Background: Plasma membrane expression is vital for the function of ASICs, which act as extracellular proton sensors.
Results: Mutations in a conserved salt bridge and its adjacent region impaired cell surface ASIC expression.
Conclusion: Surface ASIC expression involves an exposed rigid signal patch at the extracellular loop.
Significance: This finding sheds lights on new strategies to prevent excessive neuronal excitability associated with ASIC activation.
Keywords: Acid-sensing Ion Channels (ASICs), Cell Surface Receptor, Ion Channels, Neurobiology, Protein Structure, Salt Bridge, Signal Patch, Surface Expression
Abstract
Acid-sensing ion channels (ASICs) are non-selective cation channels activated by extracellular acidosis associated with many physiological and pathological conditions. A detailed understanding of the mechanisms that govern cell surface expression of ASICs, therefore, is critical for better understanding of the cell signaling under acidosis conditions. In this study, we examined the role of a highly conserved salt bridge residing at the extracellular loop of rat ASIC3 (Asp107-Arg153) and human ASIC1a (Asp107-Arg160) channels. Comprehensive mutagenesis and electrophysiological recordings revealed that the salt bridge is essential for functional expression of ASICs in a pH sensing-independent manner. Surface biotinylation and immunolabeling of an extracellular epitope indicated that mutations, including even minor alterations, at the salt bridge impaired cell surface expression of ASICs. Molecular dynamics simulations, normal mode analysis, and further mutagenesis studies suggested a high stability and structural constrain of the salt bridge, which serves to separate an adjacent structurally rigid signal patch, important for surface expression, from a flexible gating domain. Thus, we provide the first evidence of structural requirement that involves a stabilizing salt bridge and an exposed rigid signal patch at the destined extracellular loop for normal surface expression of ASICs. These findings will allow evaluation of new strategies aimed at preventing excessive excitability and neuronal injury associated with tissue acidosis and ASIC activation.
Introduction
Acid-sensing ion channels (ASICs)4 are members of the epithelial sodium channel/degenerin family. In mammals, four asic genes have been found to encode at least six ASIC subunits (ASIC1a, -1b, -2a, -2b, -3, and -4) (1, 2), which form either homo- or heterotrimeric channels. ASICs are mainly expressed in neurons and activated by extracellular acidosis associated with pain pathogenesis (3–6), mechanosensation (7–9), neurotransmission (10), ischemia (11–13), and epilepsy (14). Recently, the first low pH crystal structure of chicken ASIC1 (cASIC1) has been resolved (15, 16), revealing a unique subunit topology: short cytoplasmic tails (N and C termini), two transmembrane (TM) helices (TM1 and TM2), and a large extracellular loop (Fig. 1A). The extracellular loop can be subdivided into “palm, knuckle, β-ball, finger, and thumb” domains, using a clenched hand for analogy (Fig. 1A). Accumulating evidence has implicated these subdomains in a variety of functions, most of which are related to channel activation (15–17). Nevertheless, the functional roles of many conserved structural elements revealed by the crystal structure still remain largely unexplored.
FIGURE 1.
Three-dimensional structure and sequence alignment of the Asp-Arg interaction region at the edge of the finger domain of ASICs. A, stereo view of the crystal structure of cASIC1 (Protein Data Bank code 3HGC) with key residues (displayed in gray sticks for emphasis) involved in forming the Asp108-Arg161 salt bridge enlarged. Red broken lines indicate hydrogen bonding among polar residues. B, sequence alignment of all different ASIC channels at the conserved salt bridge and adjacent residues. The numbering is based on the rASIC3 channel.
A salt bridge most commonly arises from the anionic carboxylate (R-COO−) of either aspartic acid or glutamic acid and the cationic ammonium (R-NH3+) of lysine or the guanidinium (RNHC(NH2)2+) of arginine, with the O–N distance required to be less than 4.0 Å. Salt bridges result from a combination of two noncovalent interactions: hydrogen bonding and electrostatic interactions. The electrostatic interactions have been implicated in the gating of ion channels, such as inwardly rectifying potassium channels 1.1 (18) and 3.1/3.4 (19), voltage-gated potassium channel 7 (20), and ATP-gated purinergic receptor 2 (21), as well as conformational transitions of other bioactive proteins (22–25). With a strong attractive force, the salt bridge stabilizes and restricts the conformation change that favors channel gating (18, 19, 21, 26–28) or binding with other bioactive substances (22–25).
We have observed considerable Asp(Glu)-Arg(Lys) interaction pairs in the extracellular loop of the crystal structure of cASIC1, of which functions are not fully explored. Using sequence alignment and alanine-scanning mutagenesis strategies, we systemically examined the roles of these putative salt bridges in ASIC gating. Here, we focus on a conserved salt bridge residing at the finger domain, namely the Asp108-Arg161 interaction pair on cASIC1 and the equivalent pairs, Asp107-Arg160 on human ASIC1a (hASIC1a) and Asp107-Arg153 on rat ASIC3 (rASIC3) channels. The crystal structure revealed that the acidic side chain of aspartate faces toward the basic side chain of arginine with the O–N distance stabilized at ∼3.0 Å, suggesting a strong electrostatic interaction between these Asp-Arg residues.
Using a diversity of approaches, including comprehensive mutagenesis, electrophysiological recording, charge swapping, cysteine cross-linking, Western blotting, immunocytochemistry, molecular dynamics (MD) simulations, normal mode analysis (NMA), and protein flexibility analysis, we demonstrate that this highly conserved extracellular salt bridge, together with its adjacent residues, which form a rigid signal patch, plays an essential role in normal cell surface expression of ASIC channels.
EXPERIMENTAL PROCEDURES
Solutions, Drugs, Cell Culture, and Transfection
All solutions and drugs were purchased and prepared as described previously (17). All DNA constructs were expressed in Chinese hamster ovary (CHO) cells as described previously (17). Briefly, CHO cells were cultured in F-12 medium at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Transfections of plasmids were performed using Hilymax (Dojindo Laboratories, Kumamoto, Japan) following the manufacturer's recommendation. Electrophysiological measurements were performed 24–48 h after transfection, whereas cell surface biotinylation and immunohistochemistry experiments were performed 24 h after transfection.
Site-directed Mutagenesis
As described previously (17), the cDNA of rASIC3 or hASIC1a was subcloned into the pEGFP-C3 vector. Each mutant was generated with the QuikChange® mutagenesis kit. Individual mutations were verified by DNA sequence analysis.
Electrophysiology
As described previously (17), ASIC currents were recorded with the whole cell patch-clamp method. In CHO cells, GFP-positive cells were selected for recordings of ASIC currents. Membrane currents were measured using a patch clamp amplifier (Axon 700A, Axon Instruments, Foster City, CA). The membrane potential was held at −60 mV throughout the experiment under voltage clamp conditions. All experiments were carried out at room temperature (23 ± 2 °C). Solutions were applied using a “Y-tube” method throughout the experiments (29).
