS gene fragment encoding the ymCP1-β LeuRS fragment was amplified in a 50-μl polymerase chain reaction (PCR) that contained 100 ng of pYM3-17 (13) template plasmid DNA, 125 ng each of forward (ymCP1-β-Fwd(NdeI)(-Trp-238–Leu-442)) and reverse (ymCP1-β-Rev(BamHI)(Trp-238–Leu-442)) (supplemental Table S1) primer containing the NdeI and BamHI restriction sites, respectively, 0.05 mm dNTP mix, and 0.05 units of Pfu DNA polymerase in commercial buffer. The PCR products were digested with NdeI and BamHI at 37 °C for 6 h. The restriction-digested PCR products were separated on a 1% agarose gel and gel-purified using the QIAquick gel extraction kit-250 (Qiagen, Inc.). The vector pET-14b (Novagen, Gibbstown, NJ) was also cleaved with NdeI and BamHI followed by gel extraction. Gel-purified restriction digests of the PCR-amplified gene fragment and the vector were ligated using T4 DNA ligase at 37 °C for 15 min to yield the plasmid p14MBymCP1+N+C(RDWL). The ligation reaction was used to transform E. coli strain DH5α. Plasmid DNA was isolated from a 3-ml overnight culture of a single transformant using the QIAprep Spin mini prep kit-250 (Qiagen Inc).
The plasmid p14MBymCP1+N+C(RDWL) was used as template to generate the plasmid p14JSymCP1-βext, encoding the LeuRS protein fragment ymCP1-βext. This site-directed PCR-based insertion mutagenesis used primers ymCP1-βext-Cter-Fwd-(Trp-238–Arg-449), ymCP1-βext-Cter-Fwd-(Trp-238–Tyr-452), ymCP1-βext-Nter-Fwd-(Gln-235–Tyr-452), and ymCP1-Nter-βext-Fwd-(Lys-232–Tyr-452) supplemental Table S1). Mutations were introduced into the template plasmid as described above via PCR. The final PCR mixture was restriction-digested with 0.8 units of DpnI for 4 h at 37 °C and then used to transform E. coli strain DH5α. The plasmid p14JSymCP1-βext was used as a template to introduce the W238C point mutation with primer ymCP1-βext-Fwd-(W238C) (supplemental Table S1) to generate plasmid p14JSymW238C-CP1-βext, encoding the ymCP1-βext LeuRS that contained a W238C mutation. Likewise, the wild type full-length ymLeuRS encoding plasmid pYM3-17 (13) was used as the template to generate the plasmids pEXW238A, pEXW238C, pEXW238F, and pEXW238Y, encoding the mutant proteins W238A, W238C, W238F, and W238Y full-length ymLeuRSs. Mutations were confirmed by DNA sequencing (UIUC Core Sequencing Facility, Urbana, IL or Seq Wright, Houston, TX).
For in vivo studies in yeast strains HM410 and HM402 (23, 24), the plasmid pKIRAN (25) and ymLeuRST (26) were used to express wild type E. coli LeuRS and wild type ymLeuRS proteins, respectively. Both these plasmids contain a constitutive ADH promoter along with a 5′-tag encoding a mitochondrial import sequence and also a TRP marker for their selection in the HM410 and HM402 yeast strains. The plasmid ymLeuRST was used in site-directed PCR mutagenesis procedures to generate the plasmids pMPW238A, pMPW238C, pMPW238F, and pMPW238Y.
Preparation of Yeast Mitochondrial tRNALeu and Mischarged Ile-tRNALeu
T7 RNA polymerase (27, 28) was expressed from the plasmid pGP1–2. Yeast mitochondrial tRNAUAALeu (ymtRNALeu) was transcribed from the plasmid pymtDNALeu (13) as described in Hsu et al. (18). The concentration of the tRNA was calculated spectrophotometrically on the basis of its extinction coefficient estimated by the online Ambion oligonucleotide calculator (supplemental Table S2).
Misaminoacylation of ymtRNALeu with isoleucine was carried out by incubating the following reaction mixture at 25 °C for 3 h: 60 mm Tris, pH 7.5, 10 mm MgCl2, 10 mm KCl, 1 mm dithiothreitol (DTT), 8 μm transcribed ymtRNALeu, 23 μm [3H]isoleucine (90 Ci/mmol), 1 μm editing defective E. coli LeuRS and 4 mm ATP. The reaction was quenched under acidic conditions with 0.18% acetic acid (29) to stabilize the aminoacyl ester linkage between isoleucine and the terminal adenosine (A76) on tRNALeu. Protein was removed from the reaction mixture by phenol extraction using a 125:24:1 phenol:choloroform:isoamyl alcohol mixture, pH 4.3, (Fisher). Mischarged tRNA was ethanol-precipitated in the presence of 0.34 mg/ml glycogen at −80 °C overnight. The recovered tRNA pellet was washed twice with 70% ethanol, dried, and resuspended in 50 mm KPi, pH 5.0. Previous estimates suggest that the tRNA recovered is mischarged at ∼30% yields and the final preparation contains uncharged tRNALeu.
Precursor RNA Transcription
Plasmid pM96Δhj1-3 (14) encodes the gene for the shortest bI4 intron deletion mutant, bI4Δ1168 precursor RNA (pre-RNA), that has been shown to be catalytically active in vitro. A custom made RNA ladder ranging from 250 to 50 base pairs (14) was used for analyzing the splicing products by gel electrophoresis. Plasmids pUC8MB-250, pUC8MB-200, pUC8MB-150, pUC8MB-100, and pUC8MB-50 (14) encode specific RNAs ranging from 250 to 50 base pairs. Each plasmid was isolated from E. coli strain DH5α using a Qiagen Plasmid Mega Prep kit-25 (Qiagen Inc). The isolated DNA was then restriction-digested using 50 units of BamHI for the bI4Δ1168 RNA or SalI for the ladder RNAs overnight at 37 °C. RNA was transcribed from the respective restriction-digested DNA template using the MEGAscript T7 transcription kit (Invitrogen).
