Background: Transcription factors must locate their specific sites within the genome.
Results: Using NMR, we demonstrate the existence of sparsely populated, transient states that facilitate target location in Sox2·DNA and Oct1·Sox2·DNA complexes.
Conclusion: Rapid target searching by sparsely populated states of Sox2 and Oct1 involves two pathways: one-dimensional sliding and transfer between DNA.
Significance: Protein interactions in multiprotein·DNA complexes modulate translocation pathways.
Keywords: NMR, Protein Complexes, Protein-DNA Interaction, Protein Dynamics, Transcription Regulation, DNA Sampling, Inter- and Intramolecular Translocation, Paramagnetic Relaxation Enhancement, Spectroscopically Invisible States, z-Exchange Spectroscopy
Abstract
The pathways whereby Sox2 scans DNA to locate its specific binding site are investigated by NMR in specific and nonspecific Sox2·DNA complexes and in a specific ternary complex with Oct1 on the Hoxb1 regulatory element. Direct transfer of Sox2 between nonspecific sites on different DNA molecules occurs without dissociation into free solution at a rate of ∼106 m−1 s−1, whereas one-dimensional sliding proceeds with a diffusion constant of ≥0.1 μm2·s−1. Translocation of Sox2 from one specific DNA site to another occurs via jumping, involving complete dissociation into free solution (kd ∼5–6 s−1) followed by reassociation (ka ∼5 × 108 m−1 s−1). In the presence of Oct1 bound to an adjacent specific site, kd is reduced by more than 10-fold. Paramagnetic relaxation measurements, however, demonstrate that sparsely populated (<1%), transient states involving nonspecifically bound Sox2 in rapid exchange with specifically bound Sox2 are sampled in both binary Sox2·DNA- and ternary Oct1·Sox2·Hoxb1-DNA-specific complexes. Moreover, Sox2 modulates the mechanism of translocation of Oct1. Both Sox2 and the Oct1 POUHD domain are transiently released from the specific ternary complex by sliding to an adjacent nonspecific site, followed by direct transfer to another DNA molecule, whereas the Oct1 POUS domain is fixed to its specific site through direct interactions with Sox2. Intermolecular translocation of POUHD results in the formation of a bridged intermediate spanning two DNA molecules, enhancing the probability of complete intermolecular translocation of Oct1. By way of contrast, in the specific Oct1·DNA binary complex, POUS undergoes direct intermolecular translocation, whereas POUHD scans the DNA by sliding.
Introduction
To control gene regulation, transcription factors must be able to efficiently locate their specific target site(s) within an overwhelming sea of nonspecific sites. Kinetic and theoretical studies have suggested that three mechanisms are involved in this search process: jumping, sliding, and intersegment transfer (1–6). Jumping involves the complete dissociation of the transcription factor from the DNA, followed by reassociation at a distal DNA site, either on the same DNA molecule or another DNA molecule. The transcription factor can also diffuse in one dimension along the DNA, tracking the grooves of the DNA while maintaining its orientation relative to the DNA (7), a process that has been termed rotation-coupled sliding (8). Finally, a transcription factor can undergo direct transfer from one DNA site to either a distal site on the same DNA molecule or to another DNA molecule without ever dissociating into free solution. This involves a flycast mechanism (9) whereby a tail or domain is used to search for other DNA sites, resulting in the transient formation of a bridged complex, followed by the first order transfer of the remainder of the transcription factor to the new site (4, 10, 11).
We have previously made use of NMR paramagnetic relaxation enhancement (PRE)3 and z-exchange spectroscopy to directly demonstrate the occurrence of sliding, jumping, and intersegment transfer for the homeodomain transcription factor HoxD9 (12, 13) and the bidomain transcription factor Oct1 (10, 14). In the context of PRE studies on specific complexes, these processes manifest themselves as sparsely populated (<1%), spectroscopically invisible states whose footprints are observed on the PRE profiles of the predominant (>99%) species, namely the specific complex (12). Further, residual dipolar coupling data on nonspecific complexes demonstrated that the orientations of HoxD9 and Oct1 relative to the long axis of the DNA are maintained during the sliding process (7, 10); thus, these transcription factors track the grooves on the DNA by rotation-coupled sliding. HoxD9 and Oct1 are both major groove DNA-binding proteins, but little is known of the behavior of minor groove binding proteins. In addition, many eukaryotic transcription factors form multiprotein complexes on the DNA, thereby enabling combinatorial control of transcription.
An example of a multiprotein transcription factor assembly is the ternary complex formed by Oct1 and Sox2 on the Hoxb1 regulatory element (15). Oct1 plays a critical role in neural development, cell growth, and differentiation (15–17) and comprises two major groove binding helix-turn-helix domains, POUS and POUHD, connected by a flexible (∼23-residue) linker (18, 19). Sox2 is a member of the HMG-box family of architectural factors that bind in the minor groove and bend the DNA by 50–90° (20–22). The solution structure of the ternary Oct1·Sox2·Hoxb1-DNA complex has been solved by NMR (23). Recent z-exchange NMR experiments have shown that the rate of direct transfer of Oct1 between specific sites located on different DNA molecules is reduced by over an order of magnitude in the presence of Sox2 bound to a specific site immediately adjacent to the Oct1-specific site (14). Here we make use of PRE, z-exchange, and line width NMR measurements to characterize transcription factor translocation in the Sox2·Hoxb1-DNA binary and the Oct1·Sox2·Hoxb1-DNA ternary complexes. The data illustrate how interactions between transcription factors modulate the mechanism of translocation.
EXPERIMENTAL PROCEDURES
Sample Preparation
The POU region (POUS + POUHD) of human Oct1 (residues 280–442) and the HMG-box domain of Sox2 (residues 38–121) were expressed and purified as described previously (23). Uniform 2H/15N isotopic labeling was achieved by growing E. coli BL21-CodonPlus(DE3)-RIPL cells in minimal medium dissolved in 99.9% D2O using d7-glucose and 15NH4Cl as the sole carbon and nitrogen sources, respectively. Single-stranded unmodified and EDTA- or rhodamine-conjugated DNA oligonucleotides were purchased from Invitrogen and Midland Certified Reagents, respectively, and purified by ion exchange chromatography on a Mono-Q (GE Healthcare) column with a NaCl gradient in a buffer of 50 mm Tris-HCl, pH 7.5, and 1 mm EDTA. After strand annealing, DNA duplexes were further purified by ion exchange chromatography to remove any residual single-stranded DNA (24). NMR samples were prepared in 10 mm PIPES, 94% H2O, 6% D2O, pH 6.5. Throughout this work, the concentrations of NaCl used for the various samples refer to the concentration of added NaCl and do not take into account the presence of counterions bound to the phosphate backbone. For example, 0 mm NaCl means that there is no added NaCl but does not imply that the sample is completely devoid of NaCl; thus, for a 29-bp DNA duplex at a concentration of 0.5 mm with no added NaCl, the concentration of sodium counterions can be estimated to be ∼30 mm.
NMR Spectroscopy
All NMR experiments were carried out at 303 K on Bruker 500- and 600-MHz spectrometers equipped with z-gradient triple resonance cryoprobes. Spectra were processed using NMRPipe (25), and analyzed using the programs XIPP4 and NMRView (26).
Intermolecular exchange rates for the translocation of Sox2 between two DNA duplexes bearing slightly different specific target sites were determined using transverse relaxation optimized z-exchange spectroscopy (TROSY) (27) with at least eight different mixing times ranging from 20 to 600 ms. Fitting of the time dependence of the 1HN/15N auto and exchange cross-peaks to obtain rate constants was as described (13).
Intermolecular exchange rates for the translocation of Sox2 between two nonspecific DNA duplexes of differing sequence were derived from Lorentzian line shape fitting in either the 1H or 15N dimensions of 1H-15N heteronuclear single quantum correlation (HSQC) spectra as described previously (7). The data for residues exhibiting 1HN or 15N chemical shift differences greater than 30 Hz at a 1H frequency of 500 MHz (namely Lys26, His34, and Lys40) were used for analysis.
PRE transverse 1HN-Γ2 rates were measured on Oct1·Sox2·Hoxb1-DNA ternary complexes comprising either 0.4 mm 2H/15N-labeled Oct1, 0.44 mm unlabeled Sox2 (i.e. at natural isotopic abundance), and 0.44 mm DNA-EDTA or 0.44 mm unlabeled Oct1, 0.4 mm 2H/15N-labeled Sox2, and 0.44 mm DNA-EDTA. The DNA duplexes are 29 base pairs in length and comprise the specific Oct1- and Sox2-binding sites from the Hoxb1 promoter (23). The DNA-dT-EDTA was chelated to either Mn2+ (paramagnetic) or Ca2+ (diamagnetic), and the 1HN-Γ2 rates were obtained by taking the difference between the 1HN-R2 transverse relaxation rates between the paramagnetic and diamagnetic samples. 1HN-R2 values were obtained using a two-time point measurement with a ΔT difference of 11.6 ms as described previously (28) with a TROSY-based 1H-15N correlation pulse scheme (10, 29). For the Sox2·DNA binary complex, the sample conditions were exactly the same as those employed for the ternary Oct1·Sox2·DNA complex except for the absence of Oct1 and a longer ΔT value of 18.4 ms for the two-time point measurement.
