Background: Leucine-rich repeat and WD repeat-containing protein 1 (Lrwd1) binds histone marks (H3K9me3 and H4K20me3).
Results: The recruitment of Lrwd1 to heterochromatin requires H3K9me3. Lrwd1 is required for silencing of major satellite repeats.
Conclusion: Lrwd1 binds to heterochromatin via interactions with H3K9me3 and maintains heterochromatin silencing.
Significance: Determining how Lrwd1 functions in heterochromatin silencing is important in understanding how heterochromatin is inherited during S phase of the cell cycle.
Keywords: Cell Cycle, Chromatin, Chromatin Structure, Gene Silencing, Heterochromatin, DNA Replication, Lrwd1, ORC, Epigenetic Inheritance, Heterochromatin Silencing
Abstract
Lrwd1, a protein containing a leucine-rich repeat and a WD40 repeat domain, interacts with the origin replication complex (ORC), a protein complex involved in both initiation of DNA replication and heterochromatin silencing. Lrwd1 and ORC are known to co-purify with repressive histone marks (trimethylated lysine 9 of histone H3 (H3K9me3) and trimethylated lysine 20 of histone H4 (H4K20me3)) and localize to pericentric heterochromatin. However, how the Lrwd1 is recruited to heterochromatin and the functional significance of the localization of Lrwd1 to the heterochromatin are not known. Here, we show that Lrwd1 preferentially binds to trimethylated repressive histone marks in vitro, which is dependent on an intact WD40 domain but independent of ORC proteins. The localization of Lrwd1 and Orc2 at pericentric heterochromatin in mouse cells is lost in cells lacking H3K9me3 but not in cells lacking H4K20me3. In addition, depletion of HP1α has little impact on the localization of Lrwd1 on pericentric heterochromatin. Finally, depletion of Lrwd1 and Orc2 in mouse cells leads to increased transcription of major satellite repeats. These results indicate that the Lrwd1 is recruited to pericentric heterochromatin through binding to H3K9me3 and that the association of Lrwd1 with pericentric heterochromatin is required for heterochromatin silencing and maintenance.
Introduction
Genetic information is encoded by the DNA sequence; yet, DNA alone does not account for the complexity of the mammalian genome. Instead, chromatin, comprising DNA, core histone proteins, and other regulatory proteins, regulates gene expression and maintains genome stability. The basic repeating unit of chromatin is the nucleosome, consisting of 147 bp of DNA wrapped around an octamer of four highly conserved core histone proteins (H3, H4, H2A, and H2B). Chromatin can be classified into two main types based on cytological staining: euchromatin, characterized by a loosely packed chromatin structure and enriched with genes, and heterochromatin, which is densely packed and silences gene expression via epigenetic mechanisms (1–3).
Heterochromatin can be further classified into two subtypes: facultative and constitutive heterochromatin. The formation of facultative heterochromatin is developmentally regulated. One of the classic examples of facultative heterochromatin is inactivation of one of two X chromosomes in female mammals (4–6). In contrast, constitutive heterochromatin is found at regions rich in repetitive sequences and often at the centric and pericentric regions of mammalian chromosomes, resulting in transcriptional silencing of both homologous chromosomes (7). Constitutive heterochromatin plays an important role in repressing transposable element activity and maintaining genome integrity (8).
Constitutive heterochromatin is marked with unique histone modifications and proteins (9, 10). For instance, histone H3 lysine 9 at pericentric heterochromatin is trimethylated by the lysine methyltransferases Suv39h1/h2, and histone H4 lysine 20 is trimethylated by Suv420h1/h2 (11, 12). H3K9me32 is recognized by the chromodomain of HP1α, an adapter protein for the recruitment of other heterochromatic effector proteins to pericentric heterochromatin (1, 13, 14). In addition, mouse embryonic fibroblast cells lacking both Suv39h1/h2 have a loss of both H3K9me3 and H4K20me3, whereas the loss of Suv420h1/h2 has no apparent effect on H3K9me3 (12).
The origin recognition complex (ORC) consists of six proteins essential for the initiation of DNA replication (15). In addition to its role in initiating DNA replication, ORC proteins are known to be involved in transcriptional gene silencing in yeast, Drosophila and mammalian cells (16–18). Indeed, human Orc2 is associated with HP1 and is required for the stable association of HP1 at heterochromatin (19). Thus, both H3K9me3 and ORC are required for the stable association of HP1 at pericentric heterochromatin.
Recently, Lrwd1, an ORC-interacting protein also called ORCA (20), was found co-localized with Orc2 at pericentric heterochromatin (21, 22). Lrwd1 contains a leucine-rich repeat domain and a WD40 repeat domain. In addition, both ORC and Lrwd1 co-purify with histone peptides or mononucleosomes containing either of three transcriptional repressive lysine marks (H3K9me3, H3K27me3, and H4K20me3) (21, 22). Because ORC and Lrwd1 form a complex, it is not known whether ORC, Lrwd1, or both are involved in recognition of these repressive marks. Furthermore, the functional significance of the association of ORC or Lrwd1 with pericentric heterochromatin is not known.
Through a genome-wide screen, we found that Lrwd1 and Orc2 are among the 32 genes involved in X-chromosome inactivation (23). In the present study, we focus on characterizing the role of Lrwd1 in pericentric heterochromatin silencing and maintenance. Using in vitro peptide pulldown assays, we show that Lrwd1 binds to repressive histone marks (H3K9me3, H3K27me3, and H4K20me3) independent of ORC but dependent on an intact WD40 domain of Lrwd1. Furthermore, we show that the pericentric heterochromatin localization of Lrwd1 and Orc2 is dependent on H3K9me3, but not H4K20me3. Finally, depletion of Lrwd1 or Orc2 in mouse cells results in increased transcription of major satellite repeats. Based on these results, we propose that Lrwd1 is recruited to heterochromatin through its interaction with H3K9me3 and that Lrwd1 is required for efficient heterochromatin silencing and maintenance.
