Background: Proteasome substrates are recognized by (poly)ubiquitin receptors or mediated by ubiquitin-like domain-containing shuttles.
Results: Multiple ubiquitin-like domain-containing proteins bind transiently to different sites at proteasome subunit Rpn1.
Conclusion: Rpn1 and Rpn2 coordinate substrate shuttles, ubiquitin receptors, and deubiquitination enzymes in proximity at the proteasome.
Significance: Targeting of (poly)ubiquitin conjugates to the proteasome depends as much on auxiliary factors and substrate shuttles as on direct recognition of the ubiquitin chain.
Keywords: Deubiquitination, Proteasome, Surface Plasmon Resonance (SPR), Ubiquitin, Ubiquitin-dependent Protease, Solenoid
Abstract
Substrates tagged with (poly)ubiquitin for degradation can be targeted directly to the 26 S proteasome where they are proteolyzed. Independently, ubiquitin conjugates may also be delivered by bivalent shuttles. The majority of shuttles attach to the proteasome through a ubiquitin-like domain (UBL) while anchoring cargo at a C-terminal polyubiquitin-binding domain(s). We found that two shuttles of this class, Rad23 and Dsk2, dock at two different receptor sites embedded within a single subunit of the 19 S proteasome regulatory particle, Rpn1. Their association/dissociation constants and affinities for Rpn1 are similar. In contrast, another UBL-containing protein, the deubiquitinase Ubp6, is also anchored by Rpn1, yet it dissociates slower, thus behaving as an occasional proteasome subunit that is distinct from the transiently associated shuttles. Two neighboring subunits, Rpn10 and Rpn13, show a marked preference for polyubiquitin over UBLs. Rpn10 attaches to the central solenoid portion of Rpn1, although this association is stabilized by the presence of a third subunit, Rpn2. Rpn13 binds directly to Rpn2. These intrinsic polyubiquitin receptors may compete with substrate shuttles for their polyubiquitin-conjugate cargos, thereby aiding release of the emptied shuttles. By binding multiple ubiquitin-processing factors simultaneously, Rpn1 is uniquely suited to coordinate substrate recruitment, deubiquitination, and movement toward the catalytic core. The broad range of affinities for ubiquitin, ubiquitin-like, and non-ubiquitin signals by adjacent yet nonoverlapping sites all within the base represents a hub of activity that coordinates the intricate relay of substrates within the proteasome, and consequently it influences substrate residency time and commitment to degradation.
Introduction
The ubiquitin proteasome system is the main proteolytic pathway in eukaryotic cells by which intracellular proteins are recycled and kept in check. In most cases, substrates are first conjugated to polyubiquitin chains (polyUb),3 with the resulting polyUb conjugates subsequently recognized and degraded by the 26 S proteasome, a large proteolytic complex (1). These two fundamental and nominally independent steps are subject to a multitude of regulatory interactions that determine the selectivity and efficiency of the system. The 26 S proteasome is a 2.5-MDa molecular machine built from over 33 highly conserved subunits (2) arranged into two subcomplexes as follows: the aptly named 20 S core particle (CP), which contains the protease subunits, and the 19 S regulatory particle (RP). PolyUb conjugates are recognized by the 19 S RP, which unfolds and translocates substrates through a narrow gated pore into the secluded inner chamber of the barrel-shaped 20 S CP where they are proteolyzed into short peptides. A distal Lid subcomplex caps the 19 S RP structure thereby enclosing a cavity where substrates are held as they are unfolded. The Lid also contains a metalloprotease subunit and is therefore involved in processing of polyUb conjugates (3, 4).
Proximal to the 20 S CP is a subcomplex of the 19 S RP known as the Base, which can unfold substrates on its own (5). The Base is composed of 10 integral subunits. Six members of the AAA (ATPases associated with an assortment of cellular activities) family of ATPases, Rpt1–6, are responsible for the protein unfolding, and by docking onto the α-ring, they also stabilize the interaction of the 19 S RP with the 20 S CP (5–8). The four other subunits within the Base subcomplex are all non-ATPases and are prefixed “Rpn” for “regulatory particle non-ATPase” (Fig. 1). Rpn10 and Rpn13 bind polyUb. The other two, Rpn1 and Rpn2, are the largest proteasomal subunits (at 110 and 104 kDa, respectively). These large flexible scaffolds with affinity for ubiquitin-like (UBL) domains are the focus of this study. Currently, their function is unclear, as is the case for most of the other regulatory particle non-ATPase subunits of the 19 S complex. Their sheer size makes them enticing to study and tempting to picture as fundamental, if not central, to the function of the 19 S RP. Deletion of either subunit is lethal, and mutations in yeast result in impaired proteasome function, accumulation of polyubiquitinated proteins, and improper nuclear proteasome localization (9–12).
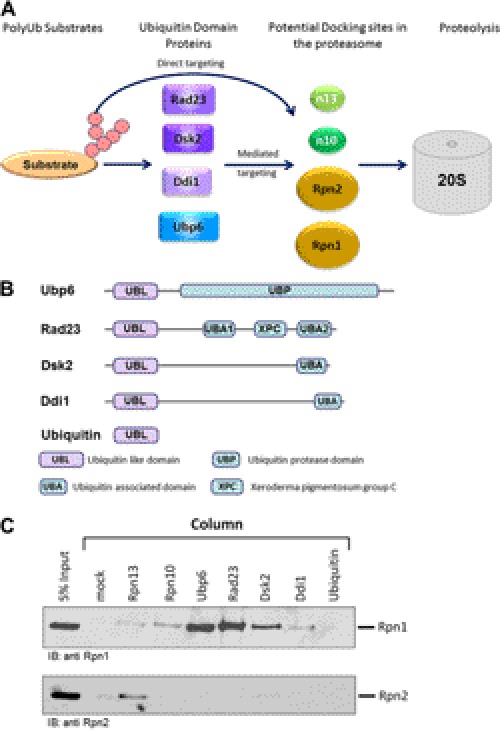
FIGURE 1.
Rpn1 and Rpn2 dock ubiquitin processing factors. A, a number of UDPs serve as bifunctional shuttles by interacting simultaneously with polyubiquitinated cargo and with receptors at the proteasome. PolyUb conjugates can also dock at the proteasome directly. Four proteasome subunits (Rpn1, Rpn2, Rpn10, and Rpn13) have been implicated in recruiting ubiquitin chains or UBLs. B, most UDPs contain an N-terminal UBL followed by a unique functional domain. Some bind polyUb through a UBA. Rad23 contains two UBA domains, UBA1 and UBA2, which share 27% sequence identity, and an XPC (xeroderma pigmentosum group C complementing protein) domain between the two (92). Dsk2 and Ddi1 contain only one UBA domain, with Ddi1 also having an internal retroviral aspartyl protease domain (93). Ubp6 associates with ubiquitin via its UBP. Protein domains are drawn roughly to scale. C, immobilized ubiquitin processing factors (Rpn13, Rpn10, Ubp6, Rad23, Dsk2, and Ddi1) as well as ubiquitin were each tested for binding Rpn1 or Rpn2 proteins. Rpn is regulatory particle non-ATPase. IB, immunoblot.
Rpn1 and Rpn2 have 40% sequence identity and are predicted to contain 11 helix-turn-helix pseudo-repeats (13), each measuring 35–40 amino acids in length. Repeats of this kind are quite widespread, and several subfamilies have been identified, including the so-called Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and PI 3-kinase TOR1 (HEAT), tetratricopeptide repeat (TPR), and Armadillo (arm) repeats (14). Crystal structures of several family members have determined them to be solenoids that vary in shape from highly curved to nearly straight (15–21), the flexibility and large surface area of which direct reversible protein-protein recognition or binding processes (22–25). The Rpn1 and Rpn2 pseudo-repeats are of a subclass associated with proteasome and cyclosome subunits and hence are named “PC repeats,” and are expected to be highly curved concave toroids (26, 27). Microscopy studies confirmed that the central PC repeats in either protein form a closed in single domain, flanked by flexible N- and C-terminal extensions (27, 28). The molecular structure of another PC repeat-like domain was determined for Blm10, a protein that binds and activates 20 S proteasomes (13, 21, 29).