To analyze the relative contributions from intra- and intermolecular translocation, mixed samples were made comprising Oct1, Sox2, and an equal mixture of two DNA duplexes, one containing the specific binding sites for Oct1 and Sox2 and the second representing a nonspecific DNA duplex for the 2H/15N-labeled protein being monitored. When the nonspecific DNA duplex is tagged with a paramagnetic label, the only PRE effects observed are those arising from intermolecular translocation, but when the paramagnetic label is attached to the DNA duplex comprising the two specific sites, both intra- and intermolecular translocation can give rise to PREs. To look at inter- and intramolecular translocation of Oct1 in the presence of Sox2, the samples comprised either 0.4 mm 2H/15N-labeled Oct1, 0.45 mm unlabeled Sox2, 0.45 mm specific DNA (containing both the Oct1- and Sox2-specific sites), and 0.45 mm nonspecific DNA or 0.4 mm 2H/15N-labeled Oct1, 0.9 mm unlabeled Sox2, 0.45 mm specific DNA, and 0.45 mm nonspecific DNA for Oct1 but containing the specific Sox2 site. To look at inter- and intramolecular translocation of Sox2 in the presence of Oct1, the samples comprised either 0.4 mm 2H/15N-labeled Sox2, 0.45 mm unlabeled Oct1, 0.45 mm specific DNA (containing both the Sox2- and Oct1-specific sites), and 0.45 mm nonspecific DNA or 0.4 mm 2H/15N-labeled Sox2, 0.9 mm unlabeled Oct1, 0.45 mm specific DNA, and 0.45 mm nonspecific DNA for Sox2 but containing the specific Oct1 site.
PRE 1HN-Γ2 rates were back-calculated from the structures of the specific Oct1·Sox2·Hoxb1-DNA ternary and Sox2·Hoxb1-DNA binary complexes (23) using a three-conformer ensemble representation for the EDTA-Mn2+ groups to account for their flexibility (30). The coordinates of the EDTA-Mn2+ moieties were optimized by simulated annealing in Xplor-NIH (31), and agreement between observed and calculated values is expressed as a PRE Q-factor given by the equation, Q = (Σ(Γ2obs(i) − Γ2calc(i))2/ΣΓ2obs(i))½, where the superscripts “obs” and “calc” represent observed and calculated values, respectively, and i denotes the residue number (30).
Fluorescence Anisotropy
The apparent equilibrium dissociation constant KD for the binding of Sox2 to nonspecific DNA at 25 °C was determined by fluorescence anisotropy using a Jobin-Yvon FluoroMax-3 spectrometer as described previously (7). The wavelengths for excitation and emission were 550 and 580 nm, respectively. Sox2 was titrated (0–26.2 μm final concentration) into a rhodamine-conjugated nonspecific 29-bp DNA duplex (1.5 μm) dissolved in 10 mm sodium phosphate buffer, 150 mm NaCl, and 1 mm EDTA at pH 7.4. The KD was calculated from the titration data as described previously (32).
RESULTS AND DISCUSSION
We first present data on the binding of Sox2 to nonspecific DNA using fluorescence anisotropy measurements and characterize intra- and intermolecular translocation of Sox2 on nonspecific DNA by PRE and line width measurements. We then present z-exchange data for the intermolecular translocation of Sox2 between specific sites and examine the impact of Oct1 on this exchange process in the context of a specific Oct1·Sox2·Hoxb1-DNA ternary complex. Finally, using PRE measurements, we examine intra- and intermolecular translocation involving transient sparsely populated states, first of Sox2 in the context of a specific Sox2·Hoxb1-DNA binary complex and then of both Oct1 and Sox2 in the context of a specific Oct1·Sox2·Hoxb1-DNA ternary complex.
Interaction between Sox2 and 29-bp Nonspecific DNA Duplex
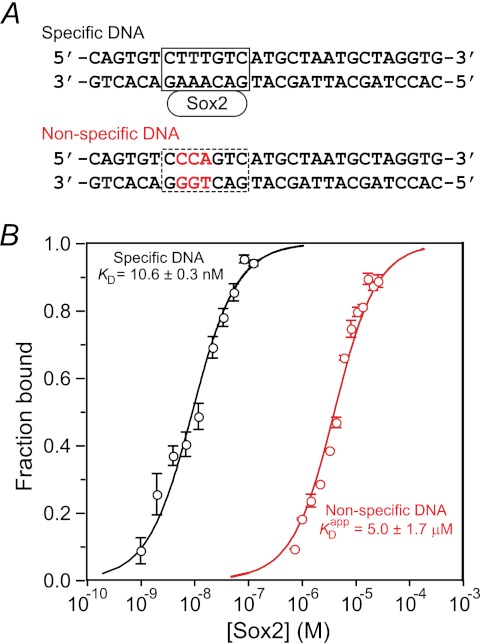
The interaction between Sox2 and a 29-bp nonspecific DNA duplex was studied by fluorescence anisotropy. The sequence of the nonspecific DNA duplex is shown in Fig. 1A. The sequence of the specific site (boxed in Fig. 1A, top) is that from the Hoxb1 regulatory element (15, 23). The nonspecific DNA duplex was derived from the specific DNA duplex by introducing three mutations in the specific binding site (displayed in red in Fig. 1A, bottom). The KD for the binding of Sox2 to the 29-bp specific DNA duplex is 10.6 nm at a salt concentration of 150 mm NaCl (14). The three DNA mutations reduce the affinity by more than 2 orders of magnitude, and the KD for the binding of Sox2 to the nonspecific 29-bp DNA duplex is 5.0 ± 1.7 μm at the same salt concentration (Fig. 1B).
FIGURE 1.
Nonspecific binding of Sox2 to DNA monitored by fluorescence anisotropy. A, specific (top) and nonspecific (bottom) DNA duplexes. The base pairs involved in specific interactions between Sox2 and DNA are located in the box (top). The bases marked in red in the nonspecific duplex indicate sites that were mutated relative to the specific duplex. B, fluorescence anisotropy titration for the binding of Sox2 to the nonspecific DNA duplex. The rhodamine fluorescent label is conjugated to the 5′-end of the bottom strand of the nonspecific 29-bp DNA duplex. The concentration of DNA is 1.5 μm in 10 mm sodium phosphate, 150 mm NaCl, and 1 mm EDTA, pH 7.4, and 25 °C. The excitation and emission wavelengths are 550 and 580 nm, respectively. The experimental data points (error bars, S.D.) are shown as open circles in red, and the best fit curve with an apparent KDapp of 5.0 ± 1.7 μm is shown as a solid red line. For comparison, the fluorescence anisotropy titration data for the binding of Sox2 to the specific DNA duplex (taken from Ref. 14) under the same conditions of buffer, salt, and temperature are shown in black with the experimental data displayed as circles and the best fit curve with a KD of 10.6 ± 0.3 nm as a solid line. The concentration of rhodamine-labeled specific DNA used in the titration experiment was 1.4 nm.
Analysis of Sox2 Sampling of Nonspecific DNA by PRE
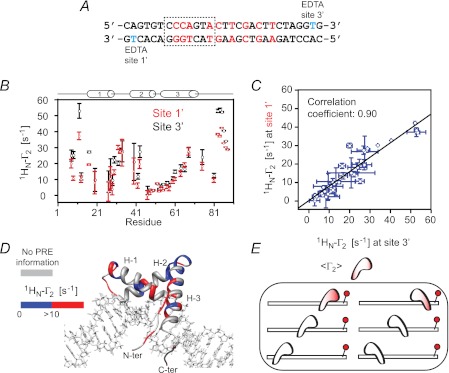
Intermolecular PRE experiments at 0 mm added NaCl were conducted using a nonspecific 29-bp DNA duplex with the paramagnetic label located at either end of the duplex, denoted as sites 1′ and 3′ (see Fig. 2A), to probe the sampling of nonspecific sites by Sox2. (Note that throughout this work, 0 mm NaCl refers to added NaCl and does not mean to imply that the sample is completely devoid of NaCl because each phosphate on the DNA will invariably be bound to a counterion; thus, for example, for a 29-bp DNA duplex at a concentration of 0.5 mm, the concentration of sodium counterions will be ∼30 mm, corresponding to one sodium ion per phosphate). The paramagnetic label comprises Mn2+ chelated to dT-EDTA (24). The transverse 1HN-Γ2 PRE relaxation rates are obtained by taking the difference in transverse 1HN-R2 relaxation rates between paramagnetic (Mn2+) and diamagnetic (Ca2+) states of the system (28). The magnitude of Γ2 is proportional to the 〈r−6〉 separation between the paramagnetic label and the proton of interest (30, 33). Because exchange of Sox2 between nonspecific sites is fast on the chemical shift time scale, the observed PRE profiles are a weighted average of all possible nonspecific configurations. The PRE profiles observed on the backbone amide protons of 2H/15N-labeled Sox2 arising from the paramagnetic label at sites 1′ and 3′ on the nonspecific DNA (Fig. 2B) are qualitatively and quantitatively very similar with a correlation coefficient of 0.90 at 0 mm NaCl (Fig. 2C). The regions on Sox2 that exhibit significant PREs (1HN-Γ2 > 10 s−1) are all located in structural elements that are in close proximity to the DNA (Fig. 2D). One can therefore conclude that Sox2 samples numerous binding sites in two orientations, related by a 180° rotation relative to the long axis of the DNA (Fig. 2E). These PRE results are very similar to those previously observed for the sampling of nonspecific DNA-binding sites by another member of the HMG-box family, namely the nonspecific architectural factor HMG-B1 (32).