Transcription reactions were carried out using 1 μg of template DNA in a 20-μl reaction mixture at 37 °C for 4 h according to the manufacturer's protocol. The transcribed RNA was radiolabeled by including 10 μCi of [α-32P]UTP in the transcription reaction. The bI4Δ1168 pre-RNA was isolated on Chroma Spin+TE-200 columns (Clontech, Mountain View, CA). RNA concentration was estimated spectrophotometrically based on its extinction coefficient, calculated by the online Ambion oligonucleotide calculator (supplemental Table S2). The bI4Δ1168 pre-RNA was folded in vitro in the presence of 1 mm MgCl2 at 50 °C for 2 min followed by cooling at 32 °C for 10 min. The ladder RNA fragments were isolated by phenol:chloroform:isomyl alcohol [125:24:1] extraction followed by ethanol precipitation. The recovered RNA pellet was washed with 70% ethanol, vacuum-dried, and then dissolved in 50–100 μl of nuclease-free water (Invitrogen).
In Vitro Protein Expression and Purification
Wild type and mutant proteins were expressed from their respective plasmids in E. coli strain BL21 (DE3) codon PLUS as described in Hsu et al. (18). The final protein concentrations were determined spectrophotometrically at 280 nm using the respective extinction coefficients, estimated by the ExPASy Protparam tool (supplemental Table S2).
Aminoacylation Assay
Each aminoacylation reaction contained 60 mm Tris, pH 7.5, 10 mm MgCl2, 10 mm KCl, 1 mm DTT, 4 μm transcribed ymtRNALeu, 21 μm [3H]leucine (150 Ci/mmol), and 1 μm enzyme and was initiated with 4 mm ATP. At various time points 10-μl reaction aliquots were quenched on Whatman filter pads that had been pre-wet in 5% trichloroacetic acid (TCA) and then dried. The pads were then subjected to the following 10 min washes: 3 times with 5% TCA, once with cold 70% ethanol, and once with anhydrous ether. The pads were then dried under a heat lamp and quantitated for radioactivity in a liquid scintillation counter (Beckman LS 6500, Beckman Coulter, Fullerton, CA). GraphPad Prism software was used to plot data.
Deacylation Assays
Deacylation reaction mixtures contained 60 mm Tris, pH 7.5, 10 mm MgCl2, 10 mm KCl, and ∼2 μm [3H]Ile-ymtRNALeu (as indicated above, the mischarged tRNA substrate contains uncharged tRNA). The reactions were initiated with 1 μm enzyme and were carried out at 25 °C. At the desired time points, reaction aliquots of 5 μl were quenched on Whatman filter pads that had been pre-wet in 5% TCA and dried. Pads were washed and then quantified for radioactivity as described above.
LeuRS Dependent in Vitro Splicing Assay
The in vitro splicing reaction (14) included 1 μm 32P-labeled bI4Δ1168 pre-RNA (0.13 μCi/μl), 1 μm LeuRS, 150 mm KCl, Stop RNase inhibitor (5 Prime, Inc., Gaithersburg, MD) in 1× binding buffer (10 mm Tris, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 1 mm EDTA, and 10% glycerol). The reaction was carried out at 37 °C and initiated with 1 mm guanosine. Aliquots of 10 μl were quenched in 10 μl of 5 m urea, 30 mm EDTA, and mixed with 5 μl of RNA loading dye for incubation at 65 °C for 10 min.
Each aliquot was electrophoresed on a 6% denaturing urea-polyacrylamide gel overnight at 10 mA in 1× Tris borate-EDTA buffer. The gel was dried and phosphorimaged using a FUJIFILM BAS Cassette 2040 (FUJIFILM Medical Systems, Stanford, CT), and products were visualized by scanning the images using a STORM 840 Molecular Dynamics scanner (Amersham Biosciences). Images were quantified using ImageQuant software. Bands corresponding to the bI4Δ1168 precursor RNA, excised intron and ligated exons, for each time point were normalized by measuring the fraction of each relative band to the sum of the intensities of the three bands for the respective time points. Background intensity was subtracted based on the zero time point. The fraction of bI4Δ1168 pre-RNA remaining and extent of reaction for both of the products, bI4 and B4-B5, were plotted using the GraphPad Prism software.
In Vivo Yeast Complementation Assay
The yeast complementation assays utilized the yeast strains HM410 and HM402 (23, 24). Both HM410 (MATα ade2-1 his-11,15 leu2-3, 112 trp1-1 ura3-1 can1-100 nam2Δ::LEU2 [777-3A]) and HM402 (MATα ade2-1 his-11,15 leu2-3, 112 trp1-1 ura3-1 can1-100 nam2Δ::LEU2 [Δ introns]) are yeast null strains that contain a genomic/allelic disruption of the nam2 gene, encoding ymLeuRS, that is replaced by a LEU marker insertion. Wild type ymLeuRS fused to mitochondrial import sequence is expressed via a maintenance plasmid YEpGMCO63 that bears a URA3 marker. The ade2 mutation enables the cells to develop a red pigment when mitochondria are functional. The strain HM410 contains all 13 mitochondrial introns. All of these introns have been deleted in HM402.
Yeast HM410 and HM402 competent cells were transformed with the plasmids pMPW238A, pMPW238C, pMPW238F, or pMPW238Y, encoding the respective mutant ymLeuRSs, or with pKIRAN, encoding the wild type E. coli LeuRS, fused to an N-terminal mitochondrial import sequence. As positive and negative controls, yeast cells were also transformed with the plasmid ymLeuRST, encoding the wild type ymLeuRS, fused to a mitochondrial import sequence and the empty vector pQB153T, respectively. Each of these plasmids encodes a TRP marker. Selection of transformants that contain both the maintenance plasmid YEpGMCO63 with a URA3 marker and the mutant or heterologous LeuRS expressing plasmid with a TRP marker was carried out on synthetic complete (SC) Leu−Ura−Trp− dropout media. The selected transformants were then grown on glucose containing 5-flouroorotic acid. This negative selection step selects for cells that have lost the maintenance plasmid YEpGMCO63 (via its URA3 marker) that expresses wild type ymLeuRS. Thus, cells that grow on glucose-5-flouroorotic acid media are Ura− segregants that contain only the plasmid with a TRP marker that expresses the mutant or heterologous LeuRSs. The 5-flouroorotic acid-resistant colonies were then grown on SC Leu− Trp− dropout media containing either 2% glucose or glycerol to test for complementation activity (26).