Multiple interactions have been reported for Rpn1 and Rpn2, including interactions with UBL-domain containing proteins (UDPs), many of which are ubiquitin-binding proteins or proteasome auxiliary factors (28, 30–39). Following ubiquitination, conjugates are directed to the 26 S complex and anchored by their polyUb chains to one of the internal ubiquitin-binding proteins in the proteasome. This is described as “direct targeting” (Fig. 1A). However, neither of the two known intrinsic ubiquitin receptors (Rpn10, Rpn13) is strictly essential (38, 40, 41) pointing to other “indirect” modes of substrate recognition. Moreover, the ubiquitination state of a substrate is a balance between ubiquitination and deubiquitination processes. If left unattended, polyUb tags may be disassembled prematurely. Auxiliary ubiquitin-binding proteins “decorate” polyUb conjugates in a chain length-dependent manner, chaperone the conjugate, and at times even trim the chain, and thus shape the proteasome recognition signal (1, 42–50). As such, delivery factors must be capable of simultaneously binding both polyUb conjugates and the proteasome. Several candidates fulfilling these properties are Rad23/hHR23, Dsk2/hPLIC/Ubiquilin, and Ddi1 (31, 51–60). Each shuttle can bind the polyUb tag on their cargo via their C-terminal ubiquitin-associated domain(s) (UBA) and associate with the proteasome by means of an N-terminal UBL domain (Fig. 1B).
In addition to substrate delivery proteins, the DUB Ubp6/USP14 also encodes a UBL domain and incorporates into the proteasome at nearly stoichiometric levels, most likely via an interaction with Rpn1 (4, 32, 61–65). This association results in a dramatic enhancement of Ubp6 and Usp14 deubiquitinating activities in vitro, although the exact mechanism of this activation remains unclear (66). Activation upon proteasome incorporation suggests that in cells some DUBs (such as Ubp6/USP14) function most effectively in the proteasome, whereas in cytosol their function is inhibited or reduced. As the case may be, Ubp6/USP14 apparently helps remove the (poly)ubiquitin tag from protein substrates, either before or during their translocation into the catalytic chamber of the proteasome for degradation (50, 67, 68), with an overall net effect of delaying proteolysis (64).
Mediated delivery implies a designated UBL receptor in the proteasome (Fig. 1A). Two proteasome subunits shown to interact with a subset of these shuttles are Rpn1 and Rpn2 (31). Using a series of deletions constructed from a recombinant GST-Rpn1 fusion, association of Rpn1 with Rad23 was roughly characterized (32). Following up on these findings, we set out to chart UBL recognition sites on Rpn1 and Rpn2, prioritize the binding partners of these platforms, and place them in context of competing/augmenting polyUb binding by neighboring subunits. Comparing binding data for UBL and (poly)ubiquitin signals unfurls a general outline of substrate relay events and points to Rpn1 and Rpn2 as the main coordinators of ubiquitin-processing factors at the proteasome.
EXPERIMENTAL PROCEDURES
Strains
Saccharomyces cerevisiae WT strain Sub62 (MATa his3-Δ200; leu2-3,112; ura3-52; lys2-801; trp1-1) was used as wild-type strain. Δubp6 strain (MATa his3-Δ200 lys2-801 leu2-3,112 trp1-1 ura3-52 YEL010c::kanMX4) was used for purification of 26 S proteasomes and in pulldown assays. A Δrpn10 mutant was used to purify Base-CP as published previously (3).
Plasmids
Full-length coding sequences of the indicated genes were amplified by PCR using Pfu DNA polymerase from genomic DNA (S. cerevisiae wild-type strain Sub62) (3, 69). The 5′ and 3′ amplification primers were designed to add restriction sites as indicated in supplemental Table S1. The fragments were cut with the indicated enzymes and subsequently cloned into a specified vector (supplemental Table S1, column 2) for expression with a His6 tag. Clones were verified by DNA sequencing and then transformed into Escherichia coli M15/BL21 strain for bacterial expression.
Antibodies
The following antibodies were used to identify proteasomal subunits: anti-Rpn1 (our laboratory), anti-Rpn2 (our laboratory), anti-20 S (BIOMOL International/Affinity), anti-Rpn10 (our laboratory), anti-Rad23 (our laboratory), anti-Dsk2 (our laboratory), anti-Rpt2 (our laboratory), anti-Rpn12 (gift from Dan Finley), anti-Ubp6 (gift from Dan Finley), anti-RGS-His6 (Qiagen), anti-GFP (Santa Cruz Biotechnology), and anti-ubiquitin (Dako). Secondary antibodies were purchased from Bio-Rad and were used according to the manufacturer's instructions.
Purification of Recombinant Proteins
Transformants for expression of protein of interest were grown in liquid LB media supplemented with 1 mm ampicillin to A600 0.6–0.8 at 37 °C, followed by 30 min of heat shock at 42 °C. Next, 0.1 mm to 1 nm isopropyl 1-thio-β-d-galactopyranoside was added for induction, and cultures were grown overnight at 16 °C, after which cells were harvested. Cells were lysed, and lysates were clarified at 16,000 rpm for 20 min at 4 °C, and the supernatant was loaded onto a Ni2+-nitrilotriacetic acid column (GE Healthcare) that was previously equilibrated with 50 mm Tris, pH 7.4, 500 mm NaCl, and 10 mm imidazole. Washes were performed with same buffer, and bound proteins were eluted using 50 mm Tris, pH 7.4, 500 mm NaCl, and 250 mm imidazole. Samples were then dialyzed for appropriate buffer, and the resulting proteins were stored at −80 °C. Purity was tested by Coomassie staining and immunoblotting. Protein concentration was determined using an ND-1000 spectrophotometer (NanoDrop). In some cases proteins were subjected to gel filtration for additional step of purification (24 ml of Superdex200).
Glycerol Gradient Fractionation
11-ml glycerol gradients were prepared manually using 10 and 40% (v/v) glycerol stock solutions supplemented with 10 mm MgCl2, 25 mm Tris, pH 7.4, 1 mm ATP, 1 mm DTT. 1 ml of whole fractionated yeast cell extract was applied on top of the gradients and resolved by centrifugation at 28,000 × g for 18 h in TH-641 rotor in ultracentrifuge. Twelve 1-ml fractions were collected and assayed for peptidase activity using N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin as was described (3). Protein samples were further resolved by 10–12% SDS-PAGE or 16.5% Tris/Tricine-PAGE and immunoblotted.
Pulldown Assays
CH-Sepharose 4B (Sigma) was swollen according to the manufacturer's recommendation. Proteins to be immobilized were first dissolved in 50 mm PBS and then were added to activated Sepharose to a final concentration of 0.6 mg/ml and incubated with agitation at 4 °C. After 2 h, the gel was washed, and its remaining active groups were blocked with 1 m ethanolamine, pH 8.0, for 2 h. Routinely, about 90% of protein was bound to the matrix. The resin was washed as recommended by the producer, equilibrated in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.1% (w/v) Triton X-100, and stored at 4 °C. A control resin was prepared in the same way, but instead of protein, the active groups were derivatized with ethanolamine. Each soluble protein was diluted to a final concentration of 100 μg/ml in incubation buffer and then incubated with 0.1 ml of slurry of immobilized proteins equilibrated in incubation buffer (50 mm Tris-HCl, pH 8.4, 150 mm NaCl, 0.1 mg/ml BSA, 10% glycerol, and 1% (w/v) Triton X-100). The mixture was agitated at 4 °C for 2.5 h and washed extensively with the equilibration buffer followed by additional five washes with equilibration buffer supplemented with 1% (w/v) Triton X-100. Bound proteins were eluted with elution buffer (incubation buffer supplemented with 1 m NaCl and/or 6 m urea) and analyzed by SDS-PAGE and immunoblotting.
26 S Proteasome Pull-out
Purified recombinant proteins were bound to nickel-nitrilotriacetic acid columns (Qiagen) according to standard procedure. The columns were incubated with 26 S proteasomes lacking Ubp6, Rad23, Dsk2, and Ddi1 proteins and tumbled for 2–4 h. The columns were washed with PBS buffer supplemented with 1% Triton and 0.5 m NaCl, and bound proteins were eluted with 250 mm imidazole. The eluates were immunoblotted with antibodies against various proteasomal subunits.
Surface Plasmon Resonance
All experiments were performed using ProteOn XPR36 instruments developed by Bio-Rad. Purified Rpn1 and Rpn2 were diluted in 10 mm sodium acetate at pH 3.0 or 4.5, respectively, and coupled to a general layer medium sensor surface activated with 37.5 mg/ml N-ethyl-N′-(3′dimethylaminopropyl) carbodiimide hydrochloride and 7.5 mg/ml N-hydroxysuccinimide. Proteins to be immobilized were injected in the vertical orientation ProteOn XPR36 fluidics for 5 min (150 μl) at 30 μl/min and then followed by blocking excess reactive esters with ethanolamine-HCl (Sigma) for 5 min. All of the SPR binding measurements were performed with PBST as the continuous running buffer at 25 °C. Rpn13, Ubp6, Rad23, Dsk2, and Ddi1 at the specified concentrations were injected in the horizontal orientation of the ProteOn XPR36 fluidics using a flow rate of 40 μl/min for 75 s (50 μl). In every separate injection, five different concentrations were injected in channels 1–5, and running buffer was injected simultaneously in the sixth channel for signal normalization. All binding sensorgrams were collected, processed, and analyzed using the integrated ProteOn Manager software (Bio-Rad).
