Background: Extracellular regions ECL2 and the N terminus of HIV coreceptor CCR5 mediate HIV-1 entry.
Results: A C-terminal CCR5 ECL2 peptide inhibits HIV-1 entry and binds to gp120 of CCR5- and CXCR4-using strains.
Conclusion: The binding site for CCR5 ECL2 is conserved in CCR5- and CXCR4-using viruses.
Significance: Our data provide new insights into HIV-1 gp120-CCR5 interactions that can be used for inhibitor design.
Keywords: G Protein-coupled Receptors, Glycoprotein, HIV-1, NMR, Peptides, HIV Coreceptor, Chemokine Receptor, Extracellular Loop
Abstract
To initiate HIV entry, the HIV envelope protein gp120 must engage its primary receptor CD4 and a coreceptor CCR5 or CXCR4. In the absence of a high resolution structure of a gp120-coreceptor complex, biochemical studies of CCR5 have revealed the importance of its N terminus and second extracellular loop (ECL2) in binding gp120 and mediating viral entry. Using a panel of synthetic CCR5 ECL2-derived peptides, we show that the C-terminal portion of ECL2 (2C, comprising amino acids Cys-178 to Lys-191) inhibit HIV-1 entry of both CCR5- and CXCR4-using isolates at low micromolar concentrations. In functional viral assays, these peptides inhibited HIV-1 entry in a CD4-independent manner. Neutralization assays designed to measure the effects of CCR5 ECL2 peptides when combined with either with the small molecule CD4 mimetic NBD-556, soluble CD4, or the CCR5 N terminus showed additive inhibition for each, indicating that ECL2 binds gp120 at a site distinct from that of N terminus and acts independently of CD4. Using saturation transfer difference NMR, we determined the region of CCR5 ECL2 used for binding gp120, showed that it can bind to gp120 from both R5 and X4 isolates, and demonstrated that the peptide interacts with a CD4-gp120 complex in a similar manner as to gp120 alone. As the CCR5 N terminus-gp120 interactions are dependent on CD4 activation, our data suggest that gp120 has separate binding sites for the CCR5 N terminus and ECL2, the ECL2 binding site is present prior to CD4 engagement, and it is conserved across CCR5- and CXCR4-using strains. These peptides may serve as a starting point for the design of inhibitors with broad spectrum anti-HIV activity.
Introduction
The chemokine receptors C-X-C chemokine receptor type 4 (CXCR4) and C-C chemokine receptor type 5 (CCR5)4 are members of a large family of seven-transmembrane G-protein-coupled receptors (1) that serve as the main coreceptors used by HIV to gain entry into a target cell in vivo. The envelope of HIV comprises a trimer of non-covalent heterodimers formed by the surface envelope glycoprotein gp120 and the transmembrane protein gp41 (2–4). HIV entry is initiated by interactions between gp120 and the target cell receptor CD4. CD4 engagement induces a structural reorganization of gp120 leading to the formation of a highly conserved coreceptor binding site (5, 6) necessary for binding to CXCR4 or CCR5. Once engaged, further conformational changes in the viral envelope trigger gp41 to mediate fusion of host and viral cell membranes (7). HIV-1 isolates can be classified according to their coreceptor usage (8). CCR5-using strains, termed R5-tropic, dominate during the early asymptomatic phase of the disease, whereas the emergence of CXCR4-using (X4-tropic) isolates or dual tropic strains (referred to as R5X4), typically coincides with transition to the symptomatic phase and loss of immune function.
Since the discovery of CCR5 and CXCR4 as obligatory coreceptors for HIV-1 entry, a major goal in HIV research has been to define the precise molecular interactions between gp120 and coreceptor. In the absence of a three-dimensional high-resolution structure of this complex, researchers have used a variety of alternative approaches to characterize gp120 binding and other functional aspects of the HIV coreceptors. Examples include theoretical modeling (9, 10), site-directed mutagenesis studies, and evaluation of chimeric constructs in which transmembrane regions and/or extracellular loops of related chemokine receptors were inserted into the HIV coreceptors (reviewed in Refs. 9 and 11). In addition, mechanistic studies using small molecules, monoclonal antibodies, and natural chemokine receptor ligands with known binding sites on the HIV coreceptors have portrayed the gp120-coreceptor interaction as a complex, multistep process involving multiple regions of the coreceptor, the most important being the N terminus and the second extracellular loop (ECL2) regions of CCR5 (highlighted in red in Fig. 1A) (12–14). Another approach that has yielded valuable insight into the molecular details of gp120-coreceptor interactions involves the use of synthetic peptides corresponding to specific coreceptor regions in biochemical or functional assays. In the case of the CCR5 N terminus, this tactic has proven especially useful for defining post-translational modifications and peptide sequences critical for function as well as inhibition of HIV-1 entry (15–18). They also facilitated determination of high-resolution structures of CCR5 N terminus peptides in complex with gp120, furthering our understanding of this interaction at an atomic level (19, 20) and making possible the discovery of small molecule CCR5 N terminus mimetics (21). Such studies demonstrate the value in using peptide fragments of G protein-coupled receptors as tools to study coreceptor interactions because appropriate peptides tend to function in part as coreceptor mimics (22, 23).
FIGURE 1.
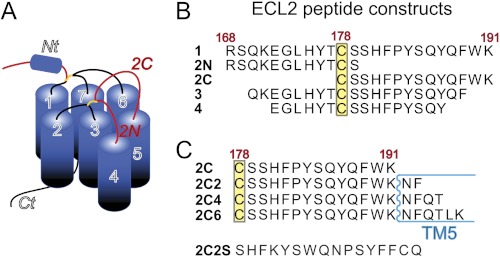
Schematic of CCR5 and ECL2 peptide constructs used in this study. A, the region of CCR5 corresponding to ECL2 is shown in red, and the helical N terminus is labeled and shown as a cylinder. B, amino acid sequences of the ECL2 peptide constructs predicted to reside in the extracellular region of ECL2. C, C-terminal ECL2 peptide sequences that contain additional residues predicted to reside in TM5.
Extending our earlier structural studies of CCR5(19, 21, 24), we sought to utilize a similar peptide fragment approach to gain insight into the molecular interactions of CCR5 ECL2 with gp120. The importance of this region in mediating viral entry has been highlighted by various studies. For example, point mutations on CCR5 identified residues in ECL2 as necessary for HIV-1 entry (25, 26). Anti-CCR5 mAbs whose epitopes include ECL2 residues potently inhibit HIV-1 fusion and entry (27, 28). Small molecule CCR5 antagonists that bind the transmembrane regions surrounding ECL2 have been suggested to distort the conformation of ECL2, thereby preventing viral entry (9). A model for the gp120-CCR5 coreceptor interaction, which involves ECL2 binding to the V3 region gp120 has been proposed (11, 29); however, there is no evidence for this direct interaction (30).