Cell Surface Biotinylation and Western Blotting Analysis
Surface biotinylation was performed on cultured CHO cells following established protocols (30). Briefly, the cells were washed in chilled PBS+/+ (containing 1 mm MgCl2 and 0.1 mm CaCl2, pH 8.0) three times and then incubated with 2 mm sulfo-EZ-Link® Sulfo-NHS-LC-biotin (Pierce) in the same buffer at 4 °C for 30 min. The reaction was terminated by further incubating the cells with 20 mm glycine in PBS. After washing with chilled PBS+/+ three times, the cells were collected and lysed with radioimmune precipitation assay buffer. Biotinylated proteins were separated from the intracellular protein fraction using agarose resin linked to NeutrAvidin (Pierce) by incubation overnight at 4 °C and subsequent centrifugation. The beads were washed five times with ice-cold PBS, and bound proteins were eluted with the boiling SDS sample buffer, whereas a 10% volume of the supernatant (unbound fraction) was diluted with the SDS sample buffer and used as the total protein fraction. Protein samples from the biotinylation assay were analyzed by Western blotting. After boiling at 100 °C for 5 min, the samples were separated by SDS-PAGE and transferred to PVDF membrane. The membrane was incubated overnight at 4 °C with anti-GFP (1:1000; Roche Applied Science) and anti-ASIC1a (1:500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-transferrin receptor (1:500; Invitrogen), or anti-GAPDH (1:4000; Kang Cheng) antibodies, followed by appropriate HRP-conjugated secondary antibodies and finally visualized using an ECL solution (Pierce) and exposure to x-ray films for 1–3 min.
Insertion of Hemagglutinin (HA) Tag
To visualize the surface expression of ASIC channels, a modified HA epitope was inserted between Glu296 and Pro297 of wild-type (WT) or mutant rASIC3. The nucleotide sequence of the HA-tagged region was TACCCATACGATGTTCCAGATTACGCT.
Immunocytochemistry
CHO cells transfected with WT and mutants of EGFP:HA-ASIC3 were used. At 24 h post-transfection, cells were fixed with 4% paraformaldehyde/PBS for 10 min at room temperature. For the permeabilized experiments, cells were treated with 0.1% Triton X-100/PBS for 5 min at room temperature. Then cells were blocked for 1 h with PBS supplemented with 10% FBS. Cells were incubated overnight at 4 °C using an anti-HA antibody (Covance, Berkeley, CA) diluted at 1:1000 in 10% FBS/PBS. After rinsing with PBS, the cells were then treated with the secondary antibody conjugated to Alexa 594 (1:2000; Invitrogen) for 1 h at 37 °C and washed three times with PBS before the coverslips were mounted onto slides for visualization by confocal fluorescence microscopy.
Homology Modeling
As described previously (17), a homology model of rASIC3 was created based on the cASIC1 structure (Protein Data Bank entry 3HGC, 3.0 Å) using Modeller 9V7 (31). The sequence of rat ASIC3 was retrieved from the UniProt (entry O35240). The alignment of the target sequence with the template was performed mainly using the method reported by Jasti et al. (15). According to the secondary structure information of the template, the sequence alignment was adjusted manually to be more reasonable. The constructed model was checked and validated by the program Procheck (32). pKa values and structures under normal or acidic pH conditions were calculated using PROPKA version 2.0 (33).
Channel Dynamics Simulations
The titrated structures of WT or mutant ASIC3 were used as the starting structures for MD simulations. All simulations used the OPLS-AA force field for proteins and ions and the simple point charge model for water. A large 1-palmitoyl-2-oleoyl-phosphatidylcholine bilayer was constructed. Redundant lipids were subsequently removed from the constructed bilayer to generate a suitable membrane system into which the TM domain of the ASIC3 could be embedded. Counterions were subsequently added to compensate for the net negative charge of the system, and additional salt (150 mm NaCl or 90 mm CaCl2) was added to mimic the microenvironment under which ASIC3 channels would be kept at desensitization or resting state (34, 35). All MD simulations were performed by using the program Desmond (36), which uses a particular “neutral territory” method called the midpoint method to efficiently exploit a high degree of computational parallelism. All MD simulations were performed using the default Desmond parameters at constant temperature (310 K) and pressure (1 bar) by using the Berendsen coupling scheme with one temperature group. As described previously (37), NMA was conducted using the Web server developed by Delarue et al. (see the NOMAD-Ref Web site). Root mean square (r.m.s.) fluctuations of each residue from MD trajectories or NMA modes were calculated according to the procedure described previously (37).
Data Analysis
Results are expressed as means ± S.E. Data were analyzed with Clampfit 10.2 (Molecular Devices). Statistical comparisons were made with Student's t test; p < 0.05 (*) and p < 0.01 (**) are considered statistically significant.
RESULTS
Highly Conserved Salt Bridge in Extracellular Loop across ASIC Family
The three-dimensional structures of cASIC1 (15, 16) revealed that residues Asp108 and Arg161, which reside in the α1 and α3 short helices of the extracellular loop, respectively, with their side chains pointing to each other, form a salt bridge (Fig. 1A). Amino acid sequence alignment showed that these two residues are highly conserved across the ASIC family, forming a Asp107-Arg160 pair in hASIC1a and a Asp107-Arg153 pair in rASIC3 (Fig. 1B). Salt bridges are generally seen in flexible structural motifs of ion channels and commonly implicated in channel dynamics (e.g. channel gating, coupling, and conformational transitions) (26, 38, 39). However, the salt bridge identified here may be less flexible, given its location in, apparently, a rigid α-helix (Fig. 1A). Further protein flexibility analysis confirmed the inflexibility of the salt bridge (see below), suggesting an unusual role in ASIC functioning.
