Background: MTCH2 and tBID proteins interact to induce apoptosis in the mitochondrial pathway.
Results: Molecular and biophysical studies of the tBID-MTCH2 complex led to two peptides, derived from the tBID-MTCH2 binding interface, which induced cell death.
Conclusion: tBID and MTCH2 have two major interaction sites. Peptides derived from the interface are anticancer leads.
Significance: The tBID-MTCH2 interaction may be a novel anticancer drug target.
Keywords: Biophysics, Peptide Arrays, Peptide Interactions, Protein Complexes, Protein-Protein Interactions
Abstract
The molecular basis of the interaction between mitochondrial carrier homologue 2 (MTCH2) and truncated BID (tBID) was characterized. These proteins participate in the apoptotic pathway, and the interaction between them may serve as a target for anticancer lead compounds. In response to apoptotic signals, MTCH2 recruits tBID to the mitochondria, where it activates apoptosis. A combination of peptide arrays screening with biochemical and biophysical techniques was used to characterize the mechanism of the interaction between tBID and MTCH2 at the structural and molecular levels. The regions that mediate the interaction between the proteins were identified. The two specific binding sites between the proteins were determined to be tBID residues 59–73 that bind MTCH2 residues 140–161, and tBID residues 111–125 that bind MTCH2 residues 240–290. Peptides derived from tBID residues 111–125 and 59–73 induced cell death in osteosarcoma cells. These peptides may serve as lead compounds for anticancer drugs that act by targeting the tBID-MTCH2 interaction.
Introduction
Protein-protein interactions (PPIs)3 are crucial for the proper function of living cells. Studying PPIs is important for understanding cell functionality and for designing drugs against diseases in which PPIs are impaired. Peptides are excellent tools for studying PPIs: They are very useful for mapping binding sites precisely, particularly by using peptide array libraries (1). They can also be used as lead compounds that stimulate or inhibit cellular processes by interfering with specific PPIs. Apoptosis (programmed cell death) is a key process in regulating physiological growth control and tissue homeostasis. Impaired regulation of apoptotic pathways is a key event in malignant transformation. Understanding how apoptotic pathways are regulated is thus essential for developing anticancer therapies. Here, the peptide approach was used to characterize the interaction between two key players in the apoptotic pathway: tBID and the mitochondrial carrier homologue 2 (MTCH2) proteins. Peptides derived from the binding interface between the two proteins were able to kill cancer cells.
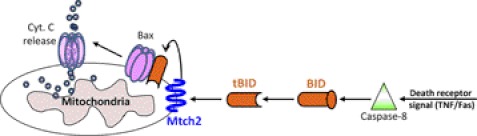
The BID protein belongs to the Bcl-2 protein family. This family consists of both proapoptotic (e.g. Bax, Bak) and antiapoptotic (e.g. Bcl-2, Bcl-XL) members, which form homo- and heterodimers that maintain the balance between cell death and survival (2–4). Proteins from the Bcl-2 family are major targets for developing anticancer therapies (2, 5, 6). BID is involved in cell proliferation and regulating genomic stability (7). However, its most important role is to induce apoptosis by interacting with other Bcl-2 family members. BID belongs to the BH3-only subfamily of the Bcl-2 family, in which only the BH3 domain is homologous. The role of BID in the apoptotic pathways is unique because it integrates both the extrinsic and intrinsic death pathways (8–10) Death signals mediated by the death receptor Fas activate caspase-8, which cleaves full-length BID at its N terminus between residues Asp-59 and Gly-60 (in mouse BID), forming a truncated BID protein (tBID). tBID contains the BH3 domain and serves as the active form of BID: It translocates to the mitochondria and activates Bax/Bak homo-oligomerization in the outer mitochondrial membrane, followed by the release of cytochrome c and apoptosis (Fig. 1). Most BH3 proteins are intrinsically disordered and probably attain structure upon binding their ligands (4). tBID shows a “Bcl-2 core” structure comprising seven amphipathic α-helices arranged around a central buried helix, that create a hydrophobic groove (4). This structure is similar to that of proteins from other pro-/antiapoptotic Bcl-2 subfamilies, e.g. Bax, Bak, and Bcl-2. Of all of the Bcl-2 family members, tBID is most similar to Bax in terms of structure and membrane-binding mechanism (11), yet the exact mechanism by which tBID translocates to the mitochondria remains unclear. A recent study revealed that tBID interacts with the MTCH2 protein, which is critical for BID-induced activation of Bax/Bak (Fig. 1) (12).
FIGURE 1.
tBID structure and mechanism of action. Death signals mediated by the death receptors Fas/TNF activate caspase-8, which cleaves BID at the N terminus and forms a truncated active BID fragment (tBID). tBID is recruited by MTCH2 to the mitochondria, enabling it to induce activation of Bax/Bak. The activation of Bax/Bak leads to cytochrome c release, resulting in an apoptotic response.
