Background: A conformational change in A1 domain of vWF is required for GPIbα binding.
Results: N-terminal flanking region of the A1 domain inhibits binding to GPIbα, stabilizes the A1A2A3 structure, and prevents platelet activation.
Conclusion: Coupling between the N-terminal flanking region of the A1 domain and the A1A2A3 complex inhibits vWF-GPIbα interaction.
Significance: This autoinhibitory mechanism prevents spontaneous thrombosis in circulation.
Keywords: Calorimetry, Platelets, Protein Domains, Protein Folding, von Willebrand Factor
Abstract
von Willebrand factor (vWF) mediates platelet adhesion and thrombus formation via its interaction with the platelet receptor glycoprotein (GP)Ibα. We have analyzed two A1A2A3 tri-domain proteins to demonstrate that the amino acid sequence, Gln1238-Glu1260, in the N-terminal flanking region of the A1 domain, together with the association between the A domains, modulates vWF-GPIbα binding and platelet activation under shear stress. Using circular dichroism spectroscopy and differential scanning calorimetry, we have described that sequence Gln1238-Glu1260 stabilizes the structural conformation of the A1A2A3 tri-domain complex. The structural stabilization imparted by this particular region inhibits the binding capacity of the tri-domain protein for GPIbα. Deletion of this region causes a conformational change in the A1 domain that increases binding to GPIbα. Only the truncated protein was capable of effectively blocking ristocetin-induced platelet agglutination. To determine the capacity of activating platelets via the interaction with GPIbα, whole blood was incubated with the N-terminal region truncated or intact tri-A domain protein prior to perfusion over a fibrin(ogen)-coated surface. At a high shear rate of 1,500 s−1, platelets from blood containing the truncated protein rapidly bound, covering >90% of the fibrin(ogen) surface area, whereas the intact tri-A domain protein induced platelets to bind <10%. The results obtained in this study ascertain the relevant role of the structural association between the N-terminal flanking region of the A1 domain (amino acids Gln1238-Glu1260) and the A1A2A3 domain complex in preventing vWF to bind spontaneously to GPIbα in solution under high shear forces.
Introduction
von Willebrand factor (vWF),2 a multimeric plasma glycoprotein, mediates platelet adhesion at sites of vascular injury and contributes to the arrest of bleeding. It is also involved in pathologic thrombus formation in diseased vessels under elevated shear stress. vWF is essential for platelet adhesion because it captures circulating platelets through an interaction with the receptor glycoprotein (GP)Ib/IX/V complex, tethers them to subendothelial collagen, and forms the initial link toward the process of clotting (1). Additionally, the vWF-GPIbα interaction also plays a role in platelet activation and adhesion to deposited fibrin (2–4).
Within each monomer of this multimeric protein, vWF contains a triplicate repeat sequence of A domains in the central portion of the 2,050-residue mature subunit (D′-D3-A1-A2-A3-D4-B-C) (5–7). The A1 domain contains contact sites for the platelet GPIbα surface receptor, heparin, sulfatides, and collagen (8–11). Its homologous A3 domain binds only collagen (12, 13). The central A2 domain contains a proteolytic site for the metalloprotease ADAMTS-13 (14, 15). These A domains are relevant to the biology of vWF, and each of them has been recombinantly characterized in its isolated form (12, 16–18).
Normally, vWF does not interact with circulating platelets unless the conformation of the A1 domain of vWF has changed as a result of the influence of hydrodynamic forces (19). This conformational change can also be induced by naturally occurring gain-of-function mutations in the A1 domain that cause type 2B von Willebrand disease by increasing the binding affinity for the platelet GPIbα receptor (20). vWF activation can also be artificially induced with the modulator, ristocetin (21).
Like multimeric vWF, a recombinantly expressed tri-domain fragment of vWF, A1A2A3 (amino acids 1238–1874), also requires gain of function mutations, or ristocetin to induce its binding to platelet GPIbα (4). The similar functional characteristics between this tri-domain protein and full-length vWF indicate that the A1A2A3 segment is the smallest size of vWF that contains the regulatory mechanism for activation of vWF in solution.
The primary structure of the mature vWF protein describes the A1 domain as being flanked by the D′D3 domains in the N-terminal region, and those domains have been described to be involved in the mechanism that inhibits the vWF-GPIbα interaction by shielding the A1 domain (22). However, the binding site for GPIbα in the A1 domain remained cryptic in our A1A2A3 protein, which excludes the D′D3 domains (4). This outcome may indicate that the amino acid sequence 1238–1247 from the C terminus of the D3 domain forms part of the mechanism that masks the binding site for GPIbα. In fact, several reports have suggested that this N-terminal segment may be associated with the mechanism that regulates binding of vWF to GPIbα (23, 24).
Because we noticed that an N-terminally truncated tri-domain (amino acids 1261–1874) resulted in apparent binding activity for GPIbα higher than that of the longer construct (25), this study has used a number of biophysical and molecular biological methods to demonstrate that the sequence 1238–1260 stabilizes the tri-domain, inhibits binding to GPIbα, and prevents activation of platelets under shear flow. These results indicate that potential coupling between the N terminus-flanking region of the A1 domain and the A domain complex forms part of the regulatory mechanism of vWF activation.