A synthetic 24-residue peptide corresponding to CCR5 ECL2168–191 (peptide 1, see Fig. 1B) has been shown to inhibit infection of R5 HIV-1 strains in cell fusion assays and block the interaction of a soluble CD4-gp120 (R5) complex with CCR5 expressing cells (9). Using this peptide as a starting point, we sought to delineate the regions of CCR5 ECL2 responsible for HIV inhibition by synthesizing various ECL2 peptide constructs and evaluating their ability to inhibit viral entry. When designing constructs we were mindful of the presence of a conserved disulfide bond present in chemokine receptors as well as most other class A G protein-coupled receptors that connects ECL2 with the extracellular end of transmembrane 3 (TM3). In CCR5, this disulfide bond occurs between TM3 Cys-101 and ECL2 Cys-178. Although not essential for mediating HIV entry (31), Cys-178 was of interest because it is located in the approximate center of ECL2 and could thus be envisioned to naturally segment this extracellular region into two structural units, namely N- and C-terminal portions (see Fig. 1B). We considered that synthesizing and evaluating these peptides independently would help narrow down the ECL2 epitopes required for HIV inhibition. In addition, constructing complementary N- and C-terminally truncated peptides would further reveal which residues on CCR5 ECL2 interact with gp120.
Here, we report the results of structure-function studies to identify the CCR5 ECL2 residues required for inhibition of HIV entry. Through screening of ECL2 truncated peptides in viral neutralization and fusion assays, we demonstrate that CCR5 residues C-terminal to Cys-178 inhibit entry of both R5 and X4 strains. Furthermore, we use co-operative infectivity assays to show that these peptides can function independently of CD4 and are non-competitive with CCR5 N terminus. We use saturation transfer difference NMR (STD NMR) experiments to validate results from our cellular assays and identify the CCR5 ECL2 residues that bind to gp120. Our results suggest the existence of a site on gp120 that is conserved across R5 and X4 strains, which binds the C-terminal end of CCR5 ECL2.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Characterization
All CCR5 ECL2 peptides were synthesized on a Liberty microwave peptide synthesizer (CEM Corp., Matthews, NC) by Fmoc (N-(9-fluorenyl)methoxycarbonyl) synthesis using CLEAR amide resins (Peptides Intl., Lexington, KY) and capping with acetic anhydride after each coupling. Peptides were cleaved from the resin by treatment with TFA:H2O:triisopropylsilane:DTT (95:2.5:1.25:1.25) for 2.5 h at room temperature and precipitated with ice-cold methyl t-butyl ether and centrifuged at 0 °C. Crude precipitated peptides were suspended in H2O and purified by RP-HPLC using a preparative Symmetry Shield RP18 column (Waters, Milford, MA) with 0.1% aqueous TFA and MeOH as eluents. Purified peptide solutions were lyophilized, and their purity (>95%) and compositions were verified by analytical HPLC and ESI-HRMS. For use in neutralization assays, fresh solutions of peptides were prepared immediately before use by dissolving lyophilized peptides in a buffer containing 20 mm Na2PO4, 70 mm NaCl, pH 6.8 (referred to as NMR buffer) to yield a final concentration of 2 mm. The pH of each solution was measured using a microelectrode and if necessary adjusted by titrating 0.1 mm aqueous NaOH or 0.1 mm aqueous HCl to yield a final pH of 6.8. At this pH, we did not observe oxidation to the dimeric form of the Cys-containing peptides as detected by HR-MS. The CCR5 N-terminal peptide comprising residues 2–18 was synthesized using previously reported procedures (20).
Generation of HIV-1 ΔV1V2 gp120 Envelope Glycoproteins
Mammalian codon-optimized genes encoding HIV-1 ΔV1V2 gp120 envelope glycoproteins from strains YU2, HxB2, and 89.6 were synthesized and cloned into mammalian expression vector pcDNA2.1 (Invitrogen), followed by transfection into FreeStyle 293F cells as described previously (32). Detailed protocols are provided in the supplemental data. After 5 days of suspension culture post-transfection, the supernatants were harvested by centrifugation, filtered through a 0.22-μm filter, and purified through an affinity column of 17b. Purified proteins were concentrated and dialyzed against PBS and characterized with SDS-PAGE.
NMR Sample Preparation
Solutions containing free peptide (800 μm), or complexes of peptide (800 μm) in the presence of gp120 (15 μm) or a stoichiometric complex of gp120-CD4 (15 μm) were prepared in 20 mm Na2PO4, 70 mm NaCl buffer, pH 6.8, with 10% D2O, and the pH of each solution was adjusted with 0.1 mm aqueous NaOH or 0.1 mm aqueous HCl. All peptides used in this study were unlabeled, with 13C present at natural abundance. Proteins were subjected to a second purification step using a Superdex 75 (16 × 600 mm) column eluting with NMR buffer. Sample concentrations were quantitated by A280 measurements.
NMR Spectroscopy
NMR data were acquired at 298 K on a Bruker Avance 600 MHz spectrometer equipped with a z-shielded gradient triple resonance cryoprobe. Water suppression was carried out using a 3-9-19 WATERGATE sequence. 1H STD NMR experiments were recorded as described previously (24, 33). Spectra were acquired with 3,072 scans, 32,768 points, and a relaxation delay of 2 s, with on- and off-resonance carrier frequencies set at −1.5 and 50 ppm, respectively. Protein saturation (1.8 s) was accomplished using a train of 50-ms Gaussian pulses followed by 1-ms delays. 1H-13C HSQC STD NMR experiments were recorded with 512 scans per increment, saturation and relaxation times of 1.5 s each, and 128 × 1024 complex points in the F1 and F2 dimensions, respectively. Difference spectra were obtained by subtracting on- from off-resonance spectra. NOESY spectra were recorded with 64 scans per increment, a τm of 150 ms, a relaxation delay of 2 s, and 512 and 1024 complex points in the F1 and F2 dimensions, respectively. 1H and 13C chemical shifts of 2C (supplemental Table S1) were assigned using standard homonuclear and heteronuclear two-dimensional NMR experiments. Data were processed using TOPSPIN (version 2.1), and peaks were integrated using NMRDraw (34) or PIPP (35). Relative intensities were obtained by normalizing peak integrals to the strongest signal/s in each spectrum. HR-MS spectra of NMR solutions containing 2C showed no evidence of oxidation to the disulfide for periods as long as 2 weeks.
Circular Dichroism
CD spectra were recorded on a Jasco J-815 CD spectrometer using a quartz cell with an optical path of 1.0 mm on 50 μm solutions of peptides in H2O or 50% trifluoroethanol/H2O at 4 °C. Three scans were performed from 190 to 260 nm at a rate of 20 nm/min, with a 1-nm bandwidth, 2 s response, and resolution of 0.2 nm. The percentage of α-helical secondary structure was calculated with the program k2d2 (36).