In general, the equilibrium dissociation constant (KD) evaluating the protein/protein binding affinity could be determined using the 1:1 Langmuir binding model or heterogeneous ligand-parallel reaction (HLPR) model (70). For both models, the association rate constant (kon) and dissociation rate constant (koff) were derived from a global fit of primary response data over the entire concentration range for each protein-ligand pair to the corresponding binding model using rate Equation 1, and the equilibrium dissociation constant KD was calculated according to Equation 2. The difference between the two binding models is that the HLPR model has two association and two dissociation reactions as schematically shown in Equation 3.
 |
In these equations, C is the concentration of the soluble ligand (referred to as analyte in the SPR literature); R represents the response unit; for the HLPR model R (observed) = R1 + R2. For generation of concentration-dependent binding curves, response levels at equilibrium were plotted against the concentration of the soluble protein (the ligand) and calculated via a nonlinear least squares fit of the Langmuir binding Equation 4,
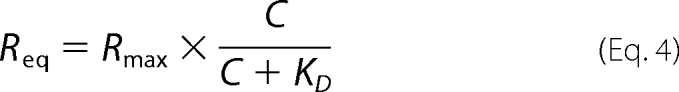 |
RESULTS
Rpn1 and Rpn2 Interact with Ubiquitin-processing Proteins
A number of ubiquitin-processing factors known from previous studies to interact with the proteasome were compared for their ability to interact directly with either Rpn1 or Rpn2. Despite high sequence and structural similarities between Rpn1 and Rpn2, the two exhibit very distinctive preferences for the potential binding partners (Fig. 1C). Although Rpn2 was found to bind only Rpn13, Rpn1 showed strong interaction with Ubp6, Rad23, and Dsk2. Additionally, in this assay, Rpn1 showed weak affinity for Rpn10 and Ddi1 (Fig. 1C). It is noteworthy that although Rpn1 bound a number of UDPs, no interaction was measured with ubiquitin, indicating that Rpn1 is able to distinguish between ubiquitin and ubiquitin-like proteins.
To achieve a deeper understanding of the differences in the binding properties of Rpn1 and Rpn2 and how they may differentiate among various ubiquitin-like signals, we quantified all pairwise binding interactions by SPR. Example titrations of single binding experiments are shown in Fig. 2, A–D. Association (kon) and dissociation (koff) rate constants were extracted by fitting binding curves over a range of concentrations using the Langmuir model for a one-to-one binding stoichiometry. Association of Ubp6 with Rpn1 was an exception to single-site binding, but we were able to model the data in Fig. 2C using the HLPR model (70). In this model, a single protein (A) interacts with two independent binding sites/states on its partner/receptor (B); each interaction is characterized by its own set of kon and koff parameters (the general scheme is shown in Equation 3 above). One way to rationalize the heterogeneous binding model with what we know of Ubp6 and the proteasome is that the single protein “A” is a bivalent molecule with two domains (depicted as A1–A2 in supplemental Scheme Si), each adapted for one of two binding sites (B1 and B2 on the receptor “B”; see Equation 3 above and supplemental Scheme Si). Initial binding of Ubp6 either via A1 or via A2 contributes to the SPR results, and hence two binding parameters were observed for the full-length Ubp6 protein attributed to two different binding modes of Ubp6-Rpn1 association (summarized in Table 1; see further analysis below and a more detailed description in supplemental Schemes Si and Sii).
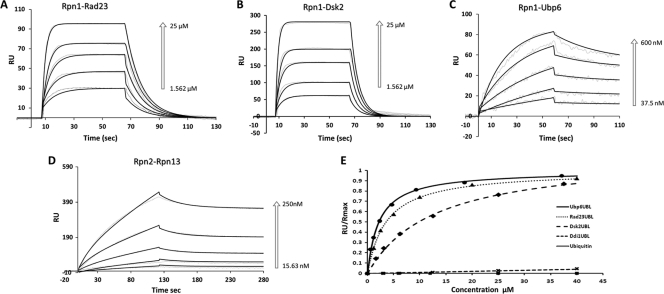
FIGURE 2.
Binding kinetics to Rpn1 or Rpn2. Rpn1 and Rpn2 were immobilized on separate channels (1700 and 1100 response units (RU)) of a ProteOn surface plasmon resonance sensor and injected with increasing concentrations of their potential binding partners followed by buffer wash. Positive associations are displayed for Rpn1-Rad23 (A), Rpn1-Dsk2 (B), Rpn1-Ubp6 (C), and Rpn2-Rpn13 (D). The response data (dashed lines) for association and dissociation are shown for a series of concentrations (arrows) overlaid with a model fit (solid line). Sensograms in A, B, and D were fit to the Langmuir model for 1:1 binding stoichiometry. Sensograms in C (Rpn1-Ubp6 response) were fit to an HLPR, assuming two-binding sites. Association (kon) and dissociation (koff) rate constants derived from a global fit of primary response data over the entire concentration range for each pair to the corresponding model are summarized in Table 1. E, binding isotherms for Ubp6UBL (circles), Rad23UBL (triangles), Dsk2UBL (diamonds), Ddi1UBL (×), and ubiquitin (squares) derived from equilibrium SPR measurements. Normalized equilibrium response is plotted as a function of soluble protein concentration; the fitting curves correspond to dissociation constants shown in Table 2.
TABLE 1.
Summary of Rpn2-Rpn13, Rpn1-Ubp6, Rpn1-Rad23, and Rpn1-Dsk2 kinetic parameters
Summary of Rpn2-Rpn13, Rpn1-Ubp6, Rpn1-Rad23, and Rpn1-Dsk2 kinetic parameters determined by SPR analysis. All response curves were fitted to a single site binding model (Langmuir kinetics) with the exception of Ubp6 for which the full-length protein required a two-site binding model (2× Ubp6 + Rpn1(site 1) + Rpn1(site 2) ↔ Ubp6-Rpn1(site 1) + Ubp6-Rpn1(site 2)). Values for dissociation constant KD were calculated as the ratio of koff over kon.
| Interaction | kon | koff | KD |
|---|---|---|---|
| m−1s−1 | s−1 | μm | |
| Rpn2-Rpn13 | 1.48 ± 0.31 × 104 | 1.80 ± 0.75 × 10−4 | 0.0118 ± 0.0028 |
| Rpn1-Ubp6 | 1.55 ± 0.36 × 104 | 9.61 ± 3.67 × 10−4 | 0.062 ± 0.004 |
| 3.53 ± 0.73 × 104 | 0.067 ± 0.006 | 1.92 ± 0.26 | |
| Rpn1-Rad23 | 2.02 ± 0.56 × 104 | 0.074 ± 0.022 | 3.65 ± 0.09 |
| Rpn1-Dsk2 | 1.37 ± 0.04 × 104 | 0.165 ± 0.007 | 12.10 ± 0.90 |
Rate constants kon and koff and the dissociation constant KD for each binding pair were calculated from at least three independent SPR experiments (Fig. 2, A–D), averaged, and summarized in Table 1. Two pairwise interactions were found to be particularly tight, displaying KD values in the nanomolar range, Ubp6-Rpn1 and Rpn13-Rpn2 (62 and 12 nm; Table 1). Rad23 and Dsk2 showed similar kinetic behavior vis-à-vis Rpn1, yielding affinities in the micromolar range (3.6 μm versus 12.1 μm, respectively). By contrast, the affinity of Ddi1 for Rpn1 was much weaker and therefore not quantifiable in the experimental concentration range. Likewise, we were unable to quantify associations of ubiquitin or Rpn10 with either Rpn1 or with Rpn2. Dissociation constants obtained by SPR (Table 1) point to a hierarchy of interactions that is completely compatible with results obtained from reciprocal binding experiments (Fig. 1). Interestingly, the proteasome interacting proteins that we assayed had similar “on” rates, yet they differed in their dissociation rates. Thus, release of Ubp6 from Rpn1 and release of Rpn13 from Rpn2 were 3 orders of magnitude slower than that of Rad23 or Dsk2 from their receptor Rpn1. These binding properties explain how transiently associated Rad23 and Dsk2 are members of the substrate delivery family, whereas proteins such as Ubp6 and Rpn13 with slow dissociation rates behave more like “sometimes proteasome subunits” (2). The fast exchange rates of shuttle proteins such as Rad23 or Dsk2 are a necessary feature if they are to unload ubiquitinated cargo and dissociate for the next round.
