Background: The Goodpasture antigen-binding protein (GPBP) and serum amyloid P component (SAP) bind to type IV collagen and are found in plasma.
Results: GPBP binds to human SAP.
Conclusion: GPBP and SAP form complexes under physiological and pathological conditions.
Significance: This interaction might be involved in protein aggregation in Alzheimer disease and the resulting innate immune response.
Keywords: Brain, Pentraxins, Protein Structure, Protein-Protein Interactions, Surface Plasmon Resonance (SPR), Amyloid-β (Aβ), Ceramide Transporter (CERT), Goodpasture Binding Protein (GPBP), Serum Amyloid P Component (SAP)
Abstract
Serum amyloid P component (SAP) is a non-fibrillar glycoprotein belonging to the pentraxin family of the innate immune system. SAP is present in plasma, basement membranes, and amyloid deposits. This study demonstrates, for the first time, that the Goodpasture antigen-binding protein (GPBP) binds to human SAP. GPBP is a nonconventional Ser/Thr kinase for basement membrane type IV collagen. Also GPBP is found in plasma and in the extracellular matrix. In the present study, we demonstrate that GPBP specifically binds SAP in its physiological conformations, pentamers and decamers. The START domain in GPBP is important for this interaction. SAP and GPBP form complexes in blood and partly colocalize in amyloid plaques from Alzheimer disease patients. These data suggest the existence of complexes of SAP and GPBP under physiological and pathological conditions. These complexes are important for understanding basement membrane, blood physiology, and plaque formation in Alzheimer disease.
Introduction
Serum amyloid P component (SAP)3 is present in the blood as single uncomplexed pentamers and decamers (1). SAP has been found to decorate amyloid deposits in different amyloid diseases, where it was first identified (2). In Alzheimer disease (AD), SAP colocalizes with amyloid-β (Aβ) deposits (3, 4) in the brain and is thought to protect the amyloid from proteolysis (5). Below we will first introduce known properties of SAP and subsequently of the Goodpasture antigen-binding protein (GPBP), which we identify and characterize as a new SAP-binding protein in this paper.
SAP is a 23-kDa glycoprotein (6) of the pentraxin family, which is characterized by Ca2+-dependent ligand binding (7–10). SAP self-association and aggregation are also influenced by Ca2+ (11).
SAP binds to proteins involved in immunological responses (12–15) and can activate the classical complement pathway through interaction with C1q (16). In addition, SAP interacts with extracellular matrix components, such as proteoglycans (8, 17), fibronectin (18), laminin (10), and collagen IV (9). Collagen molecules are heterotrimers composed of three α-chains. In collagen IV, each of these α-chains consists of an N-terminal 7S domain, a central helical structure, and a C-terminal globular non-collagenous (NC1) domain. The distribution of SAP in basal membranes (BMs) coincides with the restricted localization of the α3-α4-α5 heterotrimer of collagen IV (17).
The NC1 domain of the collagen IV α3 subunit (α3(IV)NC1) is the autoantigen in Goodpasture syndrome, an autoimmune disease in which autoantibodies are observed along glomerular and alveolar BMs, causing glomerulonephritis and lung hemorrhage (19).
GPBP binds to the α3(IV)NC1 in the glomerular basal membrane (GBM) of Goodpasture patients (20). GPBPs exist in different isoforms that are found in the extracellular compartment, either soluble or associated with the surface of the plasma membrane (21, 22), and in blood (23). A shorter splicing isoform, CERT, is located inside the cell and functions as carrier for ceramide from the ER to the Golgi apparatus (24). GPBP and CERT (also known as CERTL and GPBPΔ26, respectively) are identical in sequence with the exception of an additional 26-amino acid domain present only in GPBP (25).
Because both SAP and GPBP have been reported to self-aggregate (11, 25) and have similar properties, the question arose as to whether SAP and GPBP could bind to each other. Thus, the aim of the present study was to characterize in detail the direct interaction between these proteins using surface plasmon resonance (SPR), far Western blotting, and microscale thermophoresis technology (MST). We report, for the first time, that SAP binds to GPBP and, moreover, that SAP and GPBP are co-localized in amyloid plaques from AD patients. Our data suggest the existence of complexes of SAP and GPBP under normal and pathological conditions.
EXPERIMENTAL PROCEDURES
Purification of SAP Protein from Human Serum
SAP was purified from human plasma through an immunoaffinity procedure applying Ca2+ ion-dependent interactions of SAP with complement factor C4b-binding protein (C4BP). A monoclonal antibody against C4BP (CLB-C4BP), which is directed against the α-chains of human C4BP (a kind gift from Dr. Jan van Mourik, Sanquin Research, Central Laboratory of the Blood Transfusion Service, Amsterdam, The Netherlands) was coupled to CNBr-activated Sepharose 4B (GE Healthcare) according to the manufacturer's instructions. Human plasma was fractionated by barium citrate precipitation and eluted with 30% ammonium sulfate. After precipitation with 70% ammonium sulfate, the protein fraction was dialyzed against Tris-buffered saline (TBS; 50 mm Tris-HCl, pH 7.4, 150 mm NaCl) supplemented with 3 mm CaCl2 and applied to the anti-C4BP column. After binding of the C4BP-SAP complex, the column was washed with TBS containing 3 mm CaCl2 until A280 nm < 0.05. Subsequently, SAP was specifically eluted with TBS containing 2 mm EDTA. Purified human SAP appeared as a single band (>95% purity) on a Coomassie Blue-stained 4–15% SDS gel, and fractions were pooled and frozen at −80 °C until use. The identity of SAP was confirmed by peptide mass fingerprinting using a tandem MALDI-TOF protein analyzer (Applied Biosystems 4800).
Production of Recombinant GPBP and CERT Proteins
For recombinant expression of GPBP and CERT, we used the vector pHILD2 (Invitrogen). The expression cassettes were synthesized (GeneArt, Regensburg, Germany) using the cDNA sequences that encode the human GPBP protein (NP_005704.1) and the human CERT protein (NP_112729.1) preceded by a cassette, including an EcoRI restriction site and a standard Kozak consensus for translation initiation, followed by a sequence encoding for MAPLA and a FLAG tag peptide (DYKDDDDK). An EcoRI site was introduced after the stop codon. The plasmids were transfected and expressed in Pichia pastoris (Invitrogen). FLAG-tagged recombinant proteins were purified from the cell lysate with an anti-FLAG M2 affinity agarose gel column (A2220, Sigma) according to the manufacturer's instructions. The unbound material was washed off of the column with TBS, and FLAG-tagged proteins were eluted using 100 μg/ml FLAG peptide (F3290, Sigma).
CERT and CERT mutant constructs were synthesized by Life Technologies, GeneArt (Regensburg, Germany) in pET28b (Novagen) expression vectors through a PCR-based method. FLAG-tagged proteins were produced by overexpression in Escherichia coli BL21(DE3) pLYSs (Promega), induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. Recombinant protein was isolated using the FLAG tag as described above.
Sample Preparation and SPR Analysis
SAP self-aggregation was controlled by optimization of the buffers. To this end, native SAP was diluted at final concentrations of 25, 50, and 100 nm in 1) sodium acetate buffer, pH 4.5; 2) sodium acetate buffer, pH 4.5, with 5 mm Ca2+; 3) 25 mm HEPES buffer, pH 7.4; 4) 25 mm HEPES buffer, pH 7.4, and sonicated; and 5) 25 mm HEPES buffer with 0.01% Tween 20 at pH 7.4 and sonicated. Sonication was performed before SPR experiments using a probe (Beun-De Ronde B.V., Abcoude, The Netherlands) for three pulses of 30 s each with a 30-s rest on ice between pulses. Each sample was centrifuged at 20,000 × g for 5 min to remove protein aggregates immediately before SPR analysis.
SPR experiments were performed on a Biacore T100 apparatus (GE Healthcare) (26). The guidelines from the manufacturer were followed for the preparation of the sensor surfaces and interpretation of the sensorgrams. Purified human SAP, GPBP, and CERT (50 μg/ml in 10 mm sodium acetate buffer, pH 4.5) were covalently coupled via amine groups onto the carboxymethylated dextran surface of CM5 sensor chips (GE Healthcare), resulting in a signal of up to 15,000 resonance units (RU). Injection of specific antibodies recognizing both GPBP and CERT demonstrated the presence of the proteins immobilized in each flow cell (rabbit polyclonal anti-GPBP/CERT, epitope 1–50 of human GPBP/CERT, Bethyl Laboratories (Montgomery, TX); rabbit polyclonal anti-SAP (P-16), Santa Cruz Biotechnology, Inc.). Analytes for binding studies were prepared in 25 mm HEPES buffer, pH 7.4, 150 mm NaCl with 0.01% Tween 20. To perform binding experiments, protein samples (purified SAP, human collagen IV (Sigma), human laminin (Sigma), and bovine serum albumin (BSA; Sigma)) were injected onto the chip over a concentration range of 100 nm to 1 μm at a flow rate of 10 μl/min for 5 min at 25 °C. To test the effect of Ca2+ on protein-protein interaction, the same buffer with the addition of 5 mm Ca2+ was used for some of the experiments.
Wild type and five mutant CERT proteins were serially diluted in 25 mm HEPES, 150 mm NaCl, 0.01% Tween 20, pH 7.4, over a wide concentration range (up to 500 nm) by using 2-fold dilution steps. Samples were injected over a SAP-coated surface of CM5 sensor chip (density, 5000 RU), for 3 min at a flow rate of 30 μl/min, at 25 °C. At the end of each run, the sensor surface was regenerated (removal of bound complex) by using 25 mm NaOH before additional samples were injected. As an internal reference, a control channel was routinely activated and blocked in the absence of protein. The signals from the control channel were subtracted from the signals generated by the flow cells containing immobilized protein. Analysis was performed on the data using BIAevaluation version 3.0 software. Sensorgrams were recorded and normalized to a base line of 0 RU.