EXPERIMENTAL PROCEDURES
Cell Culture
Suv39h1h2−/− and Suv420h1h2−/− MEF cells were a kind gift from Dr. Thomas Jenuwein. Female immortalized MEF cells used to knockdown Lrwd1, Orc2 and HP1α have been described (23). HeLa and 293T cells were cultured and maintained under standard conditions.
Plasmids and shRNAs
The shRNAs used to knockdown genes in human and mouse cells were purchased from Sigma: human Lrwd1, NM_152892.1–510s1c1 and NM_152892.1–465s1c1 and Orc2, NM_006190.3–1110s1c1 and NM_006190.3–1684s1c1; mouse Lrwd1, NM_027891.1–2820s1c1 and NM_027891.1–2817s1c1; Orc2, NM_008765.2–649s1c1 and NM_008765.2–510s1c1; and HP1α, NM_007626.2–547s1c1 and NM_007626.2–356s1c1. Full-length and truncated forms of Lrwd1 cDNA were cloned into pBabe vector for expression in 293T cells.
Antibodies
Peptides with the sequence “dkvradfmr” corresponding to amino acid residues 162–170 of mouse Lrwd1 were used for rabbit immunization. Polyclonal Lrwd1 antibodies were purified using peptide-conjugated beads. The antibodies used for Orc2 and HP1α immunofluorescence were described before (23). Antibody dilutions used for Western blot are as follows: Orc1 (ab85830 Abcam), 1:1000; Orc4 (ab9641 Abcam), 1:1000; Orc5 (H-300 Santa Cruz Biotechnology) sc-20635, 1:500; HP1α (ab77256 Abcam), 1:1000; Orc2, Orc3, and Orc6 antibodies (gifts from Dr. Bruce Stillman) (24), 1:1000; and GFP (ab6556), 1:500.
In Vitro Peptide Pulldown Assay
In vitro peptide pulldown assays were performed as described (25) with some modifications. Various peptides were conjugated to SulfoLink beads from Pierce. Beads were washed three times in buffer D (20 mm HEPES, pH 7.9, 150 mm KCl, 20% v/v glycerol, 0.2 mm EDTA, 0.2% Triton X-100, and protease inhibitors) before adding to S100 extracts, nuclear extracts, or purified Lrwd1 proteins. Mixtures were incubated at 4 °C for 3 h. After extensive washing with buffer D, bound proteins were eluted by SDS sample buffer, resolved by SDS-PAGE, detected by Ponceau S staining or Western blotting. Immunoblot and RT-PCR images were quantified using ImageJ.
Co-immunoprecipitation
FLAG-tagged full-length and truncated versions of Lrwd1 were expressed in 293T cells. Cells were treated in lysis buffer (50 mm HEPES-KOH, pH,7.4, 100 mm NaCl, 1% Nonidet P-40, 10% glycerol, 1 mm EDTA, 1 mm PMSF, 1 mm DTT, and 1 mm benzamidine) and incubated at 4 °C for 1 h. Lysates were cleared by spinning at 13,000 rpm at 4 °C for 10 min. M2 beads (Sigma) were added to the lysates and incubated at 4 °C for 3 h for immunoprecipitation. Beads were washed three times with washing buffer (lysis buffer with 0.01% Nonidet P-40). Proteins that bound were boiled in SDS sample buffer and separated by SDS-PAGE for subsequent Western blotting using the indicated antibodies.
Purification of Recombinant Lrwd1 Proteins from Sf9 Cells
Full-length or truncated forms of Lrwd1 were cloned into pFastBac1HTa (Invitrogen) to generate baculovirus carrying the His6-tagged Lrwd1 transgene. Following Sf9 cell infection, cells were collected and washed with cold PBS followed by addition of lysis buffer (50 mm HEPES-KOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 5% glycerol, 0.1% Nonidet P-40, 0.1% Tween 20, 10 mm imidazole, 0.5 mm PMSF) at 4 °C for 15 min. Lysates were cleared by spinning two times at 13,000 rpm, 4 °C for 10 min. Nickel-nitrilotriacetic acid beads (Sigma) were added to the cleared lysates and incubated at 4 °C for 1 h. After washing, His-tagged Lrwd1 proteins were eluted four times with elution buffer at 16 °C for 5min. Eluted His-tagged Lrwd1 proteins were dialyzed twice with dialysis buffer (150 mm NaCl, 20 mm Tris, pH 8.0, 10% glycerol) before used for in vitro peptide pulldown assays.
Immunofluorescence
Cells grown on chamber slides (Nunc) were washed briefly with PBS followed by fixation with 3% paraformaldehyde at room temperature for 12 min. Cells were permeabilized with 0.5% Triton X-100 solution for 5 min at room temperature and then blocked in 5% normal goat serum for 1 h. Primary antibody with appropriate dilution was added to cells and incubated at 37 °C for 20 min or 4 °C overnight. Cells were then washed with PBS twice and probed with FITC/rhodamine-conjugated secondary at 37 °C for 20 min. DNA was stained with DAPI, and samples were mounted with Gold anti-fade reagents (Invitrogen) and examined using a Zeiss Axioplan fluorescence microscope. Dilution of antibodies used in immunofluorescence were as follows: anti-Orc2 (mAb920), 1:200; anti-HP1α (1:400 Millipore mAb3584, 1:100 cell signaling 2616), anti-H3K27me3 (1:80), H3K9me3, H4K20me3 (1:80). For the detection of GFP-Lrwd1 fusion protein, full-length Lrwd1 was cloned into pEGFP C1 vector to generate the GFP-Lrwd1 transgene. The transgene was subsequently cloned into pTSiN lentiviral based vector to infect WT, Suv39h1h2−/−, and Suv420h1h2−/− cells. Cells were washed with PBS and preextracted with 0.5% Triton X-100 to remove soluble proteins in the nucleus before paraformaldehyde fixation and subsequent DAPI staining and immunofluorescence detection.