Contribution of Ubiquitin-like Domains to Interaction of UDPs with Rpn1
Four UDP proteins (Rad23, Dsk2, Ubp6, and Ddi1) have very different affinities for the UBL receptor Rpn1 (Table 1). Recognition of UDPs is generally thought to occur via their structurally similar UBL domains (Fig. 1). Therefore, we compared the contribution of each UBL domain to the overall interaction with Rpn1. Association of purified Rpn1 with recombinant UBL domains from each of the four UDPs was assayed by SPR. Dissociation constants were derived from equilibrium analysis (Fig. 2E) because the on/off rates were too fast for accurate kinetic quantification. The resulting KD values are summarized in Table 2.
TABLE 2.
Comparison of Rpn1 affinities for full-length or UBL domains of UDP proteins
All soluble proteins listed in the left column were injected at different concentrations over immobilized full-length Rpn1, and response curves generated from at least six concentrations were used to generate a secondary binding plot for each pair. KD values were extracted in this case by fitting the equilibrium binding curves to a single-site binding model, except for full-length Ubp6 where the KD values were calculated as the ratio of the corresponding off- and on-rates (see Equation 3). NB means no binding was detected.
| Protein |
KD (μm) |
|
|---|---|---|
| UBL | Full length | |
| Ubp6 | 2.16 ± 0.36 | 0.062 ± 0.004 |
| 1.92 ± 0.26 | ||
| Rad23 | 3.59 ± 0.69 | 3.6 ± 0.9 |
| Dsk2 | 11.69 ± 0.87 | 11.4 ± 2.6 |
| Ddi1 | >100 | >100 |
| Ubiquitin | NB | NB |
| Ub4 (Lys-48) | NB | NB |
Three UBLs (Rad23, Dsk2, and Ubp6) bound Rpn1 with comparable micromolar range affinities; Ddi1UBL associated very weakly (>100 μm), whereas neither ubiquitin nor Lys-48-linked polyUb chains bound at detectable levels (Fig. 2E). Isolated UBLs bound Rpn1 with the same affinity as their corresponding full-length proteins (Table 2) suggesting that association is carried out through the UBL domain. A notable exception was Ubp6UBL, which bound Rpn1 weaker than Ubp6 (KD = 2 μm versus 60 nm; Table 2), yet with comparable affinity to those of Rad23UBL and Dsk2UBL (Fig. 2E). Moreover, Ubp6UBL binding to Rpn1 (Fig. 3A) was fit to a single binding-site model (Langmuir kinetics). The dissociation constant KD for Ubp6UBL corresponds within experimental error to the KD of the weaker binding site of Ubp6 (Table 2). As the net dissociation rate of this UBL domain is faster than that observed for full-length Ubp6, it seems likely that additional properties of the full-length protein slow down the apparent dissociation. In this respect, Ubp6 differs from other members of the UDP family that do not exhibit complicated association kinetics and consequently do not bind as tightly to Rpn1 receptor.
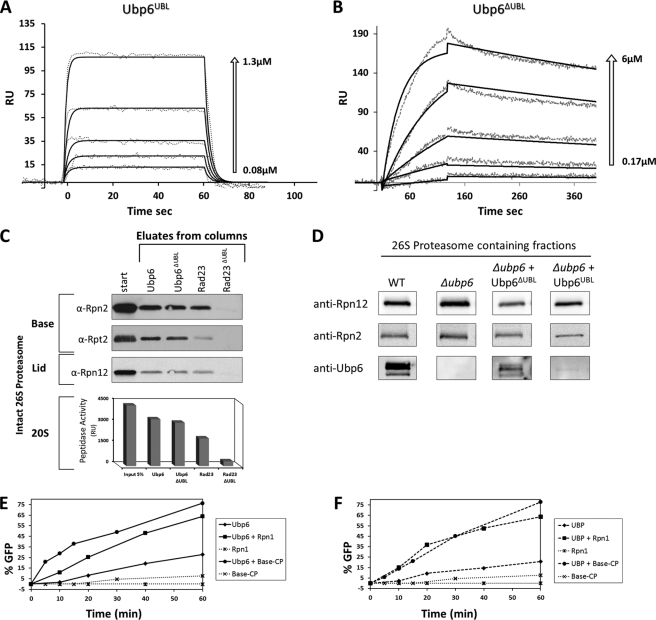
FIGURE 3.
Two-step proteasome incorporation of Ubp6. A and B, SPR sensograms of Ubp6UBL and Ubp6ΔUBL binding kinetics to Rpn1. The response data for binding and dissociation (dotted lines) are shown for a series of concentrations (arrows). Values of best fit parameters to a single bimolecular interaction model (solid lines) are summarized in Table 3. RU, response units. C, immobilized Ubp6 and Rad23 lacking their UBL domains (Ubp6ΔUBL and Rad23ΔUBL) were tested for binding to 26 S proteasome-purified free of all UDPs or other transient proteasome-interacting proteins. Association of full-length Ubp6 and Rad23 is included as positive control. Eluate from each column was tested for proteasome activity (20 S CP) as well as immunoblotted (IB) for presence of 19 S subunits from the Base and Lid subcomplexes. D, truncated Ubp6-expressing strains were molecular weight-fractionated, and proteins copurifying with peak of proteasome activity were identified by immunoblotting. E, potency of Ubp6 to remove ubiquitin from Ub-GFP was monitored over 60 min, and % reaction progression was plotted over time (diamonds). The reaction was repeated in presence of equimolar Rpn1 (squares) or purified base-CP (lidless) proteasome lacking both endogenous Ubp6 and Rpn11 (circles). Neither Rpn1 (star) nor base-CP alone (×) exhibit deubiquitination activity in the absence of added Ubp6 (dotted lines). F, experiment similar to E but for truncated Ubp6ΔUBL. Ubp6 activation is not dependent on UBL.
Coordinated Binding of Ubp6 Domains to the Rpn1 Receptor
To dissect parameters that contribute to Ubp6-Rpn1 binding, we generated truncated constructs of Ubp6. One fragment spanned just the UBL domain, and another encompassed the enzymatic UBP domain (Ubp6UBL versus Ubp6ΔUBL; Fig. 3, A and B). Whereas Ubp6UBL exhibited fast association and dissociation rates similar to those for other Rpn1-associating UDPs (Table 3 versus Table 1), the C-terminal portion of Ubp6 associated slower and dissociated much slower than other UDPs studied (Fig. 3B; Table 3). Independent binding of two proteins (A1 and A2) to two binding sites (on a platform B) can be described by the heterogeneous ligand parallel reaction model as adapted schematically in supplemental Scheme Sii. In this scenario, by measuring binding of each protein (ligand) separately, it is possible to estimate the overall equilibrium between the free state (A1 + A2 + B) and the fully occupied state (A1BA2). Coordinated binding of the same domains chemically linked in a single molecule is expected to result in a lower entropic penalty and as a result tighter apparent binding. Although most SPR measurements are limited to detecting the first step (generating intermediates A1B or BA2 in supplemental Scheme Si), similar thermodynamic considerations can provide an estimate for the overall equilibrium. Indeed, using the experimental data in Table 3, we estimated that binding between full-length Ubp6 and Rpn1 is tighter than between separate Ubp6 domains and Rpn1 (see details in supplemental Schemes Si and Sii, respectively). Moreover, we point out the similar association rate constants for Ubp6 and Ubp6UBL with Rpn1, and likewise the similar dissociation rate constants for Ubp6 and Ubp6ΔUBL. Coordinated binding of two Ubp6 domains could explain the higher affinity of the full-length protein; Ubp6 would be targeted to Rpn1 via its UBL domain and locked in place by interactions involving the C-terminal portion of the protein.
TABLE 3.
Dissecting contribution of Ubp6 domains to kinetics of Rpn1 association
Full-length Ubp6 as well as isolated truncated UBL or ΔUBL domains were injected at various concentrations over immobilized full-length Rpn1. Values of kinetic parameters and of the KD dissociation constant were extracted from a fit of response curves to a minimal binding site model (single site for the individual domains or a two-site binding model for the full length protein).
| Protein | kon | koff | KD | |
|---|---|---|---|---|
| m−1s−1 | s−1 | μm | ||
| Ubp6 | Site 1 | 1.55 ± 0.36 × 104 | 9.61 ± 3.67 × 10−4 | 0.062 ± 0.004 |
| Site 2 | 3.53 ± 0.73 × 104 | 0.067 ± 0.006 | 1.92 ± 0.26 | |
| Ubp6ΔUBL | 1.81 ± 0.04 × 103 | 6.55 ± 0.95 × 10−4 | 0.36 ± 0.05 | |
| Ubp6UBL | 5.37 ± 0.54 × 104 | 0.120 ± 0.013 | 2.23 ± 0.24 | |
To summarize, of the various UDPs in this study Ubp6 had the highest affinity for Rpn1. The overall affinity of Ubp6 for Rpn1 is comparable with that of a bona fide proteasome subunit such as Rpn13 (Fig. 2), thus placing Ubp6 in the category of occasional proteasome subunits rather than fast-exchange shuttle proteins.