FIGURE 2.
Intermolecular PRE profiles for nonspecific Sox2·DNA complexes. A, sequence of the 29-bp nonspecific DNA duplex. The two sites chosen to covalently attach EDTA to thymine (one site at a time) are shown in cyan and designated as sites 1′ and 3′. Mn2+ and Ca2+ are chelated to the EDTA moiety in the paramagnetic and diamagnetic control samples, respectively. B, PRE profiles at 0 mm NaCl and 30 °C. The concentrations of 2H/15N-labeled Sox2 and nonspecific DNA are 0.40 and 0.45 mm, respectively. C, correlation between PREs originating from sites 1′ and 3′. D, PRE profiles originating from site 3′ mapped onto the structure of the specific Sox2·DNA complex (23). The color scale for the PRE 1HN-Γ2 rates is shown to the left of the structure. E, schematic illustrating the nonspecific DNA binding states sampled by Sox2. The color coding within the schematic representation of Sox2 represents the magnitude of the PRE, and the EDTA-Mn2+ paramagnetic label is depicted by the red sphere. Two orientations of Sox2 on the DNA related by a 180° rotation relative to the long axis of the DNA are sampled.
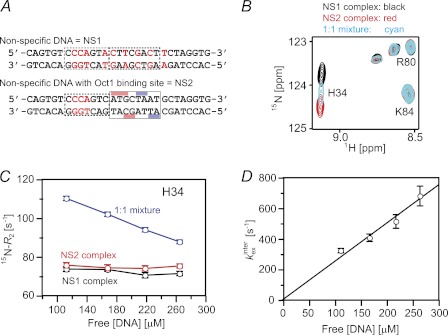
Rates of Inter- and Intramolecular Translocation of Sox2 on Nonspecific DNA
To analyze the rates of intermolecular translocation of Sox2 between nonspecific DNA duplexes, we carried out Lorentzian line shape analysis of cross-peaks in the 1H-15N HSQC spectra of three samples: two complexes with different nonspecific DNA duplexes (NS1 and NS2) and a sample comprising a 1:1 mixture of the two nonspecific DNA duplexes (Fig. 3A). An expanded region of the 1H-15N HSQC spectra for the three samples is shown in Fig. 3B. Residues exhibiting 1HN or 15N chemical shift differences of >30 Hz at a 1H frequency of 500 MHz between the two nonspecific complexes, namely Lys26, His34, and Lys40, were used for line shape analysis. The cross-peaks in the sample comprising an equimolar mixture of the two nonspecific complexes appear halfway between the corresponding cross-peaks for the two samples containing only one nonspecific DNA duplex and are broadened due to the exchange contribution from intermolecular translocation between the two DNA duplexes. The apparent intermolecular translocation rate can be readily obtained from Lorentzian line shape analysis of the cross-peaks in the three samples as described by Iwahara et al. (7). The observed transverse relaxation rate (R2mix) for a given resonance in the mixture is equal to the population-weighted average of the corresponding R2 rates for the individual nonspecific complexes plus an exchange contribution Rexinter (Fig. 3C). The apparent intermolecular translocation rate, kexinter (which is equal to the sum of the two apparent first-order rate constants for the transfer of Sox2 from NS1 to NS2 and from NS2 to NS1), is given by the equation, kexinter = 4π2pNS1pNS2|Δ|2/Rexinter, where pNS1 and pNS2 are the populations of the two nonspecific complexes (in this instance 0.5 each), and Δ is the chemical shift difference between the two nonspecific complexes (measured in Hertz). kexinter is linearly proportional to the free DNA concentration (Fig. 3D), indicative of a direct transfer mechanism from one DNA duplex to the other without involving the dissociation of Sox2 into free solution (13). The second order rate constant for direct transfer from one DNA duplex to the other is 1.3 ± 0.2 × 106 m−1 s−1. Note that a mechanism involving dissociation of Sox2 into free solution, followed by reassociation, would be independent of the free DNA concentration, and the measured exchange rate would be equal to the first order dissociation rate constant (7, 13).
FIGURE 3.
Kinetics of intermolecular translocation of Sox2 between nonspecific DNA duplexes. A, sequences of the two 29-bp nonspecific DNA duplexes denoted as NS1 and NS2. B, overlay of an expanded region of the 1H-15N HSQC spectra of 2H/15N-labeled Sox2 recorded on nonspecific complexes comprising NS1 (black), NS2 (red), and a 1:1 mixture of NS1 and NS2 (cyan) DNA duplexes. The concentration of Sox2 is 0.4 mm, the concentration of NS1 and NS2 in the individual complexes is 0.5 mm, and in the complexes comprising a 1:1 mixture of NS1 and NS2, the two DNA duplexes are at a concentration of 0.25 mm each. The NaCl concentration and temperature are 25 mm and 30 °C, respectively. C, amide 15N-R2 rates for His34 as a function of free DNA concentration. D, dependence of the overall apparent intermolecular translocation rate kexinter on the free DNA concentration. Error bars, S.D.
The R2 rates for the individual complexes include an exchange contribution from intramolecular translocation or sliding (7). Because the latter R2 rates are independent of free DNA concentration and because one can assume that the average chemical shift difference between the nonspecific sites on a single DNA duplex will be approximately equal to the average chemical difference between the complexes with the NS1 and NS2 DNA duplexes, one can conclude that the rate of intramolecular translocation is much faster than that for intermolecular translocation (7). A lower limit for this rate can be derived from the extrapolated value of kexinter obtained from the free DNA concentration at which the value of R2mix falls to the R2 values for the individual nonspecific complexes (6). From Fig. 3C, this occurs when the free DNA concentration reaches ∼360 μm, which corresponds to a kexinter value of ∼900 s−1. Because exchange between all nonspecific sites, including those located at either end of the DNA, is fast on the chemical shift time scale, the lower limit for the one-dimensional diffusion constant D1 for intramolecular translocation is given by 2kexinter, where L is the distance between the nonspecific sites at the extreme ends of the DNA, and the factor of 2 arises because there are two binding orientations. For a 29-bp DNA duplex and a binding site size of 7 base pairs, L is equal to 3.4 Å × (29 − 7) ∼ 75 Å, yielding a lower limit of ∼0.1 μm2·s−1 for D1, consistent with values of D1 derived for other proteins by single molecule experiments (5, 8).
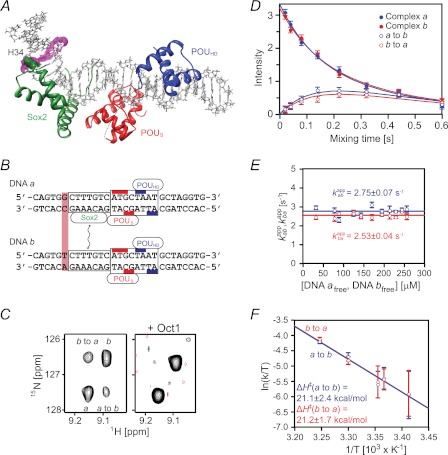
Intermolecular Translocation of Sox2 between 29-bp Specific DNA Duplexes
To measure the rate of transfer of Sox2 between two specific sites, we used an experimental design similar to that described previously (13, 14). A single base pair mutation was introduced in the Hoxb1 promoter (DNA a) adjacent to the recognition site (DNA b). This sequence alteration has no significant effect on the binding affinity for Sox2 but results in small differences in the 1HN/15N chemical shifts of several backbone amide groups (residues Met33–Ser39) between the two binary complexes (referred to as complexes a and b). Because Sox2 binds tightly to its cognate DNA sequences with a KD of 10.6 nm at 150 mm NaCl (14), the 1H-15N TROSY correlation spectrum for a 1:1 mixture of complexes a and b contains peaks from both binary complexes (Fig. 4C). To analyze the exchange kinetics for the translocation of Sox2 between the two specific DNA sites, we carried out a TROSY-based z-exchange experiment (27). The apparent exchange rates, kabapp and kbaapp, were determined by fitting the time dependence of the intensities of auto and exchange cross-peaks (Fig. 4D) to the McConnell equations (34). At 30 °C, kabapp and kbaapp are independent of the concentration of free DNA with values of 2.8 ± 0.1 and 2.5 ± 0.1 s−1 (Fig. 4E). These data indicate that transfer of Sox2 from one specific duplex to the other occurs entirely via dissociation of Sox2 into free solution followed by reassociation, and is rate-limited by the dissociation rate constants given by the equations, kaoff = 2kabapp and kboff = 2kbaapp, where the statistical factor of 2 arises from the fact that transfer of Sox2 between DNA molecules of the same sequence is of equal probability to transfer between DNA molecules of differing sequence (13). Given the measured KD of 10.6 nm, the corresponding association rate constants for the binding of Sox2 to DNA a and DNA b are 5.2 × 108 and 4.8 × 108 m−1 s−1.