Mutations at Salt Bridge and Its Adjacent Residues Decrease Current Density of ASIC3 Channels
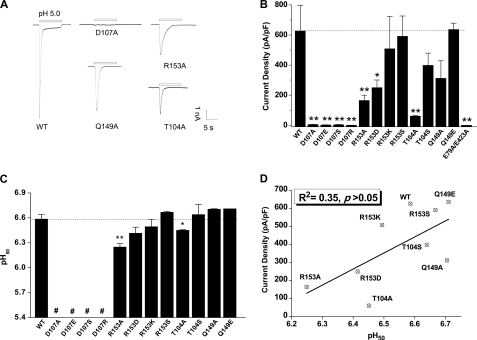
To elucidate the functional roles of the salt bridge, we first replaced the two residues with alanine (D107A and R153A) in rASIC3. CHO cells expressing the ASIC3D107A mutant showed negligible acid-induced currents. Similarly, cells expressing ASIC3R153A had markedly attenuated acid-induced currents as compared with those expressing WT channels (Fig. 2, A and B). This result suggests a crucial role of the Asp107-Arg153 pair in ASIC3 function. We then expanded mutagenesis to adjacent residues, namely Thr104 and Gln149 in rASIC3, which may play a role in stabilizing the conformation of the salt bridge via hydrogen bonding (see Fig. 1A, bottom; equivalent residues of cASIC1, Thr105 and Glu157). Indeed, alanine substitution of either Thr104 or Gln149 also reduced acid-induced currents (Fig. 2, A and B), consistent with a regulatory role of these residues. Additional mutagenesis with varying charges (D107R, R153D, and R153S) or side chain lengths (D107E, R153K, and T104S) was performed, and functional data confirmed that both steric hindrance and residue charges are important determinants of ASIC3 function because the mutations either fully abolished or tended to decrease acid-induced currents (Fig. 2B). Notably, any change with Asp107, even a minor alteration such as the D107E substitution, completely abolished acid-induced currents in CHO cells, supporting the notion that the Asp107-Arg153 pair and their adjacent residues contribute to ASIC3 function.
FIGURE 2.
Effects of mutations at the salt bridge on acid-induced currents in ASIC3 channels. A, representative pH 5.0-induced current traces of ASIC3-WT and ASIC3 mutants, D107A, R153A, Q149A, and T104A, expressed in CHO cells. B, pooled data showing acid-induced changes in current density of WT and mutations at Asp107 and Arg153 as well as adjacent residues Thr104 and Gln149. The E79A/E423A mutant was used as a loss-of-function control. Data are means ± S.E. (error bars) from 5–10 experiments. **, p < 0.01; *, p < 0.05, unpaired t test versus the WT channels (broken line). C, pH50 of WT channel and all mutants. #, pH50 was not measured due to the lack of detectable currents. **, p < 0.01; *, p < 0.05, unpaired t test versus the WT channels (broken line). D, lack of linear correlation between current density and pH50 values for the WT channel and mutations in the Asp107–Arg153 region (R2 = 0.35, p > 0.05).
Impairment of Salt Bridge Does Not Alter pH Sensing
The pH at which a protein is placed is crucial for the stability of salt bridges, largely due to alterations in electrostatic interactions. At pH extremes, two amino acids participating in a salt bridge could lose their ability to interact because one will lose its charge. Thus, the decrease in acid-induced current in the mutated ASIC3 channel could be attributed to impaired pH sensing mediated by the salt bridge and its related conformational transitions (26, 38, 39). To test this possibility, we compared the apparent proton affinities between WT and the functional mutant channels by measuring pH50 values (pH required for inducing half-maximal currents). Unexpectedly, in contrast to the marked change of current density, no change or only slight changes were observed for the pH50 (Fig. 2C). In support of such an observation, we found poor correlation between the current density and the apparent proton affinity (R2 = 0.35, p > 0.05; Fig. 2D).
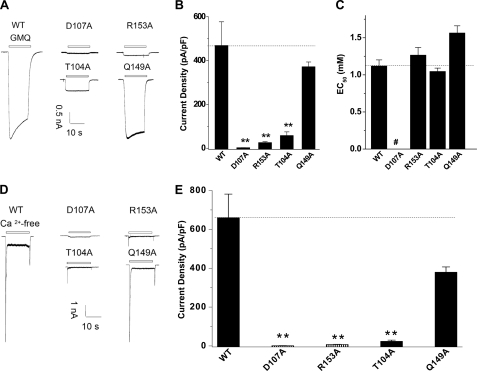
We previously reported that ASIC3 channels could be activated independent of acidosis by either a synthetic small molecule (2-guanidine-4-methylquinazoline (GMQ)) or the deprivation of extracellular Ca2+ (17). Taking advantage of these nonproton activation approaches, we further tested the activation of ASIC3 mutants in the absence of pH changes in order to rule out a role of the salt bridge in pH sensing. Indeed, we found that the mutations (D107A, R153A, and T104A) also markedly reduced GMQ- and Ca2+ deprivation-induced ASIC3 activation (Fig. 3, A, B, D, E) without affecting the EC50 for GMQ (GMQ concentration required for inducing half-maximal currents) in all functional ASIC3 mutants (Fig. 3C). Together, these data suggest that the residues forming the salt bridge do not directly sense protons. Thereby, other mechanisms may play a leading role in channel dysfunction observed in the ASIC3 mutants.
FIGURE 3.
Effects of mutations at the salt bridge on nonproton activator-induced currents in ASIC3 channels. A, representative GMQ (5 mm)-induced current traces of ASIC3-WT channel and mutants, D107A, R153A, Q149A, and T104A. B, pooled data showing GMQ-induced changes in current density of WT and all mutants. Data are means ± S.E. (error bars) from 5–7 experiments. **, p < 0.01, unpaired t test versus the WT channels (broken line). C, EC50 of GMQ for WT channel and all mutants. #, EC50 was not measured due to the lack of detectable current activation. D, representative current traces induced by removing extracellular Ca2+ for ASIC3-WT and mutants D107A, R153A, Q149A, and T104A. E, pooled data showing the Ca2+ deprivation-induced changes in current density of WT and all mutants. Data are means ± S.E. from four to five experiments. **, p < 0.01, unpaired t test versus the WT channels (broken line).
Breaking Salt Bridge Reduces Cell Surface Expression of ASIC3 Channels
Decreased protein expression level or defective trafficking to the cell surface could also account for partial or complete loss of channel function in mutated channels. Indeed, for mutants at Asp107-Arg153 and adjacent Thr104 and Gln149, although the total protein levels were comparable with the WT, cell surface expression levels were significantly decreased (Fig. 4, A, B, and D). As a control, we also tested a known mutant (E79A/E423A) with gating defects (Fig. 2B) unrelated to the salt bridge disruption (17). The expression of the E79A/E423A mutant on the cell surface was comparable with that of WT channels (Fig. 4, C and D). Thus, disruption of the salt bridge reduced ASIC3 current density predominantly through decreasing cell surface expression. This change does not seem to result from altered protein synthesis or degradation because the total ASIC3 protein levels were unchanged (Fig. 4, A–C).
FIGURE 4.
Cell surface expression of WT and mutant ASIC3 channels. A–C, representative Western blotting of the cell surface expression of WT and various ASIC3 mutants. The cell surface-expressed channels were determined by biotinylation under non-permeabilized conditions (see “Experimental Procedures”). D, pooled data showing changes in surface expression levels of mutations at the Asp107–Arg153 region. The relative levels of ASIC3 expression were quantified by densitometry. **, p < 0.01 versus WT (broken line), Student's paired t test. Error bars, S.E.
