Background: The trypanosome mitochondrion imports ∼1000 nucleus-encoded proteins. However, its protein translocation machinery remains elusive.
Results: We identified several trypanosome-specific members of the translocase of mitochondrial inner membrane (TIM) in T. brucei.
Conclusion: The TIM complex in T. brucei is significantly divergent from those of other eukaryotes.
Significance: This TIM complex could be a potential drug target in trypanosomatids.
Keywords: Mitochondria, Protein Complexes, Protein Targeting, Protein Translocation, Trypanosoma brucei, Tim17
Abstract
Translocases of mitochondrial inner membrane (TIMs) are multiprotein complexes. The only Tim component so far characterized in kinetoplastid parasites such as Trypanosoma brucei is Tim17 (TbTim17), which is essential for cell survival and mitochondrial protein import. Here, we report that TbTim17 is present in a protein complex of about 1,100 kDa, which is much larger than the TIM complexes found in fungi and mammals. Depletion of TbTim17 in T. brucei impairs the mitochondrial import of cytochrome oxidase subunit IV, an N-terminal signal-containing protein. Pretreatment of isolated mitoplasts with the anti-TbTim17 antibody inhibited import of cytochrome oxidase subunit IV, indicating a direct involvement of the TbTim17 in the import process. Purification of the TbTim17-containing protein complex from the mitochondrial membrane of T. brucei by tandem affinity chromatography revealed that TbTim17 associates with seven unique as well as a few known T. brucei mitochondrial proteins. Depletion of three of these novel proteins, i.e. TbTim47, TbTim54, and TbTim62, significantly decreased mitochondrial protein import in vitro. In vivo targeting of a newly synthesized mitochondrial matrix protein, MRP2, was also inhibited due to depletion of TbTim17, TbTim54, and TbTim62. Co-precipitation analysis confirmed the interaction of TbTim54 and TbTim62 with TbTim17 in vivo. Overall, our data reveal that TbTim17, the single homolog of Tim17/22/23 family proteins, is present in a unique TIM complex consisting of novel proteins in T. brucei and is critical for mitochondrial protein import.
Introduction
A vast majority of mitochondrial proteins are nucleus-encoded and need to be imported into mitochondria after synthesis on cytoplasmic ribosomes (1–3). Protein translocases of mitochondrial outer and inner membranes (TOMs3 and TIMs, respectively) have been extensively characterized in fungi and higher eukaryotes (3–5). TOM is a multisubunit protein complex that is responsible for import of virtually all nucleus-encoded proteins through the mitochondrial outer membrane (MOM) (6, 7). The major subunit of TOM is Tom40, a β-barrel protein that forms a channel for protein translocation. Tom20, Tom70, Tom22, and Tom5 are important in the receptor function of the translocase. Tom6, Tom7, and Tom22 are involved in structural assembly of the complex. There are two TIMs (TIM23-17 and TIM22-54) in the mitochondrial inner membrane (MIM) in fungi to mammals (3–5). The TIM23-17 complex imports N-terminal mitochondrial targeting signal (MTS)-containing proteins as well as an adjacent hydrophobic sorting signal-containing proteins destined to the mitochondrial matrix and the MIM, respectively (8, 9). The core subunits of the TIM23-17 complex are Tim23, Tim17, and Tim50. For translocation of proteins for the mitochondrial matrix, the TIM23-17 complex associates with the presequence-activated motor (PAM) complex consisting of Tim44, Tim14/Pam18, Tim16/Pam16, and Hsp70. Once the proteins reach their destination, the presequence or MTS is cleaved by a mitochondrial processing peptidase (10), and the imported proteins are folded within the matrix through the action of the Hsp60-Hsp10 chaperone system (3). In contrast, the TIM22-54 complex imports internal signal-containing proteins such as the ADP/ATP carrier or other metabolite carriers with the help of hexameric chaperone complexes consisting of small Tims (such as Tim9-Tim10 and Tim8-Tim13) located in the intermembrane space (11, 12). These small Tim proteins are also involved in the sorting and assembly of the MOM β-barrel proteins such as Tom40 and the voltage-dependent anion channel (VDAC) through a topogenesis of β-barrel protein complex (TOB), which is also known as the sorting and assembly complex (SAM) (13, 14).
Despite the elaborate discoveries of the mitochondrial protein import machinery in fungi and mammals, this essential-to-life process is poorly understood in trypanosomatids. Trypanosoma brucei belongs to a group of hemoflagellated parasitic protozoa that cause a devastating disease in humans and domestic animals (African trypanosomiasis), which is most prevalent in sub-Saharan Africa (15, 16). Trypanosomatids possess a single reticular mitochondrion with several unique characteristics essential for cell survival (17, 18). Similar to other eukaryotes, the mitochondrial genome of these parasites encodes only a limited set of proteins. About a thousand mitochondrial proteins (19, 20) in T. brucei are nucleus-encoded and therefore need to be imported into mitochondria after their synthesis in the cytoplasm to form a functional mitochondrion. Mitochondrial protein import is thus essential to the survival of T. brucei.
In fungi, plants, and mammals, the N-terminal MTS varies in length by about 20–60 amino acid residues (3–5). The amino acid sequences of MTSs are not conserved, but they all possess specific characteristics, including the presence of positively charged residues and the ability to form an amphiphilic α-helix (21). Although T. brucei possess such N-terminal MTS-containing proteins, the length of the sequence varies widely compared with other eukaryotes and can be as small as 8–9 amino acid residues (22, 23).
Mitochondrial protein import machinery also appears divergent in trypanosomatids. Extensive searches in trypanosomatid genomes (24) and the completion of mitochondrial proteomes (19, 20) have yielded only a limited number of homologs of Tims in T. brucei, including a homolog of Tim17 and a few small Tims (25, 26). In fungi and higher eukaryotes, there are three homologous proteins, Tim17, Tim22, and Tim23, which belong to the amino acid transporter family and consist of four transmembrane domains in the center of the protein that are integrated in the MIM, leaving both the N and C termini in the intermembrane space (27). Although the predicted secondary structure of T. brucei Tim17 (TbTim17) is conserved, there are no homologs of eukaryotic Tim22 and Tim23 in the trypanosome genome databases. Therefore, it is necessary to identify these unknown TbTim17-interacting partners and to investigate the functions of these proteins in T. brucei.
Here, we report that TbTim17 is present in a protein complex of about 1,100 kDa, which is much larger than the TIMs found in other eukaryotes. TbTim17 is directly involved in the import of cytochrome oxidase subunit IV (COIV), an N-terminal signal-containing protein, into mitochondria. Tandem affinity purification (TAP) of the TbTim17 protein complex identified several associated proteins that are unique for trypanosomes. Two of these unique proteins (TbTim54 and TbTim62) showed strong interaction with TbTim17 and were found to be critical for mitochondrial protein import in T. brucei both in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture
The procyclic form of T. brucei 427 cells were grown in SDM-79 medium containing 10% fetal bovine serum. T. brucei 427 procyclic double resistant cell line (Tb427 29-13) expressing the tetracycline repressor gene and T7 RNA polymerase were grown in the same medium containing 50 μg/ml hygromycin and 15 μg/ml neomycin (G418) (28). For measurement of cell growth, the procyclic cells were inoculated at a cell density of 2–3 × 106/ml in medium containing appropriate antibiotics in the presence or absence of doxycycline (1 μg/ml). Cells were harvested on different days up to 12 days, and the number of cells was counted in a Neubauer hemocytometer counter. The logs of the cumulative cell numbers were plotted versus time of incubation in culture. Large scale cultivation of the procyclic form was performed by inoculating 1 liter of medium at a cell density of 2–3 × 106/ml at 27 °C in a conical flask of 2-liter capacity with constant agitation.
Plasmid Construction, Transfection, and RNA Analysis
To generate constructs for inducible expression of C-terminal TAP-tagged TbTim17, the 459-bp open reading frame (ORF) of TbTim17 was PCR-amplified using the Tim17-TAP forward and reverse primers with HindIII and XhoI restriction sites, respectively, at their 5′-ends (supplemental Table S1). The PCR product was digested with HindIII and XhoI enzymes and cloned into pLew79-MHT vector (a generous gift from Marilyn Parsons), which contains c-myc, His6, calmodulin binding peptide, and 2X-protein A tags in that order (29–31). The calmodulin binding peptide and the 2X-protein A tags are separated by a TEV protease cleavage site. The purified plasmid DNA was linearized by NotI. The linearized plasmid was used for transfection into procyclic cells (Tb427 29-13) expressing T7 polymerase and tetracycline repressor proteins according to standard protocols (28); cells were then selected by phleomycin (2.5 μg/ml) resistance. After transfection, the plasmid was integrated into the ribosomal DNA spacer region in T. brucei. The C-terminal TAP-tagged construct for Tb927.8.1740-TAP and Tb927.6.2470-TAP was developed similarly using appropriate primers (supplemental Table S1) to amplify the corresponding ORFs. The RNAi constructs for Tb927.1.1310, Tb927.6.2470, Tb927.8.1740, and Tb927.10.3620 were generated using a tetracycline-inducible dual promoter plasmid vector, p2T7Ti-177 (32). Specific primer pairs (supplemental Table S1) containing appropriate restriction enzyme sites at the 5′-ends were used to amplify the fragments of the corresponding cDNAs. The amplified products were cloned individually into the BamHI/HindIII sites of the vector p2T7Ti-177. The constructs for RNAi were verified by sequencing. The purified plasmid DNA was linearized by NotI. The linearized plasmids were used for transfection into procyclic cells Tb427 29-13 as described above. After transfection, the plasmid was integrated into 177-bp repeat regions of the minichromosomes in T. brucei. The RNAi cell lines for TbTim17 and VDAC was similarly developed as described (25, 33).