MTCH2 was named after its single conserved mitochondrial carrier domain that was predicted at the time of its discovery (13) and was also identified as the Met-induced mitochondrial protein (MIMP) (14). MTCH2 is a conserved protein that belongs to the mitochondrial carrier protein (MCP) family. MCPs catalyze the exchange of solutes across the inner mitochondrial membrane (IMM) (15, 16). Structurally, all MCPs contain a similar mitochondrial carrier domain, composed of three tandem repeats, each containing two hydrophobic helices separated by a hydrophilic region. Both the N and the C termini face the intermembrane space, and three hydrophilic elements face the matrix (17). The adenine nucleotide translocator (ANT1) protein was the first MCP to be identified and sequenced (18). A model of the MTCH2 structure was proposed based on sequence alignment analysis and threading using the three-dimensional structure of ANT1 (19). MTCH2 differs from the other MCPs because it is exposed on the surface of the mitochondria rather than at the IMM (12). This is consistent with the interaction between MTCH2 and tBID because tBID translocates to the mitochondria but does not reach the IMM. The interaction between MTCH2 and tBID was shown by a complex formed by cross-linking of tBID in TNF-α-activated hematopoietic FL5.12 cells (20). Knockout of MTCH2 in embryonic stem cells and in mouse embryonic fibroblasts inhibited recruitment of tBID to the mitochondria and resulted in reduced Bax/Bak activation and apoptosis (12). Significantly less tBID was recruited to the mitochondria following injection of anti-Fas antibodies in mice lacking MTCH2 in the liver. This resulted in reduced Bax activation, reduced activation of caspase-3, and increased survival of the mice (12). This indicates that MTCH2 has a critical function in liver apoptosis by regulating the recruitment of tBID to the mitochondria (Fig. 1).
Here, the first detailed characterization of the interaction between tBID and MTCH2 is presented at quantitative and structural levels. The binding sites and affinities are shown for both proteins. Cancer cell death was induced by peptides derived from the binding interfaces. Targeting the specific binding interfaces between tBID and MTCH2 is a promising approach for developing anticancer lead compounds.
EXPERIMENTAL PROCEDURES
Expression and Purification of tBID
Recombinant tBid 15 was expressed and purified as described (21).
Peptide Array Screening
Peptide arrays (containing peptides derived from BID and MTCH2 sequences) were synthesized by INTAVIS Bioanalytical Instruments AG (Köln, Germany). Peptide-cellulose conjugates were synthesized and spotted on glass slides (INTAVIS). The peptide array was screened as described (23). Binding of tBID to the array was detected using rabbit anti-BID, and binding of the biotinylated MTCH2 peptides was detected using streptavidin-conjugated HRP antibody (Sigma), using a chemiluminescence blotting substrate Super Signal reagent (Beit Haemek) according to the manufacturer's instructions.
Peptide Synthesis and Purification
Peptides were synthesized on a Liberty MAPS (Microwave-Assisted Peptide Synthesizer; CEM) using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry as described (24). Peptides were purified on a Gilson HPLC (Middelton, WI) using a reverse-phase C8 semipreparative column (ACE). Peptides were analyzed using MALDI-TOF mass spectrometry on a Voyager DE-Pro instrument (Applied Biosystems). For some experiments the peptides were also labeled with biotin or fluorescein at their N terminus as described (24, 25).
Fluorescence Anisotropy
Measurements were performed as described (26) using a PerkinElmer Life Science LS-55 luminescence spectrofluorimeter equipped with a Hamilton microlaboratory 500 dispenser (26). Briefly, 1 ml of fluorescein-labeled peptide (0.1 μm in 20 mm Hepes buffer, pH 7.3, 42 mm NaCl, and 5 mm DTT) was placed in a cuvette, and the nonlabeled protein or peptides were added at 1-min intervals and stirred for 10 s. Dissociation constants (Kd) were calculated by fitting the anisotropy titration curves to a 1:1 binding model (26).
Circular Dichroism (CD) Measurements
Samples were prepared by dissolving the lyophilized peptide in 25 mm sodium phosphate buffer, pH 7.2, 100 mm NaCl. CD spectra were recorded using a J-810 spectropolarimeter (Jasco) in a 0.1-cm quartz cuvette for far-UV CD spectroscopy, collected over 190–260 nm at 25 °C.
Cell Lines
Human osteosarcoma (U2OS) cells were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 1:100 liters glutamine, 10% fetal bovine serum (FBS), and 1:100 penicillin/streptomycin antibiotic solution (10,000 units/ml penicillin, 10 mg/ml streptomycin; Biological Industries, Beit HaEmek, Israel). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 and 95% air.
Cell Penetration of Peptides
Fluorescein-labeled peptides at a final concentration of 5–60 μm in phosphate-buffered saline (PBS) were incubated with U2OS cells for 1.5 h at 37 °C. After three washes in PBS, the cells were fixed with acetone and methanol and visualized by confocal microscopy. The peptide Antennapedia helix III (43–58), sequence RQIKIWFQNRRMKWKK (also named Penetratin), was used as a positive control.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
U2OS cells were seeded in a 96-well plate at a density of 5 × 103 cells/well. After growing overnight, cells were treated with the various peptides overnight (for concentrations, see “Results”). Then supernatants were removed, and 200 μl of 0.3 mg/ml MTT was added to each well followed by incubation at 37 °C for an additional 2 h. Supernatants were removed, and 200 μl of dimethyl sulfoxide was added, followed by an additional 10-min incubation at room temperature. Absorbance at 570 nm was measured using a microplate reader.