MST Binding Analyses
MST is a new immobilization-free technique for the analysis of biomolecule interaction (27–29). The term “microscale thermophoresis” refers to the directed movement of molecules in optically generated microscopic temperature gradients. This thermophoretic movement is determined by the entropy of the hydration shell around molecules. The microscopic temperature gradient is generated by an IR laser. The readout method of the interaction analysis is based on fluorescence. In a typical MST experiment, the concentration of the labeled molecule is kept constant, whereas the concentration of the unlabeled interaction partner is varied. The MST signal will detect the binding by a quantification of the change in the normalized fluorescence (27).
MST analysis was performed on the Monolith NT.115 instrument (NanoTemper, München, Germany). In brief, a constant concentration of NT647-labeled GPBP (labeled protein concentration of 1–50 nm) was incubated for 20 min at room temperature in the dark with different concentrations of SAP (up to 5000 nm) in PBS, 0.01% Tween 20. Afterward, 3–5 μl of the samples were loaded into glass capillaries (Monolith NT Capillaries, catalog no. K002), and the thermophoresis analysis was performed (LED 40–51%, IR laser 80%). Statistical analysis was performed with Origin version 8.5 software.
Gel Filtration Chromatography
A Superose 6 column with a bed volume of 24 ml (PerkinElmer Life Sciences series 4 FPLC system) connected to an FPLC system (GE Healthcare) was washed with ethanol (20%, v/v) and 0.5 m NaOH and pre-equilibrated with 3 column volumes of 25 mm HEPES buffer (pH 7.4) with 150 mm NaCl. Samples of purified SAP were run on this column with a buffer flow rate of 0.4 ml/min. The approximate molecular mass of the fractionated proteins was calculated from peak elution volumes by comparison with molecular mass standards (thyroglobulin, 669 kDa; ferritin, 440 kDa; fibrinogen, 340 kDa; IgG, 160 kDa; BSA, 67 kDa; ribonuclease A, 13.7 kDa; GE Healthcare).
Immunoprecipitation and Western Blot
Immunoprecipitation and co- immunoprecipitation of SAP and GPBP from human serum were performed after depletion of albumin (ProteoExtract albumin removal kit, Calbiochem) to increase the resolution of the lower abundance proteins of interest. Albumin-depleted serum was centrifuged at 20,000 × g for 30 min. Pull-down of endogenous SAP and endogenous GPBP was performed with mAb 4E8 (Sigma) and mAb 3A1-C1, respectively. Rabbit polyclonal anti Dok-7 antibody (H-77, Santa Cruz Biotechnology, Inc.) and a mouse monoclonal anti syntaxin 6 (clone 3D10, Abcam, Cambridge, UK) were used as isotype controls. After incubation (1 μg of antibody per 15 μl of serum; at room temperature for 1 h), samples were centrifuged at 20,000 × g for 30 min at 4 °C. Pellets were washed three times in 50 μl of PBS and boiled in reducing sample buffer containing mercaptoethanol to dissolve immunocomplexes.
Brain tissue from 8-month-old control and Alzheimer transgenic mice (APPswe PS1ΔE9 C57BL/6) were thawed and homogenized as described (30). In brief, mouse cortex was placed in ice-cold lysis buffer containing PBS, 0.1% SDS, 0.1% Triton X-100, 1% glycerol, 1 mm EDTA, 1 mm EGTA, 30 mm NaF, and 16.7 mm sodium orthovanadate and a Complete protease inhibitor mixture tablet (Roche Applied Science) per 50 ml of buffer, following the manufacturer's recommendations. Glass beads and a bead beater (Biospec Products, Bartlesville, OK) were used for the homogenization of the tissue. The homogenization consisted of three cycles of 30 s in 1.0 ml of lysis buffer.
Total protein concentration was determined by a conventional method (BCA protein assay kit, Pierce). For immunoprecipitation, 2 mg of brain homogenates were incubated for 1 h at room temperature with 3 μg of one of the following mAbs: anti-SAP (4E8), anti-GPBP (3A1-C1), or anti-Aβ (human recombinant antibodies bapineuzumab, solanezumab, and 20C2, produced recombinantly in house, based on published sequences). Negative controls consisted of IgG mAb anti-syntaxin 6 (clone 3D10) or anti-rapsyn (clone 1234, Sigma). Next we added goat anti-mouse (Euroegentec, Maastricht, The Netherlands) during 30 min at room temperature (1 μg/sample). Samples were centrifuged at 20,000 × g for 30 min at 4 °C, and immunocomplexes were processed as described above.
Proteins were separated by SDS and native PAGE using precast Criterion Tris-HCl glycine 4–20% gradient gels (Bio-Rad) followed by electroblotting to nitrocellulose membrane (Millipore). The membranes were incubated with primary antibodies specific for SAP (4E8) or GPBP using either mAb 3A1-C1 or polyclonal rabbit anti-GPBP epitope 1–50 (Bethyl Laboratories) or specific for Aβ using anti-Aβ (6E10). After PBS washes, the membrane was incubated with goat anti-rabbit-IRDye 800 and donkey anti-mouse-IRDye 680 (Rockland Immunochemicals, Gilbertsville, PA). Finally, the membrane was washed with PBS, dried, and scanned using the Odyssey infrared imaging system (Westburg, Leusden, The Netherlands).
Far Western
For far Western experiments, SAP (80 ng) and BSA (2 μg) (Sigma) were separated by SDS-PAGE under reducing conditions and transferred to nitro cellulose membranes (Bio-Rad). For renaturation of SAP, the membranes were incubated in TBS with Tween 20 (0.05%) overnight. Renatured proteins were probed for 1 h at 37 °C with either GPBP, CERT, or CERT mutants (30 μg/ml) in the same buffer (20). Next, membranes were blocked with 5% BSA. Bound material was detected using anti-GPBP (3A1-C1), anti-SAP (4E8), and donkey anti-mouse-IRDye 680 followed by detection as described above. BSA was detected by standard Coomassie staining.
Immunohistochemistry in Kidney
Sections of monkey control kidney tissue (macaques) (Probetex, San Antonio, TX) were incubated overnight at room temperature with primary antibodies (mouse monoclonal anti-GPBP antibody (3A1-C1) and anti-SAP (4E8)), followed by the corresponding secondary antibody (donkey anti-mouse biotinylated IgG (Jackson ImmunoResearch Laboratories Europe Ltd., Newmarket, Suffolk, UK)). Subsequently, sections were incubated with the ABC kit (Vector Laboratories, Burlingame, CA) followed by 3,3-diaminobenzidine tetrahydrochloride.
Slides were mounted with 80% glycerol in TBS. Images were acquired using an Olympus AX70 microscope (Olympus, Zoeterwoude, The Netherlands) and recorded using Cell P software (Olympus).
Immunohistochemistry in Human Brain
In order to investigate the presence and localization of GPBP in human brain, post-mortem specimens from three male and three female donors were studied by immunohistochemistry. This material was obtained from the Netherlands Brain Bank (Amsterdam, The Netherlands). Staging of AD was neuropathologically evaluated according to the Braak and Braak criteria (see Table 1) (31). For immunohistochemical staining, 5-μm cryosections were mounted on coated glass slides (Menzel Gläser Superfrost PLUS, Braunschweig, Germany) and fixed in acetone for 10 min. Next, sections were incubated overnight with primary antibodies, including rabbit anti-SAP (Dako), mouse monoclonal anti-Aβ (clone 6F/3D, Dako), affinity-purified rabbit antibody specific for residues 300–350 of human GPBP/CERT (Bethyl Laboratories), or polyclonal rabbit anti-GPBP/CERT epitope 1–50. Subsequently, sections were incubated with EnVision goat anti-mouse horseradish peroxidase (HRP) or EnVision goat anti-rabbit HRP (Dako). Peroxidase labeling was visualized by EnVision DAB (EV-DAB; Dako). Sections were counterstained with hematoxylin. For co-localization studies, cryosections were incubated in thioflavin S solution to stain Aβ fibrils and washed subsequently three times in ethanol 70%. Sections were incubated with a mix of primary antibodies: anti-SAP (mAb-14) (32) in combination with anti-GPBP/CERT 1–50 or 300–350 diluted in PBS containing 1% BSA. After washing in PBS, sections were incubated with a mix of secondary antibodies: biotin-conjugated goat anti-rabbit (Dako) and EnVision goat anti-mouse HRP (Dako). Upon washing with PBS, sections were incubated with streptavidin Alexa-633; HRP signal was developed with rhodamine tyramide (in the presence of 0.01% H2O2). Slides were covered with Aqua-Poly/Mount (Polysciences Inc., Warrington, PA).
TABLE 1.
Overview of cases included for the immunohistochemical part of this study
Upon immunostaining for Aβ, the two AD cases (Braak 5C) were found to have many Aβ plaques but no classical or neuritic plaques. The AD/Lewy body variant case (case 135) did show all types of Aβ plaques, ranging from diffuse to neuritic, as did the demented case that did not fulfill the neuropathological criteria of AD (case 294). The non-demented controls (cases 47 and 115) were selected to have many cerebral Aβ deposits and had many Aβ plaques ranging from diffuse to classical type.
| Case | Diagnosisa | Braakb | Amyloidc (0, A, B, C) | Age | Genderd | PMDe | APOE genotype |
|---|---|---|---|---|---|---|---|
| years | h:min | ||||||
| 47 | Non-demented CTL | 1 | B | 98 | M | 8:40 | 3/3 |
| 115 | Non-demented CTL | 1 | B | 83 | M | 4:35 | 3/3 |
| 81 | AD | 5 | C | 78 | M | 7:45 | 4/4 |
| 264 | AD | 5 | C | 85 | F | 6:10 | 4/3 |
| 135 | AD/Lewy body variant | 4 | B | 95 | F | 5:15 | 3/3 |
| 294 | Dementia with cortical changes | 2 | B | 91 | F | 3:35 | 3/3 |
a Clinical diagnosis with neuropathological confirmation (CTL, control).
b Neuropathological staging of cases based on occurrence and distribution of neurofibrillary changes according to the criteria of Braak and Braak (52).
c Neuropathological staging of cases based on occurrence and distribution of amyloid deposits according to the criteria of Braak and Braak (52).
d M, male; F, female.
e Post-mortem delay time (hours:minutes).