HIV-1 Neutralization and Fusion Assays
Env-pseudotyped HIV neutralization assays were performed as described previously (37) using viral particles pseudotyped with HIV-1 envelopes (38). Serial dilutions of inhibitors were added to pseudovirus, followed by TZM-bl cells (which express CD4, CXCR4, and CCR5) at 37 °C. Forty-eight h post-infection, cells were lysed, and luciferase activity was measured. Representative inhibition curves are shown in supplemental Fig. S1. Post attachment assays used to determine the effect on inhibition by ECL2 peptides following CD4 binding to viral envelope were performed using synchronized infections temperature-arrested at the CD4-bound state (39, 40). HIV-1 envelope-mediated cell fusion assays were performed as described previously (41). A full list of reagents and cell lines is provided in the supplemental data.
Synergy Experiments
Neutralizing activities of multiple constant-ratio combinations of CCR5 ECL2 peptides, the small molecule CD4 mimetic NBD-556, soluble CD4 (sCD4), and the CCR5 N terminus were tested in serial dilutions in HXB2 Env-pseudotyped HIV neutralization assays. Combination effects were analyzed as described (42, 43). The dose reduction index (DRI) of inhibitor x in combination with inhibitor y is given by DRIx = (IC50)x/(IC50)x,y, where (IC50)x and (IC50)x,y are the IC50 values of x alone and in combination with y, respectively. The combination index (CI) describing the summation of effects of two inhibitors is given by CI = (DRIx)−1 + (DRIy)−1 + (DRIxDRIy)−1, where the last term, which makes a small contribution to CI, accounts for the state where both inhibitors are bound (43).
RESULTS
C-terminal Region of CCR5 ECL2 Comprising Cys-178–Lys-191 Is Sufficient for Inhibition of HIV Entry by R5 and X4 Strains
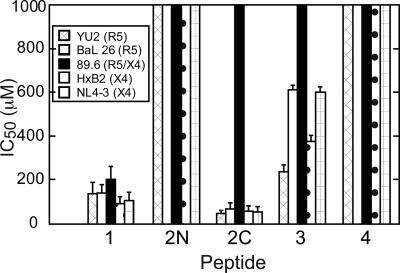
A panel of peptides corresponding to the second extracellular region (ECL2) of CCR5 was assessed for their ability to neutralize HIV-1 entry in single round infectivity assays using viral particles psuedotyped with CXCR4- and CCR5-using HIV-1 envelopes (Table 1). TZM-bl cells expressing CD4, CCR5, and CXCR4 were used as target cells. Peptide 1, comprising full-length ECL2, inhibited R5 strains YU2 and Bal26 in a dose-dependent manner with IC50 values of ∼140 μm (Table 1, see Fig. 2), a result expected for this CCR5-derived peptide. Neutralization assays also showed that peptide 1 inhibited viral entry of X4 strains HxB2 and NL4–3 with comparable potencies (IC50 values of 90–100 μm) to those observed for R5 strains (Fig. 1 and supplemental Table S1). Peptide 1 also inhibited the dual tropic strain 89.6 although with 2-fold weaker potency compared with the X4 and R5 strains (Table 1 and Fig. 2). To narrow down the region of ECL2 required for neutralization, truncations of peptide 1 lacking two and four amino acids from both the N and C termini were synthesized to give peptides 3 and 4, respectively (Fig. 1B). Peptide 3 inhibited viral cell entry in a dose-dependent manner but showed dramatically reduced potency across all strains with IC50 values >200 μm and as high as 600 μm (Table 1). Peptide 4 was inactive toward all strains tested.
TABLE 1.
IC50 values (μm) for CCR5 ECL2 peptides in neutralization assays
| Peptide | R5a |
R5/X4a (89.6)b | X4a |
||
|---|---|---|---|---|---|
| YU2b | BaL26b | HxB2b | NL4-3b | ||
| 1 | 136 ± 49 | 138 ± 36 | 203 ± 54 | 89 ± 31 | 103 ± 37 |
| 2N | NAc | NA | NA | NA | NA |
| 2C | 28 ± 7 | 65 ± 3 | NA | 54 ± 2 | 53 ± 2 |
| 3 | 237 ± 31 | 612 ± 21 | NA | 374 ± 28 | 600 ± 24 |
| 4 | NA | NA | NA | NA | NA |
a Virus tropism. R5 and X4 refer to CCR5- and CXCR4-using HIV-1 strains, respectively, and R5/X4 refers to dual tropic strains. Neutralization assays were performed as described previously (37).
b HIV strain.
c Not active, NA.
FIGURE 2.
Relative IC50 values of the extracellular ECL2 peptides (Fig. 1B) for inhibition of viral entry of R5, X4, and dual tropic HIV-1 strains. Numerical values are provided in Table 1, and representative inhibition curves are shown in supplemental Fig. S1. The legend for the HIV-1 strains used is shown in the inset in each graph. A dramatic reduction in activity can be seen readily for peptides 2N, 3, and 4.
Using Cys-178 as a midpoint, we next dissected peptide 1 into two segments, giving the N-terminal peptide 2N and the C-terminal peptide 2C. Surprisingly, 2C not only retained potency but was more potent than full-length ECL2 (peptide 1) across all X4 and R5 strains tested. 2C, however, was inactive against the dual tropic strain 89.6. Peptide 2N, on the other hand, was inactive against all five strains, including R5, X4, and one dual tropic strain (Table 1 and Fig. 2). Thus, the C-terminal segment of ECL2 alone is sufficient for inhibiting HIV-1 entry of R5 and X4 strains.
To test whether the presence of the cysteine sulfhydryl group contributed to the inhibitory activity of these peptides, we synthesized and tested two additional 2C analogs that lacked a free thiol. These included C178A and a Cys-S-acetamidomethyl variant in which the sulfur atom is protected by an acetamidomethyl group. Each of these peptides inhibited HIV-1 entry in a dose-dependent manner with similar potencies (supplemental Fig. S2), suggesting the Cys side chain is not critical for inhibitory activity. We tested several of the CCR5 ECL2 peptides against two amphotropic enveloped viruses, murine leukemia virus (MLV) and vesicular stomatitis virus. None of the peptides inhibited either of these viruses demonstrating specificity of the ECL2 peptides for HIV-1.