RNA was isolated from the procyclic trypanosomes grown for 2 days with or without doxycycline using TRIzol reagent (Invitrogen) according to the manufacturer's protocol, and Northern analysis was performed as described previously (25). Specific probes were generated using a random primer labeling protocol (Invitrogen) from the cDNA fragments generated by PCR using the same primer pairs used for preparation of RNAi constructs. The ethidium bromide-stained ribosomal RNA was used as loading controls.
MitoTracker Staining and Flow Cytometry Analysis
T. brucei cells were harvested and resuspended in fresh medium (5 × 106/ml). MitoTracker Red (Molecular Probes) was added to the cell suspension at a final concentration of 0.05 μm and incubated at 27 °C for 15 min. Following incubation, cells were washed with 10.0 ml of complete medium, resuspended in 5.0 ml of medium, and incubated at 27 °C for 30 min. After that, cells were washed with 1× PBS, fixed with 3.7% paraformaldehyde, and stored in PBS in the dark at 4 °C for further analysis. Fluorescence intensity was measured with a FACSCalibur (BD Biosciences) analytical flow cytometer using absorption at 578 nm and emission at 599 nm. CellQuest software (BD Biosciences) was used to analyze the results.
Subcellular Fractionation
Fractionation of T. brucei procyclic cells was performed as described (34). Briefly, 2 × 108 cells were resuspended in 500 μl of SMEP buffer (250 mm sucrose, 20 mm MOPS/KOH, pH 7.4, 2 mm EDTA, 1 mm PMSF) containing 0.03% digitonin and incubated on ice for 5 min. The cell suspension was then centrifuged for 5 min at 6,800 × g at 4 °C. The resultant pellet was considered a crude mitochondrial fraction, and the supernatant contained the soluble cytosolic proteins.
Isolation and Postisolation Treatment of Mitochondria
Mitochondria were also isolated from the parasite after lysis via nitrogen cavitation in isotonic buffer as described (25, 33, 35). The isolated mitochondria were stored at a protein concentration of 10 mg/ml in SME buffer (250 mm sucrose, 20 mm MOPS/KOH, pH 7.4, 2 mm EDTA) containing 50% glycerol at −70 °C. Before using, mitochondria were washed twice with 9 volumes of SME buffer to remove glycerol. Mitoplasts (i.e. mitochondrial matrix with intact MIM) were isolated after treatment of mitochondria with 3 volumes of water for 15 min on ice to disrupt MOM. Afterward, the osmolarity was restored with the addition of one-third volume of 2.4 m sucrose. Mitoplasts were recovered by centrifugation at 12,000 × g for 15 min. The supernatant and pellet fractions were analyzed for the release of Cyt c, an intermembrane space protein, as described (33).
Blue-Native PAGE (BN-PAGE) and Two-dimensional Gel Electrophoresis
Mitochondrial proteins (50 and 100 μg) were solubilized in 72 μl of ice-cold buffer N (20 mm Tris, pH 7.0, 0.1 mm EDTA, 50 mm NaCl, 10% glycerol, 1 mm PMSF, 1 μg/ml leupeptin, 1% digitonin). The solubilized mitochondrial proteins were clarified by centrifugation at 100,000 × g for 30 min at 4 °C. The supernatants were mixed with 7.5 μl of sample buffer (750 mm amino caproic acid, 5% Coomassie G-250) and electrophoresed on a linear 6–13% polyacrylamide gradient gel (33, 35, 36). Protein complexes were detected by immunoblot analysis. Molecular size marker proteins apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa) were electrophoresed on the same gel and visualized by Coomassie Brilliant Blue (CBB) staining. Gel strips representing a single lane on first dimension BN-PAGE gels were excised and electrophoresed on a second dimension using Tricine-SDS-PAGE (10%) (37). After separation, proteins were transferred to a nitrocellulose membrane for immunodetection.
SDS-PAGE and Immunoblots
Total cellular proteins and proteins from isolated mitochondria were analyzed by SDS-PAGE (10 or 15%) as described (38) and transferred to nitrocellulose membranes at 4 °C (100 V using 25 mm Tris-HCl, 192 mm glycine, 20% (v/v) methanol, pH 8.3) (39). Blots were treated with polyclonal antiserum against T. brucei VDAC (33), TbTim17 (25), T. brucei serine/threonine protein phosphatase 5 (TbPP5) (40), and T. brucei ATPase β (41). Trypanosome alternative oxidase (TAO) and β-tubulin were detected with the corresponding monoclonal hybridoma culture supernatants (42, 43) at 1:50 and 1:10,000 dilutions, respectively. Peroxidase-anti-peroxidase (PAP) mouse IgG (PAP reagent) was purchased from Sigma. Blots were treated with appropriate secondary antibodies and developed using an enhanced chemiluminescence (ECL) detection system (GE Healthcare).
Superose 12 Column Chromatography
Mitochondrial proteins were solubilized in buffer N, and the solubilized proteins were loaded on a Superose 12 gel filtration column (GE Healthcare), and size exclusion chromatography was performed on an FPLC system (GE Healthcare). The void volume of the column was ∼7.0 ml. Proteins were eluted with 30 ml of the same buffer except the digitonin concentration was reduced to 0.1%. Each fraction (0.25 ml) was analyzed by immunoblotting to detect TbTim17. Gel filtration molecular weight markers were run separately on the column and were detected in the eluted fractions by SDS-PAGE and CBB staining (supplemental Fig. S1). The size of the TbTim17 protein complex was calculated from a standard curve in which the log10 values of the molecular sizes of marker proteins were plotted against the fraction numbers.
TAP of Tim17 Protein Complex
TbTim17-TAP cells was induced for 48 h with doxycycline. Either the whole cell pellets or isolated mitochondria from 2 × 1010 cells were solubilized in PA150 buffer (150 mm KCl, 20 mm Tris-Cl, pH 7.7, 3 mm MgCl2, 0.5 mm DTT, 0.1% (v/v) Tween 20) (44, 45) containing 1% digitonin, 1 mm PMSF, and 1 μg/ml leupeptin. The soluble supernatant (4 ml) was incubated with 400 μl of IgG-Sepharose Fast Flow beads (GE Healthcare) for 4 h at 4 °C on a rotary mixer. After washing with 30 ml of PA150 buffer containing 0.125% digitonin, 1 mm PMSF, and 1 μg/ml leupeptin, beads were incubated with 300 units of TEV protease (Invitrogen) in 1.0 ml of TEV buffer (50 mm Tris-Cl, pH 8.0, 0.5 mm EDTA, 1 mm DTT) for 16 h at 4 °C with constant mixing. After collection of the TEV eluate, beads were briefly washed with 0.5 ml of TEV buffer, and the washing was combined with the eluate. The total eluate (1.5 ml) was then diluted with 3 volumes of calmodulin binding buffer (150 mm KCl, 20 mm Tris-Cl, pH 7.7, 3 mm MgCl2, 0.5 mm DTT, 1 mm CaCl2, 0.125% digitonin, 0.1% (v/v) Tween 20) and mixed with 200 μl of calmodulin resin (GE Healthcare) for 4 h at 4 °C. After washing the resin with 20 ml of calmodulin binding buffer, bound proteins were eluted four times (500 μl each) with EGTA elution buffer (5 mm Tris-Cl, 5 mm EDTA, 10 mm EGTA, 0.1% (v/v) Tween 20, 0.125% digitonin). Eluates were combined, concentrated about 2-fold by freeze drying, and mixed with 20 μl of Strataclean resins (Stratagene) for 15 min at room temperature. The bound proteins were eluted by boiling with Laemmli sample buffer (0.125 m Tris-Cl, pH 6.8, 0.004% bromphenol blue, 10% 2-mercaptoethanol, 20% glycerol, 4% SDS) and analyzed by SDS-PAGE.
Mass Spectrometry
Excised protein bands from one-dimensional SDS-PAGE were digested in-gel with trypsin (10 ng/μl). The digested peptides were loaded onto a homemade reverse phase column using a pressure bomb and further separated by liquid chromatography. Peptides were analyzed by a linear trap quadrupole (LTQ) nanospray mass spectrometer. Tandem spectra were searched against the GeneDB T. brucei database using MyriMatch and IDPicker software (46). The search database was concatenated with the reverse sequences of all proteins in the database to allow for the determination of false discovery rates. Protein matches were preliminarily filtered using a cutoff of at least two peptide spectra for each unique peptide sequence match. All proteins identified by fewer than two peptide spectra were eliminated, resulting in false discovery rates of <5%. The output was also filtered using IDPicker using a false positive identification threshold (default is 0.05 or 5% false positives) based on reverse sequence hits in the decoy database.
In Vitro Import Assay
The ORF for the COIV was amplified from T. brucei genomic DNA using forward and reverse primer pairs (supplemental Table S1). The PCR product was subcloned in pGEM-4Z vector between HindIII and EcoRI sites. The TAO-dihydrofolate reductase (DHFR) fusion construct was also generated in a pGEM-4Z vector (Promega). The TAO ORF was amplified from the TAO cDNA clone (pTAO25) (47) using forward and reverse primers (supplemental Table S1) containing HindIII and BamH1 restriction sites at the 5′-ends, respectively. The mouse DHFR ORF was PCR-amplified using pQE16 (Qiagen) as the template and the forward and reverse primers (supplemental Table S1) containing BamH1 and EcoRI restriction sites at the 5′-ends, respectively. These PCR products were then ligated and cloned into a pGEM-4Z vector between the HindIII and EcoRI sites to develop the TAO-DHFR fusion construct. Radiolabeled precursor proteins COIV and TAO-DHFR were synthesized in vitro using a coupled transcription/translation rabbit reticulocyte system (TnT® Coupled Reticulocyte Lysate Systems, Promega) according to the manufacturer's protocol using [35S]methionine. Radiolabeled precursor proteins were used for in vitro import into mitochondria isolated from T. brucei as described (35, 48). For antibody inhibition experiments, mitoplasts generated from Tb427 procyclic mitochondria were preincubated with various concentrations of either the TbTim17 antibody or normal IgG for 20 min on ice. After incubation with antibodies, an in vitro import assay was performed as described above using COIV as the substrate protein.