RESULTS
Identifying tBID Binding Sites in MTCH2 Using Peptide Array Screening
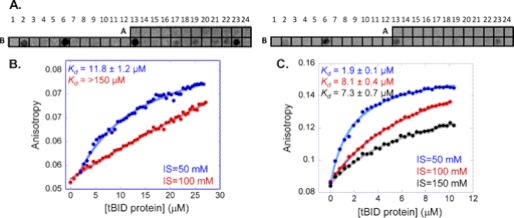
The tBID binding sites in MTCH2 were detected by screening an array consisting of 37 partly overlapping peptides derived from MTCH2. Peptide length was between 10 and 30 residues (for peptides sequences, see supplemental Table 1). Peptides were designed based on the predicted secondary and tertiary structures of MTCH2 as shown in its model (Protein Data Bank ID code 1OCK) (19). The peptide array was screened for binding recombinant tBID. The tBID protein bound eight peptides derived from MTCH2 with different signal intensities (Fig. 2 and Table 1). Four peptides gave medium-strong signal intensities. Three of these peptides were derived from the central region of MTCH2, residues 105–119, 140–161, and 210–224. One peptide was derived from the C-terminal domain between residues 276 and 290. Four additional peptides derived from the C terminus of MTCH2 bound tBID with relatively weak signal intensities (Fig. 2). These peptides were derived from MTCH2 residues 240–254, 247–264, 265–275, and 270–283. All five peptides derived from the C-terminal region were partly overlapping, and together they span the sequence of the entire MTCH2 C terminus. Although each peptide from this region showed only a weak partial interaction with tBID, the whole domain (residues 240–290) may bind tBID much more strongly.
FIGURE 2.
tBID binding sites in MTCH2: screening and quantitative analysis. A, an array consisting of partly overlapping peptides derived from MTCH2 was screened for binding tBID. Each dark spot represents binding of tBID to a specific peptide (Table 1). Left, high contrast, allowing observation of the weak binding peptides. Right, low contrast, emphasizing the different signal intensities as specified in Table 1. B and C, shown are representative binding curves for the interaction between the tBID protein and the peptides derived from MTCH2: MTCH2 240–290 (B) MTCH2 140–161 (C). Binding was quantified using fluorescence anisotropy and measured at various ionic strengths. Both peptides bound tBID with affinity at the low millimolar range.
TABLE 1.
Binding of tBID to peptides derived from MTCH2: peptide array screening
| Spot no. in arraya | Residues | Sequenceb | Signal intensity |
|---|---|---|---|
| B2 | 105–119 | GSVTVQKEYSSSFDR | Medium |
| B6 | 140–161 | PFHVITLRSMVQFIGRESKYCG | Strong |
| B13 | 210–224 | SGVSTMNEMKSYSQA | Medium |
| B17 | 240–254 | VSNLMAVNNCGLAGG | Weak |
| B19 | 247–264 | NNCGLAGGSPPYSPIYTS | Weak |
| B21 | 270–283 | CMLQKAGNMSRGNS | Weak |
| B22 | 265–275 | WIDCWCMLQK | Weak |
| B23 | 276–290 | WGNMSRGNSLFFRKVP | Medium |
a The peptide spot number in the array corresponds to Fig. 2A.
b Trp was added at the N terminus of some peptides for UV spectroscopy.
Quantitative Analysis of Interactions between tBID and MTCH2 Peptides: Selecting Lead Peptides for Further Experiments
Quantitative information about the binding of the MTCH2 peptides to tBID was obtained using the peptides found to bind by array screening. Their interaction with tBID was confirmed and quantified using fluorescence anisotropy. The four strongest binding peptides in the array were selected for further study (Fig. 2 and Table 1): MTCH2 105–119, MTCH2 140–161, MTCH2 210–224, and MTCH2 276–290, as well as the longer peptide that combined all of the binding peptides from the MTCH2 C-terminal region: MTCH2 240–290. MTCH2 140–161 and MTCH2 240–290 bound tBID, whereas the interaction of tBID with the other peptides was too weak to quantify. MTCH2 140–161 bound tBID with Kd = 11.8 ± 1.2 μm at ionic strength of 50 mm (Fig. 2B). MTCH2 240–290 bound tBID an order of magnitude tighter, with an affinity of 1.9 ± 0.1 μm at 50 mm ionic strength (Fig. 2C). The binding of both peptides to tBID was dependent on the ionic strength, indicating a partial electrostatic contribution to the interaction (Fig. 2). Based on the fluorescence anisotropy binding studies, two peptides representing two main binding sites for tBID on MTCH2 were selected for further studies: one from the central region of MTCH2 (residues 140–161) and the other from the C-terminal region of MTCH2 (residues 240–290).