Northern Blot Analysis
Total cellular RNA was extracted from yeast null strain HM410 expressing wild type ymLeuRS or the mutant ymLeuRSs (W238A or W238C). The cells were selectively grown in 25 ml of SC Leu− Trp− dropout media containing 5-flouroorotic acid until the A600 reached 1.5. The cultures were then harvested, and pellets were washed with ice-cold water. The washed pellets were resuspended in 400 μl of TES (10 mm Tris, pH 7.5, 10 mm EDTA, and 0.5% SDS) followed by hot acid phenol extraction at 65 °C for 1 h. The aqueous layer was extracted twice with chloroform. The recovered RNA was precipitated overnight with 40 μl of 3 m CH3COONa, pH 5.3, and 100% chilled ethanol (that was stored at −20 °C). The pellets were washed with 70% ethanol, dried, and resuspended in nuclease-free water (Ambion Inc., Austin, TX).
Up to 100 μg of total cellular RNA were electrophoresed on a 1% agarose gel containing 1.85% formaldehyde and 40 mm MOPS, 0.1 mm EDTA, 0.5 mm CH3COONa MOPS buffer. The RNA was transferred to a nitrocellulose membrane in 5× sodium salt citrate (SSC: 0.75 m NaCl and 0.075 m sodium citrate) overnight. RNA was UV-cross-linked to the nitrocellulose membrane for 1 min. The membrane was then prehybridized for 1 h at 42 °C with 25 ml of hybridization buffer (0.5 m NaPi, pH 7.2, 7% SDS, 1% BSA, and 0.04% EDTA).
A DNA oligomer that represents the ligated B4-B5 exon junction (5′-GAATATAGTTATCAGGATGACCTAAAGTATTAGGTGAATAG-3′) was radiolabeled in a reaction containing 0.3 μg of the oligo, 50 μCi of [γ-32P]ATP, and 10 units of T4 polynucleotide kinase (Promega). The radiolabeled probe was added to the prehybridized membrane, and hybridization was carried out overnight at 42 °C. The membrane was then washed 3 times for 10 min with 1× SSC and 0.1% SDS followed by drying. The dried membrane was exposed to FUJIFILM BAS Cassette 2040 (FUJIFILM Medical Systems), and the image was scanned using a STORM 840 Molecular Dynamics scanner.
RT-PCR Detection of Spliced Product
Denaturation of 25 μg of total cellular RNA was carried out by incubation at 95 °C for 5 min. This was followed by an annealing reaction at 48 °C for 1 h with 10 pmol of forward (bI4-F-5′-GGCATTACATATTCATGGTTCATC-3′) and reverse (bI5-R-5′-GGTGTTACTAAAGGATTACCAGGAA-3′) primers followed by reverse transcription at 37 °C for 90 min using 10 units of avian myeloblastosis virus reverse transcriptase (Promega). The PCR reaction was carried out by subsequent addition of 10 pmol of forward and reverse primers along with 5 units of Go Taq Flexi DNA polymerase (Promega). Subsequent to denaturation for 5 min at 95 °C, 30 cycles of PCR were carried out as follows: 94 °C, 1 min; 50 °C, 1 min; 72 °C for 2 min with a final extension for 10 min at 72 °C.
RESULTS
Isolated ymCP1 Domain Splices in Vitro, Unlike Its E. coli Counterpart
The isolated ymLeuRS CP1 domain is sufficient to support in vivo splicing activity of the bI4 intron RNA (17). To investigate CP1 adaptations that confer splicing activity in vitro, we designed three ymLeuRS protein fragments that contained the CP1 domain and various lengths of its flanking extensions (Fig. 1B). These constructions were based on available LeuRS crystal structures as well as biochemical investigations. Our goal was to incorporate the LeuRS protein fragments into a recently developed in vitro splicing assay for the bI4 intron (14) to distinguish adaptations of this housekeeping protein that are important to RNA splicing.
The CP1 domain is defined as a polypeptide insertion that splits the ATP binding Rossmann fold (15). However, several x-ray crystal structures show that the primary structure of CP1 folds as a discrete domain (20). The CP1 domain is linked to the LeuRS main body via the flexible N- and C-terminal β-strands (20). In ymLeuRS, the isolated CP1 domain (CP1; Fig. 1B) extends from Ile-260 to Glu-431. Previously we showed that the β-strands are required for deacylation of mischarged tRNA in the E. coli enzyme (21). Thus, we constructed a CP1-containing fragment that included the β-strands, extending from Trp-238 to Leu-446 (CP1-β; Fig. 1B). We had also determined that inclusion of short extensions from the β-strands to the aminoacylation core that contained conserved WIG and RDW motifs enhanced tRNA deacylation by the isolated E. coli CP1 domain (21). In addition to the β-strands, these extensions have been shown to be important for tRNALeu-protein interactions in E. coli LeuRS (21, 30, 31). We hypothesized that they might also be important for interacting with the bI4 intron RNA. Based on the E. coli LeuRS model (21), we designed a similar fragment for the ymLeuRS CP1 domain that includes extensions of the β-strands (CP1-βext; Lys-232 to Tyr-452; Fig. 1) to assess its contributions to RNA splicing.
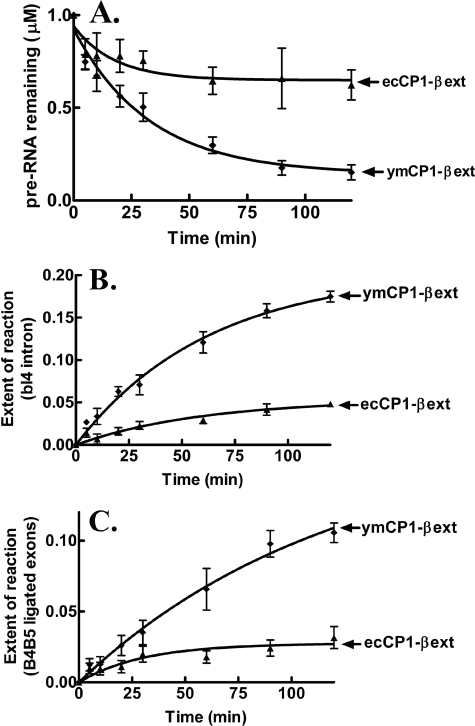
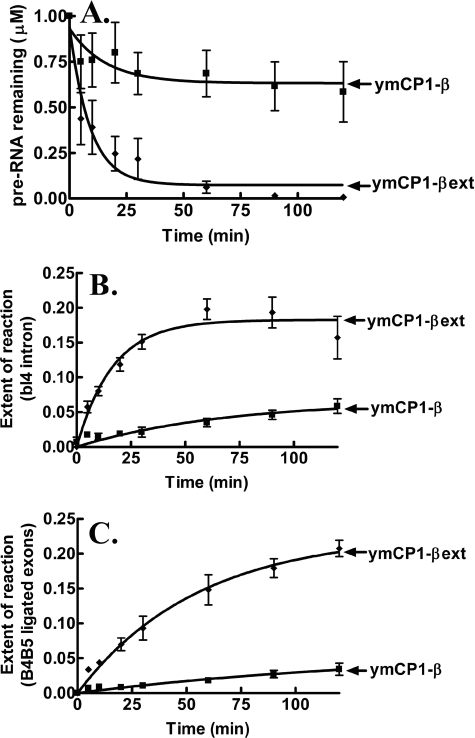
We analyzed the LeuRS CP1 domain-dependent splicing activity in vitro using a minimized bI4 intron (bI4Δ1168) (14) that was composed primarily of its catalytic core. The isolated ymCP1-βext LeuRS that contained the CP1 domain, the β-strands, and the extensions was similarly active compared with the full-length ymLeuRS (supplemental Fig. S1) in processing the bI4Δ1168 pre-RNA (Fig. 2A) and produced the expected bI4 intron and fused B4-B5 exon products (supplemental Fig. S2). The extent of the splicing reaction based on relative yields of the excised bI4 intron (Fig. 2B) and the ligated B4-B5 exons (Fig. 2C) was comparable.