Activity of ECL2 2C-TM5 Peptides
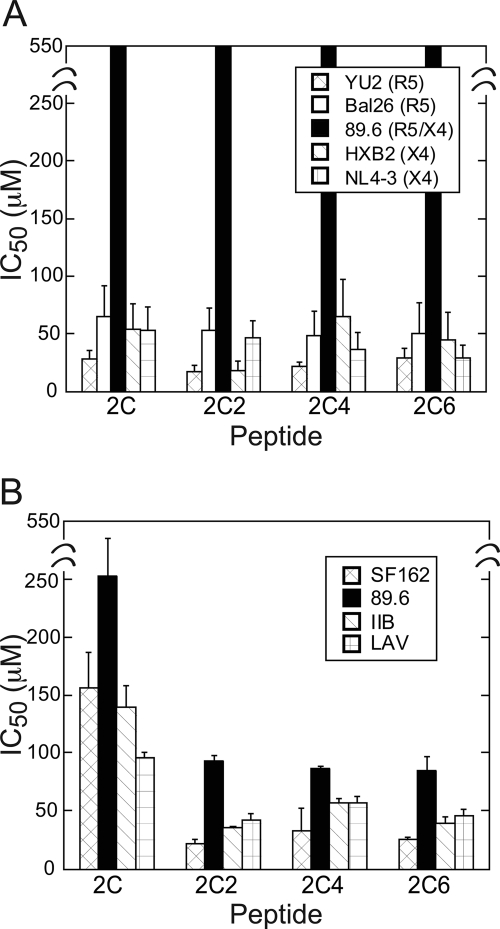
The C-terminal end of CCR5 ECL2 shown to be critical for inhibition of HIV entry directly precedes TM5. To determine whether residues extending into TM5 might contribute to HIV gp120-CCR5 binding, we used 2C as a starting point to synthesize a panel of two-residue incremented peptides following the natural sequence of CCR5 (Fig. 1C). To assess inhibition of HIV-1 entry by these 2C-TM5 peptides, both viral neutralization (38) and HIV envelope-mediated cell-cell fusion (41) assays were performed. In neutralization assays, the 2C-TM5 peptides performed similarly to 2C across both R5 and X4 strains (Fig. 3A and supplemental Table S2) with IC50 values averaging ∼20–60 μm. Following a similar trend, in the cell-cell fusion assays 2C was found to inhibit the R5 strain SF162 and X4 strains III-B and LAV, albeit with decreased potencies and IC50 values around 100–150 μm (Fig. 3B and supplemental Table S3). In contrast to the neutralization assays, however, 2C was mildly inhibitory in the cell fusion assays toward the dual tropic strain 89.6 with an IC50 value of 250 ± 30 μm. Interestingly, in the cell-cell fusion assays, 2C-TM5 peptides were found to be significantly more potent than 2C against all strains tested, with increases in potency ranging from 1.5–8× the IC50 values observed for 2C. A scrambled peptide 2C2S (Fig. 1C) was used as a control in both viral neutralization and cell-cell fusion assays and was found to be inactive against all strains tested in both assay formats.
FIGURE 3.
Histograms showing relative IC50 values for 2C-TM5 peptides in neutralization (A) and cell-cell fusion (B) assays. The legend for the HIV-1 strains used is shown in the inset in A and B. Numerical values are provided in supplemental Tables S2 and S3.
Inhibition of HIV-1 Entry by CCR5 ECL2 Peptides Occurs Independently of CD4 and CCR5 N Terminus
CD4 is the primary receptor used in HIV entry, and CD4 binding rearranges the viral envelope protein gp120 allowing for coreceptor binding. To investigate the effect of this conformational change on the ability of the C-terminal peptide 2C to inhibit HIV entry, we performed post-attachment neutralization assays as described previously (44). Briefly, virus was added to plated TZM-bl cells followed by spinoculation to aid CD4 attachment. Both steps are performed at 4 °C to prevent subsequent steps in viral entry from taking place. Unattached virus was removed and media that contained 2C at 37 °C was added to synchronize the start of infection. We found 2C to be equally potent in the post-attachment neutralization assay as in the standard neutralization assay against the R5 strain YU2 (Table 2).
TABLE 2.
IC50 values (μm) for ECL2-TM5 peptides in standard infectivity and post-attachment assays
Post-attachment assays were performed using synchronized viral infection, and temperature was arrested at 4 °C at the CD4-bound state (39, 40).
| Peptide | Post-attachment assay | Standard assay |
|---|---|---|
| 2C | 49 ± 10 | 28 ± 7 |
| 2C2 | 20 ± 7 | 17 ± 5 |
| 2C4 | 23 ± 6 | 21 ± 4 |
| 2C6 | 21 ± 8 | 29 ± 7 |
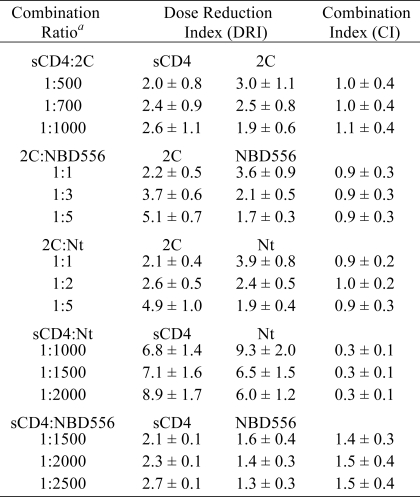
This result prompted us to test for cooperative inhibition of HIV entry between 2C and two CD4 mimetics, sCD4, and the small molecule NBD-556, both of which have been shown to induce conformational changes on gp120 similar to those induced by cell surface CD4 and to inhibit HIV-1 entry (45–48). We tested the inhibitors individually and in constant ratio combinations for their ability to inhibit YU2 HIV-1. Data were quantitatively analyzed using CI. In this method, CI values that are equal to, greater than, or less than 1 are indicative of additive, antagonistic, and synergistic effects, respectively (42, 49). The results are summarized in Table 3. When 2C was tested in combination with either sCD4 or NBD-556, CI values of 0.9–1.1 were obtained, indicating ECL2 2C is additive with each of these CD4 mimetics, and together, they inhibit HIV entry in a non-cooperative and non-competitive manner. In contrast, the CCR5 N terminus was strongly synergistic when combined with sCD4 showing a mean CI of 0.3 more than three different constant combination ratios (Table 3). We used the same method to determine the mode of inhibition of N terminus in combination with 2C. As seen for the CD4 mimetics, average CIs of 0.9–1 for the N-terminal peptide and ECL2 were obtained across three different concentration ratios indicating that these two extracellular CCR5 peptides inhibit HIV-1 entry in an additive manner (Table 2). As a control to test for antagonism in the neutralization assay, we tested NBD556 and sCD4 in combination because they should compete with one another for the CD4 binding site on gp120. Indeed, an average CI of 1.5 was measured, indicative of antagonism between these two CD4 mimetic inhibitors.
TABLE 3.
Activity of combinations of CCR5 2C, CCR5 N terminus, and CD4 mimetics sCD4 and NBD 556
All experiments were performed as described previously using HIV YU2 Env pseudotypes. The DRI is the ratio of the IC50 in the absence and presence of the second inhibitor. IC50 values for each of the four inhibitors on their own, measured in parallel with the combination experiments, were as follows: 2C; 27 ± 4 μm; sCD4, 37 ± 9 nm; NBD566, 45 ± 8 μm; CCR5 Nt, 51 ± 7 μm. The CI represents the combined effect of two inhibitors in combination where CI values of <1, 1; and >1 correspond to synergistic, additive, and antagonistic effects, respectively (42, 43). Nt, N terminus.
a The ratio of inhibitor concentrations approximates the ratio of the IC50 values of the inhibitors alone.