Identifying MTCH2 Binding Sites on tBID
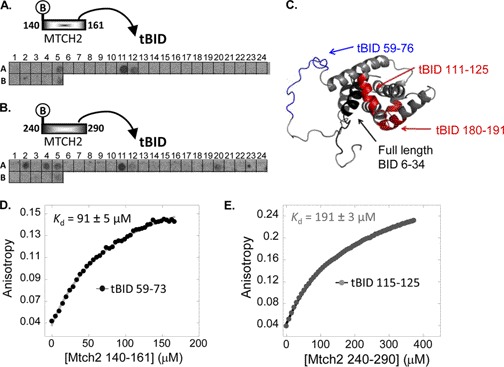
An array consisting of 30 partly overlapping peptides derived from BID was designed to identify the MTCH2 binding sites in tBID. The peptides were 10–30 residues in length (peptide sequences shown in supplemental Table 2). Peptides were designed based on the secondary and tertiary structures of the BID (Protein Data Bank ID code 2BID) (27, 28). Being a membrane protein, MTCH2 had not been successfully expressed as a recombinant protein. Thus, the tBID-binding MTCH2 peptides (MTCH2 140–161 and MTCH2 240–290) were used for screening the array. The peptides were labeled with biotin at their N termini to identify them using streptavidin-conjugated HRP antibody. Seven tBID-derived peptides bound the biotinylated MTCH2 peptides (Fig. 3, A and B, and Table 2). Two partly overlapping observed binding peptides were derived from the N terminus of tBID between residues 59–73 and 62–76. Two other peptides, BID 111–125 and BID 180–191, were derived from the central and C-terminal regions of BID, respectively. Although these peptides are distant in the protein sequence, they are spatially proximate in the folded protein (Fig. 3C), forming one binding site. The four tBID-binding peptides, together with a peptide consisting of residues 62–73 (overlapping residues between two binding peptides), were synthesized for further studies. Three additional binding peptides from the N terminus of full-length BID (residues 6–34) were observed, but because these residues are cleaved during the formation of tBID they were not pursued.
FIGURE 3.
MTCH2 binding sites in tBID: screening and quantitative analysis. A and B, an array consisting of partly overlapping peptides derived from BID was screened for binding biotinylated MTCH2 peptides MTCH2 140–161 (A) and MTCH2 240–290 (B). Each dark spot represents binding of MTCH2 peptides to a specific BID peptide (Table 2). C, MTCH2 binding sites, as discovered in the peptide array screening, are colored on the three-dimensional structure of BID (Protein Data Bank ID code 2BID). D and E, binding was quantified using fluorescence anisotropy. The interaction between the selected tBID peptides and MTCH2 140–161 (D) and MTCH2 240–290 (E). Each MTCH2 peptide bound only one peptide from tBID. MTCH2 240–290 bound tBID 111–125, and MTCH2 140–161 bound tBID 59–73.
TABLE 2.
Binding of MTCH2 140–161 and MTCH2 240–290 to peptides derived from BID: peptide array screening results
| Spot no. in arraya | Residues | Sequenceb | Signal intensity |
|---|---|---|---|
| A2 | 6–20 | NNGSSLRDECITNLL | Medium |
| A4 | 20–34 | LVFGFLQSCSDNSFR | Medium |
| A5 | 27–34 | SCSDNSFR | Medium |
| A11 | 59–73 | WTDGNRSSHSRLGRIE | Strong |
| A12 | 62–76 | WNRSSHSRLGRIEADS | Medium |
| A20 | 111–125 | WLQLRNTSRSEEDRNR | Medium |
| B5 | 180–191 | QNLRTYVRSLAR | Weak |
a The peptide number in the array corresponds to Fig. 3, A and B.
b Trp was added at the N terminus of some peptides for UV spectroscopy.
Quantitative Analysis of Interactions between Peptides from tBID and Peptides from MTCH2: Identifying Corresponding Binding Sites
The selected tBID peptides discovered in the array screening were synthesized, and their binding to MTCH2 140–161 and MTCH2 240–290 was quantified using fluorescence anisotropy. Each MTCH2 peptide bound only one peptide from tBID (supplemental Fig. 1, A and B). MTCH2 140–161 bound tBID 59–73 with Kd = 91 ± 5 μm (Fig. 3D), whereas MTCH2 240–290 bound tBID 111–125 with an affinity constant of 191 ± 3 μm (Fig. 3E). The interactions were weaker compared with the binding of MTCH2 peptides to recombinant tBID (Fig. 2), probably because peptide-peptide interactions (Fig. 3) are usually weaker than protein-peptide interactions (Fig. 2). Two peptides have much shorter interaction interfaces. The binding of both peptides was dependent on the ionic strength (supplemental Fig. 1, C and D), similar to the opposite interaction between the tBID protein and the MTCH2 peptides, further indicating the partly electrostatic mechanism. In summary, the fluorescence anisotropy binding studies revealed that tBID has two main MTCH2 binding regions. Using peptides from both proteins the positions where the interaction between the proteins occurs were identified: MTCH2 240–290 binds tBID 111–125, and MTCH2 140–161 binds tBID 59–73.