Analysis of Transcription at Major or Minor Satellites Using RT-PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen) from WT, Suv39h1h2−/−, Suv420h1h2−/− cells or MEF cells infected with viruses for knockdown of Orc2 or Lrwd1 by shRNA and the nontarget control. cDNA was synthesized using 1 μg of total RNA with random hexamer and SuperScriptIII reverse transcriptase (Invitrogen). PCR was performed in a 25-μl volume with 0.1 μm primers with cycle numbers as indicated. Primer sequences are as follows: major satellites (MajF1), 5′-GACGACTTGAAAAATGACGAAATC-3′ and (MajR1) 5′-CATATTCCAGGTCCTTCAGTGTGC-3′; minor satellites (MinF1), 5′-CATGGAAAATGATAAAAACC-3′ and (MinR1), 5′-CATCTAATATGTTCTACAGTGTGG-3′; β-actin (actin F), 5′-CCCAGCCATGTACGTAGCCATCCA-3′ and (actin R), 5′-CATGAGGTAGTCCGTCAGGTC-3′; Lrwd1 (Lrwd1 Ex3F), 5′-GTCAGTGACAACCTCAAAGTC-3′ and (Lrwd1 Ex4R), 5′-CCGCCACTGCGTGAACTCGATG-3′; Orc2 (Orc2 F3), 5′-CTGATCGSSCGCTGCAG AGG-3′ and (Orc2 R2), 5′-CTCTCTTGGACCCCAAGCC-3′. To analyze transcripts at major or minor satellites using real-time PCR, PCR primers described by Zhu et al. (26) were used.
RESULTS
Lrwd1 Binds Repressive Histone Marks Independent of ORC
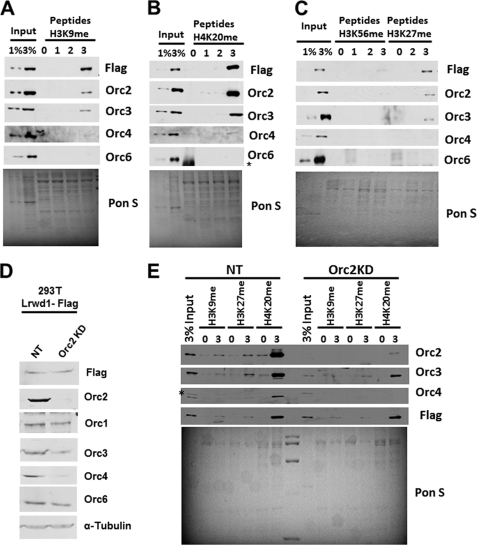
Both ORC and Lrwd1 co-purify with three repressive histone marks (H3K9me3, H3K27me3, and H4K20me3) (22). Because Lrwd1 associates with ORC (20), it is not known whether Lrwd1 or ORC is involved in direct binding to these repressive marks. To address this issue, we employed an in vitro peptide pulldown assay (Fig. 1, A–C). Briefly, unmodified peptides or peptides with varying degrees of methylation (0, 1, 2, and 3) at H3K9, H3K27, and H4K20 lysine residues were coupled to beads and used to pull down Lrwd1 from nuclear extracts of 293T cells ectopically expressing FLAG-Lrwd1. Lrwd1, Orc2, Orc3, and Orc4, but not Orc6, were preferentially pulled down with H3K9me3 (Fig. 1A), H4K20me3 (Fig. 1B), and H3K27me3 (Fig. 1C) compared with the corresponding unmodified, mono-, or dimethylated peptides. In contrast, Lrwd1, Orc2, Orc3, and Orc4 were not pulled down by unmodified, mono/di/trimethylated peptide forms of H3K56 (Fig. 1C). These results recapitulate published results showing that Lrwd1 and a subset of ORC subunits bind repressive histone marks in vitro (21, 22).
FIGURE 1.
Orc2 is not required for Lrwd1 to bind histone peptides with repressive histone marks. A–C, in vitro peptide pulldown assay was performed to assess binding of Lrwd1 and ORC proteins to peptides containing methylated histone marks. Synthetic peptides of H3K9 (A), H4K20 (B), H3K56 and H3K27 (C) with different degrees of methylation (un- (0), mono- (1), di- (2), and trimethyl (3)) were coupled to beads and used to pull down proteins in nuclear extracts prepared from 293T cells stably expressing FLAG-Lrwd1. Eluted proteins were detected by Western blotting using the indicated antibodies. Ponceau S (Pon S) staining was used to detect total proteins, which serves as loading controls. D and E, Orc2 is not required for Lrwd1 to bind H3K9me3, H3K27me3, and H4K20me3. Orc2 was depleted in 293T cells expressing FLAG-tagged Lrwd1 using shRNA, and nuclear extracts were prepared to perform the peptide pulldown assay described above using unmodified (0) and trimethylated (3) H3K9, H4K20, and H3K27 peptides. Proteins in whole cell extracts (D) as well as in pulldown (E) were detected by Western blotting using indicated antibodies. As controls, nuclear extracts were prepared from cells infected with virus for nontargeting control (NT). Each result shown is representative of three independent experiments. * indicates nonspecific band.
Using the peptide pulldown assay, we asked how depletion of Orc2, and thereby loss of an intact ORC complex, affected the ability of Lrwd1 to bind repressive histone marks. Depletion of Orc2 had no apparent effect on the ability of Lrwd1 to bind H3K9me3, H3K27me3, and H4K20me3 (Fig. 1, D and E). The association of Orc3 and Orc4 with either of these peptides was reduced, possibly due to reduced levels of Orc3 and Orc4 in Orc2-depleted cell extracts (Fig. 1D, compare the input lanes between NT and Orc2KD) (17). These results indicate that Orc2 or an intact ORC complex is not required for Lrwd1 to bind these repressive histone marks.