Incorporation of Ubp6 into Proteasomes Does Not Require the UBL Domain
It has been suggested that distribution of Ubp6 between proteasome-bound and proteasome-unbound pools reflects an equilibrium between association and dissociation (44). To test whether free Ubp6 can incorporate into preassembled proteasomes, a purified proteasome sample free of UDP proteins (lacking Ubp6, Rad23, Dsk2, and Ddi1) was assayed for recruiting Ubp6, with or without its UBL domain. Incorporation of Ubp6 or Ubp6ΔUBL into the 26 S proteasome was equally efficient. By contrast, binding to Rad23 required its UBL domain (Fig. 3C). Moreover, even in cells, proteasomes isolated from a strain expressing a construct of Ubp6 lacking the UBL domain (ubp6ΔUBL) contained Ubp6ΔUBL as a subunit (Fig. 3D). Interestingly, the UBL domain alone was not retained during the purification process (Fig. 3D, right column) supporting the in vitro observation that the UBL alone had fast on/off kinetics. Thus, we conclude that stickiness of the C-terminal UBP-containing segment contributes toward Ubp6 as a proteasome subunit.
Next, we assessed whether association with its receptor Rpn1 was the underlying cause of enzymatic activation of Ubp6 upon proteasome incorporation (66). Premixing Ubp6 with a proteasome sample devoid of all known DUBs (Lidless Base-CP lacking Rpn11 purified from a Δubp6 strain) enhanced basal Ubp6 activity to the same extent as premixing with isolated Rpn1 (Fig. 3E). In this assay, we made use of a ubiquitin-fusion substrate used previously as a model for deubiquitination (4, 71). Even a Ubp6 version lacking its N-terminal UBL domain was activated by either proteasome or Rpn1 to essentially the same extent as was full-length Ubp6 (Fig. 3F). These findings indicate that activation of Ubp6 is a direct consequence of association with Rpn1 yet does not require its UBL.
Domain Mapping of Rpn1 and Rpn2
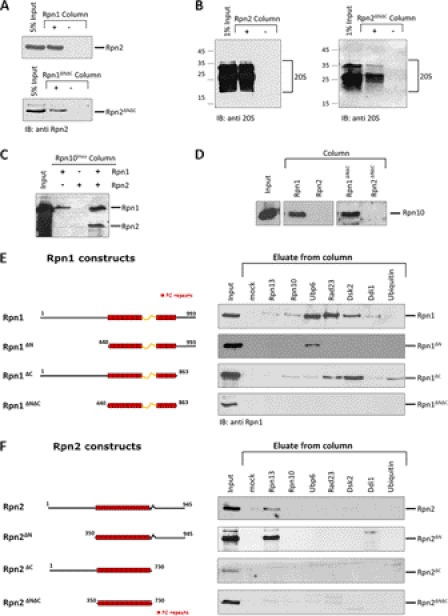
Having quantified association of Rpn1 or Rpn2 with a number of ubiquitin-processing factors, we then asked how they discern between multiple binding partners. Are there dedicated binding sites, or do UDPs compete for a single receptor? Both Rpn1 and Rpn2 proteins can be similarly dissected into three major segments. The central portions of each are composed of 11 α-helical rich repeats (also known as PC repeats) that fold into highly curved solenoids (26). Truncated versions of Rpn1 or Rpn2, encompassing only the solenoid portion, fold into stable structures (27) and are sufficient to maintain interactions with each other (Fig. 4A) or with the 20 S CP (Fig. 4B), as has been published for the full-length proteins (28). This central solenoid domain appears to be sufficient for association with Rpn10 (Fig. 4C). No other proteasome auxiliary factor or transient subunit tested in this study was found to associate stably with this region. Association of Rpn10 with Rpn1 is stabilized in presence of Rpn2 (Fig. 4D). Coupling of Rpn1 and Rpn2 stabilizes their PC folds (28) and thus may explain the stronger association of Rpn10 with a joint Rpn1-Rpn2 complex (Fig. 4C). Indeed, all three have been found in a stable complex together with the 20 S CP and a few additional factors (7, 36).
FIGURE 4.
Mapping binding sites on Rpn1 and Rpn2. Indicated proteins were immobilized on CH-Sepharose beads, mixed with various fragments of Rpn1 or Rpn2 proteins, washed, and eluates analyzed by immunoblotting (IB) for retained Rpn1 or Rpn2.
The flexible terminal extensions of Rpn1 were found to be required for UDPs to bind (Fig. 4E). Specifically, binding of either Rad23 or Dsk2 required the presence of the N-terminal region, whereas deleting this region did not abolish association of Ubp6 (Fig. 4E) suggesting that Rpn1 harbors multiple UBL-binding sites. Similarly, we mapped the interaction of Rpn13 with Rpn2 to the C-terminal segment of Rpn2 (Fig. 4F). This result is in full agreement with previously published data for Rpn2-Rpn13 association (7, 35).
Simultaneous Binding of Rad23 and Dsk2 to Rpn1
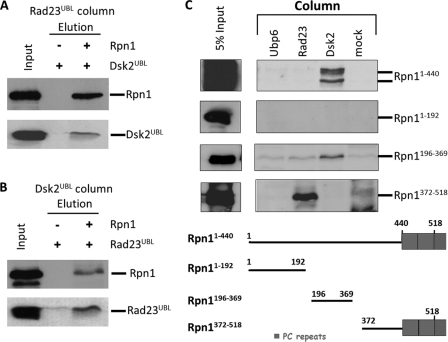
Having observed that both Rad23 and Dsk2 require the N-terminal region of Rpn1 for binding (Fig. 4E), we wished to explore whether these two shuttle proteins compete for the same site. We immobilized Rad23UBL and monitored its ability to retain pre-bound Rpn1-Dsk2UBL. The results provided evidence for a ternary complex of Rad23UBL-Rpn1-Dsk2UBL (Fig. 5A), indicative of separate binding sites on Rpn1. The reverse experiment in which immobilized Dsk2UBL retained pre-bound Rpn1-Rad23UBL also generated the same conclusion (Fig. 5B).
FIGURE 5.
Dsk2 and Rad23 bind to two different sites at Rpn1. A and B, immobilized Rad23UBL was incubated with either Dsk2UBL or a premixed Rpn1 + Dsk2UBL mixture and washed, and bound proteins were eluted and immunoblotted for the presence of Dsk2. The reciprocal experiment was repeated with Dsk2UBL immobilized and incubated with either Rad23UBL or a Rpn1 + Rad23UBL mixture. The formation of these Rad23-Rpn1-Dsk2 ternary complexes demonstrates that Rad23 and Dsk2 can bind simultaneously to Rpn1, via at least two discrete binding sites. C, purified recombinant N-terminal fragments of Rpn1 were washed over immobilized binding partners, washed, and elution immunoblotted for presence of Rpn1. A Dsk2-binding site is located between residues 196 and 369 of Rpn1, whereas Rad23 binds tighter to an Rpn1 fragment spanning residues 372–518.
The two UBL-binding sites may differ in their binding specificities as each may show preference for one UBL over the other, thereby clarifying why a 50-fold molar excess of Dsk2 was utilized to out-compete Rad23 bound to Rpn1 (40). In search of a minimally required binding region for each UBL, we tested N-terminal segments of Rpn1 for pairwise interactions. Truncating the first 440 residues abolished association with both Rad23 and Dsk2 (Fig. 4E); however, the detached segment alone was competent to bind Dsk2 but not Rad23, confirming that different sets of residues are required to anchor either of these two UBLs (Fig. 5C, top). Using the meta Protein DisOrder prediction System (PrDOS (72)), we dissected the N-terminal region of Rpn1 into three segments separated by natively disordered linkers (Fig. 5C). The first segment (Rpn1(1–192)) did not display affinity for any UDP in our assay, whereas a Dsk2-binding site was positively identified in the second segment (Rpn1(194–360)). None of these N-terminal segments bound Rad23 (Fig. 5C),and neither did the immediately downstream PC repeat region (residues 440–863, see Fig. 4E). Therefore, we hypothesized that contributions from residues in flanking regions may be required in tandem. An earlier study identified Rad23 as associating with a region of Rpn1 spanning the N terminus as well as the first two of 11 PC repeats (32). Therefore, we generated an additional fragment starting at the second disordered interceding linker and extending into the flanking PC region (Rpn1(372–518)) that spans the first two “covert” PC repeats predicted to differ in their properties from the subsequent nine PC repeats (26). This segment (entirely distinct from the Dsk2-binding region; Fig. 5C) was sufficient to associate with Rad23. The tertiary fold of Rpn1 may provide the necessary contact points for Rad23 binding. These contact points may be contributed by relatively few amino acid residues from different structural domains positioned far apart in the primary sequence.