EXPERIMENTAL PROCEDURES
Antibodies and Proteins
Human fibrinogen was obtained from Calbiochem. The recombinant A1A2A3 variants were expressed in mammalian (HEK293) cells, purified from the conditioned medium, and subjected to gel electrophoresis and gel filtration chromatography to verify its purity and monomeric state as we described previously (4, 25). A monospecific polyclonal antibody against the A1 domain, A108 (against sequence 1444–1452), was commercially prepared using synthetic peptides in rabbits.
Preparation of Protein-coated Coverslips
Plates coated with fibrinogen were prepared as we described previously (4). Human fibrinogen was diluted to 100 μg/ml in 65 mm sodium phosphate buffer, pH 6.5, and added to 35-mm culture dishes and incubated for 1 h at 37 °C. After washing twice with phosphate-buffered saline (PBS), pH 7.4, the plates were blocked with 3% BSA in PBS.
Flow Assays
To obtain blood, approval was attained from the Baylor College of Medicine institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki. Perfusion assays were carried out as we described elsewhere (26). One ml of citrated whole blood containing either the A1A2A3 protein or buffer (TBS) was perfused over the analyte surface-coated plate followed by TBS or PBS. Tethered platelets were observed with phase contrast objectives, recorded by video microscopy, and analyzed as described previously (4). Experiments were performed in duplicate using different blood donors.
Platelet Binding Assay
This assay was performed similarly to what we described elsewhere (18, 21). Microtiter wells were coated with 75 μl of a suspension with fixed platelets. To examine binding of A1A2A3 proteins to GPIbα, increasing concentrations of each of the protein were mixed with ristocetin (0.5 mg/ml) or TBS and incubated in the platelet-coated wells for 1 h at 37 °C. A polyclonal anti-vWF-horseradish peroxidase conjugated antibody was used for detection of the A1A2A3 proteins. Net specific binding was determined by subtracting optical density (OD) values from wells coated only with BSA from the total binding values obtained in wells coated with the corresponding protein. The software, KaleidaGraph 4.0, was used for curve fitting and to determine approximate half-maximal binding values using the equation Absorbance = Bmin*KD/(KD + [A1A2A3]) + Bmax*X/(KD + [A1A2A3]). [A1A2A3] is the tri-domain concentration, Bmin and Bmax are the minimal and maximal absorbance, and the dissociation constant is equal to the inverse of the apparent binding affinity, KD = 1/KB.
Ristocetin-induced Platelet Agglutination (RIPA)
RIPA was carried out in siliconized glass cuvettes at 37 °C with constant stirring at 1,200 rpm in an eight-channel aggregometer (Bio/Data Corp., Horsham, PA). A suspension of platelet-rich plasma containing different concentrations of each WT A1A2A3 protein was prepared. After a 5-min incubation at 37 °C, agglutination was initiated by the addition of ristocetin (Helena Laboratories, Beaumont, TX) to a final concentration of 1 mg/ml.
Protein Unfolding
Thermal denaturation of the 1238-A1A2A3 and the N-terminally truncated 1261-A1A2A3 tri-domains was monitored by circular dichroism using an AVIV Circular Dichroism Spectrophotometer model 420C (AVIV Biomedical) and by differential scanning calorimetry using a VP-DSC scanning microcalorimeter (MicroCal; GE Healthcare). All CD thermal scans and spectra were background-corrected by subtracting the contribution of the buffer (25 mm Tris-HCl, 150 mm NaCl, pH 7.4). All DSC traces were background-corrected with an irreversible scan that was used as the base line. All thermal transitions were irreversible.
Antibody Binding Assays
Monospecific polyclonal antibody A108 or irrelevant rabbit IgG was diluted to 5 μg/ml with 50 mm carbonate buffer, pH 9.6, and used to coat wells of a microtiter plate. Coating was carried out overnight at 4 °C. The wells were washed with TBS-0.05% Tween 20 (TBS-T) and blocked with 3% BSA in TBS-T for 60 min at 37 °C. A1A2A3 variants (250 nm) or purified plasma vWF (5 μg/ml), with and without ristocetin (0.5 mg/ml), were incubated in these wells for 60 min at 37 °C. After incubation, wells were then washed with TBS-T, and the bound A1A2A3 or vWF was detected by ELISA using a polyclonal anti-vWF-horseradish peroxidase conjugate (Dako). The wells were washed again, and the substrate (o-phenylenediamine; Sigma) was added. Substrate conversion reactions were stopped with 0.025 ml of 2 n H2SO4, and the plates were read at 490 nm. Net specific binding was determined by subtracting OD values from wells coated only with BSA from the total binding values obtained.
