Background: Tumor cell invasion is important in cancer progression, and the sigma-2 receptor (S2RPgrmc1) contributes to invasion.
Results: S2RPgrmc1-knockdown cells had diminished levels of NGAL protein and RNA, corresponding with decreased active NFκB.
Conclusion: We propose a model in which S2RPgrmc1 elevates NGAL expression via EGFR and NFκB.
Significance: S2RPgrmc1 may be a potent target for inhibiting tumor invasion.
Keywords: Epidermal Growth Factor Receptor (EGFR), Lung Cancer, NF-κB (NF-κB), Protease, Receptors
Abstract
Tumor invasion is a critical step in the spread of cancer. S2R (sigma-2 receptor)/Pgrmc1 (progesterone receptor membrane component 1) is a cytochrome b5-related drug-binding orphan receptor essential for tumor formation and invasion. Secretory proteins drive these processes, so we screened for S2RPgrmc1-dependent secreted proteins using antibody arrays. S2RPgrmc1 markedly regulated the expression of NGAL/LCN2 (neutrophil gelatinase-associated lipocalin/lipocalin 2), a secreted glycoprotein that binds to MMP-9 (matrix metalloproteinase 9) and protects it from degradation. S2RPgrmc1 knock-down blocked NGAL/LCN2 expression at the protein and RNA levels and decreased MMP9 activity. NGAL expression was required for MMP-9 activity and tumor formation. S2RPgrmc1 associates with EGFR and increases EGFR levels at the plasma membrane, and the EGFR inhibitors erlotinib and AG1478, as well as Akt and ERK inhibitors, suppressed the NGAL/LCN2 RNA and protein levels. NGAL is transcriptionally regulated by NFκB, and S2RPgrmc1 knock-down decreased the NFκB subunit p65/RelA acetylation, phosphorylation, and activation. In S2RPgrmc1 knock-down cells, p65 acetylation was reversed by inhibitors of histone deacetylase 1, and the inhibitors partially restored NGAL levels. Our results are consistent with a model in which S2RPgrmc1 increases NGAL/LCN2 levels by activating NFκB via EGFR.
Introduction
Cancers spread to distant sites by tumor invasion, which is driven by the combined action of proteases, adhesion factors, and signaling proteins. Neutrophil gelatinase-associated lipocalin/lipocalin 2 (NGAL/LCN2)2 is an iron-binding protein (1, 2) that complexes and stabilizes the matrix metalloproteinase MMP9 (3, 4), promoting survival (5, 6), and invasion (4, 7, 8). The NGAL-MMP9 complex is expressed in tumors and is detectable in the blood and urine of cancer patients (3, 9). NGAL expression is driven, at least in part, by a pathway consisting of the HER2/neu-phosphotidylinositol 3-kinase-NFκB pathway in breast cancer cells (7, 10), where it profoundly increases tumor formation and invasion (7). NFκB exists in an inactive complex in the cytoplasm, and is activated by acetylation and phosphorylation (11), although the mechanism through which HER2/neu activates NFκB to elevate NGAL is unknown.
S2RPgrmc1 was originally identified as a putative hormone receptor (12–15), but S2RPgrmc1 is related to cytochrome b5 and binds to heme (14, 16–21). In normal tissues, S2RPgrmc1 increases lipid synthesis via P450 proteins (20, 22–24), while in tumor cells, it has a profound effect on cell signaling (25–27). Specifically, S2RPgrmc1 associates with EGFR in lung and breast cancer cells, where it elevates plasma membrane levels of the receptor (28). Recently, S2RPgrmc1 was also identified as the putative sigma-2 receptor, an intracellular drug-binding protein (29). S2RPgrmc1 also associates with PAIR-BP1 in ovarian cells (30, 31), although the mechanism of this complex is unknown.
S2RPgrmc1 expression is elevated in a broad spectrum of tumors (32–34), where it is variously associated with survival, tumor stage (34), hormone receptor status (25), and hypoxia (35). In lung cancer cell lines, we have shown that S2RPgrmc1 increases proliferation, invasion, tumor growth, and metastatic colonization (26). In ovarian cancer, S2RPgrmc1 promotes tumor formation and suppresses apoptosis (30, 34, 36). The ligand-binding domain of S2RPgrmc1 is attractive as a therapeutic target, and an S2RPgrmc1 inhibitor, AG-205 (26, 37), inhibits cancer cell proliferation (26), destabilizes EGFR (28) and reverses S2RPgrmc1 agonist binding (29).
In the present study, we report that S2RPgrmc1 drives the transcription of NGAL and the activation of the NGAL-MMP9 complex. We show that NGAL is required for tumor formation in lung cancer cells; NGAL transcription requires EGFR, and both proteins are activated by S2RPgrmc1. The results suggest a model in which S2RPgrmc1 promotes metalloproteinase activity by activating receptor signaling to NFκB.
EXPERIMENTAL PROCEDURES
Cell Lines and Treatments
A549 and NCI-H226 cells were obtained from the ATCC, cultured under the suggested conditions, and their identity was verified by Genetica LLC (Cincinnati, OH). Cells were maintained in DMEM containing 10% serum supreme and antibiotics, except where described. The AG-205 inhibitor (26) has been described. Short hairpin RNA (shRNA) lentiviruses (Sigma-Aldrich) containing shRNAs for NGAL (clone D: TRCN0000060540 and clone E: TRCN0000060549) were transduced and selected in puromycin. The Pgrmc1 expression plasmid pRC40 has been described (27), and EGFR was expressed from the plasmid pcDNA3-EGFR, which was a kind gift from Drs. Penni Black of the University of Kentucky and William Pao of Vanderbilt University. Erlotinib (LC Laboratories, Woburn, MA), LY294002 (Sigma), PD98059 (Sigma), AG1478 (Sigma), SAHA (suberoylanalide hydroxamic acid, Biomol, Plymouth Meeting, MA) and sodium butyrate (Sigma) were used as indicated. Conditioned media was generated by incubating cells in serum-free DMEM media and concentrating the media 10-fold using an Amicon Ultracel 10 kDa molecular mass cut-off filter unit (Millipore, Billerica, MA). For xenografts, athymic nude female mice (5–6 weeks old, Harlan Laboratories, Indianapolis, IN) were injected subcutaneously with either A549/con or A549/shNGAL cells (3 × 106 cells in 100 μl of PBS) into the upper flank. After 21 days, tumor volume (mm3) was calculated by using the following formula V = (W2 × L)/2, where W is width (small diameter) and L is length (long diameter).