In Vivo Import Assay
An inducible dual vector assay system was developed to study the effect of RNAi on in vivo targeting of nucleus-encoded mitochondrial protein such as mitochondrial RNA-binding protein 2 (MRP2) (49). To generate the MRP2–2X-myc expression construct, the MRP2 coding region was PCR-amplified from T. brucei genomic DNA using MRP2 forward and MRP2–2X-myc reverse primers (supplemental Table S1). The nucleotide sequence that codes for 2X-myc epitope (EQKLISEEDL) was included at the 5′-end of the reverse primer. The PCR product was cloned in between HindIII and BamHI sites of the pLew-Bsd vector (a generous gift from George Cross). After transfection of Tb427 29-13 cells with this construct and selection with blasticidin (10 μg/ml), cells were individually transfected with different RNAi constructs generated using p2T7Ti-177 vector and selected with phleomycin (2.5 μg/ml). These cell lines were then induced with doxycycline for expression of MRP2–2X-myc as well as RNAi for the selected genes. At different time points after induction with doxycycline, cells were lysed, and the mitochondrial and cytosolic fractions were analyzed by SDS-PAGE and immunoblotting to assess the level of MRP2–2X-myc localized in mitochondria.
RESULTS
TbTim17 Is Present in ∼1,100-kDa Mitochondrial Membrane Protein Complex
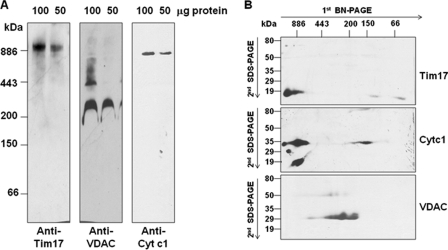
To understand whether TbTim17 is present in a multisubunit protein complex, T. brucei mitochondrial membrane proteins were solubilized with a mild non-ionic detergent (i.e. 1% digitonin), and soluble protein complexes were analyzed by BN-PAGE followed by immunoblot with anti-TbTim17 antibody. The antibody recognized a band of molecular mass greater than 886 kDa (Fig. 1A). Reprobing the same blot with anti-VDAC antibody recognized a major homo-oligomeric complex of ∼212 kDa as reported previously (32). Higher order oligomers were also detected in minor amounts when more protein was loaded (Fig. 1A). T. brucei Cyt c1 antibody was used to detect cytochrome-bc1 reductase complex. The size of this complex was ∼672 kDa as has been reported earlier (49). Therefore, mitochondrial membrane protein complexes were separated by BN-PAGE as anticipated. Digitonin (1%) gave us the best separation of T. brucei mitochondrial membrane complexes by BN-PAGE. Use of other detergents such as lauryl-d-maltoside or Triton X-100 generated a trailing phenotype, indicating the fragmentation of the protein complexes.
FIGURE 1.
Analysis of mitochondrial membrane protein complexes by two-dimensional gel electrophoresis. A, T. brucei mitochondrial proteins (100 or 50 μg) were solubilized in buffer N containing 1% digitonin. The soluble supernatant was subjected to BN-PAGE. Proteins were transferred to a nitrocellulose membrane, and the blot was developed with anti-TbTim17, -TbVDAC, and -TbCyt c1 antibodies. Apoferritin dimer (886 kDa) and monomer (443kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa) were used as molecular mass markers. B, gel strips representing individual lanes were excised from the first dimensional gel and subjected to 10% Tricine-SDS-PAGE. Proteins were transferred to a nitrocellulose membrane, and the blot was sequentially probed with anti-TbTim17, -TbCyt c1, and -TbVDAC antibodies.
To further resolve the protein complexes detected with the above antibodies, a gel strip from the first dimension BN-PAGE was subjected to Tricine-SDS-PAGE and sequentially immunoblotted with anti-TbTim17, anti-TbVDAC, and anti-Cyt c1 antibodies. The results clearly indicated that a higher molecular mass complex recognized by the anti-TbTim17 antibody on the first dimension BN-PAGE contained TbTim17, which was resolved as an ∼19-kDa protein band by second dimension SDS-PAGE (Fig. 1B). The presence of Cyt c1 (∼33 kDa) and VDAC monomer (∼29 kDa) was also verified in the respective complex. It had been reported in fungi that Tim23 and Tim17 are present primarily in a 90-kDa complex; minor amounts are also found in 140- and 250-kDa complexes (27). The T. brucei TbTim17 protein complex observed here is therefore much larger than that seen in fungi.
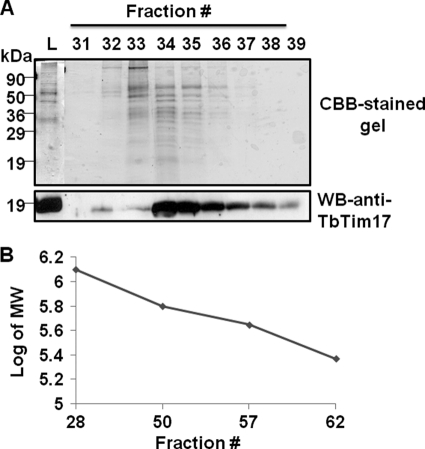
Next, we determined the size of this complex by Superose 12 size exclusion chromatography. The column fractions were assayed for the presence of Tim17 by immunoblot analysis. We found that the majority of the TbTim17 was eluted at an elution volume of 9.0–10.0 ml (fractions 34–39) (Fig. 2A). Based on calibration of the column with known molecular mass protein standards (Fig. 2B), we calculated that the molecular size of the protein complex eluting in fraction 34 was about 1,100 kDa, which supported the results found with BN-PAGE.
FIGURE 2.
Superose 12 size exclusion chromatography of T. brucei mitochondrial proteins. A, a digitonin (1%)-solubilized mitochondrial supernatant was run on a Superose 12 column. Fractions (0.25 ml each) were eluted with 30 ml of buffer containing 0.125% digitonin. Fractions were analyzed by immunoblot for detection of TbTim17. The column was run four consecutive times, and respective fractions were pooled, precipitated by TCA (10%), and analyzed by SDS-PAGE with CBB staining. The results from the positive fractions (fractions 31–39) are shown. The input lane (L) represents mitochondrial supernatant equivalent to 2% of the amount of protein loaded on the column. B, the column was calibrated with proteins of known molecular size, thyroglobulin (669 kDa), ferritin (440 kDa), and catalase (232 kDa). A pUC plasmid DNA (6.2 kb), which eluted at peak fraction 28, was used to measure the void volume. A standard curve was drawn by plotting the fraction number eluted versus the log of the molecular size of the marker. WB, Western blot.
Role of TbTim17 in Mitochondrial Protein Import
To investigate the role of TbTim17 for mitochondrial import, we performed import analysis of an N-terminal MTS-containing protein, COIV, which possesses a predicted MTS of 40 amino acids, into T. brucei mitochondria. In vitro import of this substrate protein was performed in mitochondria isolated from the uninduced control and TbTim17 knockdown cells. As reported earlier, TbTim17 protein levels were reduced about 50% within 2 days after induction of expression of the TbTim17 double-stranded RNA (supplemental Fig. S2). For in vitro protein import assays, we isolated mitochondria at day 2 after induction of TbTim17 RNAi because at this time point cell growth and mitochondrial membrane potential were minimally affected (25). In addition, during our assay, isolated mitochondria were re-energized in import buffer containing 5 mm NADH, 2 mm ATP, 10 mm creatine phosphate, and 0.1 mg/ml creatine kinase.
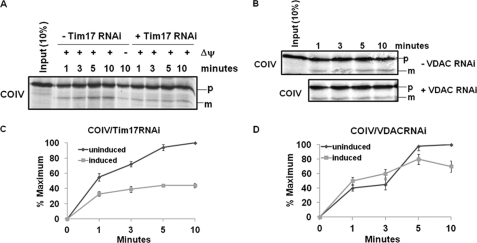
The presequence-containing precursor COIV was imported and processed to its mature form as anticipated in mitochondria from uninduced cells (Fig. 3A). COIV import was also inhibited when mitochondria were pretreated with valinomycin and carbonyl cyanide m-chlorophenylhydrazone, which are uncouplers that disrupt mitochondrial membrane potential (ΔΨ). Analysis of the import of COIV into TbTim17-depleted mitochondria clearly showed an inhibition of import of about 60% within 10 min of incubation (Fig. 3, A and C), indicating the involvement of TbTim17 in the import of COIV into mitochondria as reported (25). As a control, in vitro import of COIV was performed using mitochondria isolated from VDAC RNAi cells grown for 48 h in the presence or absence of doxycycline. The level of VDAC was depleted more than 50% at this time point after induction of the double-stranded RNA (supplemental Fig. S2). However, no significant inhibition of import was observed due to depletion of VDAC (Fig. 3, B and D).
FIGURE 3.