tBID-derived Peptides Induced Cancer Cell Death
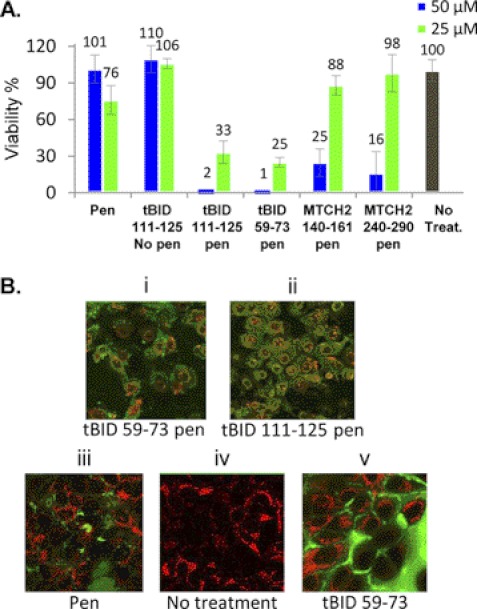
The interaction between tBID and MTCH2 is at the heart of the mitochondrial death pathway. Thus, targeting the interaction interfaces that were identified by inhibiting peptides derived from these sites can interfere with the apoptotic pathway regulation and induce cancer cell death. The ability of the four selected peptides derived from the MTCH2-tBID binding interface (MTCH2 240–290, MTCH2 140–161, tBID 111–125, and tBID 59–73) to induce cancer cell death was tested. The peptides were fused to Penetratin at their N termini to enable cell permeability. They were labeled with fluorescein at their N termini, and their cell penetration was followed by confocal fluorescence microscopy. All peptides penetrated osteosarcoma (U2OS) cells (supplemental Fig. 2). The effect of peptides on cancer cells was tested in U2OS using the MTT assay. The MTCH2- and tBID-derived peptides were incubated with the cells for 24 h, and the cells were tested for viability compared with untreated cells. The tBID peptides induced significant death of cancer cells. Treating cells with 50 μm tBID 111–125 conjugated to Penetratin or tBID 59–73 conjugated to Penetratin almost completely eliminated cell survival (Fig. 4A). Both peptides reduced cell viability in a concentration-dependent manner (Fig. 4A). The MTCH2-derived peptides had a much weaker effect on cell survival, which was observed only at 50 μm peptide but not at 25 μm. Penetratin only or peptides lacking the Penetratin sequence did not induce cell death. The MTT assay was performed also using SV40 mouse embryonic fibroblasts (supplemental Fig. 3). The tBID-derived peptides induced significant death of the mouse embryonic fibroblasts, similar to the case with the U2OS cells. tBID 111–125 conjugated to Penetratin had the strongest effect. MTCH2 140–161 conjugated to Penetratin also induced cell death at a concentration of 90 μm.
FIGURE 4.
tBID-derived peptides induce cancer cell death. A, representative MTT assay to assess cell viability. U2OS cells were treated with tBID/MTCH2-derived peptides (concentrations of 25 and 50 μm, respectively). The number of control cells, i.e. viable cells, not exposed to any treatment, was defined as 100%. MTT assay was preformed as described under “Experimental Procedures.” The two tBID peptides (tBID 59–73 and tBID 111–125) induced significant cancer cell death. B, morphologic evidence of cell death. U2OS cells were treated with fluorescent mitochondria stain RPA-C (red), and fluorescein-labeled peptides (green) in a final concentration of 25 μm. Morphologic evidence of cell death was observed among the cells treated with tBID 59–73 penicillin (Pen) (i), tBID 111–125 penicillin (ii). Cells treated with penicillin (iii) and tBID 59–73 without penicillin (v) showed no morphologic change as the nontreated cells (iv).
Following the effect of the tBID peptides using live confocal fluorescence microscopy showed significant changes in cell morphology. Cells were also treated with a fluorescent mitochondria stain RPA-C (rhodamine B-[(1,10-phenanthrolin-5-yl)aminocarbonyl] benzyl ester), to examine whether the mitochondria stay intact. Cells treated with the tBID-derived peptides fused to Penetratin demonstrated a morphology characteristic of apoptosis, including reduction in cell volume, nuclear condensation, and membrane blebbing (Fig. 4B, i and ii). Treating cells with peptides lacking the Penetratin sequence (Fig. 4Bv) or peptides containing the Penetratin sequence alone (Fig. 4Biii) had no effect on cell morphology or viability, demonstrating that the observed cell killing activity was tBID-dependent and required internalization.
DISCUSSION
In vitro molecular studies of the interaction between MTCH2 and tBID using the full-length proteins is very difficult because it is impossible to express and purify MTCH2 as it is a membrane protein. Instead, the peptide approach was used with peptides corresponding to domains of this protein. Using a combination of peptide array screening, biophysical methods, and cell viability assays, the interaction between MTCH2 and tBID was characterized, and the binding sites in both proteins were mapped. Quantification of the interactions at the peptide level showed where each peptide bound in the partner protein. The binding studies demonstrated a partial electrostatic nature for the interaction. MTT experiments in cells highlighted two tBID-derived peptides (tBID 111–125 and tBID 59–73), which induced cell death in U2OS cells. These peptides are potential anticancer lead compounds that act by a novel mechanism.
Molecular Basis of MTCH2-tBID Interaction
Our results suggest that MTCH2 140–161 binds tightest to tBID (compare peak intensities in Table 1 and Fig. 2). This is in good agreement with the fluorescence anisotropy binding studies, where from all of the observed peptides on the array only MTCH2 140–161 demonstrated significant binding to tBID with Kd = 11.8 ± 1.2 μm (Fig. 2B). MTCH2 240–290, designed according to the sequences of five partly overlapping weak binding peptides that were observed by the peptide array screening, also bound tBID in the anisotropy studies with an affinity of 1.9 ± 0.1 μm (Fig. 2C). This peptide represents the whole C-terminal tail of MTCH2. We conclude that the C-terminal region of MTCH2 binds tBID as one domain, and dividing it into shorter peptides reduces its affinity. A shorter peptide from this region (residues 276–290) gave a tBID binding interaction that was too weak to quantify, indicating that the whole surface of the C-terminal region participates in the interaction.
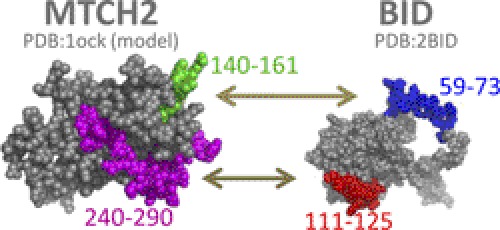
Fluorescence anisotropy binding studies of the tBID-derived peptides to the MTCH2 peptides (140–161 and 240–290) indicated a weak binding affinity at the hundred micromolar range. The affinity of MTCH2-derived peptides to the tBID protein was in the low micromolar range. This difference is probably because in the case of the tBID peptides peptide-peptide binding experiments were performed, compared with peptide-protein binding studies with the tBID protein and MTCH2 peptides. In addition, peptides are mainly unfolded in solution (supplemental Fig. 1E) and often undergo induced fit upon ligand binding and gain a folded structure. This folding process is accompanied by loss of entropy, and therefore the overall affinity of the binding is lower compared with binding a protein that is already folded before binding. An exception is the peptide MTCH2 240–290, which is relatively long and can be considered a protein domain. This peptide adopts a significant α-helical structure (supplemental Fig. 2E). This is also consistent with the results demonstrating that MTCH2 240–290 binds the tBID protein tighter than MTCH2 140–161 (Fig. 2, B and C). To improve the activity of the peptides, modified derivatives such as shortened and/or cyclic peptides should be designed. The experiments at the peptide level, described herein, provided the basis for designing such peptides by identifying the lead sequences derived from the binding interface between the two proteins. tBID residues 59–73 bound MTCH2 residues 140–161, and tBID residues 111–125 bound MTCH2 residues 240–290 (Fig. 5).
FIGURE 5.
Sites in MTCH2 and tBID that mediate their interaction. The main binding sites from both MTCH2 and tBID are highlighted on the three-dimensional structures. These binding sites were found based on peptide array screening results along with anisotropy binding studies.
Implications for MTCH2-tBID Mechanism of Action
The experiments in cancer cells (U2OS) revealed that the observed tBID-derived peptides induced substantial cancer cell death, whereas the observed MTCH2-derived peptides had a much smaller effect. It was previously found that approximately 45% of MTCH2 liver-deficient mice injected with anti-Fas antibody were resistant to the injection (12) and that approximately 80% of the BID-deficient mice were resistant (29). These results suggest that the absence of MTCH2 is less cytoprotective than the absence of BID. Results that demonstrate activity mainly for the tBID-derived peptides, and not MTCH2-derived peptides, also support this hypothesis.
The understanding of BID-MTCH2 mechanism of action is still at a very early stage. It is known that MTCH2 recruits tBID to the mitochondria, and consequently tBID activates the Bax protein to initiate apoptosis. Yet the mechanism by which tBID activates Bax is still not clear. Several models have been proposed (30, 31), and the most popular model suggests that tBID binds Bax via its BH3 domain (tBID residues 90–98) and activates Bax also at its BH3 domain. These results were surprising in the sense that the tBID peptides found were not part of the tBID-BH3 domain and were located before the BH3 domain (residues 59–73) or after it (residues 111–125). Nevertheless, these tBID-peptides demonstrated induction of cancer cell death. One possible explanation is that the mechanism of Bax activation by tBID is more complex than currently known. Perhaps the tBID-derived peptides (59–73, 111–125) are part of an extended site of tBID that activates Bax, and therefore these peptides themselves could activate Bax and consequently cause cancer cell death. This is in agreement with a recent study that presented a novel model for Bax activation by tBID (32). The model was predicted by simulations and docking of tBID with Bax. According to this model, tBID binds Bax via its N-terminal part (residues 61–78), which overlaps the region. This was identified as a binding site for MTCH2 (residues 59–73).