FIGURE 4.
Kinetics of intermolecular translocation of Sox2 between specific DNA complexes. A, ribbon diagram of the NMR structure (Protein Data Bank code 1O4X) (23) of the Sox2·Oct1·Hoxb1-DNA ternary complex with Sox2 in green, and the POUS and POUHD domains of Oct1 in red and blue, respectively; the 29-mer DNA is shown in an all heavy atom representation. The intermolecular exchange rates for Sox2 were determined by monitoring the auto and exchange 1HN/15N cross-peaks of His34, whose atoms are displayed as green spheres. The purple mesh identifies the base pair changed between DNA a and DNA b. B, sequences of Hoxb1 DNA fragments, DNA a and DNA b, differing by 1 base pair (light purple box). The boxes represent the Sox2 and Oct1 DNA recognition sites (with the bases contacted by the POUS and POUHD domains indicated by the red and blue bars, respectively). C, example of z-exchange data for His34 seen in the 1H-15N TROSY-based z-exchange spectrum (27) at a mixing time of 220 ms, 30 °C, and 150 mm NaCl for the Sox2·Hoxb1-DNA binary (left) and Oct1·Sox2·Hoxb1-DNA ternary complexes (right). The concentrations of 2H/15N-labeled Sox2, DNA a, DNA b, and Oct1 (at natural isotopic abundance) are 0.55, 0.35, 0.35, and 0.7 mm, respectively. D, time dependence of auto (closed circles) and exchange (open circles) cross-peak intensities at 30 °C and 150 mm NaCl together with the best fit curves (solid lines). The concentrations of 2H/15N-labeled Sox2, DNA a and DNA b are 0.55, 0.35, and 0.35 mm, respectively. E, apparent intermolecular exchange rates as a function of free DNA concentration. The rates are independent of free DNA concentration, indicating that the rate-limiting step is governed by dissociation of Sox2 from the DNA. F, Erying plots of the apparent intermolecular exchange rates at 20, 24, 25, 30, and 35 °C and 150 mm NaCl. Linear fits for the exchange rates kabapp and kbaapp are shown as blue and red lines, respectively. Thermodynamic parameters derived from the data are shown in Table 1. Error bars, S.D.
These results are distinct from those obtained with the homeodomain HoxD9 and Oct1, where the apparent first order exchange rate constants for intermolecular translocation between specific sites were linearly dependent on the concentration of free DNA, indicative of the presence of a direct transfer pathway between DNA sites without dissociation into free solution. In the case of HoxD9, the dissociation/reassociation pathway was too slow to detect, whereas in the case of Oct1, the dissociation rate constants could be determined by extrapolation of the apparent intermolecular translocation rates to zero free DNA concentration. The dissociation rate constants for Oct1 at 30 °C range from 3.7 to 5.1 s−1, comparable with the values determined here for Sox2 under the same experimental conditions.
Erying plots of the temperature dependence of the apparent intermolecular translocation rates for Sox2 (Fig. 4F) yield estimates of ∼21 and ∼4 kcal·mol−1 for the activation enthalpy (ΔH‡) and entropy (TΔS‡ at 30 °C), respectively (Table 1). Thus, the energy barrier for the dissociation of Sox2 from its specific DNA site is primarily enthalpic.
TABLE 1.
Apparent activation enthalpies, entropies, and free energies for intermolecular translocation of Sox2 between two specific DNA-binding sites located on separate DNA molecules
Values were derived by least squares fitting of the Erying plots shown in Fig. 4F. ΔG‡ is calculated from the equation, ΔG‡ = ΔH‡ − TΔS‡ at 303 K.
| Transfer process | ΔH‡ | ΔS‡ | ΔG‡ |
|---|---|---|---|
| kcal·mol−1 | cal·mol−1·K−1 | kcal·mol−1 | |
| DNA a to DNA b | 21.1 ± 2.4 | 13.5 ± 8.0 | 17.1 ± 3.4 |
| DNA b to DNA a | 21.2 ± 1.7 | 13.6 ± 8.0 | 17.0 ± 3.0 |
In our previous study (14), we showed that Sox2 dramatically decreases the rate of intermolecular translocation of Oct1 between specific DNA-binding sites. The same phenomenon is observed for intermolecular translocation of Sox2 in the presence of Oct1. No exchange cross-peaks are detectable for intermolecular translocation of Sox2 between Oct1·Sox2·Hoxb1-DNA ternary complexes (Fig. 4C, right). Even at higher ionic strength and temperature (260 mm NaCl at 40 °C), exchange cross-peaks in the ternary complexes could not be detected. This can be accounted for by the decrease in KD from 10.6 nm for the binary Sox2·Hoxb1-DNA complex to 0.6 nm for the ternary Oct1·Sox2·Hoxb1-DNA ternary complex (14). Given a 15N-T1 relaxation rate of ∼1 s, the minimum exchange rate measurable by z-exchange spectroscopy is ∼0.2 s−1. Thus, the intermolecular translocation rate of Sox2 between specific sites in ternary complexes is at least an order of magnitude slower than in the binary complexes.
Translocation of Sox2 in Specific Binary Complexes Probed by PRE
Given the slow dissociation of the specific Sox2·DNA complex, intra- and intermolecular translocation of Sox2 bound to its specific DNA target site must involve short lived, sparsely populated (<1%) states in which nonspecific DNA-binding sites are sampled. These states can be detected by PRE measurements, provided that exchange between the major and minor species is fast on the PRE relaxation time scale (10, 12, 33, 35, 36). Under these conditions, the observed PRE is a population weighted average of the PREs in the major and minor states, and the minor species will leave their footprint on the PRE profiles observed for the specific complex provided that the distances between the paramagnetic label and the protons of interest are shorter in the minor species than in the major one.
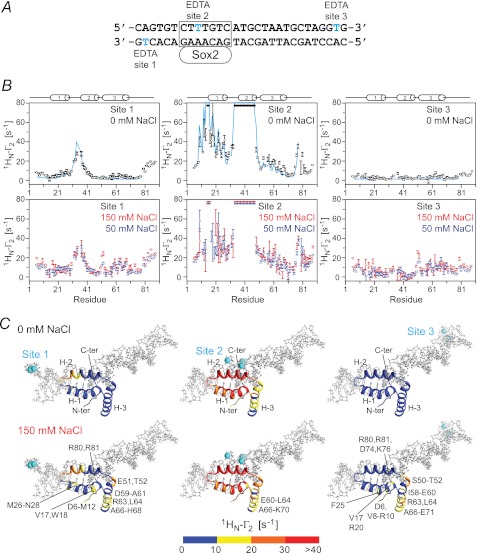
The 29-bp specific DNA duplex was paramagnetically labeled at three sites separately, on either end of the DNA and in the Sox2 recognition site (Fig. 5A). Note that EDTA is attached to T on the major groove side of the DNA, and therefore the presence of the paramagnetic label in the middle of the Sox2 recognition site does not perturb the binding of Sox2 in the minor groove. PREs were recorded at 0 mm NaCl (Fig. 5B, top panels) and at higher salt concentrations (50 and 150 mm NaCl; Fig. 5B, bottom panels). The PRE profiles mapped onto the structure of the specific Sox2·DNA complex are shown in Fig. 5C, and a summary of PRE Q-factors (30) reporting the agreement between observed and calculated PRE profiles is provided in Table 2.
FIGURE 5.
Intermolecular PRE profiles for the specific Sox2·Hoxb1-DNA complex. A, sequences of the 29-bp DNA duplex employed with the paramagnetic label (dT-EDTA-Mn2+) located at the end of the duplex (sites 1 or 3) or in the middle of the Sox2-specific site (site 2). Note that the EDTA-Mn2+ label is located in the major groove and therefore does not perturb the binding of Sox2 in the minor groove. B, intermolecular PRE profiles for the paramagnetic label at sites 1 (left panels), 2 (middle panels), and 3 (right panels) at 0 mm NaCl (top panels, black circles) and at 50 (blue circles) and 150 mm NaCl (red circles) (bottom panels). Error bars, S.D. The concentrations of 2H/15N-labeled Sox2 and DNA are 0.40 and 0.44 mm, respectively. The back-calculated PREs are displayed as solid lines (cyan). The PREs are back-calculated from the coordinates of the specific Sox2·DNA complex (23) using Xplor-NIH (37) with an ensemble representation for the paramagnetic label (30). C, experimental PRE profiles at 0 (top) and 150 mm (bottom) NaCl mapped onto the structure of the specific Sox2·DNA complex (23). The color scale for the PRE 1HN-Γ2 rates is shown at the bottom.