Effect of TbTim17 knockdown on import of COIV into mitochondria. A, in vitro import of radiolabeled COIV into isolated mitochondria from T. brucei Tim17 RNAi cells grown in the absence (− Tim17 RNAi) or presence (+ Tim17 RNAi) of doxycycline for 48 h was performed as described under “Experimental Procedures.” At different time points during import, mitochondria were reisolated, and proteins were analyzed by SDS-PAGE and autoradiography. The precursor (p) and mature (m) products are indicated. Some mitochondria were also pretreated with valinomycin and carbonyl cyanide m-chlorophenylhydrazone for disruption of the mitochondrial membrane potential (ΔΨ). These pretreated mitochondria were also used for import assays to show that import of COIV was ΔΨ-dependent. B, in vitro import of COIV into isolated mitochondria from T. brucei VDAC RNAi cells grown in the absence (− VDAC RNAi) or presence (+ VDAC RNAi) of doxycycline for 48 h was performed in parallel as a control. C and D, the intensity of mature COIV imported into mitochondria from Tim17 RNAi and VDAC RNAi, respectively, was quantitated from three independent experiments for each set using densitometry. The intensity of the mature proteins in the respective uninduced control mitochondria at the longest time point was used as maximum (100%), and the calculated percentage of maximum import at other time points into mitochondria from uninduced and induced cells was plotted versus time. Standard errors are calculated from three independent experiments.
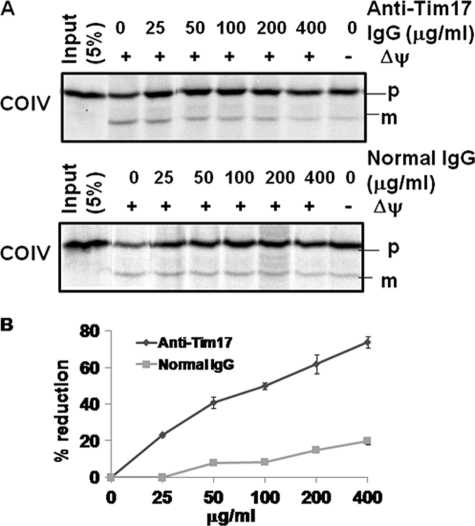
To confirm that TbTim17 plays a direct role in import of COIV, we performed antibody inhibition experiments (Fig. 4). Mitoplasts isolated from the T. brucei parental cell line were preincubated with various concentrations of anti-TbTim17 IgG and used for in vitro import assays. Functional blocking of TbTim17 with specific antibodies showed inhibition of import of COIV into mitochondria (Fig. 4A). Import efficiency was quantitated by measuring the intensity of the mature protein bands. A steady increase in inhibition of import of COIV was noted with increasing concentrations of anti-TbTim17 IgG (Fig. 4B). A maximum inhibition of about 80% was found using an anti-TbTim17 IgG concentration of 400 μg/ml. Preincubation of mitoplasts with a similar concentration of IgG purified from preimmune sera did not show any significant inhibitory effect on in vitro import, although a 10–20% inhibition was observed at the highest concentration of normal IgG (400 μg/ml), possibly due to nonspecific blocking of import.
FIGURE 4.
Effect of TbTim17 antibody on in vitro import of COIV into mitoplast. A, in vitro import of COIV into isolated T. brucei mitoplasts was performed in the presence of various concentrations of anti-TbTim17 IgG and IgG purified from preimmune serum. The radiolabeled substrate, T. brucei COIV, an N-terminal signal-containing protein, was synthesized using a coupled transcription/translation system as described under “Experimental Procedures.” The precursor (p) and mature (m) products are indicated. B, the intensity of the imported protein bands was quantified, and the percent reductions relative to no-antibody controls were calculated from three independent experiments and plotted against IgG concentrations.
C-terminal TAP-tagged TbTim17 Is Expressed and Assembled in Endogenous Protein Complex
For purification of the TbTim17 protein complex, a C-terminal TAP-tagged TbTim17 was expressed ectopically from a tetracycline-inducible pLew79-MHT vector in the procyclic form of T. brucei. The TAP-tagged protein was detected by Western blot analysis and probed with PAP reagent (Fig. 5A). The TAP-tagged TbTim17 was expressed only after induction with doxycycline. As expected, the apparent size of the protein was about 40 kDa. The expression level was maximal at about 48 h after induction. Therefore, we harvested cells at this time point for subsequent experiments. The subcellular fractionation followed by immunoblot analysis using PAP reagent showed that TbTim17-TAP is enriched in the mitochondrial fraction, similar to other endogenous mitochondrial proteins such as VDAC and TbTim17 (Fig. 5B). TbPP5, a cytosolic protein (40), was present exclusively in the cytosolic fraction. Therefore, the C-terminal TAP tag did not hamper the localization of TbTim17.
FIGURE 5.
Expression, targeting, and assembly of C-terminal TAP-tagged TbTim17 in T. brucei mitochondria. C-terminal TAP-tagged-TbTim17 was expressed from the inducible vector pLew79-MHT. A, immunoblot analysis of total proteins from cells grown in the presence or absence of doxycycline for 48 and 96 h using antibodies against TbTim17. PAP reagent was used to detect TAP-tagged-Tim17. B, proteins from subcellular fractions were analyzed by immunoblot with antibodies against mitochondrial proteins VDAC and Tim17 and cytosolic protein PP5 and with PAP reagent. T, total fraction; C, cytosolic fraction; M, mitochondrial fraction. Ten micrograms of protein were loaded per lane. C, assembly of the C-terminal TAP-tagged TbTim17 into mitochondrial membrane protein complexes. Mitochondrial proteins from the parental, TbTim17 RNAi, and TbTim17-TAP cells grown for 48 h were solubilized with digitonin (1.0%) and analyzed by BN-PAGE and immunoblot using anti-TbTim17 and anti-VDAC antibodies. PAP reagent was used to detect TbTim17-TAP-containing protein complexes. Protein complexes and molecular size markers are indicated.
For TAP of the TbTim17 protein complex, it was necessary that the tagged bait was assembled in the endogenous TIM complex. To investigate this, mitochondrial membrane protein complexes from Tb427 (parental strain), TbTim17-RNAi, and TbTim17-TAP procyclic forms were analyzed by BN-PAGE and probed with anti-TbTim17 antibody and PAP reagent (Fig. 5C). The TbTim17 protein complex was recognized in the parental and the TbTim17-TAP mitochondrial samples at the expected size. The size of the complex was slightly larger in TbTim17-TAP mitochondria, indicating that the tagged TbTim17 is assembled in this complex. In contrast, the level of this protein complex was significantly decreased in TbTim17-RNAi mitochondria, further confirming the identity of this complex. Probing a duplicate blot with the PAP reagent clearly showed the presence of the TAP-tagged TbTim17 in a protein complex of a size similar to that recognized by anti-TbTim17, indicating that the TbTim17-TAP is present in the endogenous protein complex. The PAP reagent did not recognize any protein bands from the parental and TbTim17-RNAi mitochondrial samples as expected. Reprobing both of these blots with anti-VDAC antibody recognized the major oligomeric complex of TbVDAC around 212 kDa as described previously (33). Together, these results confirmed that the TbTim17-TAP is targeted to mitochondria and is assembled in the endogenous complex in T. brucei.
TAP of TbTim17 Protein Complex Revealed That TbTim17 Is Associated with Several Novel and a Few Conserved Proteins in T. brucei
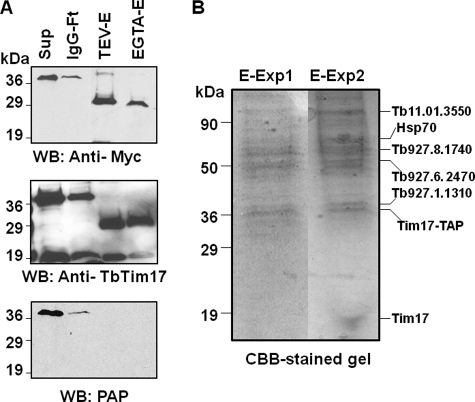
TAP of TbTim17 was performed using isolated mitochondria (10 mg of protein) as well as a whole cell (2 × 1010) lysate (Fig. 6). Samples collected from each step of the purification process were analyzed by SDS-PAGE and CBB staining and by immunoblot analysis with anti-TbTim17, anti-myc, and the PAP reagent. Immunoblot results clearly showed that TbTim17-TAP (∼40 kDa) was present in the solubilized supernatant and efficiently bound to IgG-Sepharose. The protein A domain was cleaved off after elution with TEV protease, and the eluted protein (∼29 kDa) was recognized by anti-myc antibody but not by the PAP reagent as expected. The TEV protease-eluted proteins were then bound to calmodulin-Sepharose and finally eluted with EGTA. Anti-TbTim17 antibody recognized the tagged protein as well as the endogenous TbTim17 in all samples, including the final eluate, further indicating an association of TbTim17-TAP with endogenous TbTim17 (Fig. 6A). The protein profiles of the final eluates obtained from mitochondrial and total cellular extract were compared after analysis of proteins by SDS-PAGE with CBB staining (Fig. 6B). The patterns of protein bands in these two experiments were quite comparable, although some additional protein bands were observed when the TAP was performed using total cellular extracts. Each of the visible protein bands from both experiments was excised from the gel and used for protein identification by mass spectrometry. As a control, TAP was performed using mitochondrial extracts obtained from the parental strains of T. brucei, but no protein was purified (not shown).
FIGURE 6.
Tandem affinity purification of TbTim17 protein complex. Digitonin-solubilized proteins from TbTim17-TAP cells were sequentially purified using affinity matrices, i.e. IgG-Sepharose and calmodulin-Sepharose, as described under “Experimental Procedures.” A, immunoblot analysis of the samples collected at each step of purification using anti-myc, anti-TbTim17, and PAP reagent. Sup, solubilized proteins; IgG-Ft, flow-through from IgG-Sepharose column; TEV-E, TEV eluate; EGTA-E, final eluate from the calmodulin-Sepharose column. B, samples from the final eluate (80%) were analyzed by SDS-PAGE and CBB staining to visualize the protein bands. E-Exp1, final eluate from experiment 1 (from mitochondrial lysate); E-Exp2, final eluate from experiment 2 (total cellular lysate). Identified proteins are indicated by name or GeneIDs by the side of the corresponding protein bands. WB, Western blot.