Immunohistochemistry Confirms Functional Role of Salt Bridge in Surface Expression
The reduced surface expression was further verified using immunohistochemistry. For this purpose, we inserted an HA epitope into the extracellular loop of ASIC3 (Fig. 5A). We selected the site of insertion based on alignment of the ASIC family, which revealed an extra segment specific to ASIC3 in the extracellular loop just below the thumb domain (Fig. 5A; see “Experimental Procedures”). This construct (EGFP:HA-ASIC3) was functional when expressed in CHO cells, with current kinetics and density comparable with the WT channels (Fig. 5B).
FIGURE 5.
Construction and function of the extracellularly HA-tagged ASIC3 channel. A, sequence alignment of ASIC family members at the area where an HA tag was inserted. The rectangle and arrows indicate the location of the inserted HA tag immediately after Glu296 in the extracellular loop of the ASIC3 channel. An EGFP sequence was fused to the N terminus of the same construct (inset diagram). B, representative pH 5.0-induced current traces (top) and pooled data (bottom) from CHO cells expressing EGFP-ASIC3 and extracellularly HA-tagged EGFP-ASIC3 (EGFP:HA-ASIC3). Pooled data are means ± S.E. (error bars) from five independent experiments.
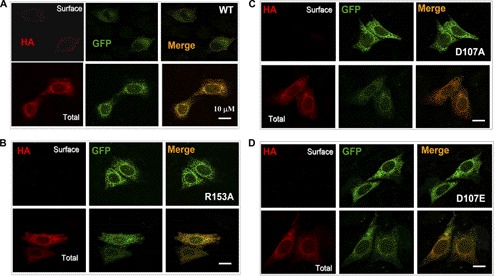
To visually measure differences between WT and mutant channels in surface expression, we constructed three additional mutants (HA-ASIC3D107A, HA-ASIC3D107E, and HA-ASIC3R153A) based on EGFP:HA-ASIC3 (with a GFP tag). Immunolabeling of non-permeabilized cells expressing the HA-tagged WT channel using the anti-HA antibody revealed a pattern of HA labeling (red) around the cell periphery that overlapped with the GFP (green) fluorescence only at the outside margin of cells (orange) (Fig. 6A, top). In permeabilized cells, the GFP and HA fluorescence colocalized perfectly (Fig. 6A, bottom). In contrast, cells expressing GFP:HA-tagged ASIC3D107A, ASIC3D107E, or ASIC3R153A showed undetectable or very weak background-like fluorescence with HA labeling under non-permeabilized conditions, despite the prominent labeling and colocalization of the HA signal with GFP in permeabilized cells (Fig. 6, B–D). These results together with the Western blotting assay (Fig. 4) demonstrate an essential role of the salt bridge in supporting cell surface expression of ASIC3 channels.
FIGURE 6.
Immunocytochemistry analysis of CHO cells expressing mutated EGFP:HA-ASIC3 channels. A–D, immunocytochemistry analysis of HA staining of CHO cells transfected with EGFP:HA-ASIC3, EGFP:HA-ASIC3R153A, EGFP:HA-ASIC3D107A, and EGFP:HA-ASIC3D107E, with (total) or without (surface) permeabilization. HA, immunofluorescence of HA labeling with a mouse monoclonal anti-HA antibody followed by anti-mouse IgG-Alexa594. GFP, GFP fluorescence of the transfected cells. Merge, overlay of GFP and HA fluorescence. Scale bars, 10 μm.
Equivalent Salt Bridge (Asp107-Arg160) Contributes to Surface Expression of ASIC1a
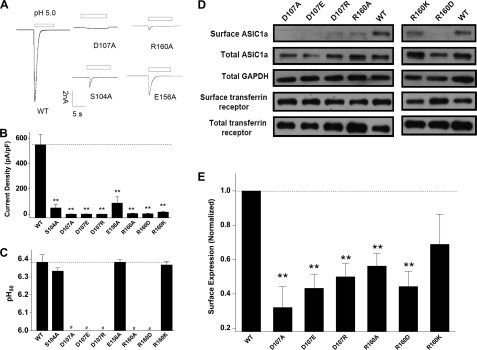
To determine how common among ASIC channels the extracellular salt bridge is in supporting cell surface expression, we analyzed the role of Asp107-Arg160 in hASIC1a (Fig. 1B). As with the mutations of ASIC3, equivalent mutations at ASIC1a (Asp107, Arg160, Ser104, and Glu156) profoundly decreased acid-induced current density of ASIC1a (Fig. 7, A and B). Also similar to the ASIC3 mutants, pH50 values of the functional ASIC1a mutants did not have an apparent change (Fig. 7C). In addition, surface expression levels of Asp107 and Arg160 mutants were significantly reduced although to a less extent compared with the equivalent ASIC3 mutants (Fig. 7, D and E). These results suggest a common mechanism governing ASIC surface expression that requires a conserved salt bridge at the extracellular loop.
FIGURE 7.
Current density and surface expression levels of WT and mutant hASIC1a channels. A, representative pH 5.0-induced current traces of ASIC1a-WT channel and mutants D107A, R160A, S104A, and E156A. B, pooled data showing the acid-induced changes in current density of the WT channel and all mutants. Data are means ± S.E. (error bars) from 5–10 experiments. **, p < 0.01, unpaired t test versus the WT channels (broken line). C, pH50 of the WT channel and all mutants. #, pH was not measured due to negligible response. D, representative images of Western blots of the cell surface expression of ASIC1a-WT channels and Asp107 and Arg160 mutants. E, pooled data showing changes in the surface expression levels of mutant ASIC1a channels. Data (mean ± S.E.; n = 3–5) were normalized to the surface expression level of the WT channel. **, p < 0.01 versus WT (broken line), by Student's paired t test.
Salt Bridge Is Stable under both Acidic and Neutral pH Conditions during MD Simulations
To obtain further insights into the structural characteristics of the salt bridge competent for the surface expression of ASIC3 channels, we created a three-dimensional homology model of ASIC3 (35) (Fig. 8A, middle), based on the crystal structure of cASIC1, which is ∼50% identical to rASIC3 at the amino acid level (see “Experimental Procedures”). We then measured pKa values of the salt bridge (pKa = 2.19, 1.82, and 2.07 for Asp107 of subunit A, B, and C, respectively; pKa = 11.87, 11.31, and 11.80 for Arg153 of subunit A, B, and C, respectively; see “Experimental Procedures”). These calculated pKa values suggest that the salt bridge should exhibit an excellent stability in response to acidosis. Given that physiological variations in pH are not acidic enough to deprive the negative charges of Asp107, these data further support the notion that Asp107-Arg153 may not directly mediate pH sensing (Fig. 2, C and D).
FIGURE 8.
Stable Asp107-Arg153 interactions during MD simulations. A, stereo view of the salt bridge and adjacent residues in the homology model of rASIC3 (middle) and top view of MD simulations under the conditions as indicated (left and right). Broken lines in the middle panel indicate hydrogen bonding between polar residues. In the left and right panels, subunits A, B, and C are colored orange, red, and green, respectively. Blue, purple, and cyan balls indicate Na+, Ca2+, and Cl−, respectively. B and C, distance of the charged atoms (O−–N+) of the salt bridge during MD simulations at pH 5.0 (B) and pH 7.4 (C) conditions. D, total number of hydrogen bonds (H-bonds) between Asp107 and Arg153 detected in trimetric ASIC3 channels during MD simulations.
To better predict the stability of the salt bridge, two sets of MD simulations were carried out at either acidic or neutral pH (Fig. 8A). Titrated ASIC3 channels at pH 5.0 and 7.4 were placed in normal (150 mm Na+) and high Ca2+ (90 mm) solutions, respectively, to mimic the microenvironments under which ASIC3 was kept stable at the desensitized or resting states (34, 35) (see “Experimental Procedures”). Both MD simulations and salt bridge interaction analysis indicate that the salt bridge is rigid, with distances between negatively (O−) and positively (N+) charged atoms varying from 3.0 to 3.5 Å during the simulation (Fig. 8, B and C), suggesting a persistent electrostatic interaction between Asp107 and Arg153. Moreover, although minor fluctuations of hydrogen bonding between Asp107 and Arg153 were observed, the total number of hydrogen bonds was limited to 5–6 (Fig. 8D) in the trimeric channel, further indicating the high stability of the salt bridge. These results, together with the results of pKa calculations, consistently suggest that the Asp107-Arg153 pair restrained by hydrogen bonding and ionic interaction is very stable at either desensitized or resting states.
Rigid Salt Bridge Is Important for Regional Conformational Stability of ASIC Channels
To better understand the functional significance of the high stability of the salt bridge, we performed MD simulations for the ASIC3D107A mutant, in which the salt bridge is disrupted. r.m.s. fluctuation of Cα of adjacent residues was calculated to check the structural fluctuations of the WT and mutant channels. The results revealed that residues adjacent to the salt bridge were more flexible in ASIC3D107A than in WT channels (Fig. 9A). Moreover, the virtual mutation ASIC3D107E, which added a methylene at the side chain compared with WT (supplemental Fig. S1A) caused an obvious bending at D107E (supplemental Fig. S1C) and thus broke the original salt bridge. These results implied that the regional conformation of the salt bridge is extremely precise, and the presence of a stable salt bridge may be important for restraining the conformations of adjacent residues that are critical for ASIC3 surface expression.
FIGURE 9.
Stable Asp107-Arg153 interaction is critical for surface expression of ASIC3 channels. A, r.m.s. fluctuations of ASIC3D107A and ASIC3-WT channels. B–E, representative Western blotting (B and D) and pooled data (C and E) of the cell surface expression of WT, cysteine cross-link mutation (B and C), and charge swap mutation (D and E). Data (mean ± S.E.; n = 4–5) were normalized to the surface expression level of WT ASIC3. *, p < 0.05 versus WT, Student's paired t test. F, total number of hydrogen bonds between Asp153 and Arg102 detected in trimetric ASIC3D107R/R153D channels during MD simulations. G, r.m.s. fluctuations of ASIC3D107C/R153C and ASIC3D107R/R153D channels.
To test this idea, we designed two strategies (i.e. cysteine cross-linking and charge swap) to mimic and/or restore the restraints conferred by the salt bridge. As expected, the double cysteine substitutions partially restored the ASIC3 surface expression, whereas single substitutions (ASIC3D107C and ASIC3R153C) did not (Fig. 9, B and C). Cysteines can automatically connect to each other when their distance is less than 5 Å (27). MD simulations in supplemental Fig. S2A show that the two cysteines are able to form a strong covalent interaction, although the resulting structure does not overlap well with the WT at the finger domain, which suggested that an obvious conformational change still exists (supplemental Fig. S2C). For charge-swapped mutant (ASIC3D107R/R153D), the surface abundance showed no significant improvement compared with that of single mutants (ASIC3D107R and ASIC3R153D; Fig. 9, D and E). The result of MD simulations indicates that the side chains of Asp153 do not face directly toward Arg107; instead, Asp153 interacts with both Arg102 (their number of hydrogen bonds is shown in Fig. 9F) and Arg107 (supplemental Fig. S2B), with a much weaker attraction force and an incorrect orientation compared with that of the salt bridge in the WT channel (supplemental Fig. S2B). Furthermore, the charge-swapped salt bridge exhibited a weaker restraint on the fluctuations of adjacent residues when compared with that of the cysteine-cross-linking mutant during MD simulations (Fig. 9G). Yet, the degree of overlap with the WT for ASIC3D107C/R153C in the finger domain is better than that for ASIC3D107R/R153D (supplemental Fig. S2C, blue arrow), although there are still regions where ASIC3D107C/R153C differs more from the WT than ASIC3D107R/R153D (supplemental Fig. S2C, yellow arrow). Together, these analyses suggest that the conserved salt bridge located at the extracellular loop is stringently precise, and it is crucial for the normal surface expression of ASIC3 channels.
Salt Bridge Separates Adjacent Rigid Signal Patch from Flexible Finger Domain
The finding that only the stabilized extracellular salt bridge is able to support ASIC surface expression prompted us to look into its detailed structural basis. For this purpose, NMA simulations, a computational approach that can effectively predict the extensive collective dynamics and inherent flexibilities in biological macromolecules (37, 40), were carried out on the rASIC3 three-dimensional model. NMA results and r.m.s. fluctuation analysis uncovered an extremely rigid region, which is surrounded by domains, including the finger, thumb, β-ball, knuckle, and upper palm domains (Fig. 10, A and B). This region is formed by Asp107, Arg153, Arg99, Glu159, Asp160, Asp216, Gln218, Gln219, Glu220, Glu221, and adjacent residues (Fig. 10B), most of which exhibited low r.m.s. fluctuation values (Fig. 10C). Any single mutation at this rigid region, designated as the “signal patch” (e.g. E159L, D160L, D216A, Q218L, Q219L, E220L, E221L, or R99A) markedly reduced ASIC3 surface abundance (Fig. 11), whereas mutations on identical or adjacent residues in hASIC1a (Q225C, Q226C, I224C, and N96C) also led to significant decreases in cell surface expressions (41). In contrast, the channel gating-related finger domain is the second most flexible domain in the entire extracellular loop, after loop-β9 (Fig. 10C). Most notably, the Asp107-Arg153 interaction pair is situated in the interface between the flexible finger domain and the rigid signal patch (Fig. 10, B and C).
FIGURE 10.
Separation of a flexible gating domain from a rigid region by the Asp107-Arg153 salt bridge. A, side view of a rigid region surrounded by multiple domains. The knuckle, finger, β-ball, thumb, loop-β9 and upper palm domains are colored pink, gold, purple, blue, yellow, and green, respectively. The residues involved in forming different domains are summarized in the table to the right. B, structure and key residues (displayed in gray sticks for emphasis) of the rigid region. The subunits A, B, and C are colored purple, yellow, and green, respectively. The two residues that form the salt bridge are indicated in red. C, r.m.s. fluctuations of ASIC3 channels in NMA analysis. Pink, orange, purple, and yellow columns highlight the fluctuations of residues in the knuckle, finger, β-ball, and loop β9 domains, respectively.
FIGURE 11.
Effects of mutations in the rigid region on the surface expression of ASIC3 channels. A, representative Western blotting of the cell surface expression of WT ASIC3 and mutants as indicated. B, pooled data showing the surface expression levels of mutant ASIC3 channels normalized to that of the WT after quantification by densitometry. Data are means ± S.E. (n = 3–5). **, p < 0.01 versus WT (broken line), Student's paired t test.
DISCUSSION
As the key extracellular proton sensors, ASICs are characterized by a large extracellular loop, within which many conserved residues with unknown functions have been identified. High resolution crystal structures of the ASIC1 channel (15, 16) caught in the desensitized state informed us of the presence of a salt bridge, lying at the edge of the finger domain of the large extracellular loop between two helices, formed by side chains of two residues that are highly conserved among ASIC family members. We used a range of complementary techniques to demonstrate that the salt bridge and a structurally rigid signal patch adjacent to it are essential for normal cell surface expression of ASIC3 and ASIC1a channels. Along with the resolution of the crystal structures, extensive investigations at the finger domain have demonstrated its role in channel gating. As the most flexible portions, the motion of the thumb and finger domains is vital to channel gating. Shaikh et al. (42) proposed that protons bind to an acidic pocket between the finger and thumb domains, whereas Yang. et al. (37) suggested that the attraction between the thumb and finger is the initial driving force of extracellular loop movement. These previous studies support that the finger domain mainly functions as a gating element. Here we demonstrate the presence of a patch of residues near the finger domain that are structurally rigid in the tertiary structure of ASICs (Fig. 10). This signal patch forms a firm region exposed to the protein surface as revealed in MD and NMA simulations, resisting structural fluctuations potentially exerted by the nearby flexible finger domain, thereby allowing properly folded ASICs to be recognized by the trafficking machinery for export to the cell surface.
It would be speculative, yet interesting, to find out whether this rigid structure is a potential recognition site for binding different auxiliary proteins that regulate ASIC trafficking to the plasma membrane. Indeed, a number of studies have focused on various interacting proteins involved in regulating cell surface expression of ASICs. For example, ASICs contain PDZ (PSD-95, Drosophila Discs-large protein, zonula occludens protein-1) binding motifs at the C termini. The interaction with CIPP (channel-interacting PDZ domain protein) (43), NHERF-1 (Na+/H+ exchanger regulatory factor-1) (44), and Lin-7b (45) increased, whereas that with PSD-95 decreased the cell surface expression of ASIC3 (45). The surface expression of ASIC1a is also regulated by its interaction with the PDZ domain of PICK-1 (protein interacting with C kinase-1) (46). In addition to PDZ domain proteins, the surface delivery of ASIC1a is also potentiated by binding of its N terminus with the light chain of annexin II (47). However, the site of interaction of ASICs with the above proteins is cytoplasmic, which is opposite from the region we have identified in the current study. Before reaching the plasma membrane, the designated extracellular signal patch is exposed to the lumen of endoplasmic reticulum, Golgi apparatus, and exocytic vesicles. However, little is known about interacting proteins involved in cell surface delivery of ASICs inside these compartments at the current stage. Therefore, it remains to be clarified in which step of the transporting process the ASICs interact with any auxiliary proteins at the identified rigid signal patch as well as how this interaction regulates the surface expression of ASIC channels.
Generally, interaction forces that maintain a protein's tertiary structures include van der Waals forces, hydrophobic interactions, disulfide bonds, hydrogen bonds, salt bridges, etc. Among them, salt bridges are very strong interactions, assembled by both hydrogen bonds and interionic attractions. NMA results and r.m.s. fluctuation analysis revealed that the region important for surface expression, composed of residues Arg99, Glu159, Asp160, Asp216, Gln218, Gln219, Glu220, and Glu221, is extremely rigid. Adjacent to this region, the finger (except for loop-β9) domain is the most flexible element. Thus, a strong separation at the interface between the flexible finger domain and the rigid region would be necessary in order to overcome the conflict generated by the distinct motion modes between the two domains. The Asp107-Arg153 salt bridge happens to situate at the interface between the two domains, and it may serve at least two distinct functions. First and most importantly, it appears to help maintain the structural stability of the rigid region, reducing its structural fluctuations and consequently enabling normal cell surface expression of the channel. Second, although we have demonstrated that the salt bridge may not be involved in pH sensing, it may coordinate with the gating domain to undergo the proper conformational change in the gating process. As predicted by MD simulations, for ASIC3D107A, ASIC3D107E, and ASIC3R153A, their α-helixes in the finger domain were all shifted compared with WT (supplemental Fig. S1, B and D), suggesting a role of the salt bridge in gating function despite its major role in controlling surface expression. Given that the salt bridge is situated between the rigid signal patch and the flexible gating domain and Asp107 is at the side of the latter structural motif, the impacts of R153A and D107A substitutions on channel gating are asymmetric (Figs. 2 and 3). Asymmetry of a salt bridge is a common feature that has been shown for other receptors, such as the GABAA receptor (26, 27). Moreover, the difference in the impact on channel function resulting from disruption of a salt bridge is entirely predictable because the protein undergoes different conformational changes, depending on which charged residue is substituted, due to the specific environment each residue is exposed to, which also differs greatly between the two residues.
In support of a crucial role for the Asp107-Arg153 interaction in determining cell surface density of ASIC3, we found that defective surface expression caused by the single mutation D107C or R153C could be partially restored by the double mutation D107C/R153C (Fig. 9). That rebuilding the Cys107-Cys153 interaction via cysteine cross-linkage only partially, and not fully, restored ASIC3 surface expression suggests that the signal patch in ASICs is a stringent structure. In fact, charge swap was ineffective in reconstructing the interaction pair to support ASIC3 surface expression. Even minor structural alterations, such as the Asp to Glu substitution (e.g. D107E) resulted in a complete loss of function of ASIC3 activity. Based on these observations, we conclude that the properly formed salt bridge at this region is critical for the tertiary structure of the signal patch important for ensuring normal surface expression of ASICs.
In summary, the present investigation provides significant insights into the mechanisms that govern ASIC surface expression. These results implied that in the tertiary structure of ASICs, a highly conserved rigidifying salt bridge plays a central role in conformational restriction that separates a rigid region critical for cell surface delivery from the flexible finger domain involved in channel gating. We suggest that the rigid region important for surface expression may be a potential binding site for associated proteins. Our finding highlights the importance of specialized tertiary structure located in the designated extracellular loop in regulating surface abundance of ion channels. Given that elevated activity of ASICs leads to a diversity of cellular dysfunctions, including ischemia (11–13), neurodegenerative diseases (48–50), and pain (3–6), the present findings allow the evaluation of new strategies aimed at preventing excessive excitability and neuronal injury associated with tissue acidosis and ASIC activation.
Supplementary Material
Acknowledgments
We thank all groups that provided ASIC cDNAs. We also thank Dr. James Celentano for help in English writing of the manuscript.
This work was supported National Basic Research Program of China Grant 2011CBA00408; National Natural Science Foundation of China Grants 91132303, 30830035, 31170787, and 20932003; Key National S&T Program “Major New Drug Development” Grant 2012ZX09504001-003; and the State Key Laboratory of Neuroscience.

This article contains supplemental Figs. S1 and S2.
- ASIC
- acid-sensing ion channel
- cASIC1
- chicken ASIC1
- hASIC1a
- human ASIC1a
- rASIC3
- rat ASIC3
- TM
- transmembrane
- MD
- molecular dynamics
- NMA
- normal mode analysis
- EGFP
- enhanced green fluorescent protein
- GMQ
- 2-guanidine-4-methylquinazoline
- r.m.s.
- root mean square.
REFERENCES
- 1. Kellenberger S., Schild L. (2002) Epithelial sodium channel/degenerin family of ion channels. A variety of functions for a shared structure. Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 2. Waldmann R., Lazdunski M. (1998) H+-gated cation channels. Neuronal acid sensors in the NaC/DEG family of ion channels. Curr. Opin. Neurobiol. 8, 418–424 [DOI] [PubMed] [Google Scholar]
- 3. Mazzuca M., Heurteaux C., Alloui A., Diochot S., Baron A., Voilley N., Blondeau N., Escoubas P., Gélot A., Cupo A., Zimmer A., Zimmer A. M., Eschalier A., Lazdunski M. (2007) A tarantula peptide against pain via ASIC1a channels and opioid mechanisms. Nat. Neurosci. 10, 943–945 [DOI] [PubMed] [Google Scholar]
- 4. Chen C. C., Zimmer A., Sun W. H., Hall J., Brownstein M. J., Zimmer A. (2002) A role for ASIC3 in the modulation of high intensity pain stimuli. Proc. Natl. Acad. Sci. U.S.A. 99, 8992–8997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sluka K. A., Price M. P., Breese N. M., Stucky C. L., Wemmie J. A., Welsh M. J. (2003) Chronic hyperalgesia induced by repeated acid injections in muscle is abolished by the loss of ASIC3 but not ASIC1. Pain 106, 229–239 [DOI] [PubMed] [Google Scholar]
- 6. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., Lingueglia E. (2008) ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Price M. P., McIlwrath S. L., Xie J., Cheng C., Qiao J., Tarr D. E., Sluka K. A., Brennan T. J., Lewin G. R., Welsh M. J. (2001) The DRASIC cation channel contributes to the detection of cutaneous touch and acid stimuli in mice. Neuron 32, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 8. Page A. J., Brierley S. M., Martin C. M., Price M. P., Symonds E., Butler R., Wemmie J. A., Blackshaw L. A. (2005) Different contributions of ASIC channels 1a, 2, and 3 in gastrointestinal mechanosensory function. Gut 54, 1408–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Price M. P., Lewin G. R., McIlwrath S. L., Cheng C., Xie J., Heppenstall P. A., Stucky C. L., Mannsfeldt A. G., Brennan T. J., Drummond H. A., Qiao J., Benson C. J., Tarr D. E., Hrstka R. F., Yang B., Williamson R. A., Welsh M. J. (2000) The mammalian sodium channel BNC1 is required for normal touch sensation. Nature 407, 1007–1011 [DOI] [PubMed] [Google Scholar]
- 10. Wemmie J. A., Chen J., Askwith C. C., Hruska-Hageman A. M., Price M. P., Nolan B. C., Yoder P. G., Lamani E., Hoshi T., Freeman J. H., Jr., Welsh M. J. (2002) The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron 34, 463–477 [DOI] [PubMed] [Google Scholar]
- 11. Xiong Z. G., Zhu X. M., Chu X. P., Minami M., Hey J., Wei W. L., MacDonald J. F., Wemmie J. A., Price M. P., Welsh M. J., Simon R. P. (2004) Neuroprotection in ischemia. Blocking calcium-permeable acid-sensing ion channels. Cell 118, 687–698 [DOI] [PubMed] [Google Scholar]
- 12. Sutherland S. P., Benson C. J., Adelman J. P., McCleskey E. W. (2001) Acid-sensing ion channel 3 matches the acid-gated current in cardiac ischemia-sensing neurons. Proc. Natl. Acad. Sci. U.S.A. 98, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yagi J., Wenk H. N., Naves L. A., McCleskey E. W. (2006) Sustained currents through ASIC3 ion channels at the modest pH changes that occur during myocardial ischemia. Circ. Res. 99, 501–509 [DOI] [PubMed] [Google Scholar]
- 14. Ziemann A. E., Schnizler M. K., Albert G. W., Severson M. A., Howard M. A., 3rd, Welsh M. J., Wemmie J. A. (2008) Seizure termination by acidosis depends on ASIC1a. Nat. Neurosci. 11, 816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 16. Gonzales E. B., Kawate T., Gouaux E. (2009) Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu Y., Chen Z., Li W. G., Cao H., Feng E. G., Yu F., Liu H., Jiang H., Xu T. L. (2010) A nonproton ligand sensor in the acid-sensing ion channel. Neuron 68, 61–72 [DOI] [PubMed] [Google Scholar]
- 18. Sackin H., Nanazashvili M., Li H., Palmer L. G., Walters D. E. (2009) An intersubunit salt bridge near the selectivity filter stabilizes the active state of Kir1.1. Biophys. J. 97, 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dibb K. M., Rose T., Makary S. Y., Claydon T. W., Enkvetchakul D., Leach R., Nichols C. G., Boyett M. R. (2003) Molecular basis of ion selectivity, block, and rectification of the inward rectifier Kir3.1/Kir3.4 K+ channel. J. Biol. Chem. 278, 49537–49548 [DOI] [PubMed] [Google Scholar]