Mapping gp120 Binding Site on Peptide 2C Using STD NMR
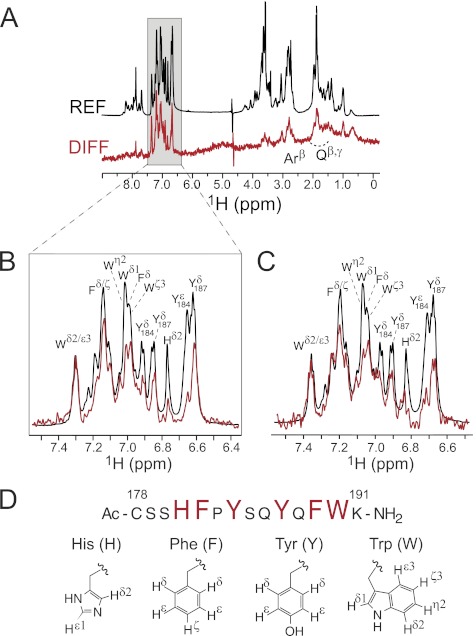
To identify which amino acid residues on 2C are critical for binding to HIV gp120, we used one-dimensional and two-dimensional STD NMR. STD NMR is a powerful technique employed to characterize the binding surface used by a ligand when binding its macromolecular receptor. It is especially useful because it can be used to characterize binding of different classes of ligands, including small molecules, peptides and carbohydrates, which have a wide range of Kd values from nm to mm (33), and does not require a 15N- or 13C-labeled ligand or receptor. When presented as a difference spectrum, signals displaying the highest intensity are in closest proximity to the receptor, allowing for a detailed description of the binding epitope used by the ligand. Using standard two-dimensional homonuclear and heteronuclear NMR experiments, we made full chemical shift assignments for 2C (supplemental Table S1), and recorded one-dimensional STD NMR spectra of 2C in the presence of YU2 g120 (Fig. 4, A and B) and HxB2 gp120 (Fig. 4C). The strongest signals observed in the difference spectra corresponded to the aromatic side chains of Phe-182, Tyr-184, Tyr-187, Phe-189, and Trp-190, and to a lesser extent His-181. STD enhancements were also observed for β- and γ-protons of Gln-186 and Gln-188, and the β-protons of several of the aromatic amino acids, but these were much weaker in comparison (Fig. 4A). Thus, interactions with gp120 and ECL2 peptide 2C are mediated primarily through the side chains of aromatic residues. Of note, the STD NMR spectra for 2C in the presence of gp120 from both R5 and X4 strains appeared very similar, consistent with its neutralization profiles against R5 and X4 strains.
FIGURE 4.
STD NMR 1H spectra of peptide 2C in the presence of soluble gp120s. A, difference (DIFF) and reference (REF) spectra for 2C in the presence of R5 strain YU2. Expansions of the aromatic regions of 2C in complex with YU2 and X4 strain HXB2 are shown in B and C, respectively. Reference and difference spectra are colored black and red, respectively. Proton signals showing enhancements and therefore involved in gp120 binding are labeled. Weak enhancements from the β- and γ-protons of Gln and the β-protons of several of the aromatic amino acids appear as an envelope in the full spectra and are labeled collectively as Qβ/γ and Arβ. Spectra were normalized to the Trp ϵ3/ζ2 signal. D, amino acid sequence of 2C with residues involved in binding displayed as large red letters.
Knowing that CD4 binding induces conformational rearrangements in gp120 to give rise to a CD4-activated conformation that is necessary for binding to CCR5, we investigated the binding of 2C to a CD4-YU2 gp120 complex. This was prepared by combining soluble YU2 gp120 with a 0.5-fold excess of two-domain CD4 and separating the complex and CD4 by gel filtration chromatography as described previously (24). NMR spectra were recorded on solutions containing 2C in the presence of the CD4-YU2 gp120 complex. As seen in supplemental Fig. S3, the STD NMR spectrum for this sample appeared very similar to that of 2C in the presence of YU2 gp120 alone. Although surprising at first, these independent NMR data are entirely consistent with neutralization data from our combination experiments where ECL2 and sCD4, or ECL2 and NBD-556, inhibited HIV entry in an additive manner. Furthermore, the STD NMR spectrum of a sample containing 2C and 89.6 gp120 was devoid of signals (supplemental Fig. S4), in agreement with our observation that 2C does not inhibit 89.6 in neutralization assays.
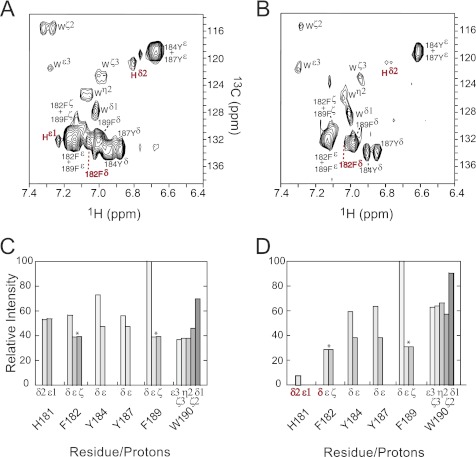
Analysis of the one-dimensional STD NMR spectra revealed which residues are involved in 2C binding to gp120, but due to spectral overlap among 1H chemical shifts, definitive assignments for a number of the protons could not be made. We resolved this issue by recording two-dimensional 1H-13C HSQC STD NMR spectra of both complexes. Spectra for 2C:YU2 gp120 and 2C:HxB2 gp120 are shown in Fig. 5, A and B, respectively. These data allowed for a detailed assessment of the contributions of each proton involved in gp120 binding. Cross-peak intensities were normalized relative to the strongest cross-peak in each of the spectra (maximum peak volume) to give a percent enhancement for each proton involved in gp120 binding as seen in Fig. 5, C and D. As in the one-dimensional spectra, the strongest signals in the HSQC difference spectra corresponded to the side chains of many (but not all) of the aromatic residues. The two-dimensional HSQC spectra revealed that Phe-189 Hδ displayed the strongest enhancements for both R5 and X4 peptide complexes, a result that was not interpretable from the one-dimensional spectra due to overlap of 1H signals. The one-dimensional STD spectra indicated similar contact trends across peptide 2C for both gp120 strains. However, several differences can be seen for the two complexes in the two-dimensional data sets. Although Phe-182 Hδ shows a strong enhancement (57%) for YU2 gp120 (R5), this signal is not seen in the complex with X4-using strain HxB2 (Fig. 5B). Also, STD intensities for Trp-190 range from 37–69% for YU2 (R5) compared with 63% to ∼100% in spectra for HxB2, indicating that Trp-190 is in closer contact to gp120 of HXB2 than for YU2. This finding agrees with neutralization results where the loss of Trp-190 in peptide 3 had a stronger deleterious effect for inhibition of HxB2 compared with YU2.
FIGURE 5.
Two-dimensional 1H-13C HSQC STD NMR spectra for 2C in the presence of soluble gp120 from YU2 (A) and HXB2 (B) HIV-1 strains. Cross-peaks are labeled according to Fig. 4C. Histograms showing the relative peak intensities (normalized to Hδ of Phe-189) for individual aromatic side chain protons involved in binding to gp120s from YU2 (C) and HXB2 (D) strains. Peak intensities were obtained by integrating peak volumes using the program PIPP (35) and correspond to cross-peaks within individual spectra. Peaks that remained overlapping in the two-dimensional spectra are labeled as pairs in A and B; and the average of their combined intensities is plotted in C and D and denoted with asterisks. Cross-peaks for His-181 and Phe-182 are labeled red in C and D to point out the changes in intensity when binding YU2 and HXB2. All spectra were recorded on solutions containing unlabeled, synthetic peptides.