Invasion was determined by adding 1 × 105 A549 cells to the inner chamber of a Matrigel-coated invasion chamber (BD Biosciences, 8 μm pore size). 600 μl of culture medium containing 5% FBS was added to the bottom chamber, and the cells were incubated for 16 h 37 °C in 5% CO2. Cells on the upper surface of the inner chamber were then removed, and cells adhering to the lower surface of the membrane were fixed, stained with 1% toluidine blue in 1% Borax and counted.
Protein Analysis
Human soluble receptor arrays were utilized according to manufacturer's instructions. Briefly, membranes were probed with 100–250 μl of conditioned media and detected with chemiluminescence. Quantification of pixels was performed by densitometry using Adobe CS2 software. Western blots were performed using previously published techniques. The antibodies used were NGAL (MAB1757, R&D Systems, Minneapolis MN; 3819-100, Bio-Vision, Mountain View, CA), ku70 (sc-5309, Santa Cruz Biotechnology, Santa Cruz, CA), S2RPgrmc1 (PGR-UK1 (26)), HA (HA11, Covance), MMP9 (3852, Cell Signaling, Danvers, MA), cathepsin D (R-20, Santa Cruz Biotechnology), Timp2 (89025, R&D), calnexin (C-20, Santa Cruz Biotechnology), p53 (Transduction Laboratories, Franklin Lakes, NJ), E2F5 (sc-999, Santa Cruz Biotechnology), NFκB p50 (sc-7178, Santa Cruz Biotechnology), NFκB p65 (Cell Signaling), NFκB p65-pSer536 (Cell Signaling), NFκB p65-Lys310 (Cell Signaling), 14–3-3β (K-19, Santa Cruz Biotechnology), Hsp90 (F-8, Santa Cruz Biotechnology), HDAC1 (10E2, Cell Signaling) and MMP9 (#3852, Cell Signaling). For zymography, equal amounts of 10-fold concentrated conditioned media were resolved on a 10% Novex Zymogram Gel (Invitrogen, Carlsbad, CA) and developed per the manufacturer's instructions, except that the developing step was increased to 24–48 h.
Nuclear fractionation was performed using the NE-PER nuclear and cytoplasmic reagent (Thermo Pierce, Fremont, CA). Thermo-Pierce p65/RelA activation was measured using the NFkB p65 chemiluminescent transcription factor assay kit according to the manufacturer's instructions. Nuclear lysates were prepared using the NE-PER reagent, and antibody incubations were for 1 h, as indicated.
RNA Analysis
For RT-PCR, total RNA was isolated by TRIzol Reagent (Invitrogen; 15596-026) according to manufacturer's protocols. cDNA synthesis from 2 μg of RNA was carried out using SuperScript II (Invitrogen) with random hexamers as described (23). Semi-quantitative RT-PCR was performed as described (38). Triplicate samples for quantitative PCR were run in an iCycler (Bio-Rad) using the SYBR Green I system (Bio-Rad). ΔCt for each gene was determined after normalization to β-actin, and ΔΔCt. was calculated relative to the control. Gene expression values were then expressed as a fold change, calculated by 2−ΔΔCt. The primer sequences were NGAL-F, 5′-TGAGCACCAACTACAACCAG-3′; NGAL-R, 5′-AGAGATTTGGAGAAGCGGATG-3′; β-actin-F, 5′-CCTTCCTGGGCATGGAGTCCT-3′; β-actin-R, 5′-GGAGCAATGATCTTGATCTTC-3′.
RESULTS
NGAL Expression Is Dependent on S2RPgrmc1
S2RPgrmc1 promotes tumor cell migration and metastatic colonization (26), and we probed conditioned media from S2RPgrmc1-knockdown A549 NSCLC cells (26) for secreted proteins associated with invasion. Using two arrays that included 81 embedded antibodies to secreted proteins, we detected numerous altered proteins (supplemental Fig. S1). NGAL was essentially absent in S2RPgrmc1-knockdown cells (Fig. 1A, plotted in Fig. 1B), and loss of NGAL in S2RPgrmc1-knockdown cells was confirmed by Western blots in conditioned media (Fig. 1C) and cell lysates (Fig. 1D). NGAL binds covalently to the MMP9 matrix metalloproteinase, forming a 125 kDa complex (3). In S2RPgrmc1-knockdown cells, the 125 kDa complex decreased 8.5-fold in zymography gels containing gelatin (Fig. 1E), while the lower MMP9 band decreased 4.1-fold. MMP9 levels were essentially unchanged in the same conditioned media (Fig. 1E, middle panel) and lysates (Fig. 1F) from S2RPgrmc1-knockdown cells, suggesting that decreased MMP9 activity in media is not due to changes in MMP9 levels.
FIGURE 1.