RESULTS
Binding of SAP and GPBP to Type IV Collagen
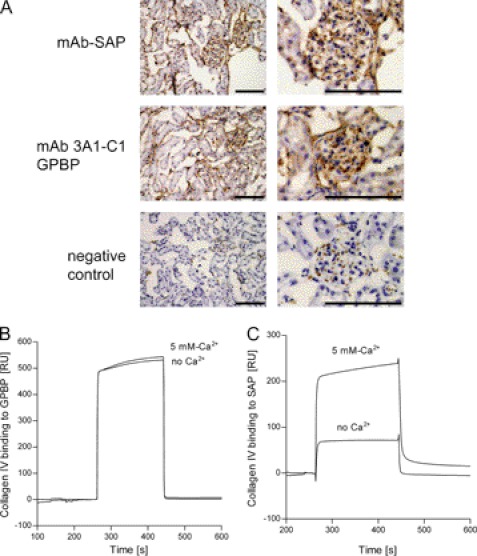
Previously, it has been reported that both SAP and GPBP specifically bind to α3(IV)NC1 monomer (20, 33). In order to analyze if this binding occurs in vivo, we studied whether SAP and GPBP colocalize in GBM, which expresses collagen IV. To this end, frozen monkey kidney sections were stained by immunohistochemistry. SAP showed a linear staining pattern along the GBM as described (34) (Fig. 1A, top). A monoclonal antibody that recognizes GPBP specifically (mAb3A1-C1) also resulted in a strong staining of the GBM (Fig. 1A, middle). Thus, the reported immunoreactivity of polyclonal antibodies against both GPBP and CERT to tubules and glomerulus in human kidney (20) can be at least partially attributed to the expression of the longer isoform GPBP.
FIGURE 1.
Collagen IV binds to SAP and to GPBP. A, representative monkey kidney sections with glomeruli stained for GPBP and SAP, SAP with mAb, and GPBP with mAb 3A1-C1. No staining was observed when the primary antibodies were omitted. Scale bars, 100 μm. B and C, the detection of the protein-protein interaction was performed by SPR technology. B, collagen IV was filtered (0.45-μm Millipore filter) and then injected (100 nm) over immobilized GPBP (101 RU; flow rate, 15 μl/min; injected volume, 60 μl) in 25 mm HEPES buffer, pH 7.4, 150 mm NaCl (either without Ca2+ or with 5 mm Ca2+). C, collagen IV was injected at 100 nm over immobilized SAP (101 RU; flow rate, 15 μl/min; injected volume, 60 μl) in 25 mm HEPES buffer, pH 7.4, 150 mm NaCl (either without Ca2+ or with 5 mm Ca2+).
We further examined the binding of SAP and GPBP to collagen IV by SPR technology. Collagen IV bound to immobilized human GPBP and human SAP (Fig. 1, B and C).
Binding of SAP to several ligands is dependent on the presence of Ca2+ (7, 8, 35). Binding of collagen IV to SAP was enhanced 3–4-fold by the addition of 5 mm Ca2+ (Fig. 1C). In contrast, binding of collagen IV to GPBP was independent of the presence of Ca2+ (Fig. 1B). Binding of BSA to immobilized SAP or GPBP was minimal (data not shown).
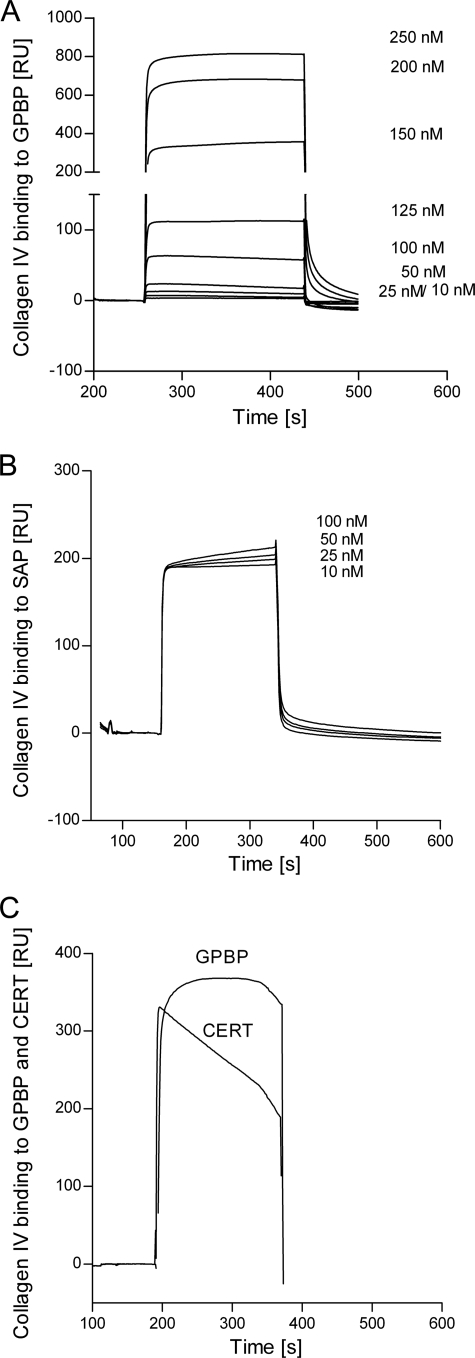
Kinetic analysis of the binding of soluble collagen IV to immobilized SAP and to GPBP was performed by SPR. Collagen IV (concentrations ranging between 10 and 250 nm) bound to GPBP strongly; increasing the concentration of collagen IV up to 250 nm did not saturate this binding (Fig. 2A). As expected, collagen IV also bound to immobilized SAP (Fig. 2B), and in this case, binding was already almost saturated at 10 nm collagen IV.
FIGURE 2.
A and B, overlay of sensorgrams resulting from the injection of different concentrations (10–250 nm) of collagen IV over immobilized GPBP (96 RU; flow rate, 15 μl/min; injected volume, 60 μl) (A) and over immobilized SAP (96 RU; flow rate, 15 μl/min; injected volume, 60 μl) (B). C, binding of collagen IV (100 nm) to immobilized GPBP and CERT (50 μg/ml in 10 mm sodium acetate buffer, pH 4.5). The sensorgrams were corrected for the signal in the empty cell (i.e. calculated as the difference between the signal in Fc2 (flow channel 2) that contained the immobilized ligand and the signal in Fc1 (empty channel), which includes injection noise, instrument drift, and nonspecific binding).
These data suggest that once collagen IV has bound to immobilized GPBP, additional collagen IV can bind by forming GPBP-collagen-collagen complexes. In contrast, SAP binding to collagen IV does not induce formation of larger collagen aggregates.
Binding of collagen IV to CERT was also studied and compared with GPBP (Fig. 2C). Collagen IV rapidly associated with both immobilized proteins. However, collagen IV dissociated faster from CERT than from full-length GPBP.
Binding of Laminin to Immobilized GPBP
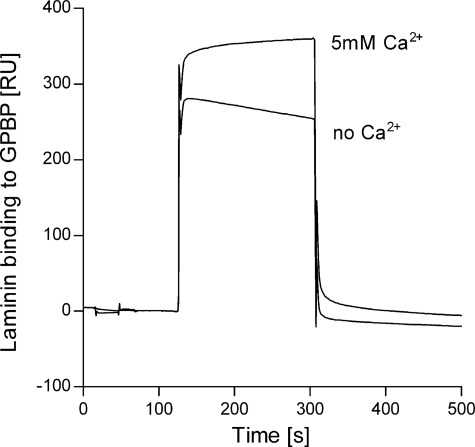
SAP has been shown to bind laminin, another important component of basement membranes (10). However, the binding of GPBP to laminin has yet to be reported. By SPR binding experiments, we found that laminin binds to immobilized GPBP (Fig. 3). Binding of laminin to GPBP was enhanced by ∼25% with the addition of 5 mm Ca2+ (Fig. 3).
FIGURE 3.
Laminin binds to GPBP. Shown is binding of laminin (100 nm) to immobilized GPBP (215 RU; flow rate, 15 μl/min; injected volume, 60 μl) in buffer containing 5 mm Ca2+ or buffer without Ca2+. The binding was first recorded in the absence of added Ca2+. The experiments were performed by SPR technology.
Binding of SAP to Immobilized GPBP
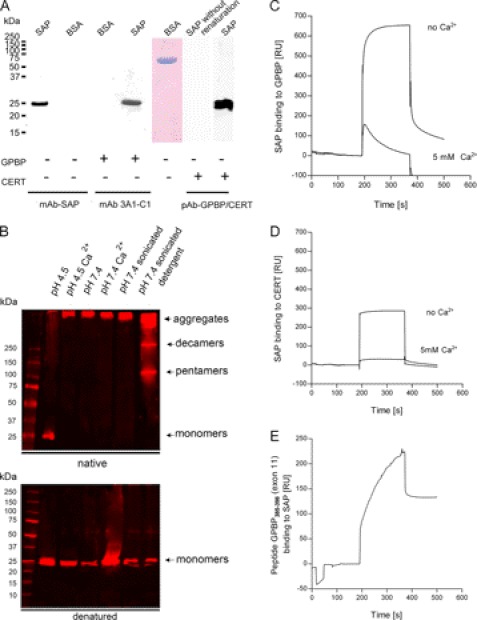
Because SAP and GPBP bind to collagen IV and colocalize at GBM, we asked whether they could interact with each other. We measured the binding of GPBP to immobilized SAP by far Western (Fig. 4A) (36, 37). For this purpose, membranes containing SAP (80 ng) and BSA (2,000 ng) as negative control were incubated in renaturation buffer overnight and subsequently probed with GPBP. Our experiment showed that GPBP strongly bound to SAP but not to BSA, suggesting that SAP and GPBP interact. This interaction is dependent on renaturation of SAP on the membrane because Western blot separation of SAP followed by immediate membrane incubation with GPBP without renaturation did not lead to significant binding of GPBP to SAP (Fig. 4A).