Orc6 was not detected in pulldown assays using H3K9me3, H3K27me3, or H4K20me3 (Fig. 1, A–C) (21, 22), suggesting that Orc6 is not required for stable association of Lrwd1 and Orc2 to the repressive histone marks. To test this idea, we determined how depletion of Orc6 by shRNA affects the association of Lrwd1 and Orc2 with repressive marks. Depletion of Orc6 did not affect the binding of Lrwd1 or Orc2 to each of the three trimethylated peptides (supplemental Fig. S1). These results suggest that Lrwd1 may form with a subset of ORC subunits a complex that binds to repressive histone marks.
WD40 Repeat Domain Is Important for Lrwd1 Binding to Repressive Histone Marks
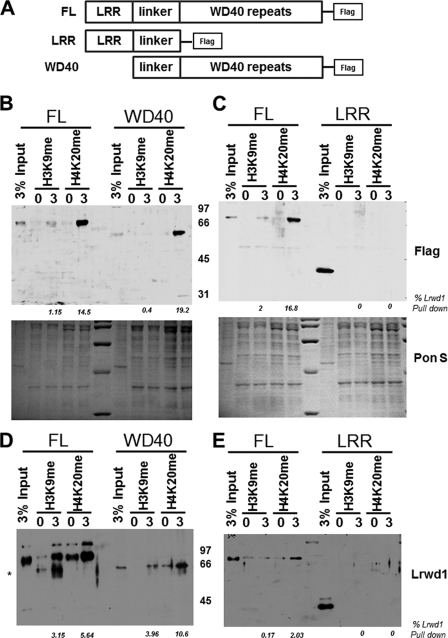
Lrwd1 contains both a leucine-rich repeat (LRR) domain and a WD40 repeat domain (Fig. 2A). Whereas the WD40 repeat domain is known to bind ORC (20), the Lrwd1 domain involved in association with methylated histones is not known. We, therefore, set out to determine which domain(s) of Lrwd1 was required for Lrwd1 association with the three repressive histone marks. Nuclear extracts were prepared from 293T cells expressing FLAG-tagged full-length (FL) Lrwd1 or Lrwd1 mutant proteins containing WD40 or LRR domains and used for peptide pulldown assays (Fig. 2A). The WD40 domain bound to these methylated peptides as efficiently as FL Lrwd1 (Fig. 2B), whereas the LRR domain failed to bind H3K9me3 and H4K20me3 (Fig. 2C). These results suggest that the WD40 repeat domain of Lrwd1 is directly involved in binding to these repressive histone marks. To test this idea further, we purified recombinant Lrwd1, the Lwrd1 WD40 domain, and LRR domain from Sf9 cells and determined how each binds to H3K9me3 and H4K20me3. Recombinant WD40 and FL Lrwd1 bound equally well to H3K9me3 and H4K20me3 (Fig. 2D), whereas recombinant LRR was not able to bind to any of these peptides (Fig. 2E). These results show that the WD40 domain of Lrwd1 is necessary and sufficient for binding to repressive histone marks in vitro.
FIGURE 2.
Lrwd1 WD40 domain is required and sufficient for Lrwd1 to bind repressive histone marks. A, schematic diagram shows FL Lrwd1 and two Lrwd1 mutants with either the LRR domain or WD40 repeat domain used for in vitro peptide pulldown assay. B and C, WD40 repeat domain, but not LRR domain of Lrwd1, binds H3K9me3 and H4K20me3. Nuclear extracts were prepared from 293T cells expressing FLAG-tagged full-length and truncated forms of Lrwd1, and peptide pulldown assays were performed as described in Fig. 1. D and E, recombinant full-length Lrwd1 and WD40 domain bind to methylated peptides. Recombinant full-length and truncated forms of Lrwd1 proteins were expressed and purified from Sf9 cells and used to incubate beads with unmodified or trimethylated form of H3K9 and H4K20 peptides for the peptide pulldown assay described in Fig. 1. Full-length and truncated forms of Lrwd1 recombinant proteins were detected by immunoblotting using antibodies prepared against the linker region of Lrwd1. Percentages of Lrwd1 pulldown in B and E were determined by normalization to input. Each result shown is a representative of three independent experiments. * indicates nonspecific band.
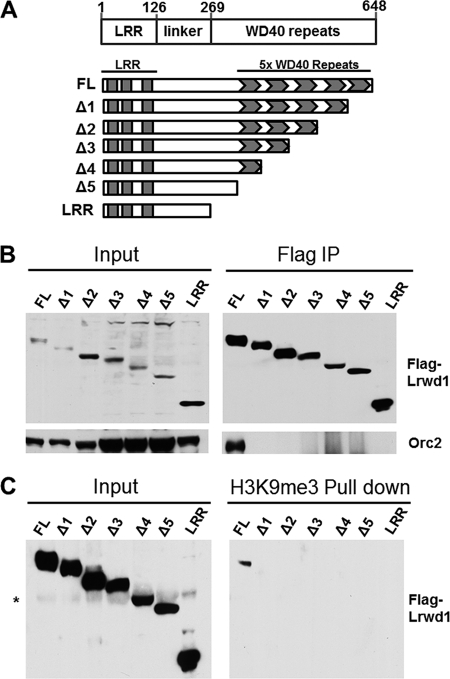
Intact WD40 Domain Is Required for Lrwd1 to Bind to Orc2 and H3K9me3
The WD40 domain of Lrwd1 comprises five blade-like small WD40 repeats (20) (Fig. 3A). To gain insight into how the WD40 domain of Lrwd1 interacts with both ORC and repressive histone marks, we systematically deleted each of the WD40 repeats, expressed the FLAG-tagged mutant proteins in 293T cells (Fig. 3A), and determined how each interacts with ORC and methylated histone peptides. FL Lrwd1 co-immunoprecipitated Orc2, whereas none of the Lrwd1 mutants lacking a WD40 repeat interacted with Orc2 (Fig. 3B). In addition, no significant association between the WD40 repeat deletion mutants with H3K9me3 peptides was detected (Fig. 3C). These results suggest that an intact Lrwd1 WD40 repeat domain is required for proper interaction with both ORC and repressive histone marks.