RESULTS
Amino Acid Sequence 1238–1260 in N-terminal Flanking Region of A1 Domain Regulates Binding to GPIbα
Previously, we noticed that an N-terminally truncated A1A2A3 (1261–1874) protein from our laboratory resulted in increased binding activity for GPIbα (25). These observations suggested a potential regulatory role for the amino acid sequence 1238–1260 in vWF, and therefore, we performed comparative studies between the 1261-A1A2A3 and 1238-A1A2A3 tri-domain fragments of vWF to ascertain how this sequence is involved in vWF-platelet interactions.
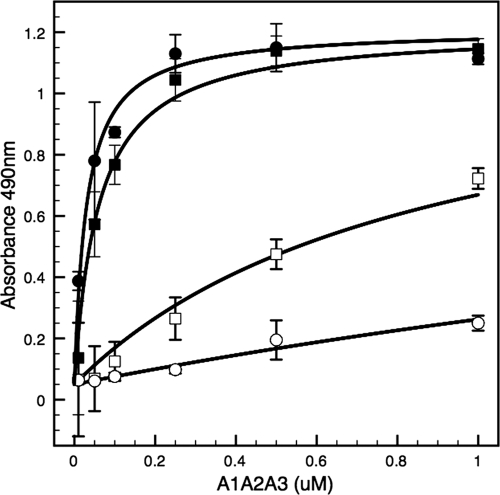
To determine the effect of the amino acid sequence 1238–1260 on platelet GPIbα binding, an ELISA was used to quantify binding to immobilized lyophilized platelets as shown in Fig. 1. Binding affinity of 1261-A1A2A3 (KB = 1.15 μm−1) to platelet GPIbα in the absence of ristocetin increased relative to 1238-A1A2A3 (KB = 0.22 μm−1). These binding affinities translate to dissociation constants of KD = 0.87 μm and KD = 4.5 μm, respectively. In the presence of ristocetin, binding affinities of 1261-A1A2A3 and 1238-A1A2A3 were 1 or 2 orders of magnitude larger (17 and 33 μm−1, respectively), corresponding to KD = 60 nm and KD = 30 nm, respectively.
FIGURE 1.
Binding of recombinant A1A2A3 proteins to platelet GPIbα. Increasing concentrations of recombinant 1261-A1A2A3 (circles) or 1238-A1A2A3 (squares) tri-domain proteins were incubated with immobilized fixed platelets in the presence (filled symbols) or absence (open symbols) of ristocetin. Bound protein was determined by ELISA as described under “Experimental Procedures.” The graphs are representative of two separate experiments. Each point represents the mean ± S.D. (error bars) of values obtained from a triplicate assay.
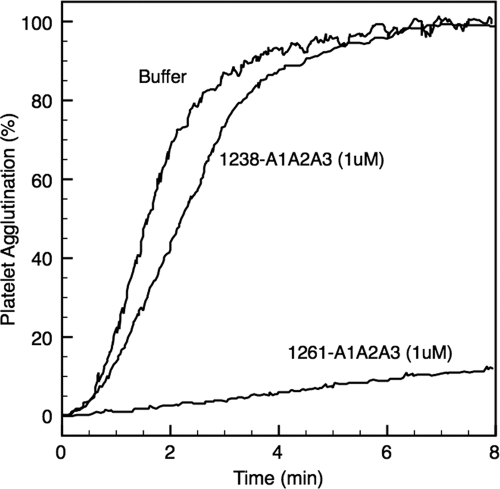
The ability of these two tri-domains to compete effectively with plasma vWF for platelet GPIbα is demonstrated with RIPA, an assay commonly used in clinics to determine clotting efficiency. In Fig. 2, the capacity of each tri-domain to block platelet agglutination induced by ristocetin correlates with their binding affinity for GPIbα. As demonstrated in Fig. 1, the increased binding activity of the 1261-A1A2A3 protein for GPIbα effectively blocked >85% RIPA at concentration of 1.0 μm, whereas in sharp contrast 1238-A1A2A3 failed to inhibit RIPA.
FIGURE 2.
Effect of A1A2A3 proteins in RIPA. 1261-A1A2A3 protein (0.5 μm) or 1238-A1A2A3 protein (1.0 μm) was incubated with platelet-rich plasma for 2 min at 25 °C. Platelet agglutination was then initiated with the addition of ristocetin (1.0 mg/ml). This figure represents two separate experiments using different two blood donors.
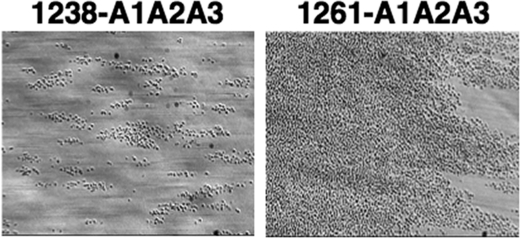
Recently, we described a method to analyze the effect of A1A2A3-GPIbα binding on platelet activation as monitored by flowing whole blood over a surface coated with fibrinogen at high shear (4). As shown in Fig. 3, 1 ml of whole blood mixed with either the 1238-A1A2A3 or 1261-A1A2A3 protein to a final concentration of 250 nm was perfused over a surface coated with fibrinogen at flow rates equivalent to 1,500 s−1 shear rate. Approximately 400 ± 100 platelets/field were observed in the blood containing 1238-A1A2A3 whereas numerous platelet deposition and thrombus formation were observed when blood was incubated with 1261-A1A2A3. This result was similar to that observed with a gain-of-function mutation in the A1 domain (R1450E) (4) and demonstrates a potential role of the N-terminal flanking region of A1 in modulating not only binding to GPIbα, but also platelet activation.