FIGURE 2.
Species-specific LeuRS CP1 domain-dependent in vitro splicing of the bI4Δ1168 precursor RNA. The processing of the bI4Δ1168 pre-RNA was evaluated with respect to substrate pre-RNA processing (A), fraction of excised bI4 intron formed (B), and fraction of ligated B4-B5 exons formed (C). Calculations were based on the intensity of the phosphorimaged bands for the substrate and products bI4 and B4-B5 as well as other alternate bands that emerged during the reaction. Splicing reactions incorporated 1 μm pre-RNA and 1 μm LeuRS and were initiated with 1 mm guanosine. ▴, ecCP1-βext; ♦, ymCP1-βext. Error bars for each time point are the result of each reaction repeated in triplicate.
LeuRSs from diverse origins are active in splicing (26), and because this includes LeuRSs that do not encounter group I introns in their natural environment, we also tested the homologous CP1-βext (Val-216 to Ala-430) construct from E. coli LeuRS. These two CP1 domain protein fragments from ymLeuRS and E. coli LeuRS share ∼34% identity. The E. coli-based ecCP1-βext LeuRS protein fragment, in contrast to its counterpart from ymLeuRS, showed significantly reduced processing of the bI4Δ1168 intron (Fig. 2). Thus, even though it has been suggested that LeuRS uses its broadly conserved evolutionarily features to function in RNA splicing (26), the ymLeuRS CP1 domain, as we hypothesized, is more highly adapted for RNA splicing. Because key sites for editing are maintained, it is possible that a combination of editing-specific sites and regions outside the editing site are important for LeuRS splicing activity.
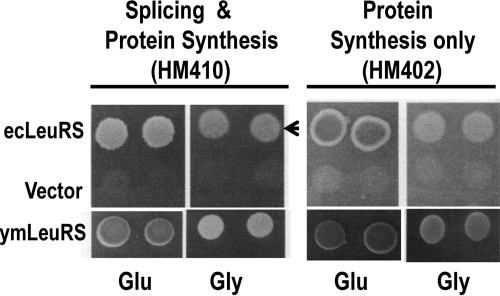
Full-length E. coli LeuRS Supports Splicing of bI4 Intron RNA in Vivo
Because the CP1 domain of E. coli LeuRS failed to support splicing of the bI4 intron in vitro, we asked whether the full-length E. coli LeuRS could substitute for the yeast enzyme in RNA splicing, similar to other LeuRSs that originated from Mycobacterium tuberculosis or human mitochondria (26). We transformed yeast null strains, HM410 and HM402, which have an allelic disruption of the nuclear encoded gene for the endogenous ymLeuRS, with the plasmid expressing full-length E. coli LeuRS fused to an N-terminal mitochondrial import sequence. The strain HM410 contains all 13 mitochondrial introns and would require both the splicing and aminoacylation functions of LeuRS. In contrast, the intronless strain HM402 would require only aminoacylation activity for growth. The full-length E. coli LeuRS supported growth of both HM410 and HM402 on glycerol media, indicating that the enzyme was active in both aminoacylation and splicing (Fig. 3). The full-length E. coli protein was also active when incorporated in the in vitro splicing assay (supplemental Fig. S3). Because the isolated E. coli LeuRS CP1 domain failed to support splicing in vitro, we hypothesize that the ymLeuRS contains evolutionary adaptations that are specific to the CP1 domain. This would allow its CP1 domain to be more robust in splicing in the absence of the full-length protein compared with the E. coli LeuRS CP1 domain.
FIGURE 3.
In vivo RNA splicing activity of full-length E. coli LeuRS. Complementation assays were performed using yeast null strains HM410 (with all 13 mitochondrial introns) and HM402 (intronless) (23). Complementation is indicated by darker colonies grown on glucose (Glu) media and by growth on glycerol (Gly) media. The arrow indicates that the full-length E. coli LeuRS supports growth of HM410 on Gly media. Both strains were transformed with the parent vector pQB153T or with plasmids expressing the wild type full-length ymLeuRS (pymLRST) or E. coli LeuRS (pKIRAN).