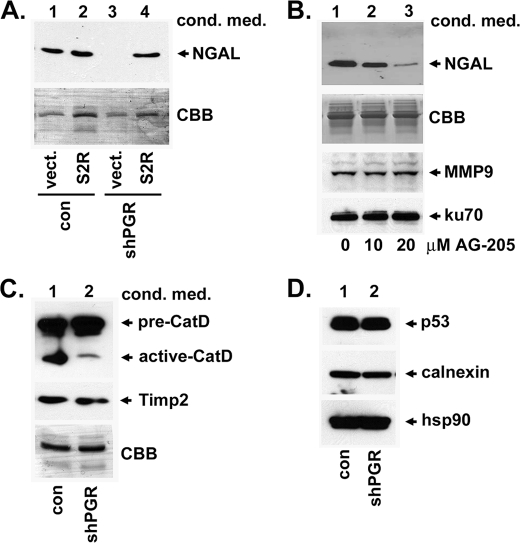
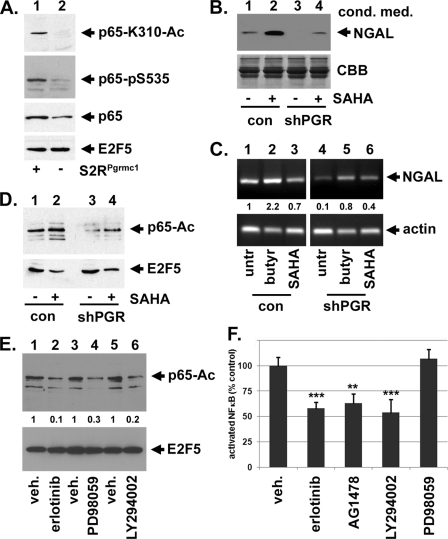
NGAL protein levels are dependent on S2RPgrmc1. A, proteome array containing antibodies to secreted proteins was probed with conditioned media from control A549 NSCLC cells (top, con) or S2RPgrmc1-knockdown cells (bottom, shPGR). NGAL is indicated. B, quantitation of NGAL spots in duplicate from a pair of arrays probed with control (top) or S2RPgrmc1-knockdown (bottom) conditioned media. C and D, Western blots of control (c, odd lanes) or S2RPgrmc1-knockdown (kd, even lanes) cells that were serum-starved for the indicated times. C, Western blot of NGAL in conditioned media with Coomassie brilliant blue-stained media (CBB) as a loading control. D, Western blot of NGAL in cellular lysates (upper panel) with a Western blot of 14-3-3 protein as a loading control (lower panel). E, zymography of control and S2RPgrmc1-knockdown conditioned media showing decreased gelatinase activity in S2RPgrmc1-knockdown cells. MMP9 expression (Western blot) in the same media samples is shown in the middle panel. Loading controls are analogous to those in panel C. F, Western blot of MMP9 (upper panel) in lysates from control and S2RPgrmc1-knockdown cells. Ku70 (lower panel) is shown as a loading control.
The shRNA targeting S2RPgrmc1 binds a sequence in the 3′-untranslated region of the S2RPgrmc1 transcript, and we expressed an exogenous S2RPgrmc1 transcript lacking this sequence. Exogenous S2RPgrmc1 restored NGAL levels in conditioned media (Fig. 2A, lane 4) to levels approximating the control cells (Fig. 2A, lanes 1 and 2). NGAL expression also decreased after treatment with AG-205, an S2RPgrmc1 small molecule inhibitor (26, 28) in a dose-dependent manner (Fig. 2B). MMP9 was unchanged (Fig. 2B, third panel). While we focused this study primarily on Ngal, other proteins were altered in the condition media, including activated cathepsin D (Fig. 2C), although Timp2 was unchanged (Fig. 2C).
FIGURE 2.
Restoration of S2RPgrmc1 elevates NGAL levels. A, A549 control (lanes 1 and 2) and S2RPgrmc1-knockdown (lanes 3 and 4) cells were transfected with a control plasmid (lanes 1 and 3) or a plasmid encoding S2RPgrmc1 (lanes 2 and 4). Conditioned media were analyzed by Western blot for NGAL (top) and by SDS-PAGE and Coomassie Brilliant Blue staining (CBB, bottom). Lane 4 shows elevated NGAL levels when S2RPgrmc1 was re-expressed. B, NGAL levels in conditioned media after treatment with increasing doses of the S2RPgrmc1 inhibitor AG-205 for 72 h. While NGAL levels decreased (upper panel), MMP9 levels were unchanged (lower panel). Ku70 served as a loading control. C, Western blot of conditioned media from control and S2RPgrmc1-knockdown cells showed decreased levels of activated cathepsin D (top panel) but no change in Timp-2 (second panel) in S2RPgrmc1-knockdown cells. D, Western blot analysis of control and S2RPgrmc1-knockdown cells revealed no change in levels of the stress response proteins p53, calnexin, and hsp90.
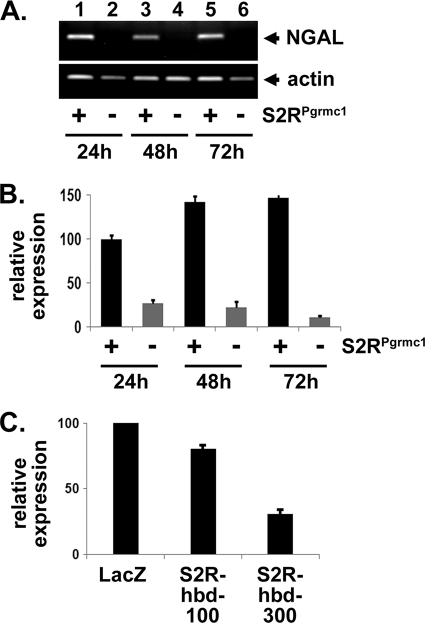
One potential mechanism governing NGAL levels in S2RPgrmc1-knockdown cells is the activation of a general stress response. However, levels of the stress response genes p53, calnexin and hsp90 (39–41) were unchanged in S2RPgrmc1-knockdown cells (Fig. 2D). Instead, the decline in NGAL protein levels was reflected in decreased NGAL RNA levels in S2RPgrmc1-knockdown cells by RT-PCR (Fig. 3A) or real-time quantitative PCR (Fig. 3B). Decreased NGAL RNA levels were also detected after expression of a heme-binding-deficient (S2R-hbd) S2RPgrmc1 mutant (Fig. 3C).
FIGURE 3.
S2RPgrmc1 increases NGAL RNA levels. A, RT-PCR for NGAL (top) and actin (bottom) in A549 control (odd lanes) and S2RPgrmc1-knockdown cells (even lanes) at increasing times after serum starvation. The figure is an agarose gel. B, real-time PCR of NGAL cDNA. The sample pattern is the same as in panel A. C, real-time PCR of NGAL in A549 cells infected with a control adenovirus (LacZ) or increasing doses of the Ad-S2R-hbd adenovirus (see Fig. 2A).