FIGURE 3.
Effect of 1261-A1A2A3 protein in flow-dependent platelet adhesion to immobilized fibrinogen. Whole blood mixed with 1238- or 1261-A1A2A3 (250 nm) was perfused over a surface coated with fibrinogen at 1,500 s−1 shear rates. After a 2-min perfusion, the plates were washed with buffer, and several frames of attached platelets were recorded. The photomicrographs represent two separate assays using two different blood donors.
N-terminal Peptide Region, Amino Acids Gln1238-Glu1260, Stabilizes A1A2A3 Tri-domain
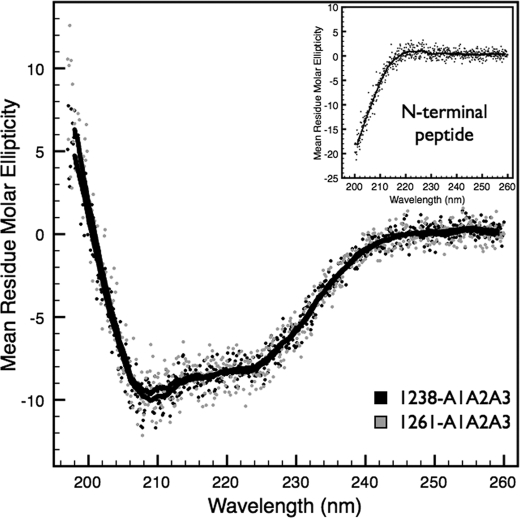
Fig. 4 shows the CD spectra of the 1238-A1A2A3 and 1261-A1A2A3 tri-domains at 5 °C. This comparison indicates that deletion of amino acid residues in the N-terminal region does not significantly perturb the overall secondary structure of the A1A2A3 complex at low temperatures. In addition, the spectra of a synthesized peptide encompassing the sequence of the N-terminal residues Gln1238-Glu1260 is unstructured due to the minima at 200 nm (inset of Fig. 4).
FIGURE 4.
Circular dichroism spectra. CD of 1238-A1A2A3 (black) and the N-terminally truncated 1261-A1A2A3 (gray) tri-domains were acquired at 5 °C. Mean residue molar ellipticity (deg cm2/dmol per residue) data are an average of three spectral scans of 1 μm protein taken in 0.1-nm intervals from 200 to 260 nm. Inset, spectral scan of the N-terminal peptide at 20 μm. Solid black lines are a 20-data point (2-nm window) smoothing of the data.
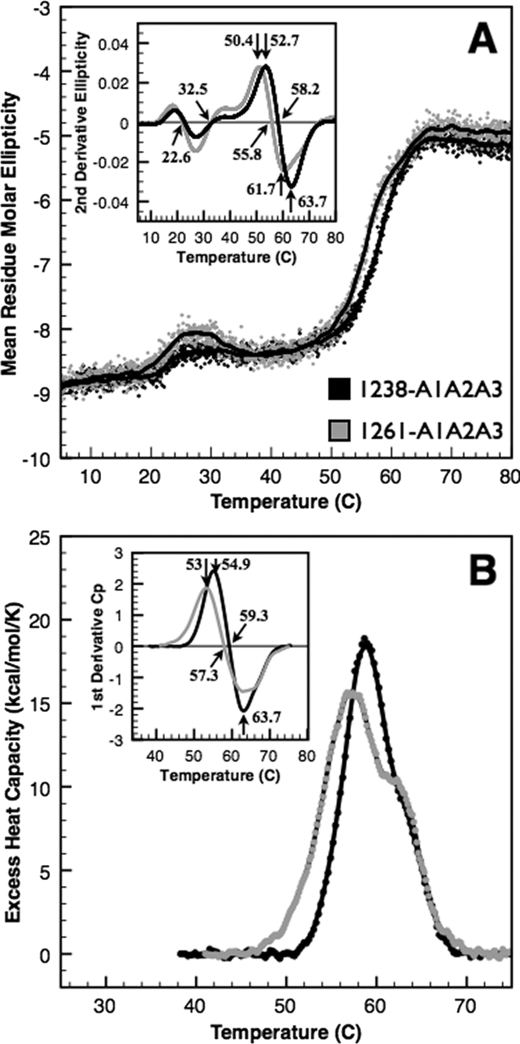
To assess the effect of the N-terminal region on the structural stability of the A1A2A3 domains, we performed thermal unfolding using both CD (Fig. 5A) at 222 nm and DSC (Fig. 5B). Both CD and DSC capture the major unfolding transitions between 50 °C and 70 °C, but these transitions are not symmetrical and contain at least two overlapping transitions that we previously assigned to the A1 and A3 domains (4). To approximate the transition temperatures, we took the second derivative of the ellipticity as a function of temperature (inset of Fig. 5A). 1238-A1A2A3 and 1261-A1A2A3 have a major transition centered at 58.2 °C and 55.8 °C assessed by the temperature at which the second derivative is equal to zero. This indicates that the N-terminal residues stabilize the tri-domain by ∼2.4 °C. However, because of the asymmetry of these transitions, the maxima and minima immediately prior to and after the temperature at which the second derivative is equal to zero identify the two transition temperatures that compose the overall transition. By this analysis, we approximate 52.7 °C and 63.7 °C transitions for 1238-A1A2A3 and 50.4 °C and 61.7 °C transitions for 1261-A1A2A3; a net stabilization of both transitions by ∼3 °C due to the N-terminal region.