Functional Divergence of LeuRS CP1 Domain to Accommodate RNA Splicing and Protein Synthesis in Yeast Mitochondria
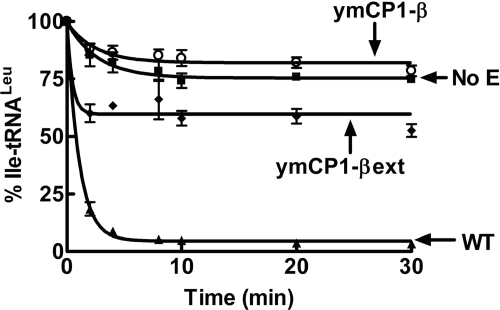
The CP1 domains from isoleucyl-tRNA synthetase (IleRS), valyl-tRNA synthetase (ValRS), and E. coli LeuRS that are independent from the rest of the enzyme effectively clear misacylated tRNAs (21, 32). The β-strands of the E. coli LeuRS CP1 domain are required for hydrolytic editing activity (21). As highlighted above, short extensions of the β-strands into the main body significantly enhance the hydrolytic activity of the E. coli LeuRS CP1 domain. They are hypothesized to bind and position the 3′-end tRNALeu for editing (21). We tested whether the β-strands and extensions of the ymLeuRS CP1 domain were similarly important to deacylation activity. In contrast to the E. coli LeuRS CP1 domain, inclusion of the β-strands in the ymCP1-β LeuRS failed to confer significant deacylation activity to the CP1 domain (Fig. 4). The kobs for deacylation by the ymCP1-β LeuRS was 0.3 ± 0.07 min−1 compared with 1.0 ± 0.11 min−1 for the full-length wild type ymLeuRS (supplemental Table S3). The addition of the extensions (ymCP1-βext LeuRS) only hydrolyzed misacylated Ile-tRNALeu at low levels (kobs = 0.3 ± 0.1 min−1 for 1 μm enzyme) (supplemental Table S3) compared with the full-length ymLeuRS (Fig. 4), albeit the extension additions increase the extent of the reaction. Increasing enzyme concentrations up to 5 μm did not show any significant increase in the observed rate constants for deacylation (supplemental Table S3 and Fig. S4).
FIGURE 4.
Deacylation activity of isolated ymLeuRS CP1 domain. Deacylation reactions included ∼2 μm [3H]Ile-tRNALeu and 1 μm enzyme. ▴, wild type; ○, ymCP1-β; ♦, ymCP1-βext; ■, no enzyme. Error bars for each time point are the result of each reaction repeated in triplicate.
We hypothesize that the ymLeuRS has functionally diverged at the CP1 domain to facilitate RNA splicing. This may have occurred at a cost to the enzyme CP1-based editing activity, as the full-length ymLeuRS clears misacylated tRNALeu via its CP1 domain (25) (Fig. 4). Interestingly, editing defects in full-length ymLeuRS had minimal effects on cell viability and mitochondrial function, although they are lethal to E. coli (25). It is possible that the cell as well as the mitochondrial LeuRS have adapted to accommodate both translational fidelity requirements and RNA splicing, which is essential to mitochondrial function.
Although the β-strands failed to enable amino acid editing by the isolated CP1 domain from ymLeuRS, we wondered if they would be sufficient in stimulating RNA splicing in vitro. As shown in Fig. 2, the ymCP1-βext LeuRS fragment, which includes the β-strands as well its extensions, is active in processing the bI4Δ1168 pre-RNA in vitro. To determine whether the addition of the β-strands alone to the CP1 domain can render the ymCP1 domain active in RNA splicing, we tested the ymCP1-β LeuRS protein fragment that includes only the β-strands but not the extensions (Fig. 1). It exhibited significantly reduced processing of the bI4Δ1168 pre-RNA as compared with the ymCP1-βext LeuRS fragment that includes both the β-strands and the extensions (Fig. 5).
FIGURE 5.
LeuRS CP1 domain extension-dependent splicing activity. In vitro bI4Δ1168 pre-RNA splicing was determined with respect to processing of the substrate pre-RNA (A), fraction of the excised bI4 intron formed (B), and fraction of the ligated B4-B5 exons formed (C). Splicing reactions incorporated 1 μm pre-RNA and 1 μm LeuRS and were initiated with 1 mm guanosine. ■, ymCP1-β; ♦, ymCP1-βext. Error bars for each time point are the result of each reaction repeated in triplicate.
It is noteworthy that the ymCP1-β LeuRS protein fragment contains the conserved WIG and RDW motifs at the end of the N- and C-terminal β-strands, respectively (Fig. 1). Thus, although these motifs provide critical interactions with the tRNA (30), they are not sufficient to confer optimal splicing activity to the ymLeuRS CP1 domain, isolated from the rest of the enzyme. We hypothesize that short extensions of the β-strands into the enzyme catalytic core domain are essential not only to confer hydrolytic editing activity but also to confer RNA splicing activity to the isolated CP1 domain of LeuRS.
WIG Peptide Extension of ymLeuRS Is Adapted for RNA Splicing
Genetic rescue experiments originally implicated Gly-240 as a critical factor in ymLeuRS splicing activity (23). This site is located in a peptide that contains a conserved WIG motif (supplemental Fig. S5) (16) in the extension of the N-terminal β-strand (Fig. 1) that links the CP1 domain of LeuRS to its aminoacylation core. Mutation of Gly-240 to serine suppresses splicing defects when the bI4 maturase is inactivated (4, 5, 23). Mutation of the Trp-238 to cysteine in the WIG motif abolishes splicing but retains aminoacylation activity in the ymLeuRS (23).
The comparison of multiple crystal structures indicates that the WIG-containing extension is flexible and assumes different conformations as LeuRS transitions through various substrate-bound states (20, 33). We predict that conformational changes within the WIG extension correlate to movements of the LeuRS CP1 domain and facilitate RNA-protein interactions. We mutationally investigated the molecular role of the WIG extension at its splicing sensitive site Trp-238. Substitutions included W238A, W238C, W238F, and W238Y in the full-length ymLeuRS.