- 20. Nakajo K., Kubo Y. (2008) Second coiled-coil domain of KCNQ channel controls current expression and subfamily-specific heteromultimerization by salt bridge networks. J. Physiol. 586, 2827–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang R., Martz A., Gonin S., Taly A., de Carvalho L. P., Grutter T. (2010) A putative extracellular salt bridge at the subunit interface contributes to the ion channel function of the ATP-gated P2X2 receptor. J. Biol. Chem. 285, 15805–15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoersch D., Otto H., Joshi C. P., Borucki B., Cusanovich M. A., Heyn M. P. (2007) Role of a conserved salt bridge between the PAS core and the N-terminal domain in the activation of the photoreceptor photoactive yellow protein. Biophys. J. 93, 1687–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan C. H., Yu T. H., Wong K. B. (2011) Stabilizing salt bridge enhances protein thermostability by reducing the heat capacity change of unfolding. PLoS One 6, e21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J. M., Altenbach C., Kono M., Oprian D. D., Hubbell W. L., Khorana H. G. (2004) Structural origins of constitutive activation in rhodopsin. Role of the Lys-296/Glu-113 salt bridge. Proc. Natl. Acad. Sci. U.S.A. 101, 12508–12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lam S. Y., Yeung R. C., Yu T. H., Sze K. H., Wong K. B. (2011) A rigidifying salt bridge favors the activity of thermophilic enzyme at high temperatures at the expense of low temperature activity. PLoS Biol. 9, e1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Venkatachalan S. P., Czajkowski C. (2008) A conserved salt bridge critical for GABAA receptor function and loop C dynamics. Proc. Natl. Acad. Sci. U.S.A. 105, 13604–13609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kash T. L., Jenkins A., Kelley J. C., Trudell J. R., Harrison N. L. (2003) Coupling of agonist binding to channel gating in the GABAA receptor. Nature 421, 272–275 [DOI] [PubMed] [Google Scholar]
- 28. Laha K. T., Wagner D. A. (2011) A state-dependent salt bridge interaction exists across the β/α intersubunit interface of the GABAA receptor. Mol. Pharmacol. 79, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y. F., Wu L. J., Li Y., Xu L., Xu T. L. (2003) Mechanisms of H+ modulation of glycinergic response in rat sacral dorsal commissural neurons. J. Physiol. 552, 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mammen A. L., Huganir R. L., O'Brien R. J. (1997) Redistribution and stabilization of cell surface glutamate receptors during synapse formation. J. Neurosci. 17, 7351–7358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sánchez R., Sali A. (2000) Comparative protein structure modeling. Introduction and practical examples with Modeller. Methods. Mol. Biol. 143, 97–129 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski R., MacArthur M., Moss D., Thornton J. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 33. Li H., Robertson A. D., Jensen J. H. (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 34. Immke D. C., McCleskey E. W. (2003) Protons open acid-sensing ion channels by catalyzing relief of Ca2+ blockade. Neuron 37, 75–84 [DOI] [PubMed] [Google Scholar]
- 35. Yu Y., Li W. G., Chen Z., Cao H., Yang H., Jiang H., Xu T. L. (2011) Atomic level characterization of the nonproton ligand sensing domain of ASIC3 channels. J. Biol. Chem. 286, 24996–25006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shaw D. E. (2005) A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J. Comput. Chem. 26, 1318–1328 [DOI] [PubMed] [Google Scholar]
- 37. Yang H., Yu Y., Li W. G., Yu F., Cao H., Xu T. L., Jiang H. (2009) Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLoS Biol. 7, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Craven K. B., Zagotta W. N. (2004) Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J. Gen. Physiol. 124, 663–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang Y., Pico A., Cadene M., Chait B. T., MacKinnon R. (2001) Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron 29, 593–601 [DOI] [PubMed] [Google Scholar]
- 40. Bahar I., Lezon T. R., Bakan A., Shrivastava I. H. (2010) Normal mode analysis of biomolecular structures. Functional mechanisms of membrane proteins. Chem. Rev. 110, 1463–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bargeton B., Kellenberger S. (2010) The contact region between three domains of the extracellular loop of ASIC1a is critical for channel function. J. Biol. Chem. 285, 13816–13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shaikh S. A., Tajkhorshid E. (2008) Potential cation and H+ binding sites in acid sensing ion channel-1. Biophys. J. 95, 5153–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anzai N., Deval E., Schaefer L., Friend V., Lazdunski M., Lingueglia E. (2002) The multivalent PDZ domain-containing protein CIPP is a partner of acid-sensing ion channel 3 in sensory neurons. J. Biol. Chem. 277, 16655–16661 [DOI] [PubMed] [Google Scholar]
- 44. Deval E., Friend V., Thirant C., Salinas M., Jodar M., Lazdunski M., Lingueglia E. (2006) Regulation of sensory neuron-specific acid-sensing ion channel 3 by the adaptor protein Na+/H+ exchanger regulatory factor-1. J. Biol. Chem. 281, 1796–1807 [DOI] [PubMed] [Google Scholar]
- 45. Hruska-Hageman A. M., Benson C. J., Leonard A. S., Price M. P., Welsh M. J. (2004) PSD-95 and Lin-7b interact with acid-sensing ion channel-3 and have opposite effects on H+-gated current. J. Biol. Chem. 279, 46962–46968 [DOI] [PubMed] [Google Scholar]
- 46. Jin W., Shen C., Jing L., Zha X. M., Xia J. (2010) PICK1 regulates the trafficking of ASIC1a and acidotoxicity in a BAR domain lipid binding-dependent manner. Mol. Brain. 3, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donier E., Rugiero F., Okuse K., Wood J. N. (2005) Annexin II light chain p11 promotes functional expression of acid-sensing ion channel ASIC1a. J. Biol. Chem. 280, 38666–38672 [DOI] [PubMed] [Google Scholar]
- 48. Vergo S., Craner M. J., Etzensperger R., Attfield K., Friese M. A., Newcombe J., Esiri M., Fugger L. (2011) Acid-sensing ion channel 1 is involved in both axonal injury and demyelination in multiple sclerosis and its animal model. Brain 134, 571–584 [DOI] [PubMed] [Google Scholar]
- 49. Xiong Z. G., Pignataro G., Li M., Chang S. Y., Simon R. P. (2008) Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr. Opin. Pharmacol. 8, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Friese M. A., Craner M. J., Etzensperger R., Vergo S., Wemmie J. A., Welsh M. J., Vincent A., Fugger L. (2007) Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat. Med. 13, 1483–1489 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.