Mass spectrometric analysis of the TbTim17 TAP samples identified 20 proteins with high confidence (Table 1 and supplemental Table S2). Among these, we identified seven novel proteins with unknown functions. GeneIDs for these are Tb09.160.0670, Tb927.1.1310, Tb927.6.2470, Tb927.8.1740, Tb11.01.3290, Tb11.02.5660, and Tb927.5.2330. Except for Tb09.160.0670 and Tb927.5.2330, all of these newly identified candidates encode mitochondrial proteins as predicted by the “MitoCarta” program (51) (Table 1). In addition, the proteins encoded by Tb927.1.1310 and Tb927.10.3620 possess N-terminal mitochondrial targeting signals of 42 and 22 amino acid residues, respectively, as analyzed by the MitoProt II program (52). When analysis was done using TMPred software (53), we found that the proteins encoded by Tb927.1.1310, Tb927.6.2470, Tb927.10.3620, Tb927.8.1740, Tb11.01.3290, and Tb11.02.5660 each possessed multiple transmembrane domains, suggesting that these are membrane proteins. These novel proteins did not show any homology with known Tims found in other eukaryotes. However, BLAST analysis revealed that the protein encoded by the gene Tb927.1.1310 (47 kDa) showed a distant similarity to a bacterial outer membrane lipoprotein of the resistance-nodulation-cell division (RND) efflux system (54). Furthermore, the protein product of Tb927.6.2470 (54 kDa) has three predicted transmembrane domains that can be grouped with a major facilitator superfamily of proteins. The Tb927.8.1740 and Tb11.01.3290 encode two multitopic mitochondrial proteins of 62 and 69 kDa, respectively. These proteins possess several tetratricopeptide repeat or ankyrin type motifs. The tetratricopeptide repeat motif is composed of two antiparallel α-helices of 34 amino acid residues and is involved in protein-protein interactions (55). The ankyrin motif is structurally similar and is involved in protein interaction. These motifs are found in various cellular proteins in different subcellular locations (55, 56). Two mitochondrial protein import receptors in fungi and animals, Tom70 and Tom20, are known to possess several tetratricopeptide repeat motifs (1–3). Tb11.02.5660 (46 kDa) has previously been found in mitochondrial proteomes and is possibly a subunit of the cytochrome oxidase complex. Tb927.5.2330 is a large protein (485 kDa) containing armadillo type folding, which is a type of superhelical structure of α-helices and is known to interact with many proteins (57). We also identified several other mitochondrial proteins that had been characterized previously. These include ATP synthase β (Tb927.3.1380), mitochondrial processing peptidase (Tb927.5.1060), Hsp60 (Tb10.70.0430), and Hsp70 (Tb927.6.3740). Hsp70 is a known component of the TIM23-17-PAM complex in other eukaryotes (8, 9). Mitochondrial processing peptidase and Hsp60 are known to be required for removal of MTS and postimport folding of translocated proteins, respectively. Therefore, it is likely that these proteins are also associated with the TbTim17 protein complex. Other proteins that have been found in the TAP eluate include Hip, an Hsc70-interacting protein (Tb927.3.5340) and a laminin-like protein (Tb927.10.3620). The list of known proteins in the TAP eluate also included several metabolic enzymes such as oxoglutarate dehydrogenase components (Tb11.01.3550 and Tb11.47.0004), acyl-CoA dehydrogenase (Tb927.8.1420), hexokinase (Tb10.70.0430), glycerol kinase (Tb09.211.3540), and tryparedoxin peroxidase (Tb927.8.1900). These could possibly be contaminants in the TbTim17-TAP eluates. However, we cannot exclude the possibility that these proteins also play a role in mitochondrial protein import besides their metabolic activities.
TABLE 1.
Proteins identified by TbTim17-TAP and mass spectrometry analysis
Proteins identified by at least two peptides are listed. Gene ID numbers are from the GeneDB T. brucei database. Estimated molecular masses and brief descriptions of proteins that have been characterized previously are included. Hypothetical proteins are trypanosome-specific, and no match has been found in other eukaryotes.
| GeneID | Molecular mass | Description | Mitochondriala | TMDb |
|---|---|---|---|---|
| kDa | ||||
| Tb09.160.0670 | 329 | Hypothetical, Hect domain E3 ubiquitin ligase | ||
| Tb927.1.1310 | 47 | Hypothetical, similarity to bacterial outer membrane lipoprotein | + | 2 |
| Tb927.6.2470 | 54 | Hypothetical | + | |
| Tb927.8.1740 | 62.14 | Hypothetical | + | |
| Tb11.01.3290 | 69 | Hypothetical | + | |
| Tb11.02.5660 | 46.2 | Hypothetical, putative complex | + | 4 |
| Tb927.5.2330 | 484.6 | Hypothetical | ||
| Tb927.3.5340 | 42.03 | Hip, Hsc70-interacting protein | 3 | |
| Tb927.10.3620 | 34 | Laminin-like protein | + | 2 |
| Tb927.3.1380 | 55.7 | ATP-synthase β | + | |
| Tb927.5.1060 | 54.1 | MPP, mitochondrial processing peptidase | + | |
| Tb10.70.0430 | 59.5 | Hsp60 | + | |
| Tb927.6.3740 | 71.4 | Hsp70 | + | |
| Tb11.01.3550 | 41.14 | Oxoglutarate dehydrogenase E2 component | + | |
| Tb927.10.2900 | 95.07 | Importin β | ||
| Tb11.47.0004 | 113.39 | Oxoglutarate dehydrogenase E1 component | + | |
| Tb927.8.1420 | 55.9 | Acyl-CoA dehydrogenase IV subunit | + | |
| Tb10.70.5820 | 51.3 | Hexokinase | ||
| Tb09.211.3540 | 56.3 | Glycerol kinase | ||
| Tb927.8.1900 | 25.6 | Tryparedoxin peroxidase |
a Mitochondrial localization of the hypothetical proteins was predicted after analysis by the MitoCarta as well as by the MitoProt program.
b Probable transmembrane domains (TMD) were predicted by TMPred server.
Evaluation of Function of Trypanosome Proteins in Mitochondrial Protein Import in T. brucei
Next, we selected four newly identified proteins encoded by genes Tb927.1.1310, Tb927.6.2470, Tb927.10.3620, and Tb927.8.1740 for further assessment. They are predicted to be mitochondrial membrane proteins and possess distinct structural motifs. The T. brucei inducible RNAi cell lines for Tb927.1.1310 (1310-RNAi), Tb927.6.2470 (2470-RNAi), Tb927.10.3620 (3620-RNAi), and Tb927.8.1740 (1740-RNAi) were developed as described under “Experimental Procedures.” The effect of RNAi on cell growth was measured after induction of expression of the corresponding double-stranded RNA. The Tim17 RNAi cell line was used as the positive control. Analysis by Northern blot clearly showed that the induction of RNAi effectively reduced the transcript of each of these genes below the detection level (supplemental Fig. S3). We found a moderate to significant growth inhibition of all these cell lines upon induction of RNAi by doxycycline (supplemental Fig. S3), suggesting that these proteins are required for cell growth. However, growth inhibition was stronger for TbTim17-RNAi than for the others.
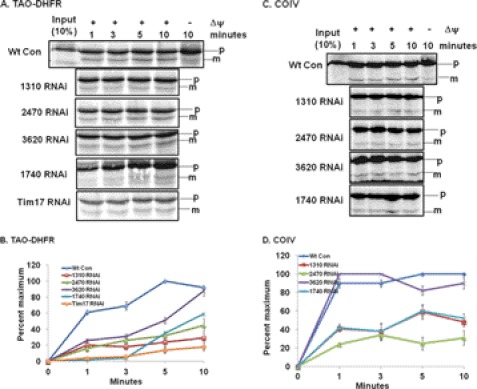
To determine whether these newly identified proteins play roles in mitochondrial protein import, in vitro protein import assays were performed using TAO-DHFR fusion protein as the substrate along with COIV that was used previously (Fig. 7). The TAO-DHFR fusion protein was radiolabeled at a much stronger intensity due to the presence of more methionine residues, which helped us see our results after a shorter exposure period than when using endogenous proteins like COIV. Similar to COIV, the TAO-DHFR fusion protein was imported into T. brucei mitochondria in a time-dependent manner and processed into the mature form as expected. The import of TAO-DHFR was also dependent on the mitochondrial membrane potential. The rate of import of TAO-DHFR into mitochondria isolated from RNAi-induced cell lines was compared with the rate of import into mitochondria from the wild type T. brucei cells (Fig. 7, A and B). Mitochondria were isolated from the RNAi cells at 48 h of growth in the presence of doxycycline. This early time point of induction of the double strand RNA was chosen to minimize any secondary effect that may be caused by RNAi. At this time point, the growth inhibition was at the minimum level (supplemental Fig. S3). We also did not see any effect of RNAi on the steady state levels of other mitochondrial proteins such as Tim17 and VDAC (supplemental Fig. S4). Mitochondrial membrane potential was also not affected by induction of 1740-, 1310-, and 2470-RNAi (supplemental Fig. S5). Our in vitro import results clearly showed a strong inhibition of import of TAO-DHFR into mitochondria from Tim17 RNAi cells. Inhibition was about 80% in comparison with that in wild type control. Import of TAO-DHFR into mitochondria isolated from doxycycline-induced 1310-RNAi, 2470-RNAi, and 1740-RNAi cells was also significantly inhibited. The percentage of inhibition of import for these RNAis was within the range of 60–80% (Fig. 7B). Although a 70% inhibition of import of TAO-DHFR was observed for 3620-RNAi within the first 3 min, import of TAO-DHFR was increased in later time points and reached a level similar to that of wild-type controls within 10 min. We also observed similar effects when COIV was used as the import substrate (Fig. 7, C and D). For this substrate, import inhibition was about 70–80% for 2470-RNAi and 50–70% for 1740- and 1310-RNAi in comparison with the wild type control. Import of TAO-DHFR into mitochondria from 3620-RNAi cells was almost unaffected, correlating somewhat with the results observed for TAO-DHFR import.