FIGURE 4.
Solid phase interaction of SAP with GPBP. A, far Western blot experiments. Human SAP and BSA were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and renatured. As a negative control for the renaturation, SAP was kept in 6 m urea (SAP without renaturation). After blocking, membranes were incubated with either GPBP or with CERT as a probe using immobilized SAP as bait. Bound proteins were detected with mAb anti-SAP, anti-GPBP (3A1-C1), or a polyclonal antibody against GPBP/CERT. The presence of BSA was confirmed by Coomassie staining. Both GPBP and CERT bound specifically to renatured SAP but not to BSA or denatured SAP. B, SAP aggregation is influenced by pH, the composition of the buffer, and the presence or absence of Ca2+. The treatment of SAP before the Western blot was performed as follows. SAP was diluted at 10 ng/μl in 10 mm sodium acetate buffer, pH 4.5 (lane 1); in sodium acetate buffer, pH 4.5, in the presence of 5 mm calcium (lane 2); in 25 mm HEPES buffer, pH 7.4 (lane 3); in HEPES buffer, pH 7.4, in the presence of 5 mm calcium (lane 4); in HEPES buffer, pH 7.4, followed by sonication with a probe sonicator for three pulses of 30 s each with a 30-s rest on ice between each pulse (lane 5); or in 25 mm HEPES buffer, pH 7.4, followed by sonication as aforementioned in the presence of 0.01% Tween 20 (lane 6). Different percentages of Tween 20 (1, 0.1, and 0.001%) were tested before choosing the optimal at 0.01%. SAP was separated using native PAGE and SDS-PAGE 4–20% gradient gels. The proteins were transferred to nitrocellulose membranes and incubated with anti-SAP antibody. 10 ng/μl SAP diluted in sodium, 10 mm acetate buffer, pH 4.5, and run in native conditions separated unique SAP species of 25 kDa (monomers). 10 ng/μl SAP diluted in HEPES buffer, pH 7.4, sonicated in the presence of 0.01% Tween, and run in native PAGE separated SAP species corresponding to high molecular aggregates of >250 kDa, 250 kDa (decamers), and 100 kDa (pentamers). As expected, independently of the buffer in which SAP has been diluted, in SDS-PAGE, SAP separated as a band of 25 kDa (monomers). C, binding of SAP to immobilized GPBP (215 RU; flow rate, 15 μl/min; injected volume, 60 μl). The binding was recorded in 25 mm HEPES buffer, pH 7.4, 150 mm NaCl. The presence of 5 mm Ca2+ decreased the binding of SAP to immobilized GPBP. D, binding of SAP to immobilized CERT (215 RU; flow rate, 15 μl/min; injected volume, 60 μl). The binding was first recorded in the absence of added Ca2+ in 25 mm HEPES buffer, pH 7.4, 150 mm NaCl. The presence of 5 mm Ca2+ decreased the binding of SAP to immobilized CERT. The general shape of the curves revealed that SAP had very fast association rates with both proteins, although SAP remained bound to GPBP for a longer period at the end of the injection. E, binding of a peptide containing amino acids 385–398 from GPBP exon 11 to immobilized SAP (215 RU; flow rate, 15 μl/min; injected volume, 60 μl) in 25 mm HEPES buffer, pH 7.4, 150 mm NaCl.
We further investigated the molecular characteristics and the kinetics of this binding by SPR. SAP is a highly interactive protein prone to extensive self-aggregation (38). We found that by careful selection of buffer conditions, such as pH and presence/absence of Ca2+ and of detergent, it was possible to control the process of self-association (Fig. 4B). Human SAP was found to be highly aggregated (>250 kDa) at pH 7.4 (with or without Ca2+) and did not migrate into a 4% polyacrylamide gel under native and non-reducing conditions. However, it was found that sonication of SAP in HEPES buffer, pH 7.4, with 0.01% Tween 20 to a large extent prevented this protein aggregation and precipitation; sonication disrupted aggregates, and the presence of the detergent stabilized SAP in physiological pentameric and decameric species. High temperature treatment, high salt, and organic solvent were also tested but resulted in irreversible denaturation (data not shown).
We studied the binding of fluid phase SAP to immobilized GPBP and CERT by SPR (Fig. 4, C and D). Interestingly, we found that SAP (stabilized as pentamers and decamers, as described above) binds both GPBP and CERT in the absence of Ca2+. Conversely, in the presence of 5 mm Ca2+, this binding was reduced 5–7-fold. SAP interaction with both GPBP and CERT was characterized by a rapid association rate; however, SAP binding to GPBP was stronger than to CERT.
To explore in more detail the different affinity of SAP for the two protein isoforms, we immobilized SAP on the sensor chip and applied as analyte a peptide containing 14 (residues 385–398) of the 26 amino acids encoded by exon 11 of GPBP. Exon 11 encodes a short domain that is absent in CERT. This peptide was shown to bind immobilized SAP (Fig. 4E), which supports the notion that this region of GPBP participates in the interaction with SAP.
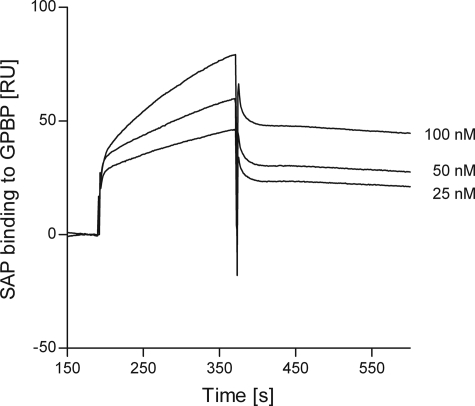
Kinetic analysis of SAP binding to immobilized GPBP was performed by varying the concentrations of SAP added (ranging from 25 to 100 nm) (Fig. 5). We found that the binding of SAP to GPBP does not saturate, suggesting that GPBP-SAP-SAP complexes can be formed, in analogy to what is described for type IV collagen binding above (Fig. 2, A and B). Therefore, these data indicate that SAP binding to GPBP in solid phase is non-saturable, making it difficult to reliably estimate the binding constant. In this respect, the SPR data should be regarded as qualitative evidence, demonstrating the direct binding of proteins involved.
FIGURE 5.
Overlay of sensorgrams resulting from the injection of different concentrations (25–100 nm) of SAP over immobilized GPBP (96 RU, flow rate, 15 μl/min; injected volume, 60 μl). The sensorgrams were corrected for the signal in the empty cell (i.e. calculated as the difference between the signal in Fc2 and the signal in Fc1, which includes injection noise, instrument drift, and nonspecific binding).
Binding of SAP to GPBP in Solution
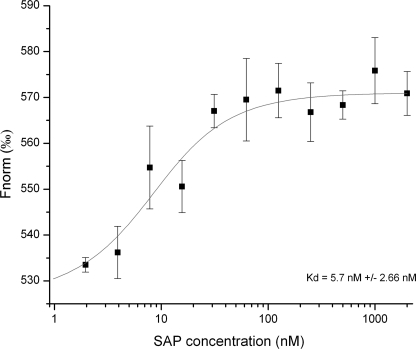
Microscale thermophoresis (MST) was performed in order to determine the dissociation constant of SAP to fluorescently labeled GPBP in fluid phase. 10 nm NT-647-labeled GPBP was mixed with increasing SAP concentrations. After a short incubation time, the samples were loaded into glass capillaries, and a thermophoretic analysis was performed on the Monolith.NT115 using 51% LED power and 80% IR laser power. The normalized fluorescence Fnorm is plotted for different concentrations of SAP. An apparent KD of 5.7 ± 2.66 nm was determined for this interaction (Fig. 6).
FIGURE 6.
SAP binds to GPBP in solution. MST was performed in order to determine the dissociation constant of SAP to fluorescently labeled GPBP. 10 nm NT-647-labeled GPBP was mixed with increasing SAP concentrations. The normalized fluorescence Fnorm is plotted for different concentrations of SAP. A Kd value of 5.7 ± 2.66 nm was determined for this interaction. Error bars, S.D.
Binding of Different Molecular Weight Species of SAP to Immobilized GPBP
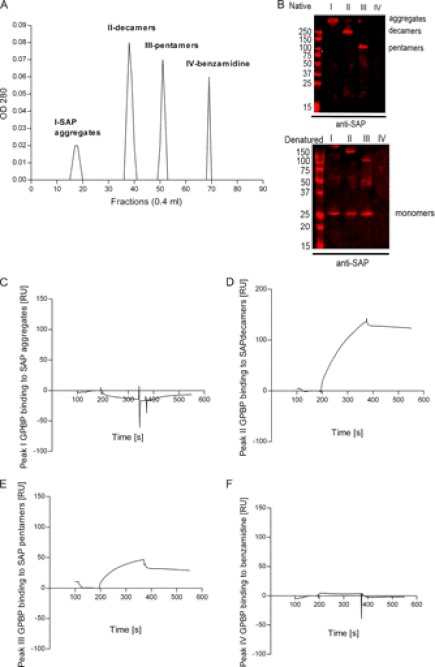
To further study the capacity of the different SAP species to interact with GPBP, we isolated fractions of SAP by size exclusion chromatography for immediate SPR analysis. Three peaks were separated by gel filtration chromatography from SAP (sonicated in HEPES buffer, pH 7.4, with 0.01% Tween 20) (Fig. 7A). SAP peaks I–III corresponded to high molecular aggregates (>250 kDa), decamers (250 kDa), and pentamers (125 kDa) as judged from native polyacrylamide gels (Fig. 7B, Native). SAP peaks separated by SDS-PAGE under reducing conditions corresponded to 25 kDa (monomers) (Fig. 7B, Denatured). SAP decamers and pentamers (peaks II and III, respectively) bound immobilized GPBP, whereas SAP aggregates (peak I) did not (Fig. 7, C–E). Likewise, peak IV, which consists of benzamidine, was found not to bind to GPBP (Fig. 7F).