Identification of UBL receptors at the previously poorly characterized N terminus of Rpn1 illuminates how Rpn1 may serve as a scaffold for ubiquitin shuttles at the proteasome. Moreover, the existence of two distinct binding sites within this region, each dedicated for a preferred ligand (Fig. 5C), suggests that Rpn1 may coordinate binding of multiple ubiquitin-processing factors simultaneously (Fig. 5, A and B), each with its own affinity for UBLs targeting the proteasome (Tables 1 and 2). Nonoverlapping binding sites within the Base, each with different specificities, raise the possibility that multiple proteasome-interacting proteins co-reside on a single proteasome (supplemental Fig. S1).
DISCUSSION
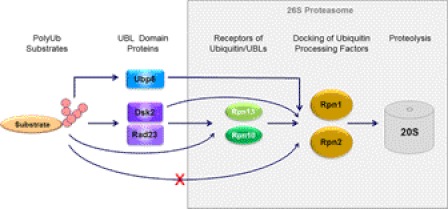
There are multiple levels of redundancy in targeting ubiquitinated substrates to the proteasome (Fig. 6). Substrate shuttles can mediate recruitment by simultaneously binding both conjugates and the proteasome. Although some of these ubiquitin-binding proteins show selectivity for certain ubiquitin-chain linkages, they appear to be generally redundant. Moreover, the few intrinsic ubiquitin receptors that have been identified at the proteasome also seem to be dispensable, suggesting functional overlap. Moreover, proteasomes lacking most, or all, known associated ubiquitin-binding subunits are functional, at least on some substrates (41, 43). Even categorizing ubiquitin-binding proteins neatly into shuttles versus receptors may be an oversimplification, as some ubiquitin receptors at the proteasome apparently also bind ubiquitin-like domains (supplemental Table S1). In this study, we identified Rpn1 as a definitive UBL receptor in the proteasome, able to distinguish UBL from ubiquitin/polyUb and with the capacity to bind multiple UDPs simultaneously. This feature of Rpn1 is distinct from the binding preferences of Rpn2, a paralog also situated within the Base of the 19 S RP, yet together these two large subunits coordinate the activity and placement of multiple ubiquitin-processing factors at the proteasome. Most of the data for this study was collected on isolated Rpn1 and Rpn2 subunits; nevertheless, by quantifying the interactions of Rpn1 and Rpn2 with their binding partners, we provide a possible outline of early steps in substrate processing by the proteasome (Fig. 6 and supplemental Fig. S1).
FIGURE 6.
Schematic description of substrate relay. Polyubiquitin-binding proteins such as Dsk2 or Rad23 are transiently associated with proteasomes and thus may serve as shuttles or delivery proteins for polyubiquitin conjugates. These proteins contain an N-terminal UBL (ubiquitin-like) domain through which they dock at the proteasome. The primary UBL receptors for Rad23 and Dsk2 was found to be proteasome subunits Rpn1 (Tables 1–3), although these two UDPs (UBL-domain containing proteins) were also capable of associating directly with two neighboring subunits, Rpn10 and Rpn13 (supplemental Table ST1). Ddi1 apparently also associates with Rpn1, although in this study we were unable to quantify the intensity of their mutual association. Polyubiquitin chains could also bind to Rpn10 and Rpn13 directly (supplemental Table ST1), thereby bypassing the need for shuttles. The relative affinities of these receptors for the different (poly)ubiquitin or UBL signals influence efficiencies of substrate targeting and coordinate their entry into the proteasome. Another UDP, the deubiquitinating enzyme Ubp6, bound only to Rpn1 yet much tighter than transiently associated shuttle proteins, thus behaving more akin to proteasome subunits Rpn10 and Rpn13. The proximal localization of Ubp6 to these polyubiquitin-chain-receptors may also coordinate trimming, processing, or shaving of conjugated chains with substrate preparation and the proteolytic mechanism (see additional details in supplemental Fig. S1).
One proteasome-associated UDP that is not a substrate shuttle but rather harbors a distinct enzymatic activity is Ubp6. Association of Ubp6 with Rpn1 activates the deubiquitinase activity of Ubp6 suggesting that proteasome-incorporated Ubp6 is functionally distinct from its free form. One explanation may be that rearrangement of the UBL domain upon proteasome binding frees up access to the active site cleft in the catalytic core (the UBP domain) (32, 73). Such an auto-inhibitory mechanism has been concluded for another DUB, USP4, also with an embedded UBL domain that can compete with substrate by acting as a ubiquitin mimic at the catalytic site (74, 75). A somewhat similar model implied conformational changes related to the UBL domain of other UDPs upon proteasome incorporation (76, 77). However, as this study observed Ubp6 lacking UBL incorporated into proteasomes, such confrontational changes are probably more subtle and do not necessitate the UBL domain for activation. The distinction between a transient shuttle, whose role is to upload cargo, depart, and make way for subsequent rounds, and a subunit that needs to be properly incorporated is evident from the different on/off kinetics (Table 2). In contrast to the UDPs that serve as delivery proteins by hovering in and out of proteasomes, subunits seem to require more than their UBL to keep them anchored in place.
The most striking feature of Rpn1 and Rpn2 is their toroidal, almost ring-like, structure, at least in the absence of other associated subunits (27). These solenoid portions are responsible for their mutual interaction as well as for association with the ubiquitin receptor, Rpn10 (Fig. 4), and possibly with regulatory particle triple nucleotidase ATPases (30). In absence of additional binding partners, the flanking N- and C-terminal extensions in each protein are likely to be unstructured or flexible (27). This study goes a considerable way to show that these extensions carry the primary proteasome docking sites for substrate delivery proteins and auxiliary ubiquitin processing subunits (Figs. 4 and 5). More specifically, it is the N-terminal region of Rpn1 that anchors UDPs at the proteasome either near to, or partially overlapping with, the central solenoid PC repeat region (Fig. 6), whereas the C-terminal regions of Rpn2 and Rpn1 are required for interaction with the ubiquitin receptor Rpn13 and the deubiquitinating enzyme Ubp6, respectively. Independently, these flexible extensions may also contribute to stability of 19 S RP-20 S CP association (37) or to proper nuclear localization of proteasome (11).
All told, Rpn2 displays specific binding to at least five partners as follows: 20 S CP, Rpn13, Rpn1, Rpt4, and Rpt6 (see this work and Refs. 31, 33, 78, 79). Rpn1 associates with an assortment of ubiquitin processing factors, Rad23, Dsk2, Rpn10, Ddi1, and Ubp6, and neighboring proteasome subunits, Rpn2, Rpt2 and Rpt5 (this work and Refs. 7, 30–32, 36, 54, 60, 78–80). In contrast, the adjacent ubiquitin receptors Rpn10 and Rpn13 also exhibit propensity for UBL domains (41, 46, 80, 81), although they do seem adapted for polyUb (supplemental Table S1). Thus, Rpn1 emerges as the primary UBL receptor at the proteasome with a marked preference for UBLs over ubiquitin. Properties of the transported cargo, such as hydrophobic/unstructured patches, or the nature of the polyubiquitin tag (linkage/length) may significantly alter the relationships identified in this study, cargo transported by these shuttles. Nevertheless, the proximity of their potential biding sites at the Base may allow for efficient funneling of polyubiquitinated substrates from shuttle proteins that dock at Rpn1 to downstream polyUb receptors, and ultimately toward the 20 S proteolytic core.
In its capacity as a UBL-binding platform, Rpn1 not only binds multiple UBLs, but does so simultaneously by utilizing specific sites for each UBL. As Rpn1 has no sequence homology to any known ubiquitin-binding domain or any protein known to bind UBLs (such as UIM, UBA, CUE, etc. (82, 83)), we believe that we have uncovered a totally new class of receptors critical for proteasome function. Many UDPs are substrate-delivery proteins (84); therefore, finding a pluripotent docking site for UDPs within the 19 S RP is a significant step toward dissecting a proteasome mechanism and the multistep trajectory of substrates within. Placement of Rpn1 at the Base, in the vicinity of proteasomal ATPases, supports a model whereby conjugated substrates may be unloaded from transitory shuttles to the ATPase unit for unfolding, whereas the polyUb tag is anchored by one of the nearby intrinsic ubiquitin receptors.