FIGURE 3.
Intact WD40 domain of Lrwd1 is required for Lrwd1 association with Orc2 and trimethylated peptides. A, schematic diagram shows FLAG-tagged Lrwd1 and deletion mutants used for immunoprecipitation in B and in vitro peptide pulldown assays in C. B, FLAG-tagged full-length and truncated versions of Lrwd1 were expressed in 293T cells and immunoprecipitated (IP) using FLAG-M2 beads. Proteins in whole cell extracts (Input, left) and IP were analyzed by Western blotting using antibodies against Orc2 and Lrwd1. C, full-length but not truncated forms of Lrwd1 bind to H3K9me3. Nuclear extracts were prepared from cells expressing FLAG-tagged full-length or truncated versions of Lrwd1 and used for in vitro peptide pulldown assays using H3K9 and H3K9me3 peptides. Proteins in whole cell extracts (Input, left) and H3K9me3 pulldown were analyzed by Western blotting. * indicates nonspecific band.
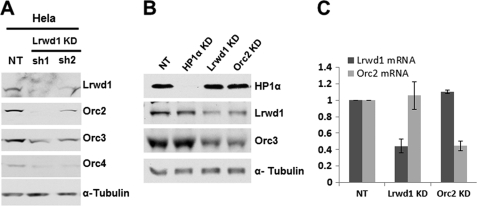
Depletion of Lrwd1 Affects Protein Levels of Subset of ORC Subunits
In an attempt to determine whether Lrwd1 is required for ORC to bind methylated histone peptides, we observed that the levels of four ORC subunits (Orc1, Orc2, Orc3, and Orc4) were reduced dramatically in Lrwd1-depleted 293T cells (data not shown). This observation seems to contradict the results of others showing that depletion of Lrwd1 had no apparent effect on the ORC protein levels (20). To confirm our observation, Lrwd1 was depleted using two independent shRNA(s) in HeLa cells (Fig. 4A). The protein levels of Orc2, Orc3, and Orc4 were reduced concomitant with the reduction in Lrwd1 level. Similar results were obtained when Lrwd1 was depleted from MEF cells (Fig. 4B). Depletion of Lrwd1 did not affect transcription of Orc2 (Fig. 4C). In addition, cells treated with proteasome inhibitor MG132 after Lrwd1 depletion did not alter the protein levels of Orc2, Orc3, and Orc4 to a detectable degree (supplemental Fig. S2). These results suggest that Lrwd1 is required for the protein stability of Orc2, Orc3, and Orc4 at least in some human cell lines and MEF in a manner that may be independent of proteasome-mediated degradation.
FIGURE 4.
Lrwd1 is required for the stability of a subset of ORC proteins in vivo. A, depletion of Lrwd1 in HeLa cells using two independent shRNA(s) results in reduced protein levels of Orc2, Orc3, and Orc4. HeLa cells were infected with two different shRNA viruses for Lrwd1 and nontargeting (NT) control. Cell lysates were prepared 72 h after infection, and ORC proteins were detected by Western blotting. B, Orc3 protein level is reduced in Lrwd1-depleted MEF cells. HP1α, Lrwd1, and Orc2 were depleted by shRNAs in MEF cells. Orc3, HP1, and Lrwd1 in whole cell extracts were analyzed by Western blotting. Note: no antibody is available to detect mouse Orc2, so Orc3 was used to monitor Orc2 protein level. C, depletion of Lrwd1 does not affect the mRNA level of Orc2. Lrwd1 or Orc2 was depleted in MEF cells as described in B. Total RNA was isolated, and the expression of Orc2 or Lrwd1 in nontargeting (NT), Lrwd1 knockdown (KD), or Orc2 knockdown (KD) cells was analyzed by real-time RT-PCR. Data and error bars indicate the mean values ± S.D. of two independent experiments.
Localization of Lrwd1 and Orc2 to Pericentric Heterochromatin Depends on H3K9me3
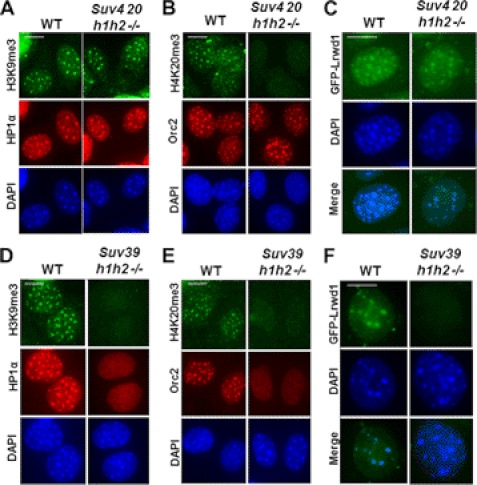
To determine the functional significance of the interactions between Lrwd1 and H3K9me3 or H4K20me3, we asked whether H3K9me3 or H4K20me3 is required for the localization of Lrwd1 and Orc2 to pericentric heterochromatin in vivo (Fig. 5). We examined the localization of Lrwd1 and Orc2 in MEF cells lacking both Suv39h1 and Suv39h2 (Suv39h1h2−/−) or MEF cells lacking both Suv420h1 and Suv420h2 (Suv420h1h2−/−) (11, 27). Suv39h1 and Suv39h2 are two lysine methyltransferases for the methylation of H3K9 at pericentric heterochromatin, whereas Suv420h1 and Suv420h2 are lysine methyltransferases for H4K20. Consistent with published reports (12), H3K9me3 and HP1α localization at pericentric heterochromatin was unchanged in Suv420h1h2−/− cells (Fig. 5A). Interestingly, the localization of Orc2 and Lrwd1 did not change to a significant degree in these mutant cells (Fig. 5, B and C). In agreement with published results (28), HP1α localization was lost, and H4K20me3 was not detectable in Suv39h1h2−/− cells (Fig. 5, D and E). Importantly, the localization of Lrwd1 and Orc2 to pericentric heterochromatin was reduced significantly in Suv39h1h2−/− cells compared with wild-type cells (Fig. 5, D–F). The expression of Lrwd1 in Suv39h1h2−/− cells was similar to that of wild-type cells (supplemental Fig. S3). Thus, the loss of Lrwd1 localization at pericentric heterochromatin in Suv39h1h2−/− cells is not due to impaired protein expression. These results suggest that recruitment of Lrwd1 to pericentric heterochromatin in vivo depends largely on the presence of H3K9me3, despite the fact that Lrwd1 binds both H3K9me3 and H4K20me3 in vitro.