FIGURE 7.
SAP decameric and pentameric species bind to GPBP. A, purification of different SAP species was performed by Superose 6 column size exclusion chromatography. Samples diluted in 25 mm HEPES buffer (pH 7.4), 150 mm NaCl were then injected at a concentration of 0.5 mg/ml and were chromatographed at a flow rate of 0.4 ml/min. Four peaks were observed. B, fractions from peaks I–IV from the Superose 6 gel filtration column were separated using native PAGE and SDS-PAGE 4–20% gradient gels. The proteins were transferred to nitrocellulose membranes and incubated with anti-SAP antibody. Top, native electrophoresis. Bottom, denatured electrophoresis. The anti-SAP antibody detected bands of the following molecular mass: >250 kDa in peak I, 250 kDa (decamers) in peak II, and 100 kDa (pentamers) in peak III in native conditions and 25 kDa (monomers) in denatured conditions in fractions from peaks I–III. C–F, sensorgrams resulting from the injection of peaks I–IV over immobilized GPBP (96 RU, flow rate, 15 μl/min; injected volume, 60 μl). C, no binding was observed with SAP peak I, which corresponds to high molecular weight SAP aggregates. D and E, 50–150 RU were observed in SAP peak fractions II and III, which correspond to decamers and pentamers, respectively. F, peak IV corresponds to benzamidine and did not bind to immobilized GPBP.
In summary, these data indicate that SAP pentamers and decamers, the physiologically active species, bind to GPBP and that the binding observed (in the previous experiments, Figs. 4, C and D, and 6) was not a result of nonspecific binding of SAP aggregates.
SAP and GPBP Form Complexes in Blood under Physiological Conditions
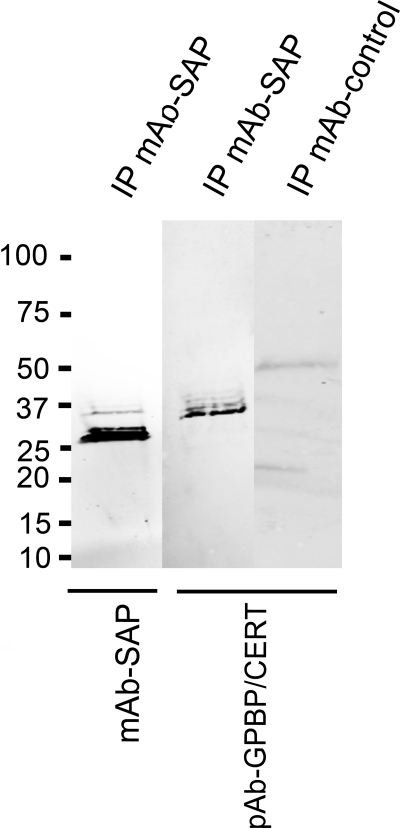
SAP is present in human serum at a concentration between 30 and 50 mg/liter (39, 40). Recently, the presence of GPBP in human serum has been described (23). To study whether SAP associates with GPBP in serum, co-immunoprecipitation experiments were performed. After precipitation with anti-SAP antibodies, both SAP and GPBP were detected by Western blot. Antibodies against GPBP epitopes 1–50 (Fig. 8) and 300–350 (data not shown) detected a GPBP fragment of ∼35–37 kDa, which has been described previously (22). This fragment was not present in a pull-down performed with an isotype control antibody when detected with the same GPBP-specific antibodies. Based on the antibody specificities, these results suggest that SAP associates in the blood with a ∼37-kDa GPBP fragment containing the N-terminal region and the middle domain.
FIGURE 8.
SAP-GPBP complexes in plasma. SAP was immunoprecipitated with mouse mAb anti-SAP (clone 4E8, Sigma), followed by immunoblotting with the same antibody to detect immunoprecipitated SAP (first lane) and a polyclonal antibody against GPBP/CERT 1–50 to detect GPBPs (second lane). The control using an isotype control mAb (mouse monoclonal anti-syntaxin 6, clone 3D10) for immunoprecipitation (IP) was negative when detected with the same polyclonal antibody against GPBP (third lane). GPBP is efficiently co-precipitated with SAP in a band of ∼37 kDa, suggesting that a part of the GPBP molecule corresponding to the N-terminal domains of the protein interacts with SAP. Results shown are representative of five experiments.
GPBP Is Present in Brain Amyloid Plaques
Because SAP is a universal component of all types of amyloid deposits, possible associations of GPBP with amyloid deposits in human brain were studied by immunohistochemistry. Stainings were performed on cryostat sections of post-mortem temporal cortex specimens from aged donors, including non-demented controls, control cases with amyloid deposits and with cortical changes (although not sufficient to be classified as AD), and AD patients (Table 1).
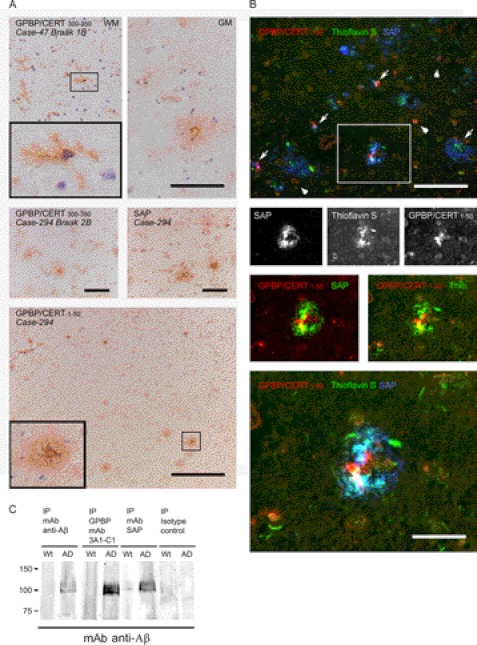
Considerable SAP immunoreactivity was found to be associated with Aβ plaques in the AD and demented cases (81, 135, and 294; Fig. 9A, middle right). SAP was less abundant but also associated with Aβ plaques in the non-demented control cases (not shown). Polyclonal antibodies reacting with GPBP/CERT stained vague, globular plaque-like structures (e.g. case 47 (Fig. 9A, top right) and case 294 (Fig. 9A, middle left)). Occasional dense deposits, possibly representing plaque cores, were observed (case 115; not shown). Plaque-associated microglia were immunoreactive in the AD cases (e.g. case 264; Fig. 9B). Additionally, microglia in the white matter were strongly immunopositive for GPBP/CERT in all cases (e.g. case 47 (Fig. 9A, top left)). In the demented control case 294, plaques and associated microglia in the gray matter were strongly immunoreactive (Fig. 9A, bottom), as were white matter microglia.
FIGURE 9.
GPBP is present in amyloid plaques. Shown are immunohistochemical stainings of GPBP/CERT and SAP in midtemporal cortex of AD and demented and non-demented control cases (cryostat sections). A, top, control case 47 (Braak 1B) white matter (WM) immunostained with anti-GPBP/CERT 300–350 shows many ramified microglia (see inset) and to some extent endothelial lining of blood vessels, whereas in the gray matter (GM), amyloid plaque staining and intense GPBP staining around the plaque core, reminiscent of clustered microglia, and in addition cytoplasmic staining of neuronal cells are seen. Bottom, demented control case (Case-294) with cortical changes but not sufficient to be classified as AD (Braak 2B). GPBP/CERT 1–50 immunostaining is seen in Aβ plaques and microglia, especially microglia in the white matter (top right) and near the core of a classical plaque (see also the higher magnification inset). Similar to anti-GPBP/CERT 1–50, plaque and microglial staining was seen with anti-GPBP/CERT 300–350, but less pronounced. With anti-SAP, in addition to plaques, also some cytoplasmic neuronal staining was observed (two central panels). B, immunofluorescence staining was performed to further investigate co-localization of GPBP and SAP. Fibrillar Aβ deposits in an AD case (case 264) were visualized with thioflavin S, SAP with monoclonal SAP-14, followed by goat anti-mouse HRP and rhodamine tyramide and GPBP with anti-GPBP/CERT 1–50- and Cy5-labeled goat anti-rabbit. Although both SAP and GPBP immunoreactivity co-localize with thioflavin positivity, their exact distribution differs (black and white images showing complete overlap in localization of thioflavin and SAP but not of GPBP). GPBP seems to be present in small cells resembling microglia clustered around Aβ and SAP deposits (arrow and lower panel (magnification)) as well as in many neuronal cells (arrowheads). Partial overlap in localization of GPBP and SAP within Aβ plaques was more clearly seen, when in the separate channels the GPBP signal was visualized as red and thioflavin or SAP as green (insets). Scale bars, 100 μm. C, Western blotting with a mAb anti-Aβ revealed that immunoprecipitation of either GPBP or SAP from brain lysates from an AD mouse model (APPswe/PS1ΔE9), but not from wild type littermates, co-isolated Aβ, which suggests the presence of Aβ-SAP and Aβ-GPBP complexes in AD model mouse brain. Aβ, GPBP, and SAP were immunoprecipitated from brain homogenates of control and Alzheimer transgenic mice using mouse mAb anti-SAP, clone 4E8 (Sigma) or mAb GPBP clone 3A1-C1, mAb β amyloid clone 6E10 (Covance). immunoprecipitation with nonspecific immunoglobulins was performed for control. Results shown are representative of three experiments.
In order to determine possible colocalization of GPBP/CERT with SAP in tissue specimens, double immunofluorescent stainings were performed. To visualize fibrillar Aβ deposits, sections were also stained with thioflavin S. In AD case 264, SAP was found to completely colocalize with fibrillar Aβ in plaques, whereas GPBP/CERT was associated with cellular structures in the center of the plaque, possibly representing recruited microglia partially colocalizing with SAP (Fig. 9B).