NGAL Requirement for Tumor Formation
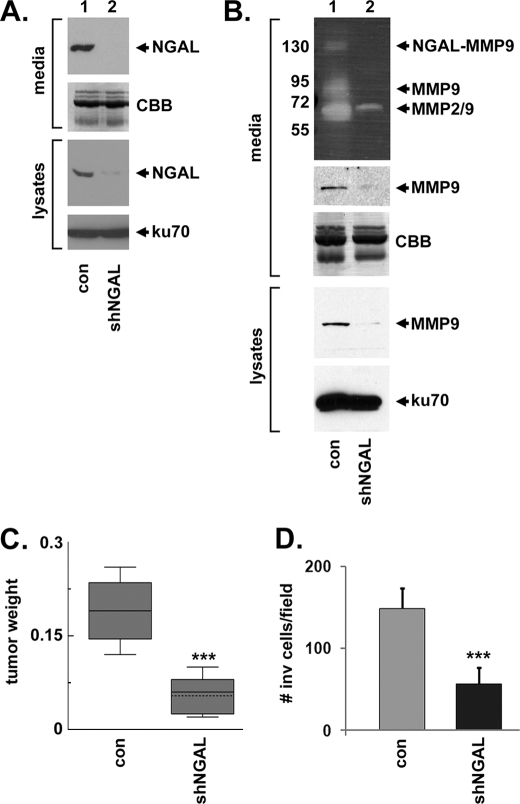
NGAL biological activity has not been reported previously for lung cancer. We inhibited NGAL expression (Fig. 4A) by introducing an NGAL-targeting shRNA into A549 NSCLC cells. Zymography of conditioned media from control and NGAL-knockdown cells indicated that both NGAL-MMP9 and MMP9 activity were decreased (26-fold and 52-fold, respectively, Fig. 4B). A 65 kDa band, which could be a cleaved form of MMP9 or uncleaved MMP2, also decreased 11-fold but was detectable in the NGAL-knockdown cells (Fig. 4B, MMP2/9). Tumor xenograft formation was sharply impaired in the mice infused with NGAL-knockdown cells (p = 0.002, t test, Fig. 4C), indicating an essential role for NGAL in lung tumor growth. NGAL also promoted motility in A549 cells in a modified Boyden chamber assay (p = 0.0002, t test, Fig. 4D), consistent with its function in regulating metalloproteinases.
FIGURE 4.
NGAL is required for A549 xenograft tumor growth. A, Western blot of NGAL (top panel) in conditioned media, concentrated 10-fold, from A549 cells infected with a control lentivirus (con, lane 1) or a lentivirus directing expression of an NGAL-targeting shRNA (shNGAL, lane 2). The second panel shows the same samples stained with Coomassie. The lower panels are Western blots of NGAL or ku70 (loading control) in lysates from control and NGAL-knockdown cells. In each case, cells were grown in the absence of serum for 72 h. B, top panel shows zymography of conditioned media from control (lane 1) or NGAL-knockdown cells (lane 2). MMP9 expression is shown in the middle panel, and Coomassie staining is shown as a loading control. MMP-9 and ku70 expression in cell lysates is shown in the lower two panels. C, box plot of tumor weight in control and NGAL-knockdown A549 tumor xenografts, showing decreased tumor growth in NGAL-knockdown cells. D, diagram of invasion assays of control and NGAL-knockdown A549 cells, showing a significant decrease in migration in NGAL-knockdown cells.
NGAL Expression Driven by an EGFR-NFκB Pathway in Lung Cancer
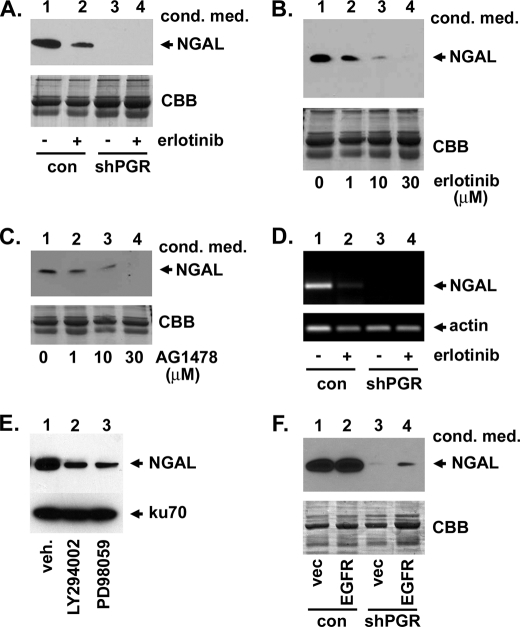
S2RPgrmc1 associates with EGFR, increasing pools of EGFR at the plasma membrane and elevating erlotinib sensitivity (28). In A549 cells, the EGFR inhibitors erlotinib and AG1478/tyrphostin inhibited NGAL protein (Fig. 5, A–C) and RNA levels (Fig. 5D), while NGAL was absent in S2RPgrmc1 knockdown cells (Fig. 5, A and D, lanes 3–4). Complete NGAL inhibition was detected at higher doses of erlotinib (Fig. 5B) and AG1478 (Fig. 5C). EGFR is upstream of Akt and ERK, two key effector kinases, and the Akt and ERK inhibitors LY294002 and PD98059, respectively, decreased NGAL levels (Fig. 5E). We then took a genetic approach and overexpressed EGFR in control and S2RPgrmc1-knockdown cells (Fig. 5F). EGFR overexpression partially restored NGAL levels in S2RPgrmc1-knockdown cells (Fig. 5F, lane 4). We conclude that EGFR signaling contributes to NGAL transcription.
FIGURE 5.
EGFR increases NGAL expression in lung cancer cells. A, NGAL levels decreased in the conditioned media of control A549 cells after treatment with 10 μm erlotinib (lanes 1–2), while S2RPgrmc1-knockdown cells had undetectable NGAL levels (lanes 3–4). For all panels, conditioned media was harvested after 72 h in serum-free media and concentrated 10-fold. B, NGAL decreased in concentrated conditioned media with increasing erlotinib doses under the same conditions. C, NGAL decreased with increasing doses of the EGFR inhibitor AG1478 under the same conditions used in panel A. D, RT-PCR showed that NGAL inhibition occurred at the RNA level in control A549 cells. RNA was harvested after 48 h of serum starvation after treatment with 10 μm erlotinib. E, Western blot of A549 cells treated with vehicle (lane 1), 15 μm LY294002 and 25 μm PD98059 for 24 h and probed for NGAL (top) or ku70 (bottom) as a loading control. F, Western blot of conditioned medium from A549 control (lanes 1 and 2) or S2RPgrmc1 knockdown (lanes 3 and 4) cells transfected with a control plasmid (vec, lanes 1 and 3) or the EGFR expression plasmid pcDNA3-EGFR (EGFR, lanes 2 and 4). The blot was probed for NGAL and showed a partial restoration of NGAL expression in S2RPgrmc1-knockdown cells following EGFR expression.