TABLE 2.
Agreement between observed and calculated intermolecular PREs for the specific Sox2·DNA complex
The position of Sox2 on the 29-bp specific Hoxb1 regulatory element is taken from the NMR structure of the Oct1·Sox2·Hoxb1-DNA ternary complex (23). The entire experimental PRE profiles are best fitted by simultaneously optimizing the positions of all three paramagnetic labels, each represented by a three-member ensemble, using simulated annealing (30).
| NaCl | Root mean square deviation between observed and calculated Γ2 rates |
PRE Q-factor |
||||
|---|---|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | Site 1 | Site 2 | Site 3 | |
| mm | s−1 | |||||
| 0 | 3.6 | 6.7 | 2.6 | 0.43 | 0.24 | 0.98 |
| 50 | 6.9 | 12.6 | 7.1 | 0.64 | 0.48 | 0.99 |
| 150 | 9.1 | 15.5 | 9.5 | 0.72 | 0.61 | 1.00 |
We first consider the PRE profiles at 0 mm added NaCl (Fig. 5B, top panels). The overall experimental PRE profiles agree well with the back-calculated profiles for the specific Sox2·DNA complex. A more detailed examination, however, shows that although the PREs for sites 1 and 2 agree well (Q-factor = 0.43 and 0.24, respectively), the agreement at site 3 is actually poor in terms of Q-factor (Q = 0.98) although not unreasonable in terms of root mean square difference between observed and calculated PRE rates (2.6 s−1) (Table 2). This is because no PREs arising from site 3 are predicted from the structure of the specific complex, whereas some small but significant PRE effects (less than 7 s−1) are in fact observed at the N terminus (residues 6–15), the N-terminal end of helix 2 (residues 34–37), and the middle of helix 3 (around residue 63) (Fig. 5B, top right panel). Thus, although Sox2 clearly spends the vast majority of its time bound to its specific DNA target site, a small population (<1%) must sample alternate configurations that come close to the paramagnetic label located at site 3. When the salt concentration is increased to 50 or 150 mm NaCl (Fig. 5B, bottom panels), the values of all the PREs arising from site 3 are increased, but the overall PRE profiles remain qualitatively similar to those obtained at 0 mm NaCl. This is a consequence of the increase in exchange rate between the specific complex and the sparsely populated states sampling nonspecific sites on the DNA duplex.
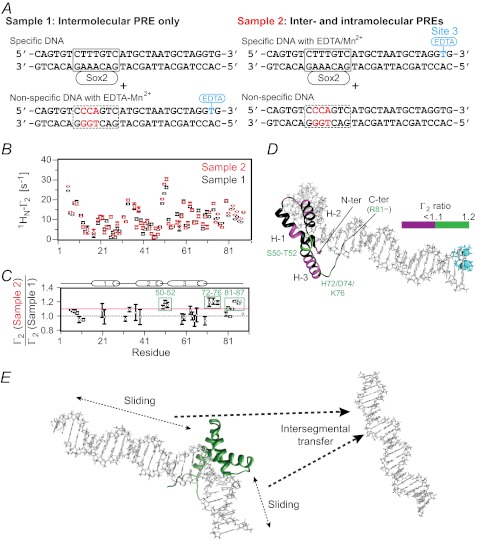
To analyze the contributions from intra- (sliding) and intermolecular translocation to the PRE profiles, we carried out a series of experiments involving samples comprising an equal mixture of specific and nonspecific DNA duplexes (12) (Fig. 6). In the first set of PRE experiments (sample 1), the paramagnetic label was placed on the nonspecific DNA (Fig. 6A, top), and therefore any observed PREs can only arise from intermolecular translocation involving transient excursions from the specific DNA to the nonspecific DNA and back to the specific DNA. In the second set of experiments, the paramagnetic label was placed on the specific DNA duplex (sample 2; Fig. 6A, bottom) with the nonspecific DNA unlabeled. The PREs observed in sample 2 will therefore comprise effects arising from both intra- and intermolecular translocation. Thus, if the PRE ratio for sample 1 to sample 2 for a particular paramagnetic center-proton vector has a value of 1, that PRE must arise predominantly from intermolecular translocation. The results of these experiments are summarized in Fig. 6B. The ratios of the sample 1 to sample 2 PREs are almost equal to 1 within experimental error with the exception of the PREs for residues 50–52 and the C-terminal tail (residues 72–76 and 81–87), indicating that the PRE effects are largely due to intermolecular translocation. In the case of residues 50–52, 72–76, and 81–87, all of which point in the direction of the paramagnetic label in the specific complex, there is a small (∼20%) contribution from sliding (with sample 1 to sample 2 PRE ratios of ∼1.2). Because dissociation of Sox2 from its specific site into free solution is slow (∼5 s−1 as measured by z-exchange spectroscopy; cf. Fig. 4), rapid intermolecular translocation must occur by Sox2 sliding out of the specific site to make a nonspecific interaction with DNA followed by intersegment transfer from the nonspecific site(s) (Fig. 6D).
FIGURE 6.
Intramolecular versus intermolecular translocation of Sox2 in the specific Sox2·Hoxb1-DNA binary complex. A, the samples comprise an equimixture of specific and nonspecific 29-bp DNA duplexes. In sample 1, the specific DNA duplex is unlabeled, and the nonspecific DNA duplex bears the paramagnetic label. In sample 2, the specific DNA duplex is paramagnetically labeled at site 3 (cf. Fig. 5A), and the nonspecific DNA duplex is unlabeled. The PRE effects observed for sample 1 arise entirely from intermolecular translocation, whereas those for sample 2 arise from both intra- and intermolecular translocation processes. The concentrations of Sox2, specific DNA, and nonspecific DNA are 0.40, 0.45, and 0.45 mm, respectively. B, PRE profiles at 150 mm NaCl from samples 1 and 2 shown as black and red circles, respectively (error bars, S.D.). C, ratio of PRE rates for sample 2 to sample 1 for residues with 1HN-Γ2 rates of >10 s−1. For Γ2(sample 2)/Γ2(sample 1) ∼ 1, the PRE effects arise predominantly from intermolecular translocation; Γ2(sample 2)/Γ2(sample 1) ratios larger than 1 are indicative of contributions from both intra- and intermolecular translocation, where the contribution from the latter is provided by the PRE profiles observed for sample 1. D, mapping of Γ2 ratios (color scale indicated on the right) onto the structure of the specific Sox2·DNA complex (23). E, schematic of intramolecular (sliding) and intermolecular (intersegment transfer) translocation events involving Sox2 bound specifically to DNA. Because bulk intersegment transfer of Sox2 between specific sites involves complete dissociation of Sox2 into free solution, followed by reassociation, the sparsely populated states arising from intermolecular translocation from the specific duplex to a nonspecific duplex and back again sampled by PRE must involve an initial sliding event from the specific site to an adjacent nonspecific site, followed by direct intersegment transfer to the nonspecific duplex and then back to the specific duplex.
Translocation of Oct1 and Sox2 on Hoxb1-DNA in Ternary Complex Probed by PRE
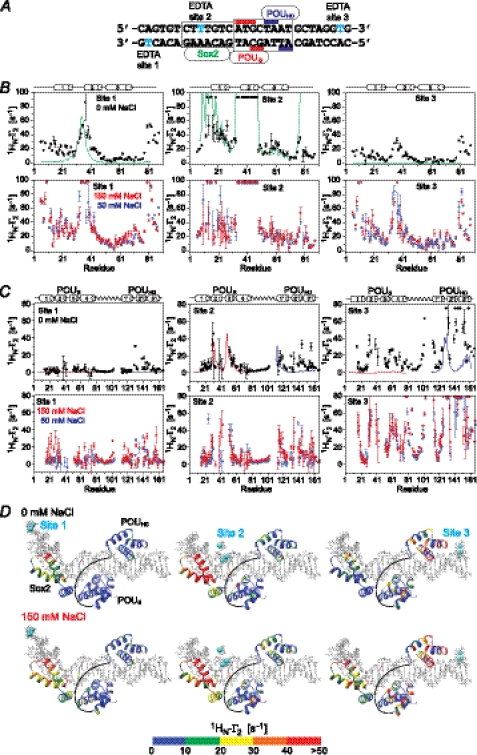
The experimental PRE profiles for the ternary Oct1·Sox2·Hoxb1-DNA complex (Fig. 7A) at 0 mm NaCl (top panels in Fig. 7, B and C) do not agree well with those back-calculated for the specific Oct1·Sox2·Hoxb1-DNA ternary complex (Table 3), in contrast to binary Sox2·DNA (Table 2) and Oct1·DNA (10) complexes. For Sox2, large PRE effects (>10 s−1) are observed at the N and C termini (residues 6–16 and 77–88) and within helix 2 (residues 34–40) from sites 1 and 3 (Fig. 7B). For Oct1, larger than expected PREs are observed within the POUHD domain from all three paramagnetic sites, and large PREs are observed within the POUS domain from the paramagnetic label at site 3 (Fig. 7C). These data indicate that although both Sox2 and Oct1 spend the vast majority of their time bound to their adjacent specific sites (Fig. 7, A and D), a small population (<1%) must sample alternate configurations that come close to the paramagnetically labeled sites. When the salt concentration is increased to 50 or 150 mm NaCl (Fig. 7, B and C, bottom panels), the magnitude of all of the PREs is increased, due to increased exchange rates between the specific complex and the sparsely populated nonspecific complexes, but the overall PRE profiles remain qualitatively the same.
FIGURE 7.
Intermolecular PRE profiles for the specific Oct1·Sox2·Hoxb1-DNA ternary complex. A, the 29-bp DNA duplexes employed with the paramagnetic label (dT-EDTA-Mn2+) located at the ends of the duplex (site 1 or 3) or in the middle of the Sox2-specific site (site 2). B, intermolecular PRE profiles observed on 2H/15N-labeled Sox2 originating from the paramagnetic label at sites 1 (left panels), 2 (middle panels), and 3 (right panels) at 0 mm NaCl (black circles, top panels) and at 50 (blue circles), and 150 mm NaCl (red circles) (bottom panels). The concentrations of 2H/15N-Sox2, Oct1 (at natural isotopic abundance) and paramagnetically labeled DNA are 0.40, 0.44, and 0.44 mm, respectively. C, intermolecular PRE profiles observed on 2H/15N-labeled Oct1 originating from the paramagnetic label at sites 1 (left panels), 2 (middle panels), and 3 (right panels) at 0 mm NaCl (black circles, top panels) and at 50 (blue circles) and 150 mm NaCl (red circles) (bottom panels). The concentrations of 2H/15N-labeled Oct1, Sox2 (at natural isotopic abundance) and paramagnetically labeled DNA are 0.40, 0.44 and 0.44 mm, respectively. The back-calculated PREs obtained from the coordinates of the Oct1·Sox2·Hoxb1-DNA ternary complex (23) are displayed as solid lines for Sox2 (green), and the POUS (red) and POUHD (blue) domains of Oct1. The PREs are back-calculated in Xplor-NIH (37) using an ensemble representation for the paramagnetic label (30). D, experimental PRE profiles at 0 (top) and 150 mm NaCl (bottom) mapped onto the structure of the specific Oct1·Sox2·Hoxb1-DNA ternary complex (23). The color scale for the PRE 1HN-Γ2 rates is shown at the bottom. Error bars, 1 S.D.
TABLE 3.
Agreement between observed and calculated intermolecular PREs for the specific Oct1·Sox2·Hoxb1-DNA ternary complex
The positions of Sox2 and Oct1 on the 29-bp specific Hoxb1 regulatory element are taken from the NMR structure of the Oct1·Sox2·Hoxb1-DNA ternary complex (23). The complete set of experimental PRE profiles are best fitted simultaneously by optimizing the positions of all three paramagnetic labels, each represented by a three-member ensemble, using simulated annealing (30).
| NaCl | Site 1 |
Site 2 |
Site 3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sox2 | POUS | POUHD | Oct1 | Sox2 | POUS | POUHD | Oct1 | Sox2 | POUS | POUHD | Oct1 | |
| mm | ||||||||||||
| PRE Q-factor | ||||||||||||
| 0 | 0.70 | 0.97 | 0.99 | 0.99 | 0.33 | 0.49 | 0.84 | 0.70 | 1.00 | 0.96 | 0.68 | 0.80 |
| 50 | 0.81 | 0.98 | 0.99 | 0.99 | 0.71 | 0.70 | 0.92 | 0.81 | 1.00 | 0.97 | 0.72 | 0.86 |
| 150 | 0.87 | 0.99 | 0.99 | 0.99 | 0.70 | 0.70 | 0.84 | 0.78 | 1.00 | 0.97 | 0.68 | 0.85 |
| Root mean square deviation between observed and calculated Γ2 rates (s−1) | ||||||||||||
| 0 | 16.8 | 3.4 | 8.0 | 5.9 | 9.8 | 6.6 | 11.2 | 9.0 | 12.7 | 17.2 | 23.4 | 20.4 |
| 50 | 28.5 | 4.3 | 9.9 | 7.4 | 21.6 | 11.2 | 13.3 | 12.3 | 31.2 | 29.2 | 30.0 | 29.9 |
| 150 | 27.7 | 12.8 | 13.0 | 12.9 | 19.9 | 11.0 | 14.7 | 13.1 | 28.5 | 32.1 | 27.2 | 30.5 |
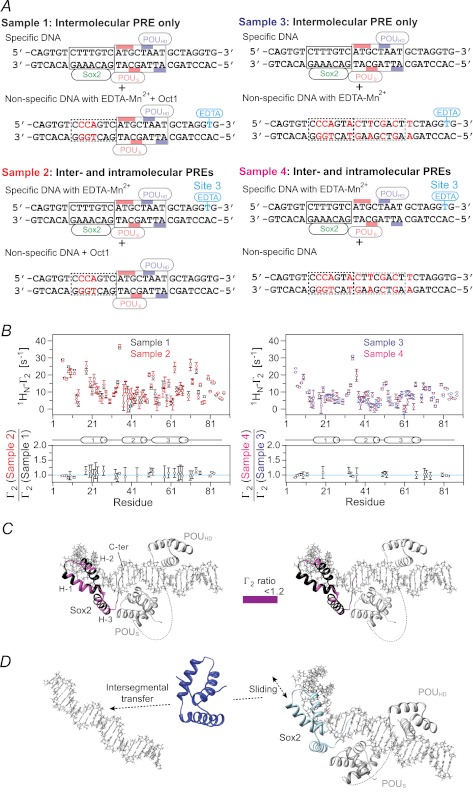
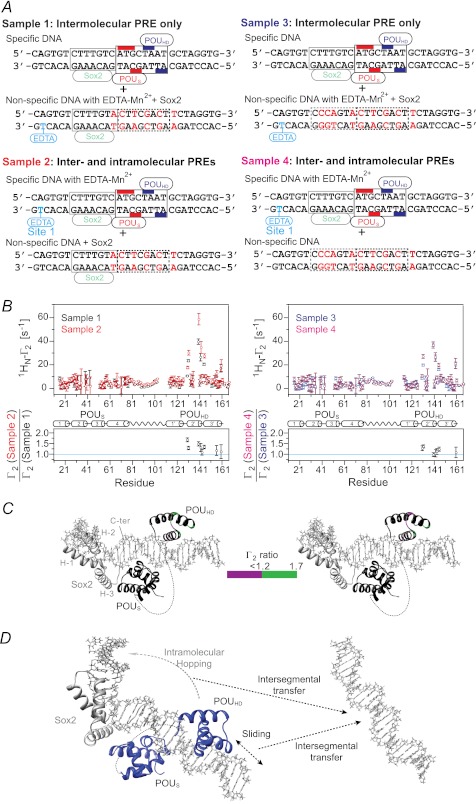
The relative contributions of intra- and intermolecular translocation were examined from a series of PRE experiments involving samples comprising an equal mixture of specific and nonspecific DNA duplexes (12). The experimental design was similar to that used for the specific Sox2·DNA binary complex (Fig. 6) except for the presence or absence of specific Oct1- or Sox2-binding sites in the nonspecific DNA used for examining translocation of Sox2 (Fig. 8A) or Oct1 (Fig. 9A), respectively. The nonspecific DNA duplex is paramagnetically labeled in samples 1 and 3, thereby reporting on intermolecular translocation, whereas the specific DNA duplex is paramagnetically labeled in samples 2 and 4 and therefore reports on both intra- and intermolecular translocation. For the experiments with 2H/15N-labeled Sox2 and Oct1 at natural isotopic abundance, the Oct1-specific site is retained in the nonspecific DNA duplex in samples 1 and 2 but removed in samples 3 and 4 (Fig. 8A). Similarly, for the experiments with 2H/15N-labeled Oct1 and Sox2 at natural isotopic abundance, the Sox2 specific site is retained in the nonspecific DNA duplex in samples 1 and 2 but removed in samples 3 and 4 (Fig. 9A).
FIGURE 8.
Intramolecular versus intermolecular translocation of Sox2 in the context of the Oct1·Sox2·Hoxb1-DNA ternary complex. A, DNA samples comprise an equimixture of specific and nonspecific 29-bp DNA duplexes. In samples 1 and 3, the specific DNA duplex is unlabeled, and the nonspecific DNA duplex bears the paramagnetic label with (sample 1) or without (sample 3) retention of the Oct1-binding site. In samples 2 and 4, the nonspecific DNA duplex with (sample 2) or without (sample 4) the Oct1-binding site is unlabeled, and the specific DNA duplex is paramagnetically labeled 3′ of the POUHD site (denoted as site 3 using the notation in Fig. 7A). The PRE effects observed for samples 1 and 3 arise entirely from intermolecular translocation of Sox2 from the specific duplex to the nonspecific duplex and back again for observation, whereas those for samples 2 and 4 arise from both intra- and intermolecular translocation processes. The concentrations of 2H/15N-Sox2, Oct1, specific DNA, and nonspecific DNA are 0.40, 0.90, 0.45, and 0.45 mm, respectively, for samples 1 and 2; for samples 3 and 4, the concentration of Oct1 is reduced to 0.45 mm because the nonspecific DNA no longer contains the specific Oct1 site. The concentrations of Oct1 were chosen to ensure that all specific Oct1-binding sites are occupied. B, intermolecular PRE profiles at 150 mm NaCl (top panels), with intermolecular PREs from samples 1 and 2 (left) and samples 3 and 4 (right) shown as black, red, blue, and magenta circles, respectively (error bars, S.D.). Bottom panels, ratio of PRE rates for sample 2 to sample 1 (left) and for sample 4 to sample 3 (right) for residues with 1HN-Γ2 rates of >10 s−1. For Γ2(sample 2 or 4)/Γ2(sample 1 or 3) ∼ 1, the PRE effects arise predominantly from intermolecular translocation; Γ2(sample 2 or 4)/Γ2(sample 1 or 3) ratios larger than 1 indicate contributions from both intra- and intermolecular translocation processes, where the contribution from the latter is provided by the PRE profile for sample 1 or 3. C, Γ2 ratios shown in B mapped onto the structure of the specific Oct1·Sox2·Hoxb1-DNA ternary complex (23). D, model for the predominant translocation processes involving Sox2 in the context of the specific Oct1·Sox2·Hoxb1-DNA ternary complex.
FIGURE 9.
Intramolecular versus intermolecular translocation of Oct 1 in the context of the Oct1·Sox2·Hoxb1-DNA ternary complex. A, DNA samples comprise an equimixture of specific and nonspecific 29-bp DNA duplexes. In samples 1 and 3, the specific DNA duplex is unlabeled, and the nonspecific DNA duplex bears the paramagnetic label with (sample 1) or without (sample 3) retention of the Sox2-binding site. In samples 2 and 4, the nonspecific DNA duplex with (sample 2) or without (sample 4) the Sox2-binding site is unlabeled, and the specific DNA duplex is paramagnetically labeled 5′ of the Sox2-specific site (site 1 in Fig. 7A). The PRE effects observed for sample 1 or 3 arise entirely from intermolecular translocation, whereas those for sample 2 or 4 arise from both intra- and intermolecular translocation processes. The concentrations of Sox2, 2H/5N-Oct1, specific DNA, and nonspecific DNA are 0.90, 0.40, 0.45, and 0.45 mm, respectively, for samples 1 and 2; for samples 3 and 4, the concentration of Sox2 is reduced to 0.45 mm. The concentrations of Sox2 were chosen to ensure that all specific Sox2-binding sites are occupied. B, intermolecular PRE profiles at 150 mm NaCl (top panels), with intermolecular PREs from samples 1 and 2 (left) and samples 3 and 4 (right) shown as black, red, blue, and magenta circles, respectively (error bars, S.D.). The ratio of PRE rates for sample 2 to sample 1 (left) and sample 4 to sample 3 (right) for residues with 1HN-Γ2 rates of >10 s−1 is displayed in the bottom panels. For Γ2(sample 2 or 4)/Γ2(sample 1 or 3) ∼ 1, the PRE effects arise predominantly from intermolecular translocation; Γ2(sample 2 or 4)/Γ2(sample 1 or 3) ratios larger than 1 indicate contributions from both intra- and intermolecular translocation processes, where the contribution from the latter is provided by the PRE profiles for sample 1 or 3. C, Γ2 ratios shown in B mapped onto the structure of the specific Oct1·Sox·Hoxb1-DNA ternary complex (23). D, model for the predominant translocation processes involving the POUHD domain of Oct1 in the context of the specific Oct1·Sox2·Hoxb1-DNA ternary complex. The POUS domain is largely fixed to its specific site as a consequence of direct interaction with the neighboring Sox2.
PRE effects within the whole of Sox2 are observed for samples 1 and 2 (Fig. 8B, left panels). Similar profiles are seen for samples 3 and 4 with some small scaling (Fig. 8B, right panels). The ratios of the sample 2 to sample 1 or sample 4 to sample 3 PREs within Sox2 are equal to 1 within experimental error (Fig. 8B, bottom panels), and there are no measurable intramolecular PRE contributions as observed in the Sox2·DNA binary complex (Fig. 6C). Thus, Sox2 largely undergoes intermolecular translocation within the context of the ternary Oct1·Sox2·Hoxb1-DNA complex, but as in the case of the binary complex, this must first involve sliding to a nonspecific site immediately adjacent to the specific one because dissociation of Sox2 from its specific site is so slow (Fig. 8D).
In the case of Oct1, sizeable PREs are observed within helices 2′ and 3′ of the POUHD domain, indicative of intermolecular translocation (Fig. 9B, top panels). However, no significant PREs (Γ2 < 10 s−1) are observed within the POUS domain for samples 1 and 3, indicating that the POUS domain is largely fixed to its specific site through interaction with Sox2. This is in direct contrast to the situation with the specific Oct1·DNA binary complex, where the POUS domain largely directs intermolecular translocation (10). For the POUHD domain, the ratios of the sample 2 to sample 1 or sample 4 to sample 3 PREs are slightly greater than 1, indicating that the PRE effects observed on the POUHD domain when the paramagnetic label is placed on the Sox2 side of the specific DNA duplex are largely due to intermolecular translocation with some contribution of intramolecular translocation. It seems likely that intramolecular translocation of the POUHD domain to nonspecific sites in close proximity to the paramagnetic label placed on the 5′ side of the Sox2-specific site (Fig. 9A) occurs by hopping rather than sliding because the presence of Sox2 and POUS domain on their respective specific sites would block direct sliding of the POUHD domain (Fig. 9D).
Because translocation of Sox2 and the POUHD domain in the context of the ternary Oct1·Sox2·Hoxb1-DNA ternary complex is primarily intermolecular, the probability of coming into close proximity to the paramagnetic label is higher when only specific DNA is present than with an equal mixture of specific and nonspecific DNA (Figs. 8 and 9). As a result, the PREs observed in Figs. 8B and 9B will be smaller than those observed in Fig. 7, B and C, respectively. For example, the sizeable PREs observed in the C-terminal tail of Sox2 when only the specific DNA is present (Fig. 7B) are much reduced in the presence of the nonspecific DNA (Fig. 8B). Similarly, the large PREs in the POUS domain seen in Fig. 7C become much smaller in Fig. 9B in the presence of nonspecific DNA. The same phenomenon is at play when comparing the magnitude of the PREs in the mixed samples 2 and 4 in Figs. 8 and 9. When the 2H/15N-labeled protein for which PREs are being measured interacts with the nonspecific DNA duplex, the presence of the unobserved protein at natural isotopic abundance (either Oct1 or Sox2) bound to its remaining specific site on the nonspecific DNA duplex reduces the number of nonspecific sites available for binding the 2H/15N-labeled protein and therefore increases the probability of close access to the paramagnetic tag. Thus, PREs observed for sample 2 (Figs. 8B and 9B, left top panel) are larger than those for sample 4 (Figs. 8B and 9B, right top panel, respectively).
Conclusions
z-Exchange experiments reveal that both Sox2 (Fig. 4) and Oct1 (14) undergo bulk intermolecular translocation between specific sites located on different DNA molecules. For Sox2, this process occurs via complete dissociation of Sox2 from its specific site on the first DNA molecule followed by a three-dimensional search in solution and reassociation, a process referred to as jumping. No evidence for direct transfer of Sox2 from one specific site to another without dissociation into free solution, a process known as intersegment transfer, was observed by z-exchange spectroscopy. For Oct1, jumping also occurs, but the predominant intermolecular translocation pathway at high concentrations of free DNA involves direct transfer from one specific site to the other (14). When the specific Oct1·Sox2·DNA ternary complex is formed on the Hoxb1 regulatory element (Fig. 4A), bulk intermolecular translocation between specific sites for both Sox2 (Fig. 4) and Oct1 (14) is slowed down by more than an order of magnitude and can no longer be detected by z-exchange spectroscopy. From the perspective of gene regulation, this implies that once the specific ternary Oct1·Sox2·Hoxb1-DNA complex is formed, the search process for other potential target sites by the Oct1 and Sox2 molecules bound to a particular regulatory element will be slowed down but, importantly, not eliminated (see below).
Intermolecular translocation between specific binding sites observed by z-exchange spectroscopy involves the major spectroscopically “visible” species (i.e. the specific complexes whose spectra can be directly observed). However, fast translocation of Sox2 and Oct1, in the context of both binary and ternary specific complexes, occurs both along the DNA via sliding (intramolecular translocation) and between DNA molecules via direct intersegment transfer (Figs. 5–9). These processes can be detected by PRE and involve sparsely populated states bound to nonspecific sites that are in fast exchange with the specific complexes on whose spectra the PRE profiles are measured. The transiently populated states reveal their footprint on the PRE profiles measured on the major species because under fast exchange conditions, the observed PREs are a population weighted average, and the 〈r−6〉 dependence of the PRE ensures that any short paramagnetic center to proton distances present in the minor species will have a large impact on the PRE rates observed on the major species, especially if the corresponding paramagnetic center to proton distances are long in the major species.
Both PRE (Fig. 2) and Lorentzian line width analysis (Fig. 3) of nonspecific Sox2·DNA complexes indicate the presence of rapid intra- and intermolecular translocation between nonspecific sites, corresponding to sliding and direct intersegment transfer, respectively. These species represent the entities whose footprints are observed in the PRE profiles measured on the specific complexes. Sliding is a first order process and is therefore independent of the concentration of nonspecific DNA. In vivo, the length of DNA over which sliding occurs will depend on two factors: 1) the competition between sliding and intermolecular translocation and 2) the presence of other proteins on the DNA that may block further ongoing sliding, thereby promoting either dissociation or intersegment transfer. The apparent rate of intersegment transfer between nonspecific DNA sites is directly proportional to the concentration of DNA and will therefore play a major role in vivo because the concentration of nonspecific DNA within the nucleus is extremely high.
The data presented here suggest the following model whereby Sox2 efficiently explores the DNA landscape to locate its specific target site. Sox2 searches for its specific target site using both direct (intersegment) transfer between nonspecific sites on different DNA duplexes and sliding along nonspecific DNA (Fig. 6E). Once Sox2 and Oct1 form a specific ternary complex with Hoxb1-DNA, further sampling of DNA sites is reduced but can still occur by initial sliding to an adjacent nonspecific site followed by intersegment transfer (Fig. 8D). Oct1 alone explores the DNA landscape by making use of the complementary interplay of intramolecular sliding by the POUHD domain and intersegment transfer of the POUS domain (10), with the latter acting as a flycast. Once the ternary complex with Sox2 is formed, however, the POUS domain is largely fixed to its specific site as a result of direct contact with Sox2. The POUHD domain, however, can still sample other DNA sites, as evidenced by the PRE data (Figs. 7 and 9), by an initial sliding event to an adjacent nonspecific site, as in the case of Sox2, followed by either intersegment transfer to another DNA molecule or by direct hopping to another nonspecific site on the same DNA molecule (Fig. 9D). In the former instance, a bridged intermediate is formed in which the POUS domain and Sox2 are located on the first DNA molecule, whereas the POUHD domain is bound to the second DNA molecule. The formation of this bridged complex can promote the completion of the release of Oct1 from the ternary complex by a first order process involving dissociation of the POUS domain from the first DNA, followed by association on to the second DNA. This contrasts to the mechanism used in the specific Oct1·DNA binary complex where the POUS domain locates the second DNA molecule by intersegment transfer, thereby promoting the subsequent transfer of the POUHD domain (10). Thus, direct interaction of Sox2 with the POUS domain of Oct1 bound to adjacent cognate sites on the DNA modulates the translocation pathway employed by Oct1 to scan alternative DNA target sites via transient, sparsely populated states.
Acknowledgments
We thank Drs. Garrett, Baber, and Ying for technical support and Dr. Carole Bewley for useful discussions.
This work was supported, in whole or in part, by the National Institutes of Health, NIDDK, Intramural Program and by the AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G. M. C.).
D. S. Garrett and G. Marius Clore, unpublished data.
- PRE
- paramagnetic relaxation enhancement
- TROSY
- transverse relaxation optimized z-exchange spectroscopy
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Berg O. G., von Hippel P. H. (1985) Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 14, 131–160 [DOI] [PubMed] [Google Scholar]
- 2. von Hippel P. H., Berg O. G. (1989) Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 [PubMed] [Google Scholar]
- 3. Halford S. E., Marko J. F. (2004) How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vuzman D., Azia A., Levy Y. (2010) Searching DNA via a “Monkey Bar” mechanism. The significance of disordered tails. J. Mol. Biol. 396, 674–684 [DOI] [PubMed] [Google Scholar]
- 5. Gorman J., Plys A. J., Visnapuu M. L., Alani E., Greene E. C. (2010) Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 17, 932–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clore G. M. (2011) Exploring translocation of proteins on DNA by NMR. J. Biomol. NMR 51, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iwahara J., Zweckstetter M., Clore G. M. (2006) NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 15062–15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blainey P. C., Luo G., Kou S. C., Mangel W. F., Verdine G. L., Bagchi B., Xie X. S. (2009) Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 16, 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levy Y., Onuchic J. N., Wolynes P. G. (2007) Fly-casting in protein-DNA binding. Frustration between protein folding and electrostatics facilitates target recognition. J. Am. Chem. Soc. 129, 738–739 [DOI] [PubMed] [Google Scholar]
- 10. Takayama Y., Clore G. M. (2011) Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc. Natl. Acad. Sci. U.S.A. 108, E169–E176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vuzman D., Polonsky M., Levy Y. (2010) Facilitated DNA search by multidomain transcription factors. Cross-talk via a flexible linker. Biophys. J. 99, 1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwahara J., Clore G. M. (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440, 1227–1230 [DOI] [PubMed] [Google Scholar]
- 13. Iwahara J., Clore G. M. (2006) Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 128, 404–405 [DOI] [PubMed] [Google Scholar]
- 14. Doucleff M., Clore G. M. (2008) Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc. Natl. Acad. Sci. U.S.A. 105, 13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Rocco G., Gavalas A., Popperl H., Krumlauf R., Mavilio F., Zappavigna V. (2001) The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 autoregulatory enhancer function. J. Biol. Chem. 276, 20506–20515 [DOI] [PubMed] [Google Scholar]
- 16. Sturm R. A., Das G., Herr W. (1988) The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeobox subdomain. Genes Dev. 2, 1582–1599 [DOI] [PubMed] [Google Scholar]
- 17. Andersen B., Rosenfeld M. G. (2001) POU domain factors in the neuroendocrine system. Lessons from developmental biology provide insights into human disease. Endocr. Rev. 22, 2–35 [DOI] [PubMed] [Google Scholar]
- 18. Herr W., Sturm R. A., Clerc R. G., Corcoran L. M., Baltimore D., Sharp P. A., Ingraham H. A., Rosenfeld M. G., Finney M., Ruvkun G. (1988) The POU domain. A large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 2, 1513–1516 [DOI] [PubMed] [Google Scholar]
- 19. Klemm J. D., Rould M. A., Aurora R., Herr W., Pabo C. O. (1994) Crystal structure of the Oct-1 POU domain bound to an octamer site. DNA recognition with tethered DNA-binding modules. Cell 77, 21–32 [DOI] [PubMed] [Google Scholar]
- 20. Kamachi Y., Uchikawa M., Kondoh H. (2000) Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 16, 182–187 [DOI] [PubMed] [Google Scholar]
- 21. Dailey L., Basilico C. (2001) Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J. Cell. Physiol. 186, 315–328 [DOI] [PubMed] [Google Scholar]
- 22. Murphy E. C., Zhurkin V. B., Louis J. M., Cornilescu G., Clore G. M. (2001) Structural basis for SRY-dependent 46-X,Y sex reversal. Modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 312, 481–499 [DOI] [PubMed] [Google Scholar]
- 23. Williams D. C., Jr., Cai M., Clore G. M. (2004) Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1·Sox2·Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 279, 1449–1457 [DOI] [PubMed] [Google Scholar]
- 24. Iwahara J., Anderson D. E., Murphy E. C., Clore G. M. (2003) EDTA-derivatized deoxythymidine as a tool for rapid determination of protein binding polarity to DNA by intermolecular paramagnetic relaxation enhancement. J. Am. Chem. Soc. 125, 6634–6635 [DOI] [PubMed] [Google Scholar]
- 25. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 26. Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 27. Sahu D., Clore G. M., Iwahara J. (2007) TROSY-based z-exchange spectroscopy. Application to the determination of the activation energy for intermolecular protein translocation between specific sites on different DNA molecules. J. Am. Chem. Soc. 129, 13232–13237 [DOI] [PubMed] [Google Scholar]
- 28. Iwahara J., Tang C., Marius Clore G. (2007) Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 184, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fawzi N. L., Doucleff M., Suh J. Y., Clore G. M. (2010) Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. U.S.A. 107, 1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwahara J., Schwieters C. D., Clore G. M. (2004) Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 126, 5879–5896 [DOI] [PubMed] [Google Scholar]
- 31. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 32. Iwahara J., Schwieters C. D., Clore G. M. (2004) Characterization of nonspecific protein-DNA interactions by 1H paramagnetic relaxation enhancement. J. Am. Chem. Soc. 126, 12800–12808 [DOI] [PubMed] [Google Scholar]
- 33. Clore G. M., Iwahara J. (2009) Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 109, 4108–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McConnell H. M. (1958) Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 28, 430–431 [Google Scholar]
- 35. Tang C., Iwahara J., Clore G. M. (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 [DOI] [PubMed] [Google Scholar]
- 36. Tang C., Schwieters C. D., Clore G. M. (2007) Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 449, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 37. Schwieters C. D., Kuszewski J. J., Clore G. M. (2006) Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 [Google Scholar]