FIGURE 7.
In vitro mitochondrial protein import assay to evaluate function of novel proteins identified by TbTim17 TAP. A and C, in vitro import of the radiolabeled TAO-DHFR fusion protein and COIV, respectively, into mitochondria isolated from wild type control (Wt Con) T. brucei and induced T. brucei 1310-RNAi, 2470-RNAi, 3620-RNAi, 1740-RNAi, and TbTim17 RNAi cells was performed as described under “Experimental Procedures.” During import, mitochondria were reisolated at the indicated time points, and proteins were analyzed by SDS-PAGE and autoradiography. The precursor (p) and mature (m) products are indicated. Some mitochondria were also pretreated with valinomycin and carbonyl cyanide m-chlorophenylhydrazone to disrupt the mitochondrial membrane potential. B and D, the intensity of the mature protein bands for TAO-DHFR and for COIV, respectively, was quantitated by densitometric scanning and plotted as a percentage of maximum of the wild type control for each experiment. Standard errors are calculated from three independent experiments. GeneIDs for RNAi targets are as follows: Tb927.1.1310 for 1310-RNAi, Tb927.6.2470 for 2470-RNAi, Tb927.10.3620 for 3620-RNAi, and Tb927.8.1740 for 1740-RNAi.
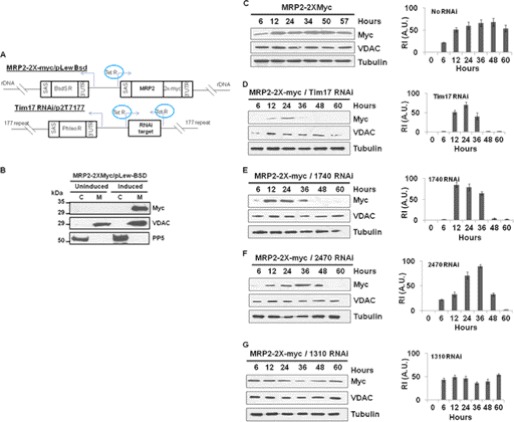
Next, we assessed the effect of these RNAis on in vivo mitochondrial targeting of the C-terminal 2X-myc-tagged MRP2. MRP2 is a T. brucei mitochondrial matrix protein (49) that has a characteristic N-terminal targeting signal. The MRP2–2X-myc was expressed from a pLew-Bsd vector after induction with doxycycline. A schematic diagram of these vectors is shown in Fig. 8A. The expressed protein was exclusively found in the mitochondrial fraction as detected by immunoblot analysis of the subcellular fractions with anti-myc antibody (Fig. 8B). The expression level of MRP2–2x-myc was detected as early as 6 h and increased about 2-fold within 12 h of induction. The expression of MRP2–2X-myc stayed at the maximum level up to 48 h, and after that, it slightly decreased (Fig. 8C). In contrast, induction of TbTim17 RNAi simultaneously with the expression of the MRP2–2X-myc showed that mitochondrial localization of this myc-tagged protein was severely hampered particularly after 24 h (Fig. 8D). At 48 h, the level of MRP2–2X-myc was below the detection limit. We also did not see any accumulation of this protein in the cytosolic fraction (not shown), possibly due to a rapid degradation of the precursor protein in the cytosol. These results indicated that depletion of TbTim17 not only inhibits the import of various mitochondrial proteins in vitro but it also inhibits targeting of a newly synthesized mitochondrial protein in vivo. This dual vector assay system was then used to investigate the effect of 1740-RNAi, 1310-RNAi, and 2470-RNAi on mitochondrial localization of the newly synthesized MRP2–2X-myc (Fig. 8, E–G). For 1740-RNAi, the maximum expression was found at 12 h; after that, it gradually decreased and went below the detection level within 48 h (Fig. 8E). Induction of 2470-RNAi also showed an inhibition of mitochondrial targeting of the MRP2–2Xmyc. However, the expression pattern of the myc-tagged protein along with 2470-RNAi was a little different from that in 1740-RNAi cells: MRP2–2X-myc was gradually accumulated in the mitochondria up to 36 h and then dropped below the detection level within 60 h (Fig. 8F). In contrast, a simultaneous induction of MRP2–2X-myc along with 1310-RNAi showed very little effect on mitochondrial localization of the newly synthesized myc-tagged protein (Fig. 8G). In contrast to the level of the newly synthesized MRP2–2X-myc, VDAC protein levels were unaffected in all these cell lines. All together, these results demonstrate that 1740-RNAi and 2470-RNAi strongly inhibit mitochondrial protein import both in vitro and in vivo. The 1310-RNAi showed strong inhibition of import of mitochondrial proteins in vitro but did not have any significant effect on in vivo targeting of a nucleus-encoded mitochondrial protein. Those proteins encoded by genes Tb927.1.1310, Tb927.6.2470, and Tb927.6.1740 are referred to here as TbTim47, TbTim54, and TbTim62, respectively. The numbers after TbTim represent the predicted molecular size of the respective protein.
FIGURE 8.
In vivo mitochondrial protein import assay to evaluate function of novel proteins identified by TbTim17 TAP. A, the schematic diagram of the MRP2–2X-myc/pLew-Bsd construct and the p2T7Ti-177 RNAi cassette. SAS, splice acceptor site; BsdS R, blasticidin resistance gene; 3′UTR, 3′-untranslated region; MRP2, MRP2 coding region; 2X-myc, double myc epitope; Phleo R, phleomycin resistance gene; rDNA, ribosomal DNA spacer region; 177 repeat, 177-base pair repeat region on minichromosome; Tet R, tetracycline repressor. Arrows indicate the direction of transcription. B, expression and mitochondrial targeting of the MRP2–2X-myc in T. brucei. The MRP2–2x-myc cell line was grown in the absence (uninduced) or presence (induced) of doxycycline for 48 h. Cells were harvested, and subcellular fractionation was performed. The cytosolic (C) and mitochondrial (M) fractions were analyzed by SDS-PAGE and immunoblotting using anti-myc antibody as the probe. VDAC and PP5 were used as the mitochondrial and cytosolic marker proteins, respectively. C–G, the single transfected cell line containing MRP2–2X-myc/pLew-Bsd cassette (C) and double transfected cell lines containing the MRP2–2X-myc/pLew-Bsd construct and either Tim17 RNAi (D), 1740-RNAi (E), 2470-RNAi (G), or 1310-RNAi (F) were induced with doxycycline (1.0 μg/ml). Cells were harvested at different time points (6–60 h) after induction. Mitochondrial fractions were analyzed by SDS-PAGE and immunoblotting using anti-myc, -VDAC, and -tubulin antibodies. The relative intensities (R.I. in arbitrary units (A.U.)) of the protein bands recognized by the anti-myc antibody were measured after normalization with the corresponding tubulin band intensity, plotted versus hours of induction, and presented by the side of the corresponding blot. Experiments are done in triplicate.
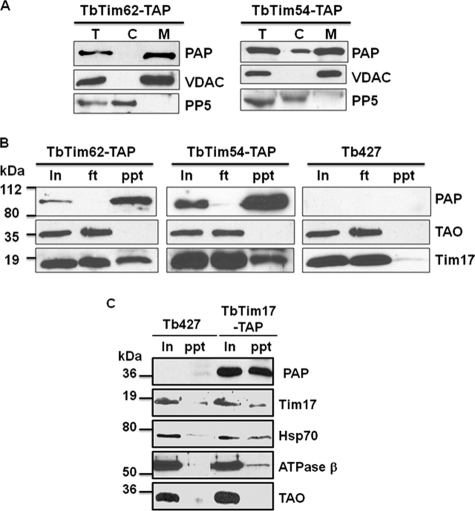
TbTim54 and TbTim62 Are Associated with TbTim17
To confirm that the TbTim54 and TbTim62 interact with TbTim17, we applied reverse TAP-pulldown experiments (Fig. 9). The C-terminal TAP-tagged TbTim54 and TbTim62 were expressed individually in T. brucei. Subcellular fractionation followed by immunoblotting clearly showed that the TAP-tagged proteins are accumulated within mitochondria (Fig. 9A). The expression level of the TbTim54-TAP was relatively higher than that for TbTim62-TAP. A small amount of TbTim54-TAP was found in the cytosolic fraction, possibly due to the effect of overexpression of this protein. Next, we used IgG-Sepharose beads to pull down proteins associated with TbTim54 and TbTim62 from the respective mitochondrial lysate. Western blot analysis of the input, flow-through (unbound), and precipitated fractions with PAP reagent clearly showed that the TAP-tagged protein was pulled down ∼100% with the beads. Reprobing these blots with anti-TbTim17 antibody demonstrated that a significant portion of Tim17 was also pulled down along with the tagged protein, indicating the association of TbTim17 with both TbTim54 and TbTim62 (Fig. 9B). We found that TAO, a mitochondrial inner membrane protein in T. brucei, was not associated with these Tim proteins, suggesting a specific interaction of TbTim54 and TbTim62 with TbTim17. As a negative control, we used mitochondrial lysate from the parental T. brucei cell line. IgG-Sepharose beads did not pull down either TAO or Tim17 from this lysate.
FIGURE 9.