NFκB has been previously implicated in NGAL transcription, and p65-RelA is activated by acetylation and phosphorylation and inhibited by histone decetylases or HDACs (11, 42–44). Acetylated p65/RelA (Ac-RelA) decreased 8-fold in the nuclear fraction of S2RPgrmc1-knockdown cells (Fig. 6A, top panel), while p65-phosphoS536 levels diminished by 5.7-fold (Fig. 6A, second panel) and total nuclear p65 by 3.9-fold (Fig. 6A, third panel). In contrast, the nuclear marker E2F5 was unchanged (Fig. 6A, lower panel). The nuclear and cytoplasmic protein separation was validated with markers for each fraction (supplemental Fig. S4). The results are consistent with a model in which p65 phosphorylation, acetylation, and nuclear transport are partially dependent on S2RPgrmc1. We detected a similar decrease in acetylated p65 in NCI-H226 cells following infection with the Ad-S2R-hbd mutant (supplemental Fig. S3C).
FIGURE 6.
S2RPgrmc1 increases NFκB activation. A, A549 control (lane 1) and S2RPgrmc1-knockdown cells (lane 2) were analyzed by Western blot for NFκB p65/RelA-K310 acetylation (top panel), Ser-536 phosphorylation (second panel), total p65 (third panel) and E2F5 as a loading control (lower panel). Nuclear fractions were analyzed after growth in serum-free media for 48 h. B, conditioned media from control (lanes 1 and 2) and S2RPgrmc1-knockdown cells (lanes 3 and 4), untreated (lanes 1 and 3) or treated with the HDAC inhibitor SAHA at 20 μm for 72 h (lanes 2 and 4), were analyzed by Western blot for NGAL. The lower panel shows loading controls for the same samples. C, A549 control (lanes 1–3) and S2RPgrmc1-knockdown cells (lanes 4–6) were analyzed by RT-PCR for NGAL (top) or actin (bottom) after treatment with vehicle (lanes 1 and 4), 5 mm sodium butyrate (lanes 2 and 5), or 20 μm SAHA (lanes 3 and 6) for 72 h. NGAL/actin ratios, relative to vehicle-treated control cells, are shown between the two panels. D, nuclear lysates from control (lanes 1 and 2) and S2RPgrmc1-knockdown cells (lanes 3 and 4), untreated (lanes 1 and 3) or treated with the HDAC inhibitor SAHA (lanes 2 and 4), were analyzed by Western blot for acetylated p65. The lower panel shows loading controls for the same samples. E, nuclear lysates from A549 cells treated with vehicle (odd lanes) or 10 μm of the EGFR inhibitor erlotinib (lane 2), 20 μm of the ERK inhibitor PD98059 (lane 4) or 20 μm of the Akt inhibitor LY294002 (lanes 2 and 4), were analyzed by Western blot for acetylated p65. The lower panel shows a loading control (E2F5) for the same samples. The numbers between the panels indicate the ratio acetylated p65 in treated samples divided by the untreated sample, relative to the loading control [(p65treated/p65vehicle)/E2F5control]. F, inhibition of activated p65/RelA levels by erlotinib, AG1478, and LY294002 (at the concentrations indicated in panel E). Surprisingly, PD98059 did not have a significant activity in this assay.
Because HDACs decrease NFκB acetylation, we posited that the HDAC inhibitor SAHA would have the opposite effect. Indeed, control cells treated with SAHA exhibited increased secreted NGAL (Fig. 6B, lane 2), and SAHA restored NGAL to near basal levels in S2RPgrmc1-knockdown cells (Fig. 6B, lane 4). Furthermore, NGAL transcription increased following treatment with the HDAC inhibitor sodium butyrate in control and shPGR cells by 2.2- and 8-fold, respectively (Fig. 6C, lanes 2 and 5). SAHA treatment slightly repressed NGAL transcription in control cells (Fig. 6C, lane 3) but induced NGAL transcription by 4-fold in S2RPgrmc1-knockdown cells. As expected, SAHA treatment had little effect on p65 acetylation in control cells (Fig. 6D, lane 2) but increased p65 acetylation by 2.8-fold in S2RPgrmc1-knockdown cells (Fig. 6D, lane 4). In contrast, E2F5 levels decreased slightly after SAHA treatment and were unchanged in control and S2RPgrmc1-knockdown cells (Fig. 6D, lower panel). The results suggest that defects in p65/RelA activation in S2RPgrmc1-knockdown cells contribute to loss of NGAL transcription.
Because inhibitors of EGFR, Akt, and ERK inhibited NGAL levels (Fig. 5), we tested whether they affect acetylated NFκB. Nuclear fractions of cells that were treated with vehicle (Fig. 6E, lanes 1, 3, and 5) had readily detectable levels of acetylated p65, while levels decreased after treatment with erlotinib, PD98059 and LY294002 by 10-fold, 3-fold, and 5-fold, respectively (Fig. 6E, lanes 2, 4, and 6). We then tested p65/RelA activation using a modified electrophoretic mobility shift assay (EMSA) modified for ELISA (enzyme-linked immunosorbent assay), and found that erlotinib, AG-1478/tyrphostin and LY294002 significantly decreased p65/RelA activation (Fig. 6F). Surprisingly, PD98059 did not significantly affect p65/RelA activation, suggesting it may inhibit NGAL through a distinct mechanism from the other inhibitors. The results suggest that the S2RPgrmc1-EGFR-Akt pathway increases nuclear p65/RelA activation.
DISCUSSION
S2RPgrmc1 is elevated in lung cancers and contributes to tumor growth, metastasis and invasion in lung cancer cells. In the present study, we demonstrate that S2RPgrmc1 elevates the transcription and protein levels of NGAL, a secreted glycoprotein that complexes with the MMP9 metalloproteinase, stabilizing the NGAL-MMP9 complex. NGAL also binds to iron (2), and the yeast S2RPgrmc1 homologue is regulated by iron (45) and is essential for iron storage (24). Our results suggest a model in which S2RPgrmc1 activates NGAL transcription. The role of NGAL in promoting or suppressing tumor growth is highly dependent on the tissue of origin, and our results support a role for NGAL in cancer cell invasion and tumor formation in lung cancer.