In prokaryotes and archaea, simplified proteasome analogs consist of a protease core complex and an ATPase regulator; few if any auxiliary factors are known to be required (85, 86). In these domains of life, substrates are not ubiquitinated, and the ATPases are sufficient for trapping substrates and transferring the unfolded polypeptide to the protease domain (87, 88). However, most substrates in eukaryotes are first conjugated to a polyUb tag. Therefore, an additional layer of factors for substrate recognition and preparation may be called for to mediate recognition, binding, anchoring, and deubiquitination, coupled with unfolding by the ATPases (65, 68). Although most observations in this study (with the exception of Ubp6) were for potential binding partners with isolated Rpn1 and Rpn2, we propose that these factors may serve to recruit ubiquitin conjugates to the proteasome. Once at the proteasome, the length of the polyUb modification could act as a threshold, with poorly ubiquitinated substrates released on account of fast dissociation rates, whereas chains of proper length and linkage are retained on the proteasome (89). As a chain-editing enzyme, Ubp6 could help define substrate residency time by trimming the chain, ubiquitin by ubiquitin (1, 50, 64, 67).
Cumulative studies have unearthed a sequence of substrate preparation steps both upstream to and within the proteasome. These include recognition, ubiquitination, delivery, recruitment by 26 S proteasome, binding, anchoring, unfolding, deubiquitination, and translocation into the 20 S CP proteolytic chamber. Curiously, many of these diverse activities have been mapped in one way or another to the Base of the 19 S RP. Nevertheless, the Lid of the 19 S RP is also required for treatment of polyUb conjugates (3), at least partially by providing (redundant) DUB activity in the form of Rpn11 (4, 90, 91). Although the ubiquitinated cargo may influence the specific properties of receptors and binding proteins measured in this study, Rpn1 and Rpn2 emerge as the primary coordinators of the ubiquitin processing machinery at the Base. As such, Rpn1 and Rpn2 appear to be the inherently eukaryotic subunits that distinguish the Base of the 26 S proteasome holoenzyme from simpler activators of prokaryotic ATP-dependent (yet ubiquitin-independent) proteases.
Supplementary Material
Acknowledgments
This manuscript was written while M. H. G. was on sabbatical at Institut Jacques Monod/Université Paris Diderot; the host laboratory of Rosine Haguenauer-Tsapisis is cordially acknowledged. We thank Noa Reis (Technion) for advice and technical knowledge.
This work was supported, in whole or in part, by National Institutes of Health Grant GM095755 (to D. F. and M. H. G.). This work was also supported by grants from the Israel Science Foundation (to M. H. G.) and by the United States-Israel Binational Science Foundation (to M. H. G. and D. F.).

This article contains supplemental Schemes Si and Sii, Table S1, Fig. S1, Plasmid, and additional references.
- polyUb
- polyubiquitin
- UBL
- ubiquitin-like
- CP
- proteasome core particle
- RP
- proteasome regulatory particle
- PC
- proteasome-cyclosome
- DUB
- deubiquitinase
- UDP
- ubiquitin-like domain-containing protein
- SPR
- surface plasmon resonance
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- HLPR
- heterogeneous ligand-parallel reaction.
REFERENCES
- 1. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Assembly, structure, and function of the 26 S proteasome. Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 4. Guterman A., Glickman M. H. (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279, 1729–1738 [DOI] [PubMed] [Google Scholar]
- 5. Bar-Nun S., Glickman M. H. (2011) Proteosomal AAA-ATPases. Structure and function. Biochim. Biophys. Acta 1823, 67–82 [DOI] [PubMed] [Google Scholar]
- 6. Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hendil K. B., Kriegenburg F., Tanaka K., Murata S., Lauridsen A. M., Johnsen A. H., Hartmann-Petersen R. (2009) The 20 S proteasome as an assembly platform for the 19 S regulatory complex. J. Mol. Biol. 394, 320–328 [DOI] [PubMed] [Google Scholar]
- 8. Peth A., Uchiki T., Goldberg A. L. (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26 S proteasome that commit to degradation. Mol. Cell 40, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeMarini D. J., Papa F. R., Swaminathan S., Ursic D., Rasmussen T. P., Culbertson M. R., Hochstrasser M. (1995) The yeast SEN3 gene encodes a regulatory subunit of the 26 S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol. Cell. Biol. 15, 6311–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsurumi C., Shimizu Y., Saeki M., Kato S., Demartino G. N., Slaughter C. A., Fujimuro M., Yokosawa H., Yamasaki M., Hendil K. B., Toh-e A., Tanahashi N., Tanaka K. (1996) cDNA cloning and functional analysis of the p97 subunit of the 26 S proteasome, a polypeptide identical to the type-1 tumor necrosis factor receptor-associated protein-2/55.11. Eur. J. Biochem. 239, 912–921 [DOI] [PubMed] [Google Scholar]
- 11. Wendler P., Lehmann A., Janek K., Baumgart S., Enenkel C. (2004) The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin αβ and proteasome functions. J. Biol. Chem. 279, 37751–37762 [DOI] [PubMed] [Google Scholar]
- 12. Wang S., Kurepa J., Smalle J. A. (2009) The Arabidopsis 26 S proteasome subunit RPN1a is required for optimal plant growth and stress responses. Plant Cell Physiol. 50, 1721–1725 [DOI] [PubMed] [Google Scholar]
- 13. Kajava A. V., Gorbea C., Ortega J., Rechsteiner M., Steven A. C. (2004) New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J. Struct. Biol. 146, 425–430 [DOI] [PubMed] [Google Scholar]
- 14. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 15. Conti E., Müller C. W., Stewart M. (2006) Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 16, 237–244 [DOI] [PubMed] [Google Scholar]
- 16. Sibanda B. L., Chirgadze D. Y., Blundell T. L. (2010) Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grinthal A., Adamovic I., Weiner B., Karplus M., Kleckner N. (2010) PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc. Natl. Acad. Sci. U.S.A. 107, 2467–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knutson B. A. (2010) Insights into the domain and repeat architecture of target of rapamycin. J. Struct. Biol. 170, 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kennedy S. A., Frazier M. L., Steiniger M., Mast A. M., Marzluff W. F., Redinbo M. R. (2009) Crystal structure of the HEAT domain from the pre-mRNA processing factor Symplekin. J. Mol. Biol. 392, 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alag R., Bharatham N., Dong A., Hills T., Harikishore A., Widjaja A. A., Shochat S. G., Hui R., Yoon H. S. (2009) Crystallographic structure of the tetratricopeptide repeat domain of Plasmodium falciparum FKBP35 and its molecular interaction with Hsp90 C-terminal pentapeptide. Protein Sci. 18, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sadre-Bazzaz K., Whitby F. G., Robinson H., Formosa T., Hill C. P. (2010) Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 37, 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aksel T., Majumdar A., Barrick D. (2011) The contribution of entropy, enthalpy, and hydrophobic desolvation to cooperativity in repeat-protein folding. Structure 19, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tao J., Petrova K., Ron D., Sha B. (2010) Crystal structure of P58(IPK) TPR fragment reveals the mechanism for its molecular chaperone activity in UPR. J. Mol. Biol. 397, 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kappel C., Zachariae U., Dölker N., Grubmüller H. (2010) An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys. J. 99, 1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kippert F., Gerloff D. L. (2009) Highly sensitive detection of individual HEAT and ARM repeats with HHpred and COACH. PLoS One 4, e7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kajava A. V. (2002) What curves α-solenoids? Evidence for an α-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J. Biol. Chem. 277, 49791–49798 [DOI] [PubMed] [Google Scholar]
- 27. Effantin G., Rosenzweig R., Glickman M. H., Steven A. C. (2009) Electron microscopic evidence in support of α-solenoid models of proteasomal subunits Rpn1 and Rpn2. J. Mol. Biol. 386, 1204–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosenzweig R., Osmulski P. A., Gaczynska M., Glickman M. H. (2008) The central unit within the 19 S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 15, 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Savulescu A. F., Glickman M. H. (2011) Proteosome activator 200. The heat is on. Mol. Cell. Proteomics 10, R110 006890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gorbea C., Taillandier D., Rechsteiner M. (2000) Mapping subunit contacts in the regulatory complex of the 26 S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 275, 875–882 [DOI] [PubMed] [Google Scholar]
- 31. Saeki Y., Sone T., Toh-e A., Yokosawa H. (2002) Identification of ubiquitin-like protein-binding subunits of the 26 S proteasome. Biochem. Biophys. Res. Commun. 296, 813–819 [DOI] [PubMed] [Google Scholar]
- 32. Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 33. Hartmann-Petersen R., Tanaka K., Hendil K. B. (2001) Quaternary structure of the ATPase complex of human 26 S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 386, 89–94 [DOI] [PubMed] [Google Scholar]