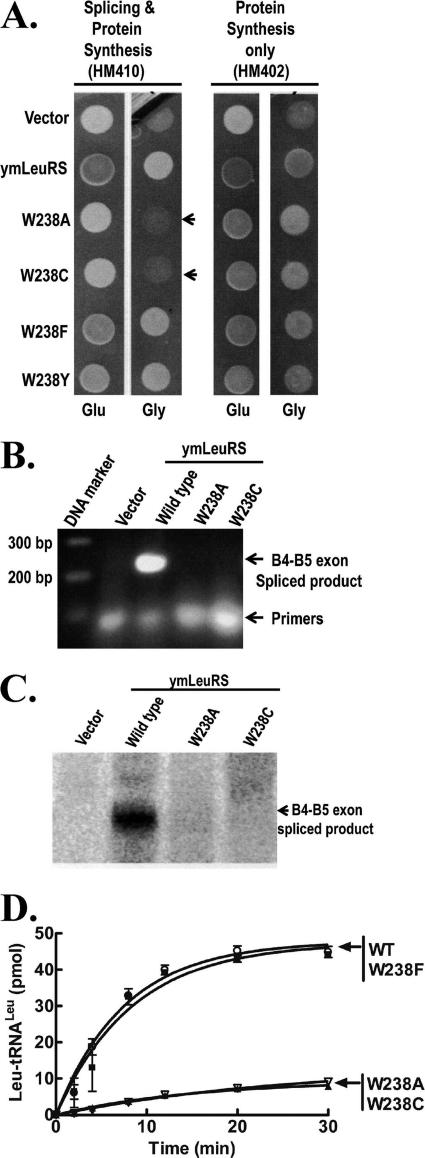
To distinguish the effects of Trp-238 on splicing and aminoacylation functions of LeuRS, we transformed wild type (HM410) and intronless (HM402) yeast strains with plasmids encoding the full-length wild type, W238A, W238C, W238F, and W238Y ymLeuRSs fused to an N-terminal mitochondrial import sequence. As expected, the full-length wild type and all the mutant ymLeuRSs complemented growth of the intronless strain HM402 on glycerol media (Fig. 6A), supporting that they are all active in aminoacylation. The full-length mutant W238F and W238Y ymLeuRSs also complemented the intron-containing HM410 strain (Fig. 6A). However, the full-length W238A and W238C ymLeuRS mutants failed to support growth of the wild type strain HM410 on glycerol media (Fig. 6A, indicated by arrows), indicating that these mutants are not active in splicing. Analysis of cell extracts from HM410 expressing full-length wild type or W238A or W238C ymLeuRSs via RT-PCR (Fig. 6B) and Northern blot (Fig. 6C) failed to detect the B4-B5 spliced product. As would be expected from the complementation assays, the full-length W238A and W238C mutants retained aminoacylation activity in vitro, albeit at significantly lower levels than the wild type enzyme (Fig. 6D). Thus, even these reduced levels of aminoacylation still meet an in vivo threshold that is necessary to support cell viability. It is also possible that the availability of appropriately modified native tRNA in the mitochondria elevates aminoacylation activity in vivo to acceptable threshold levels.
FIGURE 6.
The Trp-238 mutation in the WIG peptide abolishes splicing but maintains sufficient aminoacylation activity. A, complementation assays used yeast null strains HM410 and HM402 and is indicated by darker colonies grown on glucose (Glu) media and by growth on glycerol (Gly) media. Arrows highlight colonies expressing full-length W238A and W238C mutant ymLeuRSs support growth of HM402 but not of HM410 on Gly media. Both strains were transformed with the parent vector pQB153T or with plasmids expressing the full-length wild type (pymLRST) or W238A (pMPW238A), W238C (pMPW238C), W238F (pMPW238F), and W238Y (pMPW238Y) mutant ymLeuRSs. B, RNA amplification via RT-PCR of total cellular RNA from yeast null strain HM410 expressing full-length wild type and Trp-238 mutant ymLRSs yielded a 250 base pair (bp) amplified product representing the B4-B5 ligated exon product on a 1% agarose gel. C, shown is a Northern blot of 100 μg of total cellular RNA isolated from HM410 yeast cells expressing full-length wild type and Trp-238 mutant ymLeuRSs. Hybridization was carried out with a 32P-labeled B4-B5 exon junction probe. D, in vitro aminoacylation activity of full-length wild type and Trp-238 mutant LeuRS is shown. Reactions included 4 μm transcribed ymtRNALeu, 1 μm enzyme, and 21 μm [3H]leucine (150 μCi/ml) and were initiated with 4 mm ATP. ■, wild type; ▾, W238A; △, W238C; ○, W238F. Error bars for each time point result from each reaction repeated in triplicate.
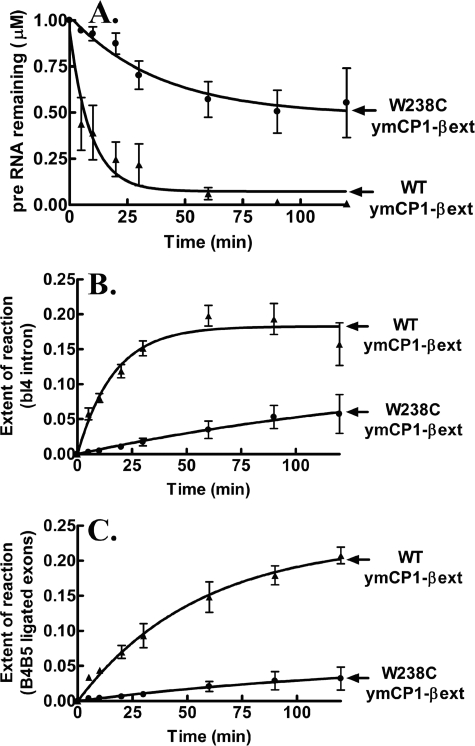
Because the full-length W238C ymLeuRS mutant failed to support mitochondrial splicing in vivo, we asked whether it would also compromise splicing in the isolated CP1 domain. We introduced the W238C mutation into ymCP1-βext LeuRS and tested its effect in our in vitro splicing assay. The W238C mutation significantly impeded processing of the bI4Δ1168 pre-RNA (Fig. 7A) and extent of formation of the products, bI4 intron (Fig. 7B) and B4-B5 ligated exons (Fig. 7C), in vitro. Thus, the W238C mutation, which abolished RNA splicing in the full-length ymLeuRS in vivo, is also critical to the isolated ymCP1 splicing domain. These combined results show that the Trp-238 site in the splicing sensitive WIG peptide requires a bulky aromatic residue for optimal interaction with the bI4 intron RNA. We hypothesize that the β-strand extension functionally diverged in ymLeuRS to adapt to its secondary alternate role in splicing while maintaining essential aminoacylation activity at sufficient levels to allow cell viability.
FIGURE 7.
In vitro splicing activity of isolated CP1 domain-based ymLeuRS Trp-238 mutants. Evaluation of in vitro bI4Δ1168 pre-RNA splicing with respect to processing of substrate pre-RNA (A), fraction of bI4 excised intron (B), and fraction of the ligated B4-B5 exons (C) is shown. Splicing reactions incorporated 1 μm concentrations of substrate pre-RNA and 1 μm LeuRS and were initiated with 1 mm guanosine. ▴, WT ymCP1-βext; ●, W238C ymCP1-βext. Error bars for each time point result from each reaction repeated in triplicate.
DISCUSSION
The bifunctional ymLeuRS is not only required for protein synthesis but also as an auxiliary protein factor in the excision of two related bI4 and aI4α group I introns for the expression of the mitochondrial cob and cox1α genes. The nuclear-encoded LeuRS and the intron-encoded mitochondrial bI4 maturase work in concert in a ternary complex formed on the bI4 intron. A direct role for LeuRS in splicing of these two introns has been established whereby LeuRS can independently bind to the bI4 intron and promote splicing (13).