In addition to increasing NGAL expression, S2RPgrmc1 elevated NGAL-MMP9 activity. Surprisingly, uncomplexed MMP9 activity was also increased by S2RPgrmc1 (Fig. 1), even though MMP9 levels were largely unchanged in S2RPgrmc1-knockdown cells. One potential mechanism is the induction of Timp1, -2, and -3 expression in S2RPgrmc1-knockdown cells (supplemental Fig. S1), which can inhibit MMP9 activation (46). However, Timp2 was not induced in conditioned media by Western blot (Fig. 2), suggesting that the proteome array results may be tightly dependent on specific epitopes recognized by the antibodies embedded in the arrays. Alternately, MMP9 can be activated by multiple proteases (46), and any of them could be altered by loss of S2RPgrmc1. Loss of S2RPgrmc1 reduced activated cathepsin D (Fig. 2), which in turn suggests alterations in cathepsins L and B (47). Interestingly, cathepsin G is important in MMP9 processing (46), and one potential model is that multiple cathepsins are activated by S2RPgrmc1. This model, which is purely speculative, is currently under investigation. The results are important because MMP9 plays a key role in tumor growth and progression (48), and NGAL is a potential mediator of tumor invasion by stabilizing MMP9.
One enigmatic finding in the present study is that S2RPgrmc1 has little effect on MMP9 levels (Fig. 1), while NGAL expression increased MMP9 levels (Fig. 4). In the simplest sense, the expected result is that both S2RPgrmc1 and NGAL inhibition should have the same phenotype, decreased MMP9, reflecting a common pathway. The results suggest that S2RPgrmc1 may have NGAL-independent functions, increasing MMP9, that are uncharacterized.
Based on previously published studies, we propose that the S2RPgrmc1-EGFR complex (28) drives NGAL expression. According to this model, NFκB p65/RelA, an essential protein in K-ras-driven lung cancer (49), is subsequently phosphorylated, acetylated and transported to the nucleus, driving NGAL transcription (Fig. 7). One caveat to this model is that the S2RPgrmc1-knockdown affected NGAL levels more potently than EGFR inhibitors (Fig. 5), suggesting that S2RPgrmc1 may utilize pathways other than EGFR in activating NGAL. Exogenous EGFR expression restored NGAL levels (Fig. 5), but only partially, likely because of the important role of S2RPgrmc1 in stabilizing EGFR pools at the plasma membrane (28). The mechanism linking S2RPgrmc1 and EGFR to p65/RelA includes Akt (for maintaining nuclear, acetylated p65/RelA and for activation) and ERK activation (for nuclear, acetylated p65/RelA, Figs. 5 and 6), but does not include altered HDAC1 expression (Fig. 6D). An alternate model is that HDAC1 can be activated by non-EGFR metabolic pathways. For example, S2RPgrmc1 ligands have been implicated in oxidative stress induction (50), suggesting that S2RPgrmc1 normally increases oxidative stress. In turn, oxidative stress inhibits HDAC activity (51), which is predicted to elevate p65/RelA acetylation. The role of S2RPgrmc1 in oxidative stress is a subject of ongoing investigation.
FIGURE 7.
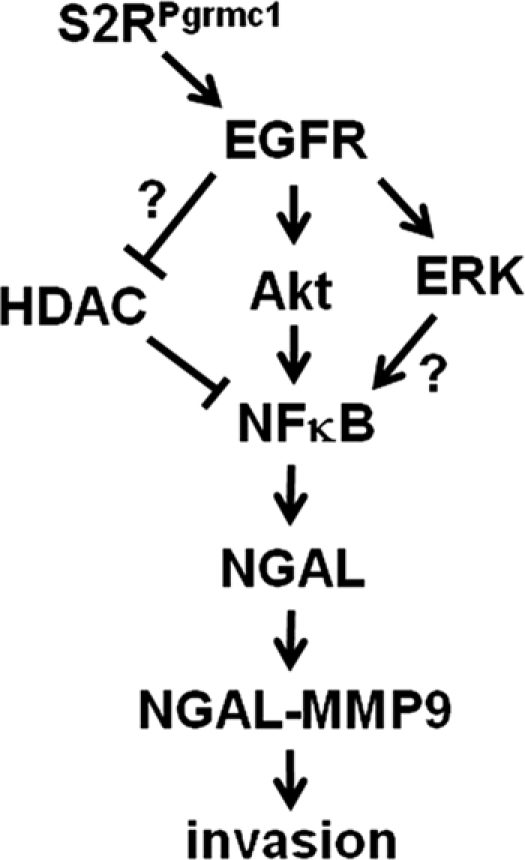
A schematic pathway linking S2RPgrmc1 to NGAL.
The significance of the findings is that tumor invasion is a hallmark of aggressive cancers and is driven, at least in part, by MMPs. Therapeutic targeting of MMPs has had mixed results (48), and the current findings suggest a new approach for targeting MMP-NGAL complexes. Indeed, a small molecule S2RPgrmc1 inhibitor suppressed NGAL expression, and other S2R ligands have activity against cancer cells (29, 52) and may indirectly inhibit MMPs. Our results suggest that strategies inhibiting S2RPgrmc1 could potentially target MMP9 activity in lung cancer and perhaps other cancer types.
Supplementary Material
Acknowledgments
We thank Sourik Ganguly for advice about zymography and Boyden chamber assays and Jim Begley for assistance with imaging.
This work was supported by the Kentucky Lung Cancer Research Program, Cycles 8 and 10, and by the Kentucky Science and Education Fund.

This article contains supplemental Figs. S1–S4.
- NGAL/LCN2
- neutrophil gelatinase-associated lipocalin/lipocalin 2
- MMP
- matrix metalloproteinase
- S2R
- sigma 2 receptor
- shRNA
- short hairpin RNA.