Another option is that the sites found in tBID also participate in mediating apoptosis via other, yet unknown, PPIs. Both of these sites are exposed in the proposed model for the interaction of tBID and Bax on the membrane (11). A third option is that tBID binds Bax via the sites mentioned above (BH3 domain) and MTCH2 via the two sites identified here. This means that tBID may bind Bax and MTCH2 via two close regions and may even create a trimeric complex (MTCH2, tBID, and Bax). A trimeric complex has already been suggested before by Grinberg et al. (20). The exact molecular details of such a trimeric complex remain to be revealed.
It is also possible that the observed death of cancer cells may be the result of inhibiting not only apoptosis, but also a different pathway in which MTCH2 participates. MTCH2 is emerging as a possible regulator of metabolism (22, 33–35). Perhaps the peptides derived from tBID that bind MTCH2 induce cancer cell death by interfering with the MTCH2 metabolic activity and inhibiting yet unknown protein interactions related to this pathway.
Our results set the interaction interfaces as targets for further development of inhibitory peptides and small molecules. tBID controls cell fate and hence is an important potential target for the development of anticancer drugs that will stimulate apoptosis by interfering with its regulation.
Supplementary Material
Acknowledgments
We thank Bioline for support and Dr. Deborah E. Shalev for critical reading of the manuscript.

This article contains supplemental Tables 1 and 2 and Figs. 1–3.
- PPI
- protein-protein interaction
- IMM
- inner mitochondrial membrane
- MCP
- mitochondrial carrier protein
- MTCH2
- mitochondrial carrier homologue 2
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- tBID
- truncated BID.
REFERENCES
- 1. Katz C., Levy-Beladev L., Rotem-Bamberger S., Rito T., Rüdiger S. G., Friedler A. (2011) Studying protein-protein interactions using peptide arrays. Chem. Soc. Rev. 40, 2131–2145 [DOI] [PubMed] [Google Scholar]
- 2. Adams J. M., Cory S. (2007) The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris M. H., Thompson C. B. (2000) The role of the Bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell Death Differ. 7, 1182–1191 [DOI] [PubMed] [Google Scholar]
- 4. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) The BCL-2 family reunion. Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang M. H., Reynolds C. P. (2009) Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin. Cancer Res. 15, 1126–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruncko M., Oost T. K., Belli B. A., Ding H., Joseph M. K., Kunzer A., Martineau D., McClellan W. J., Mitten M., Ng S. C., Nimmer P. M., Oltersdorf T., Park C. M., Petros A. M., Shoemaker A. R., Song X., Wang X., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H., Elmore S. W. (2007) Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J. Med. Chem. 50, 641–662 [DOI] [PubMed] [Google Scholar]
- 7. Yin X. M. (2006) Bid, a BH3-only multi-functional molecule, is at the cross-road of life and death. Gene 369, 7–19 [DOI] [PubMed] [Google Scholar]
- 8. Li H., Zhu H., Xu C. J., Yuan J. (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501 [DOI] [PubMed] [Google Scholar]
- 9. Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) BID, a Bcl2-interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 10. Gross A., Yin X. M., Wang K., Wei M. C., Jockel J., Milliman C., Erdjument-Bromage H., Tempst P., Korsmeyer S. J. (1999) Caspase-cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J. Biol. Chem. 274, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 11. Billen L. P., Shamas-Din A., Andrews D. W. (2008) BID: a Bax-like BH3 protein. Oncogene 27, S93–104 [DOI] [PubMed] [Google Scholar]
- 12. Zaltsman Y., Shachnai L., Yivgi-Ohana N., Schwarz M., Maryanovich M., Houtkooper R. H., Vaz F. M., De Leonardis F., Fiermonte G., Palmieri F., Gillissen B., Daniel P. T., Jimenez E., Walsh S., Koehler C. M., Roy S. S., Walter L., Hajnóczky G., Gross A. (2010) MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat. Cell Biol. 12, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Q. H., Ye M., Wu X. Y., Ren S. X., Zhao M., Zhao C. J., Fu G., Shen Y., Fan H. Y., Lu G., Zhong M., Xu X. R., Han Z. G., Zhang J. W., Tao J., Huang Q. H., Zhou J., Hu G. X., Gu J., Chen S. J., Chen Z. (2000) Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 10, 1546–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yerushalmi G. M., Leibowitz-Amit R., Shaharabany M., Tsarfaty I. (2002) Met-HGF/SF signal transduction induces mimp, a novel mitochondrial carrier homologue, which leads to mitochondrial depolarization. Neoplasia 4, 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Palmieri F. (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 16. Arco A. D., Satrústegui J. (2005) New mitochondrial carriers: an overview. Cell Mol. Life Sci. 62, 2204–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palmieri F. (1994) Mitochondrial carrier proteins. FEBS Lett. 346, 48–54 [DOI] [PubMed] [Google Scholar]