Three different regions of the ymLeuRS have been reported to be important in the protein RNA splicing activity; they are the CP1 amino acid editing domain (17), the conserved WIG signature sequence (Fig. 1B) (23), and the C-terminal domain (18, 23). The yeast mitochondrial CP1 domain, isolated from the rest of the enzyme, has been shown to independently stimulate group I intron splicing in vivo (17). We utilized the isolated CP1 domain from ymLeuRS with different lengths of its flanking β-strands to probe the adaptations of the protein that facilitate aminoacylation and RNA splicing.
The isolated ymCP1-βext LeuRS processed a minimized bI4 intron in vitro, unlike its counterpart from the E. coli enzyme. Interestingly, however, the full-length E. coli LeuRS rescued bI4 intron splicing activity in vivo, similar to LeuRSs from other origins such as M. tuberculosis and human mitochondria (26). Thus, although RNA splicing activity has been proposed to be based on LeuRS broadly conserved evolutionary features, the CP1 domain of the splicing ymLeuRS appears to be adapted to facilitate RNA processing compared with its non-splicing counterpart from E. coli LeuRS.
To accommodate its RNA splicing activity, our results suggest that the ymLeuRS CP1 domain has functionally diverged. In vitro, the isolated ymCP1-β from LeuRS failed to hydrolyze misacylated Ile-tRNALeu. Inclusion of the short β-extensions in the isolated ymCP1-βext LeuRS enabled deacylation activity but at significantly lower levels as compared with the editing active full-length protein. This contrasts with the homologous isolated CP1 construct from E. coli LeuRS (21), where inclusion of the β-strand extensions retained deacylation activity that was comparable to the full-length enzyme (21).
Previously, we showed that the robust and viable post-transfer editing activity of full-length ymLeuRS appears to be dispensable to cell viability (25). It is possible that yeast mitochondria have evolved to tolerate increased infidelity during translation. Alternatively, they may possess distinct alternate mechanisms to suppress mistakes, such as precisely controlling mitochondrial amino acid concentrations or increased discrimination between cognate and noncognate amino acids.
Interestingly and unlike TyrRS (CYT18), LeuRS has not acquired an idiosyncratic domain or peptide (34, 35) insert that is dedicated to RNA splicing. Thus, we hypothesize that evolutionary pressures on the ymLeuRS CP1 domain for dual functions in the cell compromised its post-transfer editing activity to facilitate its secondary but essential role of RNA splicing.
The β-strands and its extensions provide critical contacts between the discretely folded CP1 domain and the LeuRS main body (20). They also provide flexibility to the CP1 domain to facilitate transitions of the enzyme between the different stages of the aminoacylation reaction (33, 36). In our investigation of structure-function relationships that are responsible for LeuRS CP1-based adaptations for RNA splicing, we determined that the β-strand extensions of the CP1 domain significantly enhance in vitro RNA processing activity. Mutational analysis in the aminoacylation-critical conserved WIG peptide at the end of the N-terminal β-strand demonstrated that a bulky aromatic residue is a requirement at this site to maintain RNA splicing activity of the ymLeuRS CP1 domain.
Significantly, the mitochondrial and bacterial LeuRS conserved WIG peptide was not sufficient to confer optimal splicing to the isolated ymCP1 domain and required the extensions of the β-strands into the canonical core to generate a splicing-active construct (ymCP1-βext). In the E. coli enzyme these extensions contain flexible hinge motifs that have critical interactions with the tRNA during post-transfer editing (33) and are essential to confer deacylation activity to the isolated CP1 domain (21). In lieu of a splicing-specific peptide or domain in LeuRS, it is possible that the WIG peptide and/or hinge regions intimately contact the intron RNA in the context of the isolated CP1 domain and full-length ymLeuRS. Direct or indirect interactions may facilitate RNA folding into a catalytically competent form. Because inclusion of these short extensions had only small effects on deacylation by ymCP1-βext LeuRS yet significantly enhanced its RNA splicing activity, we hypothesize that this region of LeuRS is differentially adapted to favor RNA splicing in ymLeuRS. Significantly, this contrasts with our results with the non-splicing adapted E. coli LeuRS, which relied on these extensions for tRNA deacylation.
Previously we showed that the leucine-specific C-terminal domain differentially adapted to accommodate RNA splicing and tRNALeu aminoacylation (18). In the non-splicing LeuRS, deletion of the C-terminal domain abolishes aminoacylation. In contrast, the deletion in the splicing ymLeuRS enhances aminoacylation but results in a splicing-inactive ternary complex of the C-terminal domain deletion mutant LeuRS:maturase:bI4 intron (18). Together these results suggest that common regions of the protein, such as the β-strands, its extensions, the conserved WIG peptide, and the C-terminal domain are critical to interactions with both tRNALeu and the bI4 intron. We hypothesize that these have been the localized sites where splicing LeuRSs have undergone differential adaptations.
The only other splicing-active aminoacyl tRNA-synthetase, TyrRS, relies upon newly acquired RNA intron binding insertions to recognize highly conserved secondary and tertiary structural features of group I introns. These insertions are distinct from the regions of the protein that bind tRNATyr (35). In contrast, LeuRS has not acquired any splicing-specific domain or peptide insertion. Key sites for editing remain intact. This LeuRS splicing factor relies on canonical features such as the CP1 domain-based WIG peptide and the β-strand extensions, which are evolutionarily conserved among bacteria and mitochondrial LeuRSs (supplemental Fig. S4). It is possible that within the pre-existing domains of LeuRS, splicing-sensitive molecular determinants are distinct from the ones that the protein uses for tRNALeu-binding. Thus, to accommodate dual essential functions in aminoacylation and splicing, we propose that the ymLeuRS functionally diverged within the confines of the CP1 domain and other regions, including the C-terminal domain and the consensus WIG motif.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM063789 and GM63107.

This article contains supplemental Tables S1–S3 and Figs. S1–S5.
- ymLeuRS
- yeast mitochondrial LeuRS
- ymtRNALeu
- yeast mitochondrial tRNAUAALeu
- CP1
- connective polypeptide 1
- pre-RNA
- precursor RNA
- SC
- synthetic complete
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- TRP
- tryptophan.