FIGURE 5.
H3K9me3 is required for stable association of Lrwd1 and Orc2 with pericentric heterochromatin in vivo. A–C, localization of HP1α (A), Orc2 (B), and Lrwd1 (C) at pericentric heterochromatin in wild-type and Suv420 h1/h2−/− MEF cells. D–F, localization of HP1α (D), Orc2 (E), and Lrwd1 (F) at pericentric heterochromatin compromised in Suv39 h1/h2−/− MEF cells. DAPI stains pericentric heterochromatin. Immunofluorescence using antibodies against HP1α, GFP-Lrwd1, or H3K9me3 of wild-type, Suv39 h1/h2−/−, and Suv420 h1/h2−/− MEF cells was performed as described under “Experimental Procedures.” Scale bar, 5 μm.
Pericentric Heterochromatin Localization of Lrwd1 Does Not Depend on HP1α
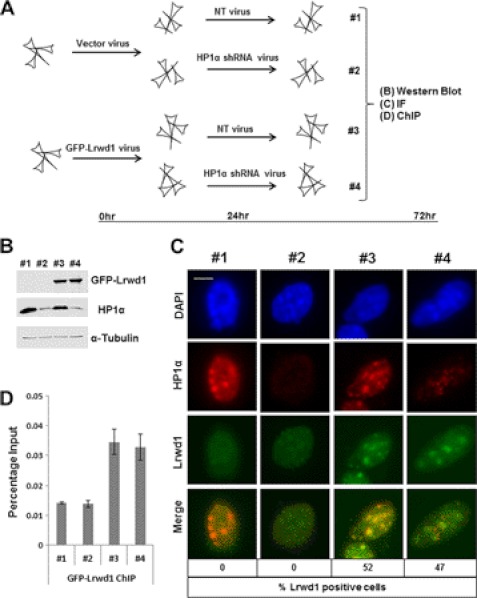
In Suv39h1h2−/− cells, the pericentric heterochromatin localization of effector proteins (e.g. HP1α) is lost. In addition, the heterochromatin localization of Orc2 is also affected in HP1α-depleted cells (19). To determine whether Lrwd1 localization is also affected by HP1α depletion, we analyzed the Lrwd1 localization in HP1α-depleted MEF cells. MEF cells were infected with virus for vector control or for expressing GFP-Lrwd1 (Fig. 6A). Twenty-four hours after infection, cells were split for infection with virus for nontargeting or shRNA targeting HP1α. Cells were collected 72 h after infection for Western blot analysis, immunofluorescence, and chromatin immunoprecipitation (ChIP). Western blot analysis revealed that GFP-Lrwd1 was expressed equally well in cells with/without HP1α depletion, and HP1α knockdown was efficient (Fig. 6B). Immunofluorescence studies revealed that the pericentric heterochromatin localization of Lrwd1 was not affected to a detectable degree in HP1α-depleted cells (Fig. 6C). In agreement with this interpretation, the binding of Lrwd1 at pericentric heterochromatin was not affected in HP1α-depleted cells as detected by ChIP assay using antibodies against GFP-Lrwd1 (Fig. 6D). Thus, the loss of Lrwd1 localization at heterochromatin in Suv39h1h2−/− cells is likely due to the loss of H3K9me3, but not the loss of HP1α at heterochromatin.
FIGURE 6.
Pericentric heterochromatin localization of Lrwd1 is independent of HP1α. A, diagram shows the experimental procedures. B, Western blotting analysis of expression of GFP-Lrwd1 and HP1α in four samples from A, C, and D is shown. C, localization of Lrwd1 is unaffected in HP1α-depleted cells compared with nontargeting cells. Cells with Lrwd1 localization at pericentric heterochromatin were counted (n = 100) in three independent experiments. Scale bar, 5 μm. D, depletion of HP1α does not affect Lrwd1 binding to pericentric heterochromatin. ChIP assays using antibodies against GFP-Lrwd1 were performed, and ChIP DNA was analyzed by real-time PCR using PCR primers amplifying major satellite repeat.