A number of GPBP-immunopositive neuronal cells were observed, also in plaques; occasionally, the endothelial cell lining as well as the basement membrane of a blood vessel were GPBP-immunopositive (Fig. 9B, top).
Additionally, we performed immunoprecipitation experiments using homogenates from control and AD mouse brains to confirm the interaction of SAP with GPBP and to distinguish between GPBP and CERT (Fig. 9C). Immunoprecipitation of either GPBP (mAb 3A1-C1) or SAP from brain lysates from the AD mouse model co-isolated Aβ (detected with 6E10) in a few bands of ∼100 kDa. The same bands were also found in a control experiment following immunoprecipitation of Aβ from AD mouse brain with a combination of recombinant human antibodies recognizing different aggregation forms of Aβ (bapineuzumab directed against plaques, 20C2 directed against fibrils, solanezumab directed against monomers). In wild type littermates, these bands were not observed. The observed Aβ bands (∼100 kDa) could correspond to Aβ aggregates that are preserved using the tissue disruption protocol and/or to amyloid precursor protein (APP). The possible presence of APP would not be surprising because it is several times up-regulated in this animal model. Lower Aβ molecular weight forms were not detected using anti-Aβ (6E10), as reported previously (41).
Western blotting with a monoclonal anti-Aβ revealed that immunoprecipitation of either GPBP (mAb 3A1-C1) or SAP from brain lysates from an AD mouse model, but not from wild type littermates, co-isolated Aβ, which suggests the presence of Aβ-SAP and Aβ-GPBP complexes in AD model mouse brain. Immunoprecipitation with nonspecific immunoglobulins performed as a control did not co-isolate Aβ.
Functional Implications of SAP for GPBP Binding and Protein-Protein Interaction Interface Prediction
In order to identify the binding site of GPBP on SAP, we used two well known ligands of SAP (C1q and PE) (16, 32) and analyzed by SPR if they could compete with GPBP. To test this, either SAP alone or SAP that had been preincubated with either PE or C1q was added as analytes and flown over a surface onto which GPBP was immobilized. Using an equimolar ratio of SAP and C1q, binding of SAP to GPBP was inhibited by 59%. In contrast, SAP preincubated with PE even at a 3-fold molar ratio of PE as compared with SAP had no effect on the binding of SAP to GPBP.
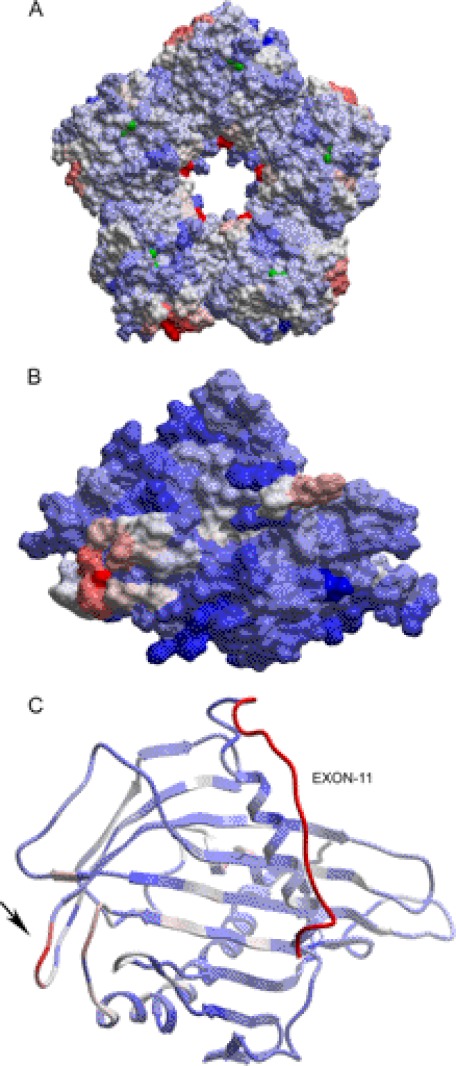
Because the crystal structures of SAP and the GPBP START domain are published (Protein Data Bank entries 2A3Y and 2E3O), we attempted to predict the possible protein-protein interaction surfaces by structural bioinformatics analysis using the ICM-pro package (Fig. 10). For SAP, our in silico analysis indicated the presence of two potential protein interaction hotspots with a 5-fold symmetry in the pentamer: one on the internal surface of the pentameric ring (Fig. 10A, red) around Glu86 and a second one more dispersed on the outer side of the ring, around residues Gly122–Phe124 (light red). In the GPBP START domain, an interactive region is located at the protein surface near Ala501 (Fig. 10, B and C, arrow).
FIGURE 10.
Protein-protein interaction interface prediction of SAP with GPBP. A, human SAP pentameric structure and predicted protein-protein interaction hot spots as obtained after structural analysis using the ICM Pro package of the published coordinates from 2A3Y.pdb (51). B, homology model for the structure of GPBP, including the region important for the interaction with SAP (shown in red). The homology model was built using the WhatIf-Yasara Twinset and ICM Pro package, whereas the coordinate file 2E3O.pdb (CERT START domain structure) was used as a template. C, GPBP structure and predicted protein-protein interaction hot spots based on the homology model as shown in B. A structural bioinformatics analysis was performed using ICM Pro to predict potential protein-protein interactions. Interaction scores are color-coded, with red representing high potential for interaction and blue indicating low potential. On the right, exon 11 containing 26 amino acids is indicated in red. The arrow indicates a hypothetical protein-protein interaction region.
Binding of CERT Mutants to SAP
In order to experimentally test the importance of the START domain for SAP binding, we generated five CERT mutants containing two amino acid substitutions each. We targeted the region near Ala501 and three additional regions with hydrophobic amino acids near the protein surface. Protein surface hydrophobicity is known to be a good determinant of protein-protein recognition (42). For consistency with the numbering used above, we will refer to the GPBP amino acid numbering throughout (see Table 2).
TABLE 2.
Mutagenesis of GPBPΔ26/CERT protein to identify amino acids relevant for SAP interaction
| Mutanta | Mutation at GPBPb protein level | Corresponding mutation at CERTc protein level |
|---|---|---|
| 1 | I439A/W588A | I413A/W562A |
| 2 | N460A/Y461A | N434A/Y435A |
| 3 | Y461A/F462A | Y435A/F436A |
| 4 | V498A/W499A | V472A/W473A |
| 5 | P500G/A501Q | P474G/A475Q |
a Mutants listed according to the position of the mutation in the cDNA.
b NP_005704.1.
c NP_112729.1.
In mutant 1, Ile439 (present in a short β-strand that flanks the N terminus of Ala591–Ala618 helix) and Trp588 (located in a loop structure that precedes the Ala591–Ala618 helix) are predicted to be arranged closely in the three-dimensional structure of GPBP. This hydrophobic pair represents a potential protein contact site that was effectively removed by substitution for two less hydrophobic Ala residues. In mutant 2, Asn460 and the adjacent Tyr461 (present in a short α-helix that is located on the opposite side of the molecule as compared with the other mutations) are substituted for two Ala residues, thereby removing a potential protein-interacting site. In mutant 3, adjacent to the changed amino acids of mutant 2, the hydrophobic Tyr461 and Phe462 residues were replaced with Ala residues. Phe462 faces the core of the protein. In mutant 4, we substituted Val498 and Trp499 with two Ala residues. These hydrophobic residues are located in an unstructured loop. In mutant 5, the adjacent Pro500 and Ala501 were substituted with Gly and Gln, respectively.
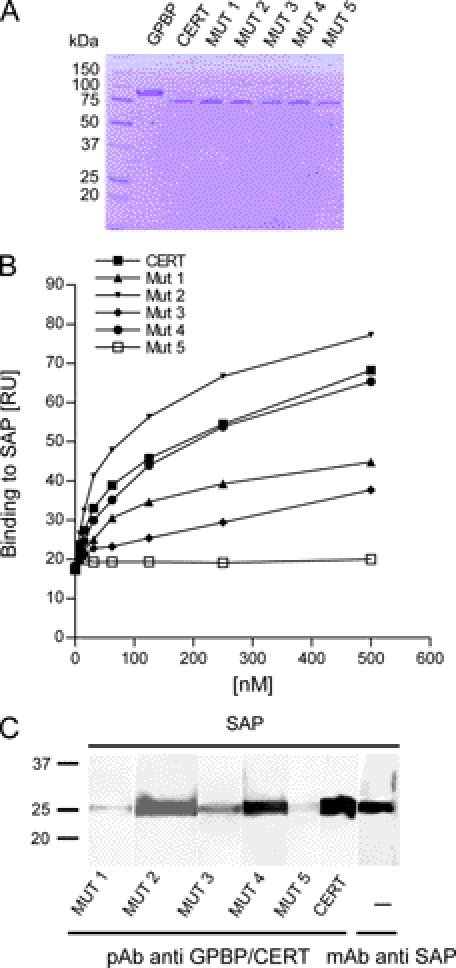
All mutants described above and the wild type protein were expressed in E. coli and purified on a FLAG column (Fig. 11A). Using SAP as ligand, the relative affinity of CERT and CERT mutant 1–5 proteins were tested by SPR (Fig. 11B). The binding of mutants 2 and 4 to SAP was similar to the wild type CERT, whereas mutants 1 and 3 showed reduced binding, and mutant 5 showed a greatly reduced binding for SAP.
FIGURE 11.
Binding of CERT and CERT mutants to SAP. A, Coomassie staining of immunopurified wild type and mutant 1–5 CERT proteins (2 μg of each sample were loaded per lane). B, 2-fold serial dilutions (3.9–500 nm) of wild type and mutant 1–5 CERT proteins in 25 mm HEPES, pH 7.4, 150 mm NaCl with 0.01% Tween 20 were tested for binding on immobilized SAP by SPR. At each concentration, the highest binding signal was measured. C, SAP was separated on a 12% SDS-polyacrylamide gel, electroblotted to nitrocellulose, and prepared for far Western analyses as described in the legend to Fig. 4A. The membrane with renatured SAP was cut into strips and probed with CERT mutants (lanes 1–5), WT (lane 6), or mAb anti-SAP (lane 7). Membrane strips incubated with CERT proteins were detected with anti-GPBP/CERT 300–350.
The relative affinity of CERT and CERT mutant 1–5 proteins was also tested by far Western blot with in blot renaturation (Fig. 11C). The results were similar to those obtained by SPR and showed reduced binding of mutants 1, 3, and 5 to SAP. In conclusion, the START domain of GPBP and CERT significantly contributes to (or might even be essential for) SAP binding.
DISCUSSION
Our results demonstrate, for the first time, that GPBP binds SAP. Both GPBP and SAP are present in amyloid plaques and co-precipitate with Aβ. Therefore, the interaction of GPBP with SAP might be involved in protein aggregation in Alzheimer disease and the resulting innate immune response.
SAP is a very compact and highly structured molecule. Its incorporation within specific types of BM may contribute to the structural conformation and consequently the correct functionality of extracellular matrix proteins. This is supported by its altered distribution in the GBM in glomerular diseases (33, 43).
Our SPR analysis has shown that GPBP binds type IV collagen, laminin, and SAP, whereas our immunohistochemical studies have shown the presence of SAP and GPBP in GBMs. Intriguingly, this raises the possibility that SAP and GPBP interact to support matrix extracellular proteins, helping to maintain their biologically active conformations in a particular group of tissues, including kidney, lungs, and choroid plexus, tissues that perform a filtering function and are therefore more vulnerable.
We also found SAP and GPBP interaction in human blood. By immunoprecipitation studies in human sera, we were able to pull down complexes of SAP and an N-terminal ∼37-kDa fragment of GPBP that has been described previously (22). However, it is worth noting that proteolysis of GPBP remains a possibility because our in vitro mutagenesis shows that the (C-terminal) START domain of GPBP is important for interaction with SAP. Moreover, full-length GPBP is the only isoform that has been described so far that is secreted and present in blood (23). Because this N-terminal domain is identical in the GPBP and CERT isoforms, our results demonstrate that both GPBP and the shorter isoform CERT could form complexes with SAP under physiological conditions. Several studies have investigated the three-dimensional structure of SAP in order to establish whether SAP stably associates with another protein or exists free in solution (44–46). Despite these efforts, it remains unclear how SAP can remain in physiological conformation (i.e. avoid aggregation) in a high Ca2+ environment. One suggestion is that SAP forms a complex with an as yet unidentified low molecular weight component that prevents self-association (6). To date, no such factor or protein has been reported. In our experiments, aggregation of SAP by Ca2+ might have reduced binding to GPBP, because GPBP specifically binds to pentamers and decamers.
Our molecular characterization of the SAP GPBP/CERT interaction suggests that residues Pro500 and Ala501 (mutant 5) of the START domain of GPBP/CERT are essential. Although we cannot rule out the possibility that the loss of SAP to CERT binding affinity seen with mutant 5 is due to drastic changes in protein conformation, our results are in line with what is known about this domain. The START domain of CERT binds one ceramide molecule in its central amphiphilic cavity and mediates ceramide transfer from donor to acceptor membranes (24). The Ω1 loop region of the CERT START domain (496KRVWPAS502 in GPBP) is an important regulatory element for the binding of ceramide (47, 48). This Ω1 loop has been proposed to function as a gate of the cavity. Specifically, the interaction between the Trp499 residue of the Ω1 loop and the membrane results in a conformational change of the protein that allows opening of the cavity. We found that substitution of either the Val498 or Trp499 had no effect on the SAP to GPBP binding, whereas mutation of the Pro500 and Ala501 abolishes SAP-GPBP interaction. This suggests that GPBP binding to SAP is independent of the ceramide transfer function. This would be consistent with the notion that ceramide function is important within the cell and that the SAP GPBP interaction described here occurs primarily extracellularly.
SAP has been observed decorating the majority of Aβ plaques in AD (49), where it has been found to enhance Aβ fibrilization. Also, SAP has been shown to hamper the uptake of Aβ by adult human microglia in vitro (50).
The binding of SAP and GPBP to plaques in AD raises the important question of whether this interaction has a role in disease progression. It may be the case that GPBP and SAP are found in the pathological plaques of AD because these plaques, that consist of amyloid deposits, are “sticky” and thus incorporate GBPB and SAP from the extracellular matrix. On the other hand, it has been reported that binding of SAP to amyloid fibrils slows proteolysis of plaque material and contributes to the pathogenesis of amyloidosis (5). Here we show that GPBPs are present in Aβ plaques, localized in the core and partially colocalizing with SAP. In agreement with this, pull-downs of extracts of AD mouse brain with antibodies against Aβ yield high molecular weight aggregates containing GPBP, indicating that GPBP, similarly to SAP, interacts with Aβ.
Additionally, we observed GPBP staining in the white matter corresponding to round and ramified microglia. Interestingly, clustering of microglia was only observed in SAP- and C1q-immunopositive Aβ deposits, and has been shown to precede neurodegenerative changes in AD brain (4).
SAP, when aggregated, can bind C1q and initiate the activation of the complement system (16). Our finding that C1q and GPBP can compete for their binding to SAP raises the question as to whether GPBP and C1q share an identical SAP binding site. This implies that when SAP binds to GPBP, the C1q binding site on the SAP molecule is blocked, and this might help to control the activation of the complement system.
These findings may then have important implications in several human diseases where complement activation plays a role and SAP and GPBP are in close proximity (e.g. in AD). Our results further raise the question of whether GPBP, similarly to SAP, could serve as an innate immune system regulator.
Acknowledgments
We are very grateful to Eline van der Esch and Marlies Jacobs for excellent technical assistance, to Anna Carrano (Pathology Department, Vrije Universiteit University Medical Center, Amsterdam) for preparing the human brain fluorescence micrographs, and to Roy L. J. Schrijver for excellent technical assistance with the SPR technology. We thank Prof. T. Hackeng (Department of Biochemistry, Cardiovascular Research Institute Maastricht, Maastricht University) for the kind gift of the purified SAP and for advice with the FPLC. We also thank Moran Jerabek-Willemsen for help with the MST technology. We are very grateful to Prof. W. Buurman for critical reading of the manuscript, advice, and encouragement.
This work was supported by Netherlands Organization for Scientific Research Grant 91107006 (to G. A. F. N.) and a grant from the Transnationale Universiteit Limburg (to G. A. F. N.).
- SAP
- serum amyloid P component
- AD
- Alzheimer disease
- Aβ
- amyloid-β
- CERT
- ceramide transporter
- SPR
- surface plasmon resonance
- GPBP
- Goodpasture antigen-binding protein
- BM
- basal membrane
- GBM
- glomerular basal membrane
- MST
- microscale thermophoresis technology
- C4BP
- C4b-binding protein
- RU
- resonance units
- APP
- amyloid precursor protein
- PE
- phosphatidyl ethanolamine.
REFERENCES
- 1. Ashton A. W., Boehm M. K., Gallimore J. R., Pepys M. B., Perkins S. J. (1997) Pentameric and decameric structures in solution of serum amyloid P component by x-ray and neutron scattering and molecular modeling analyses. J. Mol. Biol. 272, 408–422 [DOI] [PubMed] [Google Scholar]
- 2. Cathcart E. S., Comerford F. R., Cohen A. S. (1965) Immunologic Studies on a Protein Extracted from Human Secondary Amyloid. N. Engl. J. Med. 273, 143–146 [DOI] [PubMed] [Google Scholar]
- 3. Schwab C., McGeer P. L. (2008) Inflammatory aspects of Alzheimer disease and other neurodegenerative disorders. J. Alzheimers Dis. 13, 359–369 [DOI] [PubMed] [Google Scholar]
- 4. Veerhuis R., Van Breemen M. J., Hoozemans J. M., Morbin M., Ouladhadj J., Tagliavini F., Eikelenboom P. (2003) Amyloid β plaque-associated proteins C1q and SAP enhance the Aβ1–42 peptide-induced cytokine secretion by adult human microglia in vitro. Acta Neuropathol. 105, 135–144 [DOI] [PubMed] [Google Scholar]
- 5. Tennent G. A., Lovat L. B., Pepys M. B. (1995) Serum amyloid P component prevents proteolysis of the amyloid fibrils of Alzheimer disease and systemic amyloidosis. Proc. Natl. Acad. Sci. U.S.A. 92, 4299–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Painter R. H., De Escallón I., Massey A., Pinteric L., Stern S. B. (1982) The structure and binding characteristics of serum amyloid protein (9.5S α 1-glycoprotein). Ann. N.Y. Acad. Sci. 389, 199–215 [DOI] [PubMed] [Google Scholar]
- 7. Hamazaki H. (1995) Ca2+-dependent binding of human serum amyloid P component to Alzheimer β-amyloid peptide. J. Biol. Chem. 270, 10392–10394 [DOI] [PubMed] [Google Scholar]
- 8. Hamazaki H. (1987) Ca2+-mediated association of human serum amyloid P component with heparan sulfate and dermatan sulfate. J. Biol. Chem. 262, 1456–1460 [PubMed] [Google Scholar]
- 9. Zahedi K. (1996) Characterization of the binding of serum amyloid P to type IV collagen. J. Biol. Chem. 271, 14897–14902 [DOI] [PubMed] [Google Scholar]