Conformational Analysis of Peptide 2C in Complex with gp120
To study the conformation of 2C when bound to gp120, we recorded two-dimensional NOESY spectra on samples containing free peptide (800 μm) or peptide in the presence of gp120 (50-fold excess). In addition to confirming binding, the pattern of NOEs observed in a NOESY spectrum provides secondary structural information for the bound ligand. As seen in supplemental Fig. 6A, few cross-peaks are observed in the NOESY spectrum of free 2C recorded with a mixing time of 150 ms (due to its molecular mass of ∼1800 Da, falling in the range where NOE values are small or close to zero (50). In the presence of gp120, the transferred NOESY spectrum of 2C contains many more cross-peaks than that of the free peptide, and nearly all predicted intraresidue CαH/CβH(i)—NH(i) NOEs are observed (Fig. 6). Several inter-residue correlations were also observed, but each of these corresponded to sequential CαH(i)—NH(i + 1) NOEs rather than any long range NOEs that would be indicative of secondary structure. We used circular dichroism spectroscopy to assess the conformation of 2C in solution. As in the bound state, CD spectra recorded on free 2C in water or 50% trifluoroethanol showed no evidence of regular secondary structure (supplemental Fig. S5). Our combined data therefore suggest that the C-terminal ECL2 peptide 2C lacks regular secondary structure and binds gp120 in an extended or coil-like conformation.
FIGURE 6.
NOESY spectra of ECL2 peptide 2C. A, NOESY spectrum of free 2C. B, transferred NOESY spectrum of 2C in the presence of YU2 gp120. C, expansion of and assignments for cross-peaks in the NH-CαH/CβH region of the spectrum for 2C in complex with gp120. In spectra for the complex, only intraresidue (black labels) and sequential (i to (i+1)) (red labels) correlations were observed; no long range correlations were detected. Spectra were recorded on samples prepared under identical conditions (with concentrations of 800 μm peptide for free 2C, and 800 μm peptide/15 μm gp120 for the complex) and with identical acquisition parameters at 600 MHz; τm = 150 ms.
DISCUSSION
In this study, we used rationally selected synthetic peptides bearing different sequences derived from ECL2 of CCR5 to define the regions critical for binding to gp120 and inhibiting HIV entry. It was shown previously that a synthetic full-length ECL2 peptide could inhibit HIV entry of R5, but not X4 or R5X4, strains in cell-cell fusion assays at 50 μg/ml, corresponding to ∼15 μm peptide (51). In single round infectivity assays employing five different HIV isolates, we found that full-length ECL2 (1) not only inhibited R5 viruses with IC50 values around 100 μm, but 1 also inhibited X4 isolates and the dual tropic strain 89.6 with similar potency. Equally surprising, we found that a peptide comprising only the C-terminal portion of ECL2 (2C) was sufficient for inhibiting both R5 and X4 strains in neutralization and cell-cell fusion assays, and was two to five times more potent than full-length ECL2. The complete lack of inhibition by the N-terminal peptide 2N against all strains, combined with the dramatic reduction or complete loss of inhibitory activity observed with peptides 3 and 4, respectively, revealed the importance of the last four amino acids, QFWK, for inhibition of viral entry. From a structural perspective, the neutralization data suggest that the C- and N-terminal regions of ECL2 likely form separate loops within the receptor that is facilitated by the presence of the conserved disulfide bond between Cys -178 of ECL2 and Cys-101 at the top of TM3 and that the C-terminal portion is recognized by a conserved binding site on gp120 that is present in both R5 and X4 strains. Our results corroborate an elegant study by Potnow and Ratner (52) where they showed that the CXCR4-using virus HXB2 could gain entry into cells expressing a CXCR4 receptor mutated to possess the ECL2 sequence of CCR5. From their results, they proposed that CCR5 contains elements that support usage by X4 viral strains and suggested that the gp120 interaction sites of CCR5 and CXCR4 are related structurally (52).
It is known that binding to coreceptor by gp120 requires CD4 activation. Thus, most biochemical and structural studies with the CCR5 N terminus utilize a gp120-CD4 complex (19). On this basis, we did not anticipate finding that CCR5-derived 2C can interact with gp120 in the absence of CD4. This CD4 independence was established through three complimentary sets of experiments. First, when 2C was added in combination with sCD4, neutralization assays showed that the two entry inhibitors acted in an additive manner rather than synergistically as was observed for the CCR5 N terminus and sCD4 (Table 3). Second, in post attachment assays where 2C was added subsequent to gp120-CD4 engagement, no change in potency was observed indicating that the conformational change associated with CD4 binding to fusion competent HIV envelope does not affect inhibition by 2C. This is in contrast to other fusion inhibitors where CD4 engagement affects potency (44, 53). Last, we definitively show by NMR that 2C binds an R5 and an X4 strain of soluble gp120 in the absence of CD4 and that the binding epitope used by the peptide to bind gp120 does not differ whether free gp120 or a gp120-CD4 complex is present. We note that in the NMR experiments reported here gp120 constructs that lack the V1/V2 loop were used; however, results from the synergy and post-attachment neutralization assays, which must employ the fusion competent form of gp120, are consistent with the results of our NMR studies, all of which point to the ability of 2C to bind non-CD4-activated gp120.
NMR data provided a detailed view of how the C-terminal region of CCR5 ECL2 interacts with gp120. When binding gp120, 2C relies on the side chains of its aromatic amino acids, including Phe, Tyr, and Trp, and to a much lesser extent His. Slight differences are seen in the extent of these interactions among the R5 and X4 isolates. In interactions with the R5 virus YU2, all of the aromatic amino acids show strong STD enhancements indicating that a substantial portion of 2C is involved in binding. In interactions with the X4 virus HxB2, however, the binding surface shifts toward the C-terminal end of 2C, with interactions involving His-181 and Phe-182 gone or greatly diminished and those with Trp-190 having increased. Given the reduction in inhibitory activity observed with peptide 3, which lacks Trp-190, it is possible that the region of the coreceptor binding site on gp120 that recognizes the C-terminal portion of 2C is highly conserved among X4 and R5 strains. The absence of binding to, and inhibition of, 89.6 by 2C suggest that the coreceptor binding site of dual tropic strains on the other hand may require more extensive contacts with ECL2 that would include both the N- and C-terminal portions of this loop. It is important to point out that our results do not preclude the possibility that the N-terminal region of ECL2 (2N) contacts the coreceptor binding site on gp120 from R5 and X4 envelopes; rather, they demonstrate that this region is not necessary for inhibition of viral entry.
With regard to secondary structural features of 2C, although crystallography has shown that ECL2s in some G protein-coupled receptors form β-hairpins (54, 55) or contain α-helical regions (56), our NMR (transferred NOESY) data indicate that 2C binds gp120 in an extended conformation, lacking regular secondary structure even in the bound state. Although peptide 2N does not inhibit HIV entry, it will be of interest to learn whether this region is structured within the full receptor.