Lrwd1 Is Required for Silencing of Major Satellite Repeats
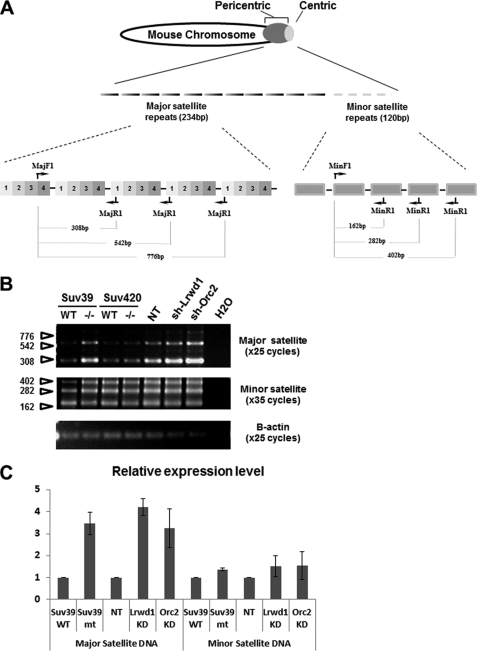
H3K9me3 is required for the silencing of the major satellite repeats in mouse ES cells (29) To determine the functional significance of the association of Lrwd1 and Orc2 with pericentric heterochromatin, we tested whether depletion of Orc2 and Lrwd1 affected silencing of major and minor satellite repeats. Total RNA was collected from MEF cells (23) infected with virus for nontargeting shRNA control, Orc2 shRNA, or Lrwd1 shRNA, and transcripts from major and minor satellites were detected by RT-PCR using primers shown in Fig. 7A. As a control, we also analyzed major and minor satellite transcripts in Suv39h1h2−/− MEF cells. Consistent with a previous report (29), major satellite transcripts, but not minor satellite transcripts, were up-regulated in Suv39h1h2−/− MEF cells (Fig. 7B). Depletion of either Lrwd1 or Orc2 by shRNA (Fig. 7B) also resulted in significant up-regulation of major satellite repeat transcripts but not the minor satellite repeats. Similar results were also observed by quantitative real-time PCR using another independent set of PCR primers (Fig. 7C), which have been recently used to detect transcripts at major and minor satellite repeats (26). Interestingly, depletion of both Orc2 and Lrwd1, while having no apparent effect on transcription of minor satellite repeats, resulted in a significant increase in transcription of major satellite repeats than either Orc2 or Lrwd1 depletion alone (supplemental Fig. S4). These results indicate that Lrwd1 and Orc2, like H3K9me3, are required for heterochromatin silencing in vivo and that Lrwd1 and Orc2 may not function in the same pathway to maintain transcriptional silencing at major satellite repeats.
FIGURE 7.
Lrwd1 is required for silencing of major satellite repeats. A, schematic diagram showing location of major and minor satellite repeats at pericentric heterochromatin and centric heterochromatin on a mouse chromosome (upper) and organization of mouse major and minor satellite repeats and primers used to detect transcripts by RT-PCR (lower). B, RT-PCR analysis of cDNA derived from wild-type, Suv39h1h2−/−, Suv420h1h2−/− MEF cells or wild-type cells infected with virus for nontargeting control (NT), shRNA for Lrwd1 or shRNA for Orc2. One representative result of three independent experiments is shown. C, experiment performed as described in B except that transcripts of major and minor satellites were detected by real-time PCR using PCR primers described under “Experimental Procedures.” β-Actin serves as loading control. The average ± S.D. (error bars) of three independent experiments are shown. KD, knockdown.
DISCUSSION
The Lrwd1 associates with ORC and both Lrwd1 and ORC co-purified with three trimethylated repressive histone marks in vitro (H3K9me3, H3K27me3, and H4K20me3) (21, 22). However, it was not known how the ORC-Lrwd1 complex recognizes these repressive histone markers and what is the functional significance of the association of Lrwd1-ORC with these repressive histone marks. We have presented the following lines of evidence to support a model that Lrwd1 is recruited to pericentric heterochromatin through its binding to H3K9me3, which in turn maintains silencing at major satellite repeats of pericentric heterochromatin. First, we show that while both Orc2 and Lrwd1 are copurified with repressive histone marks, Orc2 and Orc6 are not required for the association of Lrwd1 with these repressive marks, suggesting that the recruitment of Lrwd1 to repressive histone marks is independent of ORC proteins. Second, we show that the WD40 domain of Lrwd1 binds directly to these repressive marks in vitro. More importantly, we show that despite being able to bind to H3K9me3 and H4K20me3, the recruitment of Lrwd1 to pericentric heterochromatin depends on H3K9me3, but not H4K20me3 in vivo. Third, we show that cells with Lrwd1 or Orc2 depletion exhibit increased transcription of major satellite repeats in MEF cells. We discuss the implications of our findings below.
Lrwd1 contains a WD40 repeat domain with five small WD40 repeats. It is known that the WD40 domain of Lrwd1 binds to ORC protein (20). We show that this domain is also involved in binding methylated histone marks (Fig. 3). WD40 repeat domains have been found in many proteins, and some of them are known to bind methylated peptides. For instance, EED, a subunit of the PRC2 complex, contains seven small WD40 repeats. This seven-blade β-propeller domain forms an aromatic cage that functions as the binding site for the trimethylated repressive histone marks (30). In addition, WDR5, the core subunit of the MLL/SET1 histone methyltransferase complex (31), also contains a WD40 domain, which binds the dimethylated form of histone H3 at lysine 4, an active histone mark. These structure-function studies suggest that different forms of the WD40 domain may recognize methylated histone marks differently. How the WD40 domain of Lrwd1 binds both methylated histone marks and ORC proteins is not known. We show that deletion of a single WD40 repeat of Lrwd1 compromises its ability to bind Orc2 and H3K9me3, suggesting that an intact WD40 repeat domain structure is important for its association with ORC and methylated peptides. The x-ray crystal structure of EED reveals that the seven-blade WD40 small repeats are symmetrically arranged around a pseudosymmetric axis and form a central pocket that facilitates the interaction with trimethylated peptides and the EED-interacting partner Ezh2. Trimethylated peptides bind to the top surface, whereas Ezh2 binds to the bottom surface of the EED WD40 domain. It would be interesting to determine how the WD40 repeat domain of Lrwd1 recognizes both repressive histone marks and binds ORC proteins.