FIGURE 5.
Thermal unfolding of A1A2A3 proteins. Thermal unfolding of 1238-A1A2A3 (black) and the N-terminally truncated 1261-A1A2A3 (gray) tri-domains was monitored by CD at 222 nm (A) and by DSC (B). Mean residue molar ellipticity (deg cm2/dmol per residue) on left was obtained by continuous scanning at 1 ºC/min taking data at 3-s intervals. CD results are an average of two scans of 1.1 μm 1238-A1A2A3 and six scans of 0.6 μm 1261-A1A2A3 to minimize data scatter and maximize signal/noise ratio. Solid black lines are a 20-data point (1 ºC window) smoothing of the data. Excess heat capacity (cal/mol per K) on right was obtained by continuous scanning of ∼3 μm protein at 1 ºC/min. Low temperature transitions observed by CD were not detectable by calorimetry. Insets, second derivatives of the data identify the midpoint temperatures that comprise these overlapping transitions (see “Results”).
DSC analysis by the first derivative (inset of Fig. 5B) resulted in a major transition at 59.3 °C for 1238-A1A2A3 that resolved to 54.9 °C and 63.9 °C and a major transition for 1261-A1A2A3 at 57.3 °C that resolved to 53 °C and 63.5 °C. By DSC, there is a net stabilization of ∼2 °C of the first transition due to the N-terminal region. Despite slight differences between CD and DSC results, overall they are in good agreement.
In addition to the major unfolding transitions between 50 °C and 70 °C, we observed smaller conformational transitions between 20 °C and 35 °C by CD that were not detectable by DSC (Fig. 5A). These transitions were <1 unit of ellipticity, changing by 0.4 unit for 1238-A1A2A3 and 0.7 unit for 1261-A1A2A3. These transitions were reproducible for multiple thermal scans taken from separate samples on different days. Although the transition temperatures obtained from second derivative analysis were similar for both proteins (22.9 °C and 32.8 °C for 1238-A1A2A3 and 22.3 °C and 32.5 °C for 1261-A1A2A3), the larger change in ellipticity for 1261-A1A2A3 within this temperature region suggests a more significant conformational rearrangement of the domains that transiently affects local secondary structure.
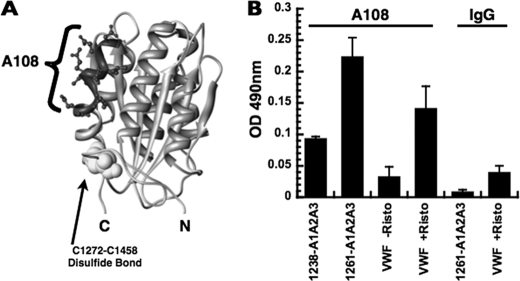
We also detected conformational differences between 1238-A1A2A3 and 1261-A1A2A3 at 37 °C using a monospecific polyclonal antibody against the A1 domain, A108 (against sequence 1444–1452) (Fig. 6A). Although the overall secondary structure of these tri-domains at 37 °C is identical, given by the 222-nm ellipticity in Fig. 5, the reactivity to these antibodies was altered by the N-terminal deletion. Compared with the 1238-A1A2A3 protein, reactivity of 1261-A1A2A3 for A108 was 2.5-fold higher (Fig. 6B). To test whether this epitope is exposed in full-length vWF, purified plasma vWF (0.5 μg/ml) was activated with 0.5 mg/ml ristocetin, which resulted in a higher reactivity for immobilized A108.
FIGURE 6.
Reactivity of the 1238-A1A2A3 and 1261-A1A2A3 tri-domains and vWF to antibody A108. A, structure of the A1 domain (1AUQ) with sequences recognized by A108 is indicated. The N-terminal sequence is not present in the crystal structure. B, tri-domains (250 nm) and purified plasma vWF (0.5 μg/ml) were incubated with immobilized antibody A108 or rabbit IgG, and the captured protein was detected by ELISA (see “Experimental Procedures”). Data (mean ± S.D. (error bars)) are representative of two separate triplicate experiments.
DISCUSSION
Previously, we described the functional similarities between a monomeric A1A2A3 protein and full-length vWF and suggested that the recombinant tri-domain fragment (amino acids 1238–1874) of vWF contains regulatory structural elements that modulate activation of vWF in solution (4). On the other hand, another recombinant A1A2A3 protein (amino acids 1261–1874) from our laboratory apparently resulted in a GPIbα binding site that was less cryptic in the A1 domain. Unlike the 1238-A1A2A3 protein, it retained moderate increased binding activity for GPIbα (25). Based on these observations, we tested the hypothesis that the amino acid sequence 1238–1260 in the N terminus of the A1 domain forms part of the mechanism that regulates vWF-GPIbα binding. A direct association between these vicinal regions in the context of full-length vWF has not been shown, but we have used recombinant proteins encompassing the natural sequence of the A1A2A3 domains to test this hypothesis.
Comparative analyses between 1238- and 1261-A1A2A3 proteins demonstrate that deletion of amino acid residues 1238–1260, which is located outside of the disulfide loop of the A1 domain structure (Fig. 6A), alters the stability of the tri-domain complex. The results obtained from thermal unfolding using both CD and DSC indicate that there is a close relationship between the N terminus flanking region of the A1 domain and the three A domains (Fig. 5). Not only are the thermal transition midpoints lowered, but the transitions are also broadened. A broadening of a thermal transition indicates that the unfolding cooperativity is decreased due to a reduced enthalpic contribution to unfolding. This reduced cooperativity may represent an impaired association between domains in the tri-domain complex. The thermal analyses demonstrate a potential association between the N-terminal flanking region (Gln1238-Glu1260) and that the tri-domain maintains stability and proper domain quaternary association in the tri-domain complex.
Deletion of the sequence Gln1238-Glu1260 increased binding activity to GPIbα. However, this deletion did not entirely terminate the regulatory mechanism of GPIbα binding under static conditions because the 1261-A1A2A3 protein required ristocetin to saturate the binding capacity for GPIbα. This observation suggests that the association between the A domains, although weak, partially inhibits the binding to GPIbα under static conditions. However, the remaining inhibitory mechanism was completely unrestricted by the influence of high shear forces (Fig. 3), confirming the crucial role of the A domains in force sensing (27). In other words, a reduced structural stability of the A1A2A3 complex increases mechanical susceptibility to shear forces, provoking the dissociation of the A domains and changing the conformation of the A1 domain to bind to GPIbα with a higher affinity (28). Thus, alterations within the structural conformation of the A domain region in vWF may positively modulate the reactivity of vWF for platelet GPIbα under hydrodynamic forces in pathological conditions. In fact, protein modifications within the A domains of vWF, such as oxidation (29), might contribute to thrombotic microangiopathies under inflammatory conditions.
The reactivity of 1261-A1A2A3 to immobilized antibody A108 was higher than that of the 1238-A1A2A3 protein (Fig. 6B). This outcome indicated that deletion of the Gln1238-Glu1260 sequence from the N-terminal region of the A1 domain changed the structural conformation of the A1 domain in the tri-A domain protein. Similarly, full-length vWF with ristocetin had higher reactivity for A108 than vWF without the modulator. It is established that ristocetin induces a conformational change in the A1 domain of vWF, enhancing binding to GPIbα (21). One way in which ristocetin activates vWF is by inducing the separation between the N-terminal flanking region of the A1 domain and the A domain complex. In fact, ristocetin recognizes part of the sequence Gln1238-Glu1260 (30, 31).
Previous studies used synthetic peptides or mutagenesis and proposed that the N terminus flanking region of the A1 domain was part of the GPIbα binding site or the regulatory element for vWF-GPIbα binding (22–24, 32, 33). The results obtained in this study have demonstrated that the amino acid sequence, Gln1238-Glu1260, plays a relevant role in modulating binding of A1 to GPIbα. The precise location of this sequence with respect to the structure of the A1A2A3 domain complex is undefined because none of the crystallographic studies with the A1 domain includes this amino acid sequence in their structures. Further studies are necessary to describe the structural basis by which the N-terminal region associates within the tri-A domain complex. Nonetheless, we propose that to terminate the mechanism that inhibits vWF-GPIbα binding, the N-terminal flanking region has to separate from the A1A2A3 complex, causing the dissociation of the A1A2A3 tri-domain, which mechanically becomes more susceptible to hydrodynamic forces.
In summary, we have used recombinant A1A2A3 domain proteins to demonstrate that the N terminus flanking region (particularly Gln1238-Glu1260) of the A1 domain stabilizes the quaternary association among the A domain complex. This structural mechanism regulates binding of vWF to GPIbα in solution, which, if terminated via hydrodynamic forces, causes platelet activation and promotes thrombus formation.
This work was supported, in whole or in part, by National Institutes of Health Grants HL72886 (to M. A. C.) and HL109109 (to M. A.). This work was also supported by the Mary Gibson Foundation and the Alkek Foundation.
- vWF
- von Willebrand factor
- DSC
- differential scanning calorimetry
- GP
- glycoprotein
- RIPA
- ristocetin-induced platelet agglutination
- TBS
- Tris-buffered saline.