In conclusion, we have identified synthetic peptides derived from the C-terminal portion of CCR5 ECL2 as inhibitors of HIV-1 entry. HIV infectivity assays and STD NMR show that these peptides inhibit viral fusion by binding to gp120 irrespective of virus tropism. The finding that these peptides bind gp120 and inhibit viral entry independent of CD4 engagement and in a non-competitive and non-synergistic manner relative to the CCR5 N terminus indicates that gp120 must have a separate binding site that recognizes ECL2, that this site is present both in the absence and presence of CD4, and is conserved among X4 and R5 strains. It is remarkable that CXCR4-using strains that do not require binding to CCR5 for entry can nevertheless recognize part of its extracellular region. These peptides should make valuable probes to study and expand understanding of conserved binding sites of gp120 and serve as a template for the design of molecules for broad spectrum inhibition of HIV-1 entry.
Supplementary Material
Acknowledgments
We thank J. R. Lloyd for HR-MS data and H. Baker for technical assistance.
This work was supported by the Intramural Research Program, National Institutes of Health (NIDDK and NIAID), and the Intramural AIDS Targeted Antiviral Program, Office of the Director, National Institutes of Health (to C. A. B. and G. M. C.).

This article contains supplemental “Experimental Procedures,” Figs. S1–S5, and additional references.
- CCR5
- C-C chemokine receptor type 5
- CXCR4
- C-X-C chemokine receptor type 4
- R5
- CCR5-using
- X4
- CXCR4-using
- X4/R5
- dual tropic
- STD NMR
- saturation transfer difference NMR
- ECL2
- second extracellular loop
- TM
- transmembrane
- sCD4
- soluble CD4
- DRI
- dose reduction index
- CI
- combination index.
REFERENCES
- 1. Zhang Y. J., Moore J. P. (1999) Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Virol. 73, 3443–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Freed E. O., Martin M. A. (1995) The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270, 23883–23886 [DOI] [PubMed] [Google Scholar]
- 3. Moore J. P., Trkola A., Dragic T. (1997) Co-receptors for HIV-1 entry. Curr. Opin. Immunol. 9, 551–562 [DOI] [PubMed] [Google Scholar]
- 4. Eckert D. M., Kim P. S. (2001) Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 [DOI] [PubMed] [Google Scholar]
- 5. Trkola A., Dragic T., Arthos J., Binley J. M., Olson W. C., Allaway G. P., Cheng-Mayer C., Robinson J., Maddon P. J., Moore J. P. (1996) CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384, 184–187 [DOI] [PubMed] [Google Scholar]
- 6. Wu L., Gerard N. P., Wyatt R., Choe H., Parolin C., Ruffing N., Borsetti A., Cardoso A. A., Desjardin E., Newman W., Gerard C., Sodroski J. (1996) CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384, 179–183 [DOI] [PubMed] [Google Scholar]
- 7. Chan D. C., Kim P. S. (1998) HIV entry and its inhibition. Cell 93, 681–684 [DOI] [PubMed] [Google Scholar]
- 8. Berger E. A., Doms R. W., Fenyö E. M., Korber B. T., Littman D. R., Moore J. P., Sattentau Q. J., Schuitemaker H., Sodroski J., Weiss R. A. (1998) A new classification for HIV-1. Nature 391, 240. [DOI] [PubMed] [Google Scholar]
- 9. Maeda K., Das D., Yin P. D., Tsuchiya K., Ogata-Aoki H., Nakata H., Norman R. B., Hackney L. A., Takaoka Y., Mitsuya H. (2008) Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: Insights into the mechanism of allosteric inhibition. J. Mol. Biol. 381, 956–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Da L. T., Wu Y. D. (2011) Theoretical studies on the interactions and interferences of HIV-1 glycoprotein gp120 and its coreceptor CCR5. J. Chem. Inf. Model. 51, 359–369 [DOI] [PubMed] [Google Scholar]
- 11. Cormier E. G., Tran D. N., Yukhayeva L., Olson W. C., Dragic T. (2001) Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75, 5541–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu L., LaRosa G., Kassam N., Gordon C. J., Heath H., Ruffing N., Chen H., Humblias J., Samson M., Parmentier M., Moore J. P., Mackay C. R. (1997) Interaction of chemokine receptor CCR5 with its ligands: Multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186, 1373–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsamis F., Gavrilov S., Kajumo F., Seibert C., Kuhmann S., Ketas T., Trkola A., Palani A., Clader J. W., Tagat J. R., McCombie S., Baroudy B., Moore J. P., Sakmar T. P., Dragic T. (2003) Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77, 5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maeda K., Das D., Ogata-Aoki H., Nakata H., Miyakawa T., Tojo Y., Norman R., Takaoka Y., Ding J., Arnold G. F., Arnold E., Mitsuya H. (2006) Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 281, 12688–12698 [DOI] [PubMed] [Google Scholar]
- 15. Farzan M., Mirzabekov T., Kolchinsky P., Wyatt R., Cayabyab M., Gerard N. P., Gerard C., Sodroski J., Choe H. (1999) Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 [DOI] [PubMed] [Google Scholar]
- 16. Cormier E. G., Persuh M., Thompson D. A., Lin S. W., Sakmar T. P., Olson W. C., Dragic T. (2000) Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U.S.A. 97, 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farzan M., Vasilieva N., Schnitzler C. E., Chung S., Robinson J., Gerard N. P., Gerard C., Choe H., Sodroski J. (2000) A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275, 33516–33521 [DOI] [PubMed] [Google Scholar]
- 18. Farzan M., Chung S., Li W., Vasilieva N., Wright P. L., Schnitzler C. E., Marchione R. J., Gerard C., Gerard N. P., Sodroski J., Choe H. (2002) Tyrosine-sulfated peptides functionally reconstitute a CCR5 variant lacking a critical amino-terminal region. J. Biol. Chem. 277, 40397–40402 [DOI] [PubMed] [Google Scholar]
- 19. Huang C. C., Lam S. N., Acharya P., Tang M., Xiang S. H., Hussan S. S., Stanfield R. L., Robinson J., Sodroski J., Wilson I. A., Wyatt R., Bewley C. A., Kwong P. D. (2007) Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317, 1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schnur E., Noah E., Ayzenshtat I., Sargsyan H., Inui T., Ding F. X., Arshava B., Sagi Y., Kessler N., Levy R., Scherf T., Naider F., Anglister J. (2011) The conformation and orientation of a 27-residue CCR5 peptide in a ternary complex with HIV-1 gp120 and a CD4-mimic peptide. J. Mol. Biol. 410, 778–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Acharya P., Dogo-Isonagie C., LaLonde J. M., Lam S. N., Leslie G. J., Louder M. K., Frye L. L., Debnath A. K., Greenwood J. R., Luongo T. S., Martin L., Watts K. S., Hoxie J. A., Mascola J. R., Bewley C. A., Kwong P. D. (2011) Structure-based identification and neutralization mechanism of tyrosine sulfate mimetics that inhibit HIV-1 entry. ACS Chem. Biol. 6, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konishi K., Ikeda K., Achiwa K., Hoshino H., Tanaka K. (2000) Synthesis of peptides mimicking chemokine receptor CCR5 and their inhibitory effects against HIV-1 infection. Chem. Pharm. Bull. 48, 308–309 [DOI] [PubMed] [Google Scholar]