Co-precipitation of TbTim17 with TbTim54, TbTim62, and Hsp70. A, expression of the C-terminal TAP-tagged TbTim62 and TbTim54. Transfected cell lines containing pLew79-MHT/TbTim62 (TbTim62-TAP) or pLew79-MHT/TbTim54 (TbTim54-TAP) were induced separately with doxycycline for 48 h. Cells were harvested, and subcellular fractions were prepared as described under “Experimental Procedures.” The total (T), cytosolic (C), and mitochondrial (M) fractions were analyzed by SDS-PAGE and immunoblot using PAP reagent for detection of the protein A tag. VDAC and PP5 were used as the markers for mitochondrial and cytosolic fractions, respectively. B, mitochondria isolated from the TbTim62-TAP and TbTim54-TAP cells induced for 48 h and from the parental T. brucei (Tb427) cells were lysed with digitonin (1%), and solubilized proteins were used for immunoprecipitation using IgG-Sepharose beads. Proteins from the input (In) 10%, flow-through (ft) 10%, and precipitated (ppt) 50% fractions were analyzed by immunoblot probed with the PAP reagent TAO and Tim17 antibodies. C, mitochondria isolated from the Tim17-TAP cells induced for 48 h and from the Tb427 cells were lysed with digitonin (1%). The soluble proteins were immunoprecipitated with IgG-Sepharose beads. Proteins from the input (In) 20% and precipitated (ppt) 80% were analyzed by immunoblot using the PAP reagent and the Tim17, Hsp70, ATPase β, and TAO antibodies. Molecular masses of the marker proteins are indicated by the side of the blots.
Interestingly, we found that induction of TbTim62 RNAi for 96 h or longer specifically reduced the level of TbTim17 protein in mitochondria (supplemental Fig. S6). However, the level of other mitochondrial proteins such as VDAC and Hsp70 remained unaffected. In contrast, induction of TbTim54 RNAi and 1310-RNAi for a similar number of days showed a minimal effect, and 3620-RNAi did not show any reduction of the TbTim17 protein. These results indicated that TbTim62 and TbTim17 protein levels are interdependent, possibly because they are associated and are present in the same protein complex.
We also demonstrated that Hsp70 is present in the TAP eluate of TbTim17 by immunoblot analysis (Fig. 8C), which confirmed the association of Hsp70 with Tim17 in T. brucei. Similarly, we demonstrated that ATPase subunit β is present in the TAP eluate by immunoblot analysis. However, this protein was precipitated at a low stoichiometric ratio with TbTim17, suggesting that this could be a loosely associated protein or a minor contaminant from the ATPase complex. All together, we identified at least three novel proteins of T. brucei, TbTim47, TbTim54, and TbTim62, that are associated with TbTim17 and play important roles in mitochondrial protein import in T. brucei.
DISCUSSION
The TbTim17 protein complex was purified, and its function was analyzed. TbTim17, a homolog of the Tim17/22/23 protein family in T. brucei, is directly involved in the import of an N-terminal MTS-containing nucleus-encoded protein into T. brucei mitochondria and is present in a multiprotein complex. Purification of this complex identified a number of trypanosome-specific proteins. Among these, TbTim54 and TbTim62, two mitochondrial proteins, interact with TbTim17 and were found to be critically important for mitochondrial protein import in T. brucei both in vitro and in vivo, suggesting that these are unique and essential components of the T. brucei TIM complex.
Despite the conservation of Toms and Tims among fungi and animals, the parasitic protozoa of the excavate group, which includes trypanosomatids, are rather unique (58, 59). The homologs of many of these known components are missing in the completed genome as well as in the mitochondrial proteomes of trypanosomatids like T. brucei (19–21). Although a MOM protein, Sam50/Tob55, is structurally and functionally conserved in T. brucei (35), a true homolog of Tom40 is not present. Recently, an archaic type, Sam50-like protein (ATOM) has been identified in T. brucei that appears to be acting as Tom40 in this organism (60). However, how ATOM works and what other proteins are associated with it need to be elucidated. Among the two TIM complexes found in fungi, plants, and animals, TIM23 appears to be evolutionarily more ancient than TIM22 (58, 59). The components of TIM22 have not been identified in trypanosomatid parasites. We previously characterized a homolog of Tim17 in T. brucei that belongs to the eukaryotic Tim17/Tim23/Tim22 family of proteins, and here, we extend our findings on TbTim17.
BN-PAGE analysis and size exclusion chromatography both showed that TbTim17 is present in a large protein complex in detergent-solubilized mitochondrial extract. The estimated size of this complex appears larger than the size of TIMs reported in other eukaryotes. However, determining whether this size difference reflects an increased number of component proteins or proteins with larger molecular masses will require further investigation. It may also be possible that the large protein complex of TbTim17 found in the detergent extracts is a supercomplex consisting of proteins of the MOM and MIM that are necessary for mitochondrial protein import in T. brucei.
A tandem affinity purification of the TbTim17 protein complex enabled us to identify a number of candidate proteins. Interestingly, none of these proteins showed a similarity to known Tims found in other eukaryotes. Using our functional assay, we found that three of these unique proteins, TbTim47, TbTim54, and TbTim62, are important for mitochondrial protein import in T. brucei under in vitro conditions. A similar result was observed when using two different substrates, TAO-DHFR and COIV, for in vitro import. However, the percentage of inhibition of import for TAO-DHFR was relatively greater than the import inhibition for COIV in TbTim47-, TbTim54-, and TbTim62-depleted mitochondria. This is possibly because TAO-DHFR is a larger protein than COIV; thus, it takes a longer time to be imported into mitochondria. As a result, the effect of depletion of important importer molecules is more prominent for TAO-DHFR than COIV. This TAO-DHFR import substrate may be useful for further analysis of the protein import process by arresting this precursor protein at different times during transit followed by chemically cross-linking it with specific importer molecules.
We also performed an in vivo mitochondrial targeting assay to confirm our results. Interestingly, we found that our in vivo import assay confirmed the importance of the last two proteins, TbTim54 and TbTim62, for translocation of nucleus-encoded proteins into mitochondria. Depletion of TbTim47 did not show significant inhibition of in vivo mitochondrial targeting of MRP2–2X-myc within 60 h. This suggests that TbTim47 may serve an auxiliary function during protein import that is critical during in vitro import of a precursor protein but not important under the in vivo condition with an intact mitochondrion and in the presence of cytosolic proteins. It is possible that depletion of TbTim47 could be functionally complemented in vivo by some cytosolic factor(s). It would be interesting to see whether longer induction of TbTim47 RNAi has an additional effect on in vivo import. However, our results clearly showed that at least two of these novel trypanosome-specific proteins are critical for mitochondrial protein import in T. brucei.
Using a reverse TAP tag approach, we confirmed the interaction of TbTim54 and TbTim62 with TbTim17. By this approach, we also showed that TbTim54 and TbTim62 are indeed mitochondrial proteins. The relative amount of endogenous TbTim17 pulled down by TbTim62-TAP and by TbTim54-TAP was higher than that by TbTim17-TAP, possibly due to the larger size of the former two proteins compared with TbTim17. Therefore, analysis of proteins pulled down by TbTim54 and TbTim62 by mass spectrometry may identify additional components of this complex. In addition, depletion of TbTim62 by RNAi showed a parallel reduction in the TbTim17 protein level, suggesting that possibly these two proteins are directly associated and are the essential core components of the TbTIM17 protein complex. Bioinformatics analysis revealed that TbTim62 is a multitopic membrane protein containing tetratricopeptide repeat or ankyrin type motifs that would support its ability for protein-protein interactions. It is possible that TbTim62 is also interacting with other Tims or the precursor proteins via these motifs.
Recently it has been reported that TbTim17 is also involved in import of cytosolic tRNAs into mitochondria (61). Because the trypanosomatid mitochondrial genome does not encode any tRNAs, import of cytosolic tRNAs is essential for translation of organellar proteins needed for important cellular functions (50). Therefore, the TbTim17 protein complex may be capable of transporting two different macromolecules, proteins and RNAs. Whether the novel proteins associated with TbTim17 reported here also play roles in tRNA import requires further investigation.
Overall, we found that T. brucei possesses a unique TIM complex. The complex function of the TbTim17 is reflected by its association with several trypanosome-specific proteins. Further investigation of the functions of these newly identified Tims and identification of other interacting partners are necessary to understand the mitochondrial protein and tRNA biogenesis in T. brucei.
Supplementary Material
Acknowledgments
We thank George Cross and Elizabeth Wirtz for pLew100 vector and Pro427 (29-13) and SM427 cell lines, Marilyn Parsons for pLew79-MHT vector, Steve Hajduk for anti-Cyt c1, Keith Gull for p2T7Ti-177 RNAi vector and β-tubulin antibody, Peter A. Weil for allowing us to use his FPLC system, and the proteomics core facility and flow cytometry core facilities at Meharry for mass spectrometry and FACS analysis, respectively. We thank Ifeanyi J. Arinze and Diana Marver for critically reviewing the manuscript.
Core facilities are supported by Grants U54RR026140 from National Center for Research Resources, U54MD07593 from National Institute for Minority Health and Health Disparities, and G12RR003032 from National Institutes of Health. V. H., E. W., and M. R. D. are supported by training Grant T32HL007737 from National Institute for Heart, Lung, and Blood. M. C. is supported by Grant 2SC1GM081146 from the National Institutes of Health.

This article contains supplemental Figs. S1–S6 and Tables S1 and S2.
- TOM
- translocase of mitochondrial outer membrane
- TIM
- translocase of mitochondrial inner membrane
- CBB
- Coomassie Brilliant Blue
- COIV
- cytochrome oxidase subunit IV
- DHFR
- dihydrofolate reductase
- Δψ
- mitochondrial membrane potential
- MOM
- mitochondrial outer membrane
- MIM
- mitochondrial inner membrane
- PAP
- peroxidase anti-peroxidase
- TAO
- trypanosome alternative oxidase
- TAP
- tandem affinity purification
- Tom
- subunits of TOM
- Tim
- subunits of TIM
- and VDAC
- voltage-dependent anion channel
- Cyt
- cytochrome
- Tb
- T. brucei
- MTS
- mitochondrial targeting signal
- PAM
- presequence-activated motor
- TOB
- topogenesis of β-barrel protein complex
- SAM
- sorting and assembly complex
- TEV
- tobacco etch virus
- BN
- blue-native
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- PP5
- phosphatase 5
- MRP2
- mitochondrial RNA-binding protein 2.