REFERENCES
- 1. Savage B., Saldívar E., Ruggeri Z. M. (1996) Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell 84, 289–297 [DOI] [PubMed] [Google Scholar]
- 2. Endenburg S. C., Hantgan R. R., Lindeboom-Blokzijl L., Lankhof H., Jerome W. G., Lewis J. C., Sixma J. J., de Groot P. G. (1995) On the role of von Willebrand factor in promoting platelet adhesion to fibrin in flowing blood. Blood 86, 4158–4165 [PubMed] [Google Scholar]
- 3. Zaidi T. N., McIntire L. V., Farrell D. H., Thiagarajan P. (1996) Adhesion of platelets to surface-bound fibrinogen under flow. Blood 88, 2967–2972 [PubMed] [Google Scholar]
- 4. Auton M., Sowa K. E., Smith S. M., Sedlák E., Vijayan K. V., Cruz M. A. (2010) Destabilization of the A1 domain in von Willebrand factor dissociates the A1A2A3 tri-domain and provokes spontaneous binding to glycoprotein Ibα and platelet activation under shear stress. J. Biol. Chem. 285, 22831–22839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sadler J. E. (1998) Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 67, 395–424 [DOI] [PubMed] [Google Scholar]
- 6. Bonthron D. T., Handin R. I., Kaufman R. J., Wasley L. C., Orr E. C., Mitsock L. M., Ewenstein B., Loscalzo J., Ginsburg D., Orkin S. H. (1986) Structure of pre-pro-von Willebrand factor and its expression in heterologous cells. Nature 324, 270–273 [DOI] [PubMed] [Google Scholar]
- 7. Verweij C. L., Diergaarde P. J., Hart M., Pannekoek H. (1986) Full-length von Willebrand factor (vWF) cDNA encodes a highly repetitive protein considerably larger than the mature vWF subunit. EMBO J. 5, 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cruz M. A., Handin R. I., Wise R. J. (1993) The interaction of the von Willebrand factor-A1 domain with platelet glycoprotein Ib/IX: the role of glycosylation and disulfide bonding in a monomeric recombinant A1 domain protein. J. Biol. Chem. 268, 21238–21245 [PubMed] [Google Scholar]
- 9. Morales L. D., Martin C., Cruz M. A. (2006) The interaction of von Willebrand factor-A1 domain with collagen: mutation G1324S (type 2M von Willebrand disease) impairs the conformational change in A1 domain induced by collagen. J. Thromb. Haemost. 4, 417–425 [DOI] [PubMed] [Google Scholar]
- 10. Christophe O., Obert B., Meyer D., Girma J. P. (1991) The binding domain of von Willebrand factor to sulfatides is distinct from those interacting with glycoprotein Ib, heparin, and collagen and resides between amino acid residues Leu-512 and Lys-673. Blood 78, 2310–2317 [PubMed] [Google Scholar]
- 11. Bonnefoy A., Romijn R. A., Vandervoort P. A., Van Rompaey I, Vermylen J., Hoylaerts M. F. (2006) von Willebrand factor A1 domain can adequately substitute for A3 domain in recruitment of flowing platelets to collagen. J. Thromb. Haemost. 4, 2151–2161 [DOI] [PubMed] [Google Scholar]
- 12. Cruz M. A., Yuan H., Lee J. R., Wise R. J., Handin R. I. (1995) Interaction of the von Willebrand factor (vWF) with collagen: localization of the primary collagen-binding site by analysis of recombinant vWF A domain polypeptides. J. Biol. Chem. 270, 10822–10827 [DOI] [PubMed] [Google Scholar]
- 13. Lankhof H., van Hoeij M., Schiphorst M. E., Bracke M., Wu Y. P., Ijsseldijk M. J., Vink T., de Groot P. G., Sixma J. J. (1996) A3 domain is essential for interaction of von Willebrand factor with collagen type III. Thromb. Haemost. 75, 950–958 [PubMed] [Google Scholar]
- 14. Dent J. A., Berkowitz S. D., Ware J., Kasper C. K., Ruggeri Z. M. (1990) Identification of a cleavage site directing the immunochemical detection of molecular abnormalities in type IIA von Willebrand factor. Proc. Natl. Acad. Sci. U.S.A. 87, 6306–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitelock J. L., Nolasco L., Bernardo A., Moake J., Dong J. F., Cruz M. A. (2004) ADAMTS-13 activity in plasma is rapidly measured by a new ELISA method that uses recombinant VWF-A2 domain as substrate. J. Thromb. Haemost. 2, 485–491 [DOI] [PubMed] [Google Scholar]
- 16. Cruz M. A., Diacovo T. G., Emsley J., Liddington R., Handin R. I. (2000) Mapping the glycoprotein Ib-binding site in the von Willebrand factor A1 domain. J. Biol. Chem. 275, 19098–19105 [DOI] [PubMed] [Google Scholar]
- 17. Cruz M. A., Whitelock J., Dong J. F. (2003) Evaluation of ADAMTS-13 activity in plasma using recombinant von Willebrand factor A2 domain polypeptide as substrate. Thromb. Haemost. 90, 1204–1209 [DOI] [PubMed] [Google Scholar]