REFERENCES
- 1. Martinis S. A., Plateau P., Cavarelli J., Florentz C. (1999) Aminoacyl-tRNA synthetases. A family of expanding functions. EMBO J. 18, 4591–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambowitz A. M., Perlman P. S. (1990) Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem. Sci. 15, 440–444 [DOI] [PubMed] [Google Scholar]
- 3. Ibba M., Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 4. Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P. (1988) The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases and are involved in mRNA splicing. EMBO J. 7, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Labouesse M. (1990) The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 224, 209–221 [DOI] [PubMed] [Google Scholar]
- 6. Labouesse M., Dujardin G., Slonimski P. P. (1985) The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell 41, 133–143 [DOI] [PubMed] [Google Scholar]
- 7. Akins R. A., Lambowitz A. M. (1987) A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell 50, 331–345 [DOI] [PubMed] [Google Scholar]
- 8. Paukstelis P. J., Lambowitz A. M. (2008) Identification and evolution of fungal mitochondrial tyrosyl-tRNA synthetases with group I intron splicing activity. Proc. Natl. Acad. Sci. U.S.A. 105, 6010–6015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Labouesse M., Herbert C. J., Dujardin G., Slonimski P. P. (1987) Three suppressor mutations which cure a mitochondrial RNA maturase deficiency occur at the same codon in the open reading frame of the nuclear NAM2 gene. EMBO J. 6, 713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Labouesse M., Netter P., Schroeder R. (1984) Molecular basis of the “box effect.” A maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. Eur. J. Biochem. 144, 85–93 [DOI] [PubMed] [Google Scholar]
- 11. De La Salle H., Jacq C., Slonimski P. P. (1982) Critical sequences within mitochondrial introns. Pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell 28, 721–732 [DOI] [PubMed] [Google Scholar]
- 12. Huang H. R., Rowe C. E., Mohr S., Jiang Y., Lambowitz A. M., Perlman P. S. (2005) The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. U.S.A. 102, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rho S. B., Martinis S. A. (2000) The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA 6, 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boniecki M. T., Rho S. B., Tukalo M., Hsu J. L., Romero E. P., Martinis S. A. (2009) Leucyl-tRNA synthetase-dependent and -independent activation of a group I intron. J. Biol. Chem. 284, 26243–26250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Starzyk R. M., Webster T. A., Schimmel P. (1987) Evidence for dispensable sequences inserted into a nucleotide fold. Science 237, 1614–1618 [DOI] [PubMed] [Google Scholar]
- 16. Hou Y. M., Shiba K., Mottes C., Schimmel P. (1991) Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 88, 976–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rho S. B., Lincecum T. L., Jr., Martinis S. A. (2002) An inserted region of leucyl-tRNA synthetase plays a critical role in group I intron splicing. EMBO J. 21, 6874–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu J. L., Rho S. B., Vannella K. M., Martinis S. A. (2006) Functional divergence of a unique C-terminal domain of leucyl-tRNA synthetase to accommodate its splicing and aminoacylation roles. J. Biol. Chem. 281, 23075–23082 [DOI] [PubMed] [Google Scholar]
- 19. Nawaz M. H., Martinis S. A. (2008) Wiley Encyclopedia of Chemical Biology, pp. 1–12, John Wiley & Sons, New York [Google Scholar]
- 20. Cusack S., Yaremchuk A., Tukalo M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 19, 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Betha A. K., Williams A. M., Martinis S. A. (2007) Isolated CP1 domain of Escherichia coli leucyl-tRNA synthetase is dependent on flanking hinge motifs for amino acid editing activity. Biochemistry 46, 6258–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li G. Y., Bécam A. M., Slonimski P. P., Herbert C. J. (1996) In vitro mutagenesis of the mitochondrial leucyl tRNA synthetase of Saccharomyces cerevisiae shows that the suppressor activity of the mutant proteins is related to the splicing function of the wild type protein. Mol. Gen. Genet. 252, 667–675 [DOI] [PubMed] [Google Scholar]
- 24. Séraphin B., Boulet A., Simon M., Faye G. (1987) Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc. Natl. Acad. Sci. U.S.A. 84, 6810–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karkhanis V. A., Boniecki M. T., Poruri K., Martinis S. A. (2006) A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J. Biol. Chem. 281, 33217–33225 [DOI] [PubMed] [Google Scholar]
- 26. Houman F., Rho S. B., Zhang J., Shen X., Wang C. C., Schimmel P., Martinis S. A. (2000) A prokaryote and human tRNA synthetase provide an essential RNA splicing function in yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 97, 13743–13748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sampson J. R., Uhlenbeck O. C. (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. U.S.A. 85, 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grodberg J., Dunn J. J. (1988) ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 170, 1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schreier A. A., Schimmel P. R. (1972) Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry 11, 1582–1589 [DOI] [PubMed] [Google Scholar]
- 30. Nawaz M. H., Pang Y. L., Martinis S. A. (2007) Molecular and functional dissection of a putative RNA binding region in yeast mitochondrial leucyl-tRNA synthetase. J. Mol. Biol. 367, 384–394 [DOI] [PubMed] [Google Scholar]
- 31. Mascarenhas A. P., Martinis S. A. (2008) Functional segregation of a predicted “hinge” site within the β-strand linkers of Escherichia coli leucyl-tRNA synthetase. Biochemistry 47, 4808–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin L., Hale S. P., Schimmel P. (1996) Nature 384, 33–34 [DOI] [PubMed] [Google Scholar]
- 33. Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 34. Cherniack A. D., Garriga G., Kittle J. D., Jr., Akins R. A., Lambowitz A. M. (1990) Function of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing requires an idiosyncratic domain not found in other synthetases. Cell 62, 745–755 [DOI] [PubMed] [Google Scholar]
- 35. Paukstelis P. J., Chen J. H., Chase E., Lambowitz A. M., Golden B. L. (2008) Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature 451, 94–97 [DOI] [PubMed] [Google Scholar]
- 36. Fukunaga R., Yokoyama S. (2005) Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 12, 915–922 [DOI] [PubMed] [Google Scholar]
- 37. Williams A. M., Martinis S. A. (2006) Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity. Proc. Natl. Acad. Sci. U.S.A. 103, 3586–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li T., Guo N., Xia X., Wang E. D., Wang Y. L. (1999) The peptide bond between Glu-292–Ala-293 of Escherichia coli leucyl-tRNA synthetase is essential for its activity. Biochemistry 38, 13063–13069 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.