REFERENCES
- 1. Schmidt O., Pfanner N., Meisinger C. (2010) Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 [DOI] [PubMed] [Google Scholar]
- 2. Endo T., Yamano K. (2009) Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 3. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 4. Bauer M. F., Gempel K., Reichert A. S., Rappold G. A., Lichtner P., Gerbitz K. D., Neupert W., Brunner M., Hofmann S. (1999) Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 289, 69–82 [DOI] [PubMed] [Google Scholar]
- 5. Lister R., Hulett J. M., Lithgow T., Whelan J. (2005) Protein import into mitochondria: origins and functions today (review). Mol. Membr. Biol. 22, 87–100 [DOI] [PubMed] [Google Scholar]
- 6. Bains G., Lithgow T. (1999) The Tom channel in the mitochondrial outer membrane: alive and kicking. BioEssays 21, 1–4 [DOI] [PubMed] [Google Scholar]
- 7. Rapaport D. (2005) How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 171, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Laan M., Hutu D. P., Rehling P. (2010) On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim. Biophys. Acta 1803, 732–739 [DOI] [PubMed] [Google Scholar]
- 9. Truscott K. N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A. J., Rassow J., Pfanner N., Wagner R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 10. Gakh O., Cavadini P., Isaya G. (2002) Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592, 63–77 [DOI] [PubMed] [Google Scholar]
- 11. Kerscher O., Holder J., Srinivasan M., Leung R. S., Jensen R. E. (1997) The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139, 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H. E., Kühlbrandt W., Wagner R., Truscott K. N., Pfanner N. (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299, 1747–1751 [DOI] [PubMed] [Google Scholar]
- 13. Becker T., Vgtle F. N., Stojanovski D., Meisinger C. (2008) Sorting and assembly of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1777, 557–563 [DOI] [PubMed] [Google Scholar]
- 14. Rehling P., Pfanner N., Meisinger C. (2003) Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J. Mol. Biol. 326, 639–657 [DOI] [PubMed] [Google Scholar]
- 15. World Health Organization (2006) Report on Infectious Diseases, World Health Organization, Geneva [Google Scholar]
- 16. Sternberg J. M., Maclean L. (2010) A spectrum of disease in human African trypanosomiasis: the host and parasite genetics of virulence. Parasitology 137, 2007–2015 [DOI] [PubMed] [Google Scholar]
- 17. Lukes J., Hashimi H., Zíková A. (2005) Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 48, 277–299 [DOI] [PubMed] [Google Scholar]
- 18. Schneider A. (2001) Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 31, 1403–1415 [DOI] [PubMed] [Google Scholar]
- 19. Panigrahi A. K., Ogata Y., Zíková A., Anupama A., Dalley R. A., Acestor N., Myler P. J., Stuart K. D. (2009) A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9, 434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Acestor N., Panigrahi A. K., Ogata Y., Anupama A., Stuart K. D. (2009) Protein composition of Trypanosoma brucei mitochondrial membranes. Proteomics 9, 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. von Heijne G., Steppuhn J., Herrmann R. G. (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180, 535–545 [DOI] [PubMed] [Google Scholar]
- 22. Schneider A., Bursać D., Lithgow T. (2008) The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol. 18, 12–18 [DOI] [PubMed] [Google Scholar]
- 23. Häusler T., Stierhof Y. D., Blattner J., Clayton C. (1997) Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur. J. Cell Biol. 73, 240–251 [PubMed] [Google Scholar]
- 24. Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E., El-Sayed N. M. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422 [DOI] [PubMed] [Google Scholar]
- 25. Singha U. K., Peprah E., Williams S., Walker R., Saha L., Chaudhuri M. (2008) Characterization of the mitochondrial inner membrane protein translocator Tim17 from Trypanosoma brucei. Mol. Biochem. Parasitol. 159, 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gentle I. E., Perry A. J., Alcock F. H., Likić V. K., Dolezal P., Ng E. T., Purcell A. W., McConnville M., Naderer T., Chanez A. L., Charrière F., Aschinger C., Schneider A., Tokatlidis K., Lithgow T. (2007) Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol. Biol. Evol. 24, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 27. Rassow J., Dekker P. J., van Wilpe S., Meijer M., Soll J. (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 286, 105–120 [DOI] [PubMed] [Google Scholar]
- 28. Biebinger S., Wirtz L. E., Lorenz P., Clayton C. (1997) Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 85, 99–112 [DOI] [PubMed] [Google Scholar]
- 29. Hashimi H., Zíková A., Panigrahi A. K., Stuart K. D., Lukes J. (2008) TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panigrahi A. K., Zíková A., Dalley R. A., Acestor N., Ogata Y., Anupama A., Myler P. J., Stuart K. D. (2008) Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 7, 534–545 [DOI] [PubMed] [Google Scholar]
- 31. Panigrahi A. K., Schnaufer A., Ernst N. L., Wang B., Carmean N., Salavati R., Stuart K. (2003) Identification of novel components of Trypanosoma brucei editosomes. RNA 9, 484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wickstead B., Ersfeld K., Gull K. (2002) Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125, 211–216 [DOI] [PubMed] [Google Scholar]
- 33. Singha U. K., Sharma S., Chaudhuri M. (2009) Downregulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot. Cell 8, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paris Z., Hashimi H., Lun S., Alfonzo J. D., Lukeš J. (2011) Futile import of tRNAs and proteins into the mitochondrion of Trypanosoma brucei evansi. Mol. Biochem. Parasitol. 176, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma S., Singha U. K., Chaudhuri M. (2010) Role of Tob55 on mitochondrial protein biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 174, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 37. Wittig I., Schägger H. (2008) Features and applications of blue-native and clear-native electrophoresis. Proteomics 8, 3974–3990 [DOI] [PubMed] [Google Scholar]
- 38. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 39. Towbin H., Staehelin T., Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chaudhuri M. (2001) Cloning and characterization of a novel serine/threonine protein phosphatase type 5 from Trypanosoma brucei. Gene 266, 1–13 [DOI] [PubMed] [Google Scholar]
- 41. Zíková A., Schnaufer A., Dalley R. A., Panigrahi A. K., Stuart K. D. (2009) The F0F1-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 5, e1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chaudhuri M., Ajayi W., Hill G. C. (1998) Biochemical and molecular properties of the Trypanosoma brucei alternative oxidase. Mol. Biochem. Parasitol. 95, 53–68 [DOI] [PubMed] [Google Scholar]
- 43. Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., Gull K. (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491–500 [DOI] [PubMed] [Google Scholar]
- 44. Günzl A., Schimanski B. (2009) Tandem affinity purification of proteins. Curr. Protoc. Protein Sci. Chapter 19, Unit 19.19 [DOI] [PubMed] [Google Scholar]
- 45. Panigrahi A. K., Schnaufer A., Stuart K. D. (2007) Isolation and compositional analysis of trypanosomatid editosomes. Methods Enzymol. 424, 3–24 [DOI] [PubMed] [Google Scholar]
- 46. Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., Tabb D. L. (2009) IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8, 3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chaudhuri M., Hill G. C. (1996) Cloning, sequencing, and functional activity of the Trypanosoma brucei brucei alternative oxidase. Mol. Biochem. Parasitol. 83, 125–129 [DOI] [PubMed] [Google Scholar]
- 48. Williams S., Saha L., Singha U. K., Chaudhuri M. (2008) Trypanosoma brucei: differential requirement of membrane potential for import of proteins into mitochondria in two developmental stages. Exp. Parasitol. 118, 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zíková A., Horáková E., Jirků M., Dunajcíková P., Lukes J. (2006) The effect of down-regulation of mitochondrial RNA-binding proteins MRP1 and MRP2 on respiratory complexes in procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 149, 65–73 [DOI] [PubMed] [Google Scholar]
- 50. Schneider A. (2011) Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 80, 1033–1053 [DOI] [PubMed] [Google Scholar]
- 51. Zhang X., Cui J., Nilsson D., Gunasekera K., Chanfon A., Song X., Wang H., Xu Y., Ochsenreiter T. (2010) The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res. 38, 7378–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Claros M. G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 53. Bagos P. G., Liakopoulos T. D., Hamodrakas S. J. (2006) Algorithms for incorporating prior topological information in Hmms: applications to transmembrane proteins. BMC Bioinformatics 7, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blair J. M., Piddock L. J. (2009) Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 12, 512–519 [DOI] [PubMed] [Google Scholar]
- 55. D'Andrea L. D., Regan L. (2003) TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 56. Al-Khodor S., Price C. T., Kalia A., Abu Kwaik Y. (2010) Functional diversity of ankyrin repeats in microbial proteins. Trends. Microbiol. 18, 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tewari R., Bailes E., Bunting K. A., Coates J. C. (2010) Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 20, 470–481 [DOI] [PubMed] [Google Scholar]
- 58. Hewitt V., Alcock F., Lithgow T. (2011) Minor modifications and major adaptations: the evolution of molecular machines driving mitochondrial protein import. Biochim. Biophys. Acta 1808, 947–954 [DOI] [PubMed] [Google Scholar]
- 59. Lithgow T., Schneider A. (2010) Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 799–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pusnik M., Schmidt O., Perry A. J., Oeljeklaus S., Niemann M., Warscheid B., Lithgow T., Meisinger C., Schneider A. (2011) Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 [DOI] [PubMed] [Google Scholar]
- 61. Tschopp F., Charrière F., Schneider A. (2011) In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 12, 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.