- 23. Tarasova N. I., Rice W. G., Michejda C. J. (1999) Inhibition of G protein-coupled receptor function by disruption of transmembrane domain interactions. J. Biol. Chem. 274, 34911–34915 [DOI] [PubMed] [Google Scholar]
- 24. Lam S. N., Acharya P., Wyatt R., Kwong P. D., Bewley C. A. (2008) Tyrosine-sulfate isosteres of CCR5 N-terminus as tools for studying HIV-1 entry. Bioorg. Med. Chem. 16, 10113–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doranz B. J., Lu Z. H., Rucker J., Zhang T. Y., Sharron M., Cen Y. H., Wang Z. X., Guo H. H., Du J. G., Accavitti M. A., Doms R. W., Peiper S. C. (1997) Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71, 6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross T. M., Bieniasz P. D., Cullen B. R. (1998) Multiple residues contribute to the inability of murine CCR-5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J. Virol. 72, 1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olson W. C., Rabut G. E., Nagashima K. A., Tran D. N., Anselma D. J., Monard S. P., Segal J. P., Thompson D. A., Kajumo F., Guo Y., Moore J. P., Maddon P. J., Dragic T. (1999) Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73, 4145–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Misumi S., Nakajima R., Takamune N., Shoji S. (2001) A cyclic dodecapeptide-multiple-antigen peptide conjugate from the undecapeptidyl arch (from Arg-168 to Cys-178) of extracellular loop 2 in CCR5 as a novel human immunodeficiency virus type 1 vaccine. J. Virol. 75, 11614–11620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cormier E. G., Dragic T. (2002) The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76, 8953–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hartley O., Klasse P. J., Sattentau Q. J., Moore J. P. (2005) V3: HIV's switch-hitter. AIDS Res. Hum. Retroviruses 21, 171–189 [DOI] [PubMed] [Google Scholar]
- 31. Blanpain C., Lee B., Vakili J., Doranz B. J., Govaerts C., Migeotte I., Sharron M., Dupriez V., Vassart G., Doms R. W., Parmentier M. (1999) Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J. Biol. Chem. 274, 18902–18908 [DOI] [PubMed] [Google Scholar]
- 32. Zhou T., Georgiev I., Wu X., Yang Z. Y., Dai K., Finzi A., Kwon Y. D., Scheid J. F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M. C., Sodroski J., Shapiro L., Nabel G. J., Mascola J. R., Kwong P. D. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayer M., Meyer B. (1999) Characterization of ligand binding by saturation transfer difference spectroscopy. Angew. Chem. Int. Ed. Engl. 38, 1784–1788 [DOI] [PubMed] [Google Scholar]
- 34. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) Nmrpipe - a multidimensional spectral processing system based on Unix pipes. J. Biol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 35. Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) A common sense approach to peak picking two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 36. Perez-Iratxeta C., Andrade-Navarro M. A. (2008) K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gustchina E., Louis J. M., Lam S. N., Bewley C. A., Clore G. M. (2007) A monoclonal Fab derived from a human nonimmune phage library reveals a new epitope on gp41 and neutralizes diverse human immunodeficiency virus type 1 strains. J. Virol. 81, 12946–12953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Doherty U., Swiggard W. J., Malim M. H. (2000) Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74, 10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reeves J. D., Gallo S. A., Ahmad N., Miamidian J. L., Harvey P. E., Sharron M., Pohlmann S., Sfakianos J. N., Derdeyn C. A., Blumenthal R., Hunter E., Doms R. W. (2002) Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U.S.A. 99, 16249–16254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nussbaum O., Broder C. C., Berger E. A. (1994) Fusogenic mechanisms of enveloped virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68, 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chou T. C., Talalay P. (1981) Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur. J. Biochem. 115, 207–216 [DOI] [PubMed] [Google Scholar]
- 43. Gustchina E., Louis J. M., Bewley C. A., Clore G. M. (2006) Synergistic inhibition of HIV-1 envelope-mediated membrane fusion by inhibitors targeting the N- and C-terminal heptad repeats of gp41. J. Mol. Biol. 364, 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gustchina E., Bewley C. A., Clore G. M. (2008) Sequestering of the prehairpin intermediate of gp41 by peptide N36Mut(e,g) potentiates the human immunodeficiency virus type 1 neutralizing activity of monoclonal antibodies directed against the N-terminal helical repeat of gp41. J. Virol. 82, 10032–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deen K. C., McDougal J. S., Inacker R., Folena-Wasserman G., Arthos J., Rosenberg J., Maddon P. J., Axel R., Sweet R. W. (1988) A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331, 82–84 [DOI] [PubMed] [Google Scholar]
- 46. Fisher R. A., Bertonis J. M., Meier W., Johnson V. A., Costopoulos D. S., Liu T., Tizard R., Walker B. D., Hirsch M. S., Schooley R. T. (1988) HIV infection is blocked in vitro by recombinant soluble CD4. Nature 331, 76–78 [DOI] [PubMed] [Google Scholar]
- 47. Zhao Q., Ma L., Jiang S., Lu H., Liu S., He Y., Strick N., Neamati N., Debnath A. K. (2005) Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 339, 213–225 [DOI] [PubMed] [Google Scholar]
- 48. Schön A., Madani N., Klein J. C., Hubicki A., Ng D., Yang X., Smith A. B., 3rd, Sodroski J., Freire E. (2006) Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45, 10973–10980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chou T. C., Talalay P. (1984) Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22, 27–55 [DOI] [PubMed] [Google Scholar]
- 50. Neuhaus D., Williamson M. (1989) The Nuclear Overhauser Effect in Structural and Conformational Analysis, 1st Ed., VCH Publishers, New York [Google Scholar]
- 51. Agrawal L., VanHorn-Ali Z., Berger E. A., Alkhatib G. (2004) Specific inhibition of HIV-1 coreceptor activity by synthetic peptides corresponding to the predicted extracellular loops of CCR5. Blood 103, 1211–1217 [DOI] [PubMed] [Google Scholar]
- 52. Pontow S., Ratner L. (2001) Evidence for common structural determinants of human immunodeficiency virus type 1 coreceptor activity provided through functional analysis of CCR5/CXCR4 chimeric coreceptors. J. Virol. 75, 11503–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shahzad-ul-Hussan S., Gustchina E., Ghirlando R., Clore G. M., Bewley C. A. (2011) Solution structure of the monovalent lectin microvirin in complex with Manα(1–2)Man provides a basis for anti-HIV activity with low toxicity. J. Biol. Chem. 286, 20788–20796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Structures of the CXCR4 chemokine G-protein-coupled receptor with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.