- 18. Aquila H., Misra D., Eulitz M., Klingenberg M. (1982) Complete amino acid sequence of the ADP/ATP carrier from beef heart mitochondria. Hoppe Seylers Z Physiol. Chem. 363, 345–349 [PubMed] [Google Scholar]
- 19. Schwarz M., Andrade-Navarro M. A., Gross A. (2007) Mitochondrial carriers and pores: key regulators of the mitochondrial apoptotic program? Apoptosis 12, 869–876 [DOI] [PubMed] [Google Scholar]
- 20. Grinberg M., Schwarz M., Zaltsman Y., Eini T., Niv H., Pietrokovski S., Gross A. (2005) Mitochondrial carrier homolog 2 is a target of tBID in cells signaled to die by tumor necrosis factor alpha. Mol. Cell. Biol. 25, 4579–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 22. Cogliati S., Scorrano L. (2010) A BID on mitochondria with MTCH2. Cell Res. 20, 863–865 [DOI] [PubMed] [Google Scholar]
- 23. Katz C., Benyamini H., Rotem S., Lebendiker M., Danieli T., Iosub A., Refaely H., Dines M., Bronner V., Bravman T., Shalev D. E., Rüdiger S., Friedler A. (2008) Molecular basis of the interaction between the antiapoptotic Bcl-2 family proteins and the proapoptotic protein ASPP2. Proc. Natl. Acad. Sci. U.S.A. 105, 12277–12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayouka Z., Rosenbluh J., Levin A., Loya S., Lebendiker M., Veprintsev D., Kotler M., Hizi A., Loyter A., Friedler A. (2007) Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. U.S.A. 104, 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coster G., Hayouka Z., Argaman L., Strauss C., Friedler A., Brandeis M., Goldberg M. (2007) The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J. Biol. Chem. 282, 32053–32064 [DOI] [PubMed] [Google Scholar]
- 26. Friedler A., Hansson L. O., Veprintsev D. B., Freund S. M., Rippin T. M., Nikolova P. V., Proctor M. R., Rüdiger S., Fersht A. R. (2002) A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proc. Natl. Acad. Sci. U.S.A. 99, 937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chou J. J., Li H., Salvesen G. S., Yuan J., Wagner G. (1999) Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell 96, 615–624 [DOI] [PubMed] [Google Scholar]
- 28. McDonnell J. M., Fushman D., Milliman C. L., Korsmeyer S. J., Cowburn D. (1999) Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell 96, 625–634 [DOI] [PubMed] [Google Scholar]
- 29. Yin X. M., Wang K., Gross A., Zhao Y., Zinkel S., Klocke B., Roth K. A., Korsmeyer S. J. (1999) BID-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400, 886–891 [DOI] [PubMed] [Google Scholar]
- 30. Leber B., Lin J., Andrews D. W. (2007) Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 12, 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Membrane binding by tBID initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 32. Veresov V. G., Davidovskii A. I. (2009) Activation of Bax by joint action of tBID and mitochondrial outer membrane: Monte Carlo simulations. Eur. Biophys. J. 38, 941–960 [DOI] [PubMed] [Google Scholar]
- 33. Ng M. C., Tam C. H., So W. Y., Ho J. S., Chan A. W., Lee H. M., Wang Y., Lam V. K., Chan J. C., Ma R. C. (2010) Implication of genetic variants near NEGR1, SEC16B, TMEM18, ETV5/DGKG, GNPDA2, LIN7C/BDNF, MTCH2, BCDIN3D/FAIM2, SH2B1, FTO, MC4R, and KCTD15 with obesity and type 2 diabetes in 7705 Chinese. J. Clin. Endocrinol. Metab. 95, 2418–2425 [DOI] [PubMed] [Google Scholar]
- 34. Zhao J., Bradfield J. P., Li M., Wang K., Zhang H., Kim C. E., Annaiah K., Glessner J. T., Thomas K., Garris M., Frackelton E. C., Otieno F. G., Shaner J. L., Smith R. M., Chiavacci R. M., Berkowitz R. I., Hakonarson H., Grant S. F. (2009) The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity 17, 2254–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willer C. J., Speliotes E. K., Loos R. J., Li S., Lindgren C. M., Heid I. M., Berndt S. I., Elliott A. L., Jackson A. U., Lamina C., Lettre G., Lim N., Lyon H. N., McCarroll S. A., Papadakis K., Qi L., Randall J. C., Roccasecca R. M., Sanna S., Scheet P., Weedon M. N., Wheeler E., Zhao J. H., Jacobs L. C., Prokopenko I., Soranzo N., Tanaka T., Timpson N. J., Almgren P., Bennett A., Bergman R. N., Bingham S. A., Bonnycastle L. L., Brown M., Burtt N. P., Chines P., Coin L., Collins F. S., Connell J. M., Cooper C., Smith G. D., Dennison E. M., Deodhar P., Elliott P., Erdos M. R., Estrada K., Evans D. M., Gianniny L., Gieger C., Gillson C. J., Guiducci C. (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.