REFERENCES
- 1. Richardson D. R. (2005) 24p3 and its receptor: dawn of a new iron age? Cell 123, 1175–1177 [DOI] [PubMed] [Google Scholar]
- 2. Devireddy L. R., Gazin C., Zhu X., Green M. R. (2005) A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 123, 1293–1305 [DOI] [PubMed] [Google Scholar]
- 3. Yan L., Borregaard N., Kjeldsen L., Moses M. A. (2001) The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J. Biol. Chem. 276, 37258–37265 [DOI] [PubMed] [Google Scholar]
- 4. Nuntagowat C., Leelawat K., Tohtong R. (2010) NGAL knockdown by siRNA in human cholangiocarcinoma cells suppressed invasion by reducing NGAL/MMP-9 complex formation. Clin. Exp. Metastasis 27, 295–305 [DOI] [PubMed] [Google Scholar]
- 5. Tong Z., Wu X., Ovcharenko D., Zhu J., Chen C. S., Kehrer J. P. (2005) Neutrophil gelatinase-associated lipocalin as a survival factor. Biochem. J. 391, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iannetti A., Pacifico F., Acquaviva R., Lavorgna A., Crescenzi E., Vascotto C., Tell G., Salzano A. M., Scaloni A., Vuttariello E., Chiappetta G., Formisano S., Leonardi A. (2008) The neutrophil gelatinase-associated lipocalin (NGAL), a NF-κB-regulated gene, is a survival factor for thyroid neoplastic cells. Proc. Natl. Acad. Sci. U.S.A. 105, 14058–14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leng X., Ding T., Lin H., Wang Y., Hu L., Hu J., Feig B., Zhang W., Pusztai L., Symmans W. F., Wu Y., Arlinghaus R. B. (2009) Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res. 69, 8579–8584 [DOI] [PubMed] [Google Scholar]
- 8. Tong Z., Kunnumakkara A. B., Wang H., Matsuo Y., Diagaradjane P., Harikumar K. B., Ramachandran V., Sung B., Chakraborty A., Bresalier R. S., Logsdon C., Aggarwal B. B., Krishnan S., Guha S. (2008) Neutrophil gelatinase-associated lipocalin: a novel suppressor of invasion and angiogenesis in pancreatic cancer. Cancer Res. 68, 6100–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolignano D., Donato V., Lacquaniti A., Fazio M. R., Bono C., Coppolino G., Buemi M. (2010) Neutrophil gelatinase-associated lipocalin (NGAL) in human neoplasias: a new protein enters the scene. Cancer Lett. 288, 10–16 [DOI] [PubMed] [Google Scholar]
- 10. Li S. H., Hawthorne V. S., Neal C. L., Sanghera S., Xu J., Yang J., Guo H., Steeg P. S., Yu D. (2009) Upregulation of neutrophil gelatinase-associated lipocalin by ErbB2 through nuclear factor-κB activation. Cancer Res. 69, 9163–9168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L. f., Fischle W., Verdin E., Greene W. C. (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293, 1653–1657 [DOI] [PubMed] [Google Scholar]
- 12. Falkenstein E., Meyer C., Eisen C., Scriba P. C., Wehling M. (1996) Full-length cDNA sequence of a progesterone membrane-binding protein from porcine vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 229, 86–89 [DOI] [PubMed] [Google Scholar]
- 13. Gerdes D., Wehling M., Leube B., Falkenstein E. (1998) Cloning and tissue expression of two putative steroid membrane receptors. Biol. Chem. 379, 907–911 [DOI] [PubMed] [Google Scholar]
- 14. Min L., Strushkevich N. V., Harnastai I. N., Iwamoto H., Gilep A. A., Takemori H., Usanov S. A., Nonaka Y., Hori H., Vinson G. P., Okamoto M. (2005) Molecular identification of adrenal inner zone antigen as a heme-binding protein. Febs. J. 272, 5832–5843 [DOI] [PubMed] [Google Scholar]
- 15. Wehling M., Schultz A., Lösel R. (2007) To be or not to be (a receptor). Steroids 72, 107–110 [DOI] [PubMed] [Google Scholar]
- 16. Mifsud W., Bateman A. (2002) Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol. 3, RESEARCH0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Min L., Takemori H., Nonaka Y., Katoh Y., Doi J., Horike N., Osamu H., Raza F. S., Vinson G. P., Okamoto M. (2004) Characterization of the adrenal-specific antigen IZA (inner zone antigen) and its role in the steroidogenesis. Mol. Cell. Endocrinol. 215, 143–148 [DOI] [PubMed] [Google Scholar]
- 18. Ghosh K., Thompson A. M., Goldbeck R. A., Shi X., Whitman S., Oh E., Zhiwu Z., Vulpe C., Holman T. R. (2005) Spectroscopic and biochemical characterization of heme binding to yeast Dap1p and mouse PGRMC1p. Biochemistry 44, 16729–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson A. M., Reddi A. R., Shi X., Goldbeck R. A., Moenne-Loccoz P., Gibney B. R., Holman T. R. (2007) Measurement of the heme affinity for yeast dap1p, and its importance in cellular function. Biochemistry 46, 14629–14637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hand R. A., Jia N., Bard M., Craven R. J. (2003) Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot Cell 2, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crudden G., Chitti R. E., Craven R. J. (2006) Hpr6 (heme-1 domain protein) regulates the susceptibility of cancer cells to chemotherapeutic drugs. J. Pharmacol. Exp. Ther. 316, 448–455 [DOI] [PubMed] [Google Scholar]
- 22. Hughes A. L., Powell D. W., Bard M., Eckstein J., Barbuch R., Link A. J., Espenshade P. J. (2007) Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 5, 143–149 [DOI] [PubMed] [Google Scholar]
- 23. Mallory J. C., Crudden G., Johnson B. L., Mo C., Pierson C. A., Bard M., Craven R. J. (2005) Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Craven R. J., Mallory J. C., Hand R. A. (2007) Regulation of iron homeostasis mediated by the heme-binding protein Dap1 (damage resistance protein 1) via the P450 protein Erg11/Cyp51. J. Biol. Chem. 282, 36543–36551 [DOI] [PubMed] [Google Scholar]
- 25. Neubauer H., Clare S. E., Wozny W., Schwall G. P., Poznanovic S., Stegmann W., Vogel U., Sotlar K., Wallwiener D., Kurek R., Fehm T., Cahill M. A. (2008) Breast cancer proteomics reveals correlation between estrogen receptor status and differential phosphorylation of PGRMC1. Breast Cancer Res. 10, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahmed I. S., Rohe H. J., Twist K. E., Mattingly M. N., Craven R. J. (2010) Progesterone receptor membrane component 1 (Pgrmc1): a heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J. Pharmacol. Exp. Ther. 333, 564–573 [DOI] [PubMed] [Google Scholar]