- 34. Davy A., Bello P., Thierry-Mieg N., Vaglio P., Hitti J., Doucette-Stamm L., Thierry-Mieg D., Reboul J., Boulton S., Walhout A. J., Coux O., Vidal M. (2001) A protein-protein interaction map of the Caenorhabditis elegans 26 S proteasome. EMBO Rep. 2, 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Murata S. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26 S proteasomes. EMBO J. 25, 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andersen K. M., Madsen L., Prag S., Johnsen A. H., Semple C. A., Hendil K. B., Hartmann-Petersen R. (2009) Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26 S proteasome. J. Biol. Chem. 284, 15246–15254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marques A. J., Glanemann C., Ramos P. C., Dohmen R. J. (2007) The C-terminal extension of the β7 subunit and activator complexes stabilizes nascent 20 S proteasomes and promotes their maturation. J. Biol. Chem. 282, 34869–34876 [DOI] [PubMed] [Google Scholar]
- 38. Chen X., Lee B. H., Finley D., Walters K. J. (2010) Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell 38, 404–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fatimababy A. S., Lin Y. L., Usharani R., Radjacommare R., Wang H. T., Tsai H. L., Lee Y., Fu H. (2010) Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26 S proteasome-mediated proteolysis. FEBS J. 277, 796–816 [DOI] [PubMed] [Google Scholar]
- 40. Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 41. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang D., Raasi S., Fushman D. (2008) Affinity makes the difference. Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 377, 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 44. Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walters K. J., Zhang N. (2008) Rpn10 protects the proteasome from Dsk2. Mol. Cell 32, 459–460 [DOI] [PubMed] [Google Scholar]
- 46. Zhang D., Chen T., Ziv I., Rosenzweig R., Matiuhin Y., Bronner V., Glickman M. H., Fushman D. (2009) Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol. Cell 36, 1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hartmann-Petersen R., Hendil K. B., Gordon C. (2003) Ubiquitin-binding proteins protect ubiquitin conjugates from disassembly. FEBS Lett. 535, 77–81 [DOI] [PubMed] [Google Scholar]
- 48. Tyrrell A., Flick K., Kleiger G., Zhang H., Deshaies R. J., Kaiser P. (2010) Physiologically relevant and portable tandem ubiquitin-binding domain stabilizes polyubiquitylated proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 19796–19801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M. S. (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee M. J., Lee B. H., Hanna J., King R. W., Finley D. (2011) Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteomics 10, R110 003871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofmann K., Bucher P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21, 172–173 [PubMed] [Google Scholar]
- 52. Schauber C., Chen L., Tongaonkar P., Vega I., Lambertson D., Potts W., Madura K. (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391, 715–718 [DOI] [PubMed] [Google Scholar]
- 53. Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. (2001) Proteins containing the UBA domain are able to bind to multiubiquitin chains. Nat. Cell Biol. 3, 939–943 [DOI] [PubMed] [Google Scholar]
- 54. Chen L., Madura K. (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22, 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Funakoshi M., Sasaki T., Nishimoto T., Kobayashi H. (2002) Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. U.S.A. 99, 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hartmann-Petersen R., Seeger M., Gordon C. (2003) Transferring substrates to the 26 S proteasome. Trends Biochem. Sci. 28, 26–31 [DOI] [PubMed] [Google Scholar]
- 57. Raasi S., Orlov I., Fleming K. G., Pickart C. M. (2004) Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J. Mol. Biol. 341, 1367–1379 [DOI] [PubMed] [Google Scholar]
- 58. Raasi S., Varadan R., Fushman D., Pickart C. M. (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12, 708–714 [DOI] [PubMed] [Google Scholar]
- 59. Kang Y., Vossler R. A., Diaz-Martinez L. A., Winter N. S., Clarke D. J., Walters K. J. (2006) UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J. Mol. Biol. 356, 1027–1035 [DOI] [PubMed] [Google Scholar]
- 60. Gomez T. A., Kolawa N., Gee M., Sweredoski M. J., Deshaies R. J. (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., Ploegh H. L. (2001) A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koulich E., Li X., Demartino G. N. (2008) Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26 S proteasome. Mol. Biol. Cell 19, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wyndham A. M., Baker R. T., Chelvanayagam G. (1999) The Ubp6 family of deubiquitinating enzymes contains a ubiquitin-like domain, SUb. Protein Sci. 8, 1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 65. Peth A., Besche H. C., Goldberg A. L. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20 S gate opening. Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 67. Guterman A., Glickman M. H. (2004) Deubiquitinating enzymes are IN/(trinsic to proteasome function). Curr. Protein Pept. Sci. 5, 201–211 [DOI] [PubMed] [Google Scholar]
- 68. Kraut D. A., Prakash S., Matouschek A. (2007) To degrade or release. Ubiquitin-chain remodeling. Trends Cell Biol. 17, 419–421 [DOI] [PubMed] [Google Scholar]
- 69. Finley D., Ozkaynak E., Varshavsky A. (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046 [DOI] [PubMed] [Google Scholar]
- 70. Cheskis B., Freedman L. P. (1996) Modulation of nuclear receptor interactions by ligand. Kinetic analysis using surface plasmon resonance. Biochemistry 35, 3309–3318 [DOI] [PubMed] [Google Scholar]
- 71. Hetfeld B. K., Helfrich A., Kapelari B., Scheel H., Hofmann K., Guterman A., Glickman M., Schade R., Kloetzel P. M., Dubiel W. (2005) The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr. Biol. 15, 1217–1221 [DOI] [PubMed] [Google Scholar]
- 72. Ishida T., Kinoshita K. (2007) PrDOS. Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu M., Li P., Li M., Li W., Yao T., Wu J. W., Gu W., Cohen R. E., Shi Y. (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054 [DOI] [PubMed] [Google Scholar]
- 74. Luna-Vargas M. P., Faesen A. C., van Dijk W. J., Rape M., Fish A., Sixma T. K. (2011) Ubiquitin-specific protease 4 is inhibited by its ubiquitin-like domain. EMBO Rep. 12, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75. Harper S., Besong T. M., Emsley J., Scott D. J., Dreveny I. (2011) Structure of the USP15 N-terminal domain. A β-hairpin mediates close association between the DUSP and UBL domains. Biochemistry 50, 7995–8004 [DOI] [PubMed] [Google Scholar]
- 76. Heessen S., Masucci M. G., Dantuma N. P. (2005) The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol. Cell 18, 225–235 [DOI] [PubMed] [Google Scholar]
- 77. Heinen C., Acs K., Hoogstraten D., Dantuma N. P. (2011) C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat. Commun. 2, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seeger M., Hartmann-Petersen R., Wilkinson C. R., Wallace M., Samejima I., Taylor M. S., Gordon C. (2003) Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J. Biol. Chem. 278, 16791–16796 [DOI] [PubMed] [Google Scholar]
- 79. Richmond C., Gorbea C., Rechsteiner M. (1997) Specific interactions between ATPase subunits of the 26 S protease. J. Biol. Chem. 272, 13403–13411 [DOI] [PubMed] [Google Scholar]
- 80. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Andersen K. M., Hofmann K., Hartmann-Petersen R. (2005) Ubiquitin-binding proteins. Similar, but different. Essays Biochem. 41, 49–67 [DOI] [PubMed] [Google Scholar]
- 83. Hurley J. H., Lee S., Prag G. (2006) Ubiquitin-binding domains. Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hartmann-Petersen R., Gordon C. (2004) Integral UBL domain proteins. A family of proteasome interacting proteins. Semin. Cell Dev. Biol. 15, 247–259 [DOI] [PubMed] [Google Scholar]
- 85. Maupin-Furlow J. A., Humbard M. A., Kirkland P. A., Li W., Reuter C. J., Wright A. J., Zhou G. (2006) Proteasomes from structure to function. Perspectives from Archaea. Curr. Top. Dev. Biol. 75, 125–169 [DOI] [PubMed] [Google Scholar]
- 86. Stadtmueller B. M., Ferrell K., Whitby F. G., Heroux A., Robinson H., Myszka D. G., Hill C. P. (2010) Structural models for interactions between the 20 S proteasome and its PAN/19 S activators. J. Biol. Chem. 285, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schrader E. K., Harstad K. G., Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang F., Wu Z., Zhang P., Tian G., Finley D., Shi Y. (2009) Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Glickman M. H., Adir N. (2004) The proteasome and the delicate balance between destruction and rescue. PLoS Biol. 2, E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yao T., Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 91. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26 S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 92. Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8, 417–422 [DOI] [PubMed] [Google Scholar]
- 93. Sirkis R., Gerst J. E., Fass D. (2006) Ddi1, a eukaryotic protein with the retroviral protease fold. J. Mol. Biol. 364, 376–387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.