- 10. Zahedi K. (1997) Characterization of the binding of serum amyloid P to laminin. J. Biol. Chem. 272, 2143–2148 [PubMed] [Google Scholar]
- 11. Nielsen E. H., Sørensen I. J., Vilsgaard K., Andersen O., Svehag S. E. (1994) Calcium-enhanced aggregation of serum amyloid P component and its inhibition by the ligands heparin and heparan sulfate. An electron microscopic and immunoelectrophoretic study. APMIS 102, 420–426 [DOI] [PubMed] [Google Scholar]
- 12. de Beer F. C., Baltz M. L., Holford S., Feinstein A., Pepys M. B. (1981) Fibronectin and C4-binding protein are selectively bound by aggregated amyloid P component. J. Exp. Med. 154, 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bristow C. L., Boackle R. J. (1986) Evidence for the binding of human serum amyloid P component to Clq and Fab γ. Mol. Immunol. 23, 1045–1052 [DOI] [PubMed] [Google Scholar]
- 14. Hutchcraft C. L., Gewurz H., Hansen B., Dyck R. F., Pepys M. B. (1981) Agglutination of complement-coated erythrocytes by serum amyloid P component. J. Immunol. 126, 1217–1219 [PubMed] [Google Scholar]
- 15. García de Frutos P., Härdig Y., Dahlbäck B. (1995) Serum amyloid P component binding to C4b-binding protein. J. Biol. Chem. 270, 26950–26955 [DOI] [PubMed] [Google Scholar]
- 16. Ying S. C., Gewurz A. T., Jiang H., Gewurz H. (1993) Human serum amyloid P component oligomers bind and activate the classical complement pathway via residues 14–26 and 76–92 of the A chain collagen-like region of C1q. J. Immunol. 150, 169–176 [PubMed] [Google Scholar]
- 17. Dyck R. F., Lockwood C. M., Kershaw M., McHugh N., Duance V. C., Baltz M. L., Pepys M. B. (1980) Amyloid P-component is a constituent of normal human glomerular basement membrane. J. Exp. Med. 152, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tseng J., Mortensen R. F. (1986) Binding specificity of mouse serum amyloid P-component for fibronectin. Immunol. Invest. 15, 749–761 [DOI] [PubMed] [Google Scholar]
- 19. Stanton M. C., Tange J. D. (1958) Goodpasture's syndrome (pulmonary hemorrhage associated with glomerulonephritis). Australas. Ann. Med. 7, 132–144 [DOI] [PubMed] [Google Scholar]
- 20. Raya A., Revert F., Navarro S., Saus J. (1999) Characterization of a novel type of serine/threonine kinase that specifically phosphorylates the human Goodpasture antigen. J. Biol. Chem. 274, 12642–12649 [DOI] [PubMed] [Google Scholar]
- 21. Mencarelli C., Losen M., Hammels C., De Vry J., Hesselink M. K., Steinbusch H. W., De Baets M. H., Martínez-Martínez P. (2010) The ceramide transporter and the Goodpasture antigen binding protein. One protein, one function? J. Neurochem. 113, 1369–1386 [DOI] [PubMed] [Google Scholar]
- 22. Revert F., Ventura I., Martínez-Martínez P., Granero-Moltó F., Revert-Ros F., Macías J., Saus J. (2008) Goodpasture antigen-binding protein is a soluble exportable protein that interacts with type IV collagen. Identification of novel membrane-bound isoforms. J. Biol. Chem. 283, 30246–30255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saus J. V., Revert F. M. (May 3, 2011) Goodpasture antigen-binding protein and its detection, U.S. Patent no. US7935492
- 24. Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 25. Raya A., Revert-Ros F., Martinez-Martinez P., Navarro S., Rosello E., Vieites B., Granero F., Forteza J., Saus J. (2000) Goodpasture antigen-binding protein, the kinase that phosphorylates the Goodpasture antigen, is an alternatively spliced variant implicated in autoimmune pathogenesis. J. Biol. Chem. 275, 40392–40399 [DOI] [PubMed] [Google Scholar]
- 26. Jönsson U., Fägerstam L., Ivarsson B., Johnsson B., Karlsson R., Lundh K., Löfås S., Persson B., Roos H., Rönnberg I. (1991) Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. BioTechniques 11, 620–627 [PubMed] [Google Scholar]
- 27. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baaske P., Wienken C. J., Reineck P., Duhr S., Braun D. (2010) Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed. Engl. 49, 2238–2241 [DOI] [PubMed] [Google Scholar]
- 29. Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S. (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100. [DOI] [PubMed] [Google Scholar]
- 30. Mencarelli C., Hammels C., Van Den Broeck J., Losen M., Steinbusch H., Revert F., Saus J., Hopkins D. A., De Baets M. H., Steinbusch H. W., Martinez-Martinez P. (2009) The expression of the Goodpasture antigen-binding protein (ceramide transporter) in adult rat brain. J. Chem. Neuroanat. 38, 97–105 [DOI] [PubMed] [Google Scholar]
- 31. Braak H., Braak E. (1995) Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol. Aging 16, 271–278; discussion 278–284 [DOI] [PubMed] [Google Scholar]
- 32. Familian A., Zwart B., Huisman H. G., Rensink I., Roem D., Hordijk P. L., Aarden L. A., Hack C. E. (2001) Chromatin-independent binding of serum amyloid P component to apoptotic cells. J. Immunol. 167, 647–654 [DOI] [PubMed] [Google Scholar]
- 33. Melvin T., Kim Y., Michael A. F. (1986) Amyloid P component is not present in the glomerular basement membrane in Alport-type hereditary nephritis. Am. J. Pathol. 125, 460–464 [PMC free article] [PubMed] [Google Scholar]
- 34. al-Mutlaq H., Wheeler J., Robertson H., Watchorn C., Morley A. R. (1993) Tissue distribution of amyloid P component as defined by a monoclonal antibody produced by immunization with human glomerular basement membranes. Histochem. J. 25, 219–227 [DOI] [PubMed] [Google Scholar]
- 35. Potempa L. A., Kubak B. M., Gewurz H. (1985) Effect of divalent metal ions and pH upon the binding reactivity of human serum amyloid P component, a C-reactive protein homologue, for zymosan. Preferential reactivity in the presence of copper and acidic pH. J. Biol. Chem. 260, 12142–12147 [PubMed] [Google Scholar]
- 36. Chan C. S., Winstone T. M., Turner R. J. (2008) Investigating protein-protein interactions by far Westerns. Adv. Biochem. Eng. Biotechnol. 110, 195–214 [DOI] [PubMed] [Google Scholar]
- 37. Burgess R. R., Arthur T. M., Pietz B. C. (2000) Mapping protein-protein interaction domains using ordered fragment ladder far Western analysis of hexahistidine-tagged fusion proteins. Methods Enzymol. 328, 141–157 [DOI] [PubMed] [Google Scholar]
- 38. Baltz M. L., De Beer F. C., Feinstein A., Pepys M. B. (1982) Calcium-dependent aggregation of human serum amyloid P component. Biochim. Biophys. Acta 701, 229–236 [DOI] [PubMed] [Google Scholar]
- 39. Pepys M. B., Dash A. C., Markham R. E., Thomas H. C., Williams B. D., Petrie A. (1978) Comparative clinical study of protein SAP (amyloid P component) and C-reactive protein in serum. Clin. Exp. Immunol. 32, 119–124 [PMC free article] [PubMed] [Google Scholar]
- 40. Bottazzi B., Doni A., Garlanda C., Mantovani A. (2010) An integrated view of humoral innate immunity. Pentraxins as a paradigm. Annu. Rev. Immunol. 28, 157–183 [DOI] [PubMed] [Google Scholar]
- 41. Jankowsky J. L., Younkin L. H., Gonzales V., Fadale D. J., Slunt H. H., Lester H. A., Younkin S. G., Borchelt D. R. (2007) Rodent A β modulates the solubility and distribution of amyloid deposits in transgenic mice. J. Biol. Chem. 282, 22707–22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Young L., Jernigan R. L., Covell D. G. (1994) A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 3, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dyck R. F., Evans D. J., Lockwood C. M., Rees A. J., Turner D., Pepys M. B. (1980) Amyloid P-component in human glomerular basement membrane. Abnormal patterns of immunofluorescent staining in glomerular disease. Lancet 2, 606–609 [DOI] [PubMed] [Google Scholar]
- 44. Hohenester E., Hutchinson W. L., Pepys M. B., Wood S. P. (1997) Crystal structure of a decameric complex of human serum amyloid P component with bound dAMP. J. Mol. Biol. 269, 570–578 [DOI] [PubMed] [Google Scholar]
- 45. Thompson D., Pepys M. B., Tickle I., Wood S. (2002) The structures of crystalline complexes of human serum amyloid P component with its carbohydrate ligand, the cyclic pyruvate acetal of galactose. J. Mol. Biol. 320, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 46. Haverkate F., Thompson S. G., Pyke S. D., Gallimore J. R., Pepys M. B. (1997) Production of C-reactive protein and risk of coronary events in stable and unstable angina. European Concerted Action on Thrombosis and Disabilities Angina Pectoris Study Group. Lancet 349, 462–466 [DOI] [PubMed] [Google Scholar]
- 47. Kudo N., Kumagai K., Matsubara R., Kobayashi S., Hanada K., Wakatsuki S., Kato R. (2010) Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer. J. Mol. Biol. 396, 245–251 [DOI] [PubMed] [Google Scholar]
- 48. Kudo N., Kumagai K., Tomishige N., Yamaji T., Wakatsuki S., Nishijima M., Hanada K., Kato R. (2008) Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc. Natl. Acad. Sci. U.S.A. 105, 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhan S. S., Veerhuis R., Kamphorst W., Eikelenboom P. (1995) Distribution of β amyloid-associated proteins in plaques in Alzheimer disease and in the non-demented elderly. Neurodegeneration 4, 291–297 [DOI] [PubMed] [Google Scholar]
- 50. Familian A., Eikelenboom P., Veerhuis R. (2007) Minocycline does not affect amyloid β phagocytosis by human microglial cells. Neurosci. Lett. 416, 87–91 [DOI] [PubMed] [Google Scholar]
- 51. Ho J. G., Kitov P. I., Paszkiewicz E., Sadowska J., Bundle D. R., Ng K. K. (2005) Ligand-assisted aggregation of proteins. Dimerization of serum amyloid P component by bivalent ligands. J. Biol. Chem. 280, 31999–32008 [DOI] [PubMed] [Google Scholar]
- 52. Braak H., Braak E. (1991) Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]