- 27. Hand R. A., Craven R. J. (2003) Hpr6.6 protein mediates cell death from oxidative damage in MCF-7 human breast cancer cells. J. Cell. Biochem. 90, 534–547 [DOI] [PubMed] [Google Scholar]
- 28. Ahmed I. S., Rohe H. J., Twist K. E., Craven R. J. (2010) Pgrmc1 (progesterone receptor membrane component 1) associates with epidermal growth factor receptor and regulates erlotinib sensitivity. J. Biol. Chem. 285, 24775–24782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu J., Zeng C., Chu W., Pan F., Rothfuss J. M., Zhang F., Tu Z., Zhou D., Zeng D., Vangveravong S., Johnston F., Spitzer D., Chang K. C., Hotchkiss R. S., Hawkins W. G., Wheeler K. T., Mach R. H. (2011) Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat. Commun. 2, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peluso J. J., Pappalardo A., Losel R., Wehling M. (2006) Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone's antiapoptotic action. Endocrinology 147, 3133–3140 [DOI] [PubMed] [Google Scholar]
- 31. Peluso J. J., Pappalardo A., Losel R., Wehling M. (2005) Expression and function of PAIRBP1 within gonadotropin-primed immature rat ovaries: PAIRBP1 regulation of granulosa and luteal cell viability. Biol. Reprod. 73, 261–270 [DOI] [PubMed] [Google Scholar]
- 32. Rohe H. J., Ahmed I. S., Twist K. E., Craven R. J. (2009) PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation, and drug binding. Pharmacol. Ther. 121, 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crudden G., Loesel R., Craven R. J. (2005) Overexpression of the cytochrome p450 activator hpr6 (heme-1 domain protein/human progesterone receptor) in tumors. Tumour Biol. 26, 142–146 [DOI] [PubMed] [Google Scholar]
- 34. Peluso J. J., Liu X., Saunders M. M., Claffey K. P., Phoenix K. (2008) Regulation of ovarian cancer cell viability and sensitivity to cisplatin by progesterone receptor membrane component-1. J. Clin. Endocrinol. Metab. 93, 1592–1599 [DOI] [PubMed] [Google Scholar]
- 35. Dressman H. K., Hans C., Bild A., Olson J. A., Rosen E., Marcom P. K., Liotcheva V. B., Jones E. L., Vujaskovic Z., Marks J., Dewhirst M. W., West M., Nevins J. R., Blackwell K. (2006) Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin. Cancer Res. 12, 819–826 [DOI] [PubMed] [Google Scholar]
- 36. Peluso J. J., Romak J., Liu X. (2008) Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology 149, 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshitani N., Satou K., Saito K., Suzuki S., Hatanaka H., Seki M., Shinozaki K., Hirota H., Yokoyama S. (2005) A structure-based strategy for discovery of small ligands binding to functionally unknown proteins: combination of in silico screening and surface plasmon resonance measurements. Proteomics 5, 1472–1480 [DOI] [PubMed] [Google Scholar]
- 38. Mallory J. C., Crudden G., Oliva A., Saunders C., Stromberg A., Craven R. J. (2005) A novel group of genes regulates susceptibility to antineoplastic drugs in highly tumorigenic breast cancer cells. Mol. Pharmacol. 68, 1747–1756 [DOI] [PubMed] [Google Scholar]
- 39. Delom F., Emadali A., Cocolakis E., Lebrun J. J., Nantel A., Chevet E. (2007) Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ. 14, 586–596 [DOI] [PubMed] [Google Scholar]
- 40. Borrás C., Gómez-Cabrera M. C., Viña J. (2011) The dual role of p53: DNA protection and antioxidant. Free Radic Res. 45, 643–652 [DOI] [PubMed] [Google Scholar]
- 41. Kroes R. A., Abravaya K., Seidenfeld J., Morimoto R. I. (1991) Selective activation of human heat shock gene transcription by nitrosourea antitumor drugs mediated by isocyanate-induced damage and activation of heat shock transcription factor. Proc. Natl. Acad. Sci. U.S.A. 88, 4825–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashburner B. P., Westerheide S. D., Baldwin A. S., Jr. (2001) The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21, 7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y., Smith P. W., Jones D. R. (2006) Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol. Cell. Biol. 26, 8683–8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen L. F., Williams S. A., Mu Y., Nakano H., Duerr J. M., Buckbinder L., Greene W. C. (2005) NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 25, 7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Puig S., Askeland E., Thiele D. J. (2005) Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120, 99–110 [DOI] [PubMed] [Google Scholar]
- 46. Okada Y., Gonoji Y., Naka K., Tomita K., Nakanishi I., Iwata K., Yamashita K., Hayakawa T. (1992) Matrix metalloproteinase 9 (92-kDa gelatinase/type IV collagenase) from HT 1080 human fibrosarcoma cells. Purification and activation of the precursor and enzymic properties. J. Biol. Chem. 267, 21712–21719 [PubMed] [Google Scholar]
- 47. Laurent-Matha V., Derocq D., Prébois C., Katunuma N., Liaudet-Coopman E. (2006) Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J. Biochem. 139, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coussens L. M., Fingleton B., Matrisian L. M. (2002) Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295, 2387–2392 [DOI] [PubMed] [Google Scholar]
- 49. Bassères D. S., Ebbs A., Levantini E., Baldwin A. S. (2010) Requirement of the NF-κB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 70, 3537–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ostenfeld M. S., Fehrenbacher N., Høyer-Hansen M., Thomsen C., Farkas T., Jäättelä M. (2005) Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 65, 8975–8983 [DOI] [PubMed] [Google Scholar]
- 51. Doyle K., Fitzpatrick F. A. (2010) Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J. Biol. Chem. 285, 17417–17424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crawford K. W., Bowen W. D. (2002) Sigma-2 receptor agonists activate a novel apoptotic pathway and potentiate antineoplastic drugs in breast tumor cell lines. Cancer Res. 62, 313–322 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.