Whereas Lrwd1 and ORC bind three repressive histone marks (H4K20me3, H3K9me3/H3K27me3), we show that the association of Lrwd1 and Orc2 to pericentric heterochromatin is not affected to a detectable degree in Suv420h1h2−/− cells, which lack H4K20me3, whereas the association of Lrwd1 and ORC with pericentric heterochromatin is compromised in cells lacking H3K9me3 (Suv39h1h2−/−) in vivo. These results indicate that H3K9me3 is the primary repressive histone mark for the recruitment of Lrwd1 to pericentric heterochromatin in vivo. H3K9me3 is known to recruit HP1α to pericentric heterochromatin via interactions between the chromodomain of HP1α and H3K9me3. We demonstrate that the recruitment of Lrwd1 to the pericentric heterochromatin in vivo is independent of HP1α but is dependent upon the Lrwd1 WD40 domain. Thus, H3K9me3 recruits multiple effector proteins, including HP1 and Lrwd1, to pericentric heterochromatin to maintain silencing. Consistent with this model, depletion of Orc2 or Lrwd1 leads to the loss of heterochromatin silencing at major satellite repeats. Recent reports indicate that increased transcription of satellite repeats contributes to genomic instability and subsequent tumorigenesis (26, 32). It would be interesting to determine whether Lrwd1 is a tumor suppressor gene and whether misregulation of Lrwd1 is linked to tumorigenesis.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Jenuwein for providing Suv39h1h2−/− and Suv420h1h2−/− cells, Dr. Bruce Stillman for ORC antibodies, and Dr. Qing Li and Rebecca Burgess for critical reading of this manuscript.
This work is supported, in whole or in part, by National Institutes of Health Grants GM99722 and GM81838.

This article contains supplemental Figs. S1–S4.
- H3K9me3
- trimethylated lysine 9 of histone H3
- FL
- full-length
- H4K20me3
- trimethylated lysine 20 of histone H4
- LRR
- leucine-rich repeat
- Lrwd1
- leucine-rich repeat and WD repeat-containing protein 1
- MEF
- mouse embryonic fibroblast
- ORC
- origin recognition complex.
REFERENCES
- 1. Grewal S. I., Jia S. (2007) Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 2. Grewal S. I., Moazed D. (2003) Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 [DOI] [PubMed] [Google Scholar]
- 3. Grewal S. I., Elgin S. C. (2002) Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12, 178–187 [DOI] [PubMed] [Google Scholar]
- 4. Augui S., Nora E. P., Heard E. (2011) Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442 [DOI] [PubMed] [Google Scholar]
- 5. Wutz A. (2011) Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12, 542–553 [DOI] [PubMed] [Google Scholar]
- 6. Tattermusch A., Brockdorff N. (2011) A scaffold for X chromosome inactivation. Hum. Genet. 130, 247–253 [DOI] [PubMed] [Google Scholar]
- 7. Probst A. V., Almouzni G. (2011) Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 27, 177–185 [DOI] [PubMed] [Google Scholar]
- 8. Beisel C., Paro R. (2011) Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12, 123–135 [DOI] [PubMed] [Google Scholar]
- 9. Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 10. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 11. Peters A. H., Mermoud J. E., O'Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T. (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30, 77–80 [DOI] [PubMed] [Google Scholar]
- 12. Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eustermann S., Yang J. C., Law M. J., Amos R., Chapman L. M., Jelinska C., Garrick D., Clynes D., Gibbons R. J., Rhodes D., Higgs D. R., Neuhaus D. (2011) Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 18, 777–782 [DOI] [PubMed] [Google Scholar]
- 14. Zeng W., Ball A. R., Jr., Yokomori K. (2010) HP1: heterochromatin binding proteins working the genome. Epigenetics 5, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bell S. P., Stillman B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357, 128–134 [DOI] [PubMed] [Google Scholar]
- 16. Ehrenhofer-Murray A. E., Gossen M., Pak D. T., Botchan M. R., Rine J. (1995) Separation of origin recognition complex functions by cross-species complementation. Science 270, 1671–1674 [DOI] [PubMed] [Google Scholar]
- 17. Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23, 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pak D. T., Pflumm M., Chesnokov I., Huang D. W., Kellum R., Marr J., Romanowski P., Botchan M. R. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91, 311–323 [DOI] [PubMed] [Google Scholar]
- 19. Prasanth S. G., Shen Z., Prasanth K. V., Stillman B. (2010) Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc. Natl. Acad. Sci. U.S.A. 107, 15093–15098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen Z., Sathyan K. M., Geng Y., Zheng R., Chakraborty A., Freeman B., Wang F., Prasanth K. V., Prasanth S. G. (2010) A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell 40, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartke T., Vermeulen M., Xhemalce B., Robson S. C., Mann M., Kouzarides T. (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 23. Chan K. M., Zhang H., Malureanu L., van Deursen J., Zhang Z. (2011) Diverse factors are involved in maintaining X chromosome inactivation. Proc. Natl. Acad. Sci. U.S.A. 108, 16699–16704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Méndez J., Zou-Yang X. H., Kim S. Y., Hidaka M., Tansey W. P., Stillman B. (2002) Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9, 481–491 [DOI] [PubMed] [Google Scholar]
- 25. Wysocka J. (2006) Identifying novel proteins recognizing histone modifications using peptide pulldown assay. Methods 40, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu Q., Pao G. M., Huynh A. M., Suh H., Tonnu N., Nederlof P. M., Gage F. H., Verma I. M. (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schotta G., Sengupta R., Kubicek S., Malin S., Kauer M., Callén E., Celeste A., Pagani M., Opravil S., De La Rosa-Velazquez I. A., Espejo A., Bedford M. T., Nussenzweig A., Busslinger M., Jenuwein T. (2008) A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22, 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 29. Lehnertz B., Ueda Y., Derijck A. A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A. H. (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 30. Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J., 3rd, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruthenburg A. J., Wang W., Graybosch D. M., Li H., Allis C. D., Patel D. J., Verdine G. L. (2006) Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ting D. T., Lipson D., Paul S., Brannigan B. W., Akhavanfard S., Coffman E. J., Contino G., Deshpande V., Iafrate A. J., Letovsky S., Rivera M. N., Bardeesy N., Maheswaran S., Haber D. A. (2011) Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.