- 18. Auton M., Cruz M. A., Moake J. (2007) Conformational stability and domain unfolding of the von Willebrand factor A domains. J. Mol. Biol. 366, 986–1000 [DOI] [PubMed] [Google Scholar]
- 19. Shankaran H., Alexandridis P., Neelamegham S. (2003) Aspects of hydrodynamic shear regulating shear-induced platelet activation and self-association of von Willebrand factor in suspension. Blood 101, 2637–2645 [DOI] [PubMed] [Google Scholar]
- 20. Federici A. B., Mannucci P. M., Castaman G., Baronciani L., Bucciarelli P., Canciani M. T., Pecci A., Lenting P. J., De Groot P. G. (2009) Clinical and molecular predictors of thrombocytopenia and risk of bleeding in patients with von Willebrand disease type 2B: a cohort study of 67 patients. Blood 113, 526–534 [DOI] [PubMed] [Google Scholar]
- 21. Azuma H., Sugimoto M., Ruggeri Z. M., Ware J. (1993) A role for von Willebrand factor proline residues 702–704 in ristocetin-mediated binding to platelet glycoprotein Ib. Thromb. Haemost. 69, 192–196 [PubMed] [Google Scholar]
- 22. Ulrichts H., Udvardy M., Lenting P. J., Pareyn I., Vandeputte N., Vanhoorelbeke K., Deckmyn H. (2006) Shielding of the A1 domain by the D'D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J. Biol. Chem. 281, 4699–4707 [DOI] [PubMed] [Google Scholar]
- 23. Mohri H., Fujimura Y., Shima M., Yoshioka A., Houghten R. A., Ruggeri Z. M., Zimmerman T. S. (1988) Structure of the von Willebrand factor domain interacting with glycoprotein Ib. J. Biol. Chem. 263, 17901–17904 [PubMed] [Google Scholar]
- 24. Nakayama T., Matsushita T., Dong Z., Sadler J. E., Jorieux S., Mazurier C., Meyer D., Kojima T., Saito H. (2002) Identification of the regulatory elements of the human von Willebrand factor for binding to platelet GPIb: importance of structural integrity of the regions flanked by the Cys1272-Cys1458 disulfide bond. J. Biol. Chem. 277, 22063–22072 [DOI] [PubMed] [Google Scholar]
- 25. Martin C., Morales L. D., Cruz M. A. (2007) Purified A2 domain of von Willebrand factor binds to the active conformation of von Willebrand factor and blocks the interaction with platelet glycoprotein Ibα. J. Thromb. Haemost. 5, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 26. Cruz M. A., Chen J., Whitelock J. L., Morales L. D., López J. A. (2005) The platelet glycoprotein Ib-von Willebrand factor interaction activates the collagen receptor α2β1 to bind collagen: activation-dependent conformational change of the α2-I domain. Blood 105, 1986–1991 [DOI] [PubMed] [Google Scholar]
- 27. Wu T., Lin J., Cruz M. A., Dong J. F., Zhu C. (2010) Force-induced cleavage of single VWFA1A2A3 tri-domains by ADAMTS-13. Blood 115, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Auton M., Zhu C., Cruz M. A. (2010) The mechanism of VWF-mediated platelet GPIbα binding. Biophys. J. 99, 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu X., Chen J., Gallagher R., Zheng Y., Chung D. W., López J. A. (2011) Shear stress-induced unfolding of VWF accelerates oxidation of key methionine residues in the A1A2A3 region. Blood 118, 5283–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujimura Y., Usami Y., Titani K., Niinomi K., Nishio K., Takase T., Yoshioka A., Fukui H. (1991) Studies on anti-von Willebrand factor (vWF) monoclonal antibody NMC-4, which inhibits both ristocetin- and botrocetin-induced vWF binding to platelet glycoprotein Ib. Blood 77, 113–120 [PubMed] [Google Scholar]
- 31. Girma J. P., Takahashi Y., Yoshioka A., Diaz J., Meyer D. (1990) Ristocetin and botrocetin involve two distinct domains of von Willebrand factor for binding to platelet membrane glycoprotein Ib. Thromb. Haemost. 64, 326–332 [PubMed] [Google Scholar]
- 32. Sugimoto M., Dent J., McClintock R., Ware J., Ruggeri Z. M. (1993) Analysis of structure-function relationships in the platelet membrane glycoprotein Ib-binding domain of von Willebrand factor by expression of deletion mutants. J. Biol. Chem. 268, 12185–12192 [PubMed] [Google Scholar]
- 33. Matsushita T., Sadler J. E. (1995) Identification of amino acid residues essential for von Willebrand factor binding to platelet glycoprotein Ib: charged-to-alanine scanning mutagenesis of the A1 domain of human von Willebrand factor. J. Biol. Chem. 270, 13406–13414 [DOI] [PubMed] [Google Scholar]