Background: The coiled-coil and C2 domain-containing 1A (CC2D1A) gene is implicated in mental retardation. Cyclic adenosine monophosphate (cAMP) pathway is critical for brain development.
Results: CC2D1A regulates the protein kinase A (PKA) activity.
Conclusion: CC2D1A is a positive regulator of the cAMP pathway in neurons.
Significance: Our findings may provide a molecular explanation why mutation in the CC2D1A gene leads to mental retardation.
Keywords: CREB, Cyclic AMP (cAMP), Neurodevelopment, Phosphodiesterases, Phospholipid, Protein Kinase A (PKA), Synapses, C2 domain, CC2D1A, DM14
Abstract
Mutation of the coiled-coil and C2 domain-containing 1A (CC2D1A) gene, which encodes a C2 domain and DM14 domain-containing protein, has been linked to severe autosomal recessive nonsyndromic mental retardation. Using a mouse model that produces a truncated form of CC2D1A that lacks the C2 domain and three of the four DM14 domains, we show that CC2D1A is important for neuronal differentiation and brain development. CC2D1A mutant neurons are hypersensitive to stress and have a reduced capacity to form dendrites and synapses in culture. At the biochemical level, CC2D1A transduces signals to the cyclic adenosine 3′,5′-monophosphate (cAMP)-protein kinase A (PKA) pathway during neuronal cell differentiation. PKA activity is compromised, and the translocation of its catalytic subunit to the nucleus is also defective in CC2D1A mutant cells. Consistently, phosphorylation of the PKA target cAMP-responsive element-binding protein, at serine 133, is nearly abolished in CC2D1A mutant cells. The defects in cAMP/PKA signaling were observed in fibroblast, macrophage, and neuronal primary cells derived from the CC2D1A KO mice. CC2D1A associates with the cAMP-PKA complex following forskolin treatment and accumulates in vesicles or on the plasma membrane in wild-type cells, suggesting that CC2D1A may recruit the PKA complex to the membrane to facilitate signal transduction. Together, our data show that CC2D1A is an important regulator of the cAMP/PKA signaling pathway, which may be the underlying cause for impaired mental function in nonsyndromic mental retardation patients with CC2D1A mutation.
Introduction
Autosomal recessive nonsyndromic mental retardation (NSMR)4 is estimated to account for a quarter of individuals with NSMR (1). Although many X- linked genes associated with NSMR have been identified (2), only three autosomal genes are found to be associated with NSMR (3). The CC2D1A gene was identified as the third autosomal recessive NSMR gene (4). Homozygosity for a deletion of 3,589 nucleotides beginning at intron 13 and ending at intron 16 of the CC2D1A gene was identified in nine affected individuals among the 10 affected and 24 unaffected individuals in the families. The deletion introduces a frameshift, creating a 30-amino acid nonsense peptide and a stop codon at position 438 of the mutant protein. Various biological processes involved in neuronal differentiation and synaptic plasticity, synaptic vesicle cycling, and gene expression regulation are considered to be important in the causation of mental retardation.
CC2D1A is the founding member of the CC2D1 gene family that contains DM14 (Drosophila melanogaster 14) and C2 domains (4). The CC2D1A gene encodes a 951-amino acid protein that contains four DM14 domains, a predicted helix-loop-helix DNA binding domain, and a protein kinase C conserved region 2 (C2)/calcium-dependent lipid-binding calcium/phospholipid binding domain. The C2 domain was originally identified as one of the two conserved regulatory domains (C1 and C2) of Ca2+-dependent protein kinase C (PKC) (5). It can act as a phospholipid binding or protein-protein interaction domain (6, 7); the DM14 motif is a conserved protein domain that was originally identified in the Drosophila genome and has no assigned function (4). The truncation mutation in NSMR patients causes deletion of one of the four DM14 domains and the C2 domain, suggesting that either or both C2 and DM14 domains are important for CC2D1A function.
CC2D1A was originally identified in a large scale overexpression screen as an activator of the NF-κB promoter (8); later, CC2D1A was shown to bind a 5′-repressor element of the serotonin-1A (5-HT1A) receptor to repress its expression in transfection experiments and was named Freud-1 (9, 10). Because 5-HT1A receptors are G-protein-coupled receptors that signal through Gi/Go proteins to inhibit adenylyl cyclase, which produces cAMP, the loss of CC2D1A may result in higher expression levels of the 5-HT1A receptor, which may inactivate adenylyl cyclase, causing inactivation of the cAMP pathway in patients (11). This overexpression study suggests that CC2D1A positively regulates the cAMP pathway through transcription that may provide a potential link between CC2D1A and neuronal function, but the function of the endogenous CC2D1A protein remains to be determined.
The cAMP-dependent protein kinase A (PKA) pathway regulates synaptic plasticity, learning, and memory and thus is an important pathway for mental retardation research (12, 13). This pathway has also served as a paradigm for signal transduction that emanates from cell membranes where receptors are activated to trigger the activation of transcription factors in the nucleus. Binding of ligands (hormones, neurotransmitters, and growth factors) to their specific receptors located in the plasma membranes activates GTP-binding proteins (G-proteins) that are coupled to the receptors; the G protein then stimulates the activation of the enzyme adenylyl cyclase, which converts ATP to cAMP. The mammalian PKA holoenzyme consists of two regulatory (R) subunits and two catalytic (C) subunits (14). Each regulatory subunit contains two tandem cAMP-binding sites (15). In the absence of cAMP, PKA is inactive. When cAMP concentration rises, sequential and cooperative binding of cAMP on the R subunits results in dissociation of the monomeric C subunits that are active in phosphorylating substrates (16–18). When the cAMP concentration is high enough, PKA eventually translocates into the nucleus and phosphorylates transcription factors to affect gene expression. One of the best studied effectors that is relevant to neuronal function is CREB (18, 19). Upon phosphorylation at Ser-133, CREB becomes competent to bind the potent co-activator CREB-binding protein and activates the transcriptional program that is essential for brain plasticity and memory (13).
Because cAMP is diffusible and is the key for PKA activation, the distribution of cAMP inside the cells dictates where PKA is activated. The flux of cAMP is also governed by phosphodiesterases (PDEs), which terminate cAMP signaling by hydrolyzing it to 5′- AMP (20). PDEs are pivotal in shaping and controlling intracellular cAMP gradients in cells (21). There are 40 different isoenzymes in the PDE superfamily. Among them, PDE4 attracts the most attention (22–24), as PDE4-selective inhibitors show considerable promise as therapeutic agents for a range of disease indications, including chronic obstructive pulmonary disease (25–28), a finding that has been strongly supported by gene knock-out studies (26, 28).
The cAMP-PKA pathway is also influenced by protein kinase A anchoring proteins (AKAPs). AKAPs specifically bind the R subunit of the PKA holoenzyme as well as PDEs and other signaling molecules to coordinate the signaling event. As cAMP is produced at the plasma membrane, for rapid and optimal activation, PKA needs to be proximal to the membrane. AKAP such as Gravin/AKAP12 has been shown to bind to membranes. Localization of AKAPs/PKA/PDEs to the plasma membrane may shape upstream events in cAMP signaling. It is commonly accepted that anchoring of PDEs to the plasma membrane through AKAPs together with PKA limits the extent and duration of PKA activation (29–32). In all these cases, the mechanism for PKA activation is through fixing AKAPs in space to scaffold activators and effectors.
CC2D1A was initially identified as a binding protein of SRC2 (steroid receptor coactivator-2) and the Tank-binding kinase 1 (TBK1) in a proteomic screen designed to identify proteins that interact with the nuclear hormone co-activator SRC2. To further explore the role of CC2D1A, we isolated CC2D1A protein complexes. We show that it associates with several components of the cAMP/PKA signaling module including AKAP8, AKAP10, PDE4D, protein kinase A catalytic subunit (PrKacb), and protein phosphatase 2 (PP2) regulatory subunit. These results were highly suggestive of the involvement of CC2D1A in the cAMP/PKA pathway.
In this study, we also show that the NSMR protein CC2D1A is a positive regulator of the cAMP-PKA pathway. CC2D1A translocates to the plasma membrane in response to cAMP signaling, facilitated by the binding of membrane phosphatidic acid (PA) with its C2 domain thus allowing maximal activation. Significantly, using a mouse model that produces a truncated form of CC2D1A that lacks the C2 domain and three of the four DM14 domains, we show that CC2D1A is required for neuronal differentiation and plasticity. CC2D1A mutant neurons are hypersensitive to stress and have a reduced capacity to form dendrites and synapses in culture. Together, our findings may provide a molecular explanation why mutation in the CC2D1A gene leads to mental retardation.
EXPERIMENTAL PROCEDURES
GFP and GST Fusion Constructs
For the N-terminal GFP and GST fusion plasmids, a BamHI-EcoRI cDNA fragment encoding full-length CC2D1A, the N-terminal 620 amino acids of CC2D1A, or the C-terminal 320 amino acids of CC2D1A was inserted into the BamHI/EcoRI site of the vector pEGFP-N1 (Stratagene) and into the BamHI/EcoRI site of the vector pGEX-4T1 (GE Healthcare). PCR was performed with CC2D1A cDNA in a V5 vector as template. The PCR product and vectors were digested with BamHI and EcoRI, gel-purified, ligated, and transformed into DH5α cells and plated on agarose gel plates with kanamycin or ampicillin antibiotic for selection.
For the expression and purification of GST-CC2D1A fusion proteins, Escherichia coli (BL-21) cells expressing GST or GST-CC2D1A, GST-CC2D1AΔN, or GST-CC2D1AΔC were induced with isopropyl 1-thio-β-d-galactopyranoside. Proteins were purified after lysing the cells by sonication. Cell debris was removed by centrifugation at 10,000 × g for 10 min. The supernatants were incubated with glutathione S-agarose beads by shaking gently for 1 h at 4 °C and then washed with PBS buffer.
Mutant Mouse Construction
A knock-out mouse for the CC2D1A gene was constructed in the laboratory of B. Su at M.D. Anderson Cancer Center. Briefly, a genomic knock-out vector was constructed in which the DNA corresponding to exons 8–14 of the CC2D1A gene was replaced with a neomycin phosphotransferase gene cassette. The construct was used to transform a mouse embryonic stem (ES) cell line, and ES clones that were resistant to neomycin were screened for a homologous recombination event at the CC2D1A locus in which the knock-out vector replaced exons 8–14 in the genome. Cells of the appropriate ES cell line were then injected into blastocysts, and the blastocysts were introduced into a pseudopregnant foster mother. Chimeric offspring that incorporate some of the CC2D1A knock-out ES cells into their germ line were crossed to make a founder line of heterozygotes. These mice were then bred to generate homozygous mutant and heterozygous mice that we used in all of the experiments described. Genotyping primer sequences are as follows: CC2D1A common forward, 5′-GGT CTT CCT AGC TAG AAT GTG TGC CCT GG-3′; CC2D1A WT reverse (+/+), 5′-GGT CCC CTT AGT AGC CGT CTG ACC TAG-3′; and CC2D1A mutant reverse (−/−), 5′-GTT GGC GCC TAC CGG TGG ATG TGG AAT G-3′.
Cell Lines and Cell Culture
For conditioned medium preparation, L929 cells were grown in Dulbecco's modified Eagle's medium (DMEM, from HyClone) plus 10% fetal calf serum (FCS) (Invitrogen). Three days after cells are at 100% confluence, medium was collected, centrifuged, filtered, and stored −20 °C for future use. For culturing macrophages, wild-type and CC2D1A homozygous E18 mice livers were disassociated in 1 ml of phosphate-buffered saline (HyClone) by trituration and plated in Eagle's modified Dulbecco's medium plus 10% FCS supplemented with 25–30% conditioned medium and incubated for 24 h. Nonadherent cells were collected and re-plated with the same culture medium for an additional 5–6 days. For culturing neurons, hippocampi were dissected from wild-type and CC2D1A homozygous P0 mice brains, and meninges were removed in a 3 °C pre cooled Ca2+- and Mg2+-free Hanks' buffered salt solution. Hippocampi were incubated with warmed 0.25% trypsin in Hanks' balanced salt solution, for 15 min at 37 °C. Tissues were washed with the Hanks' balanced salt solution and triturated by using a pipette. Dissociated cells were cultured on gelatinized plates with warmed G3 (Neurobasal medium with B27 supplement (2 ml per 100 ml of medium), l-glutamine (0.5 mm), l-glutamate (25 μm), and gentamicin (10 g/liter of medium)). After 3–5 days in culture, the medium was half-changed to G2 medium (G2 is the same as G3, minus the glutamate). To construct the GFP-CC2D1A mouse embryonic fibroblast cells (MEF), 2 × 106 CC2D1A mutant MEF cells were electroporated with 20 μg of GFP-CC2D1A plasmid and GFP vector pEGFP-N1 (Stratagene) separately and seeded in 6-cm plates and on coverslips.
Antibodies and Western Blotting
Stimulated cells with bacterial lipopolysaccharide (LPS, 1 μg/ml, Sigma) or forskolin (50 μm, Sigma) were collected and lysed for 15 min by NETN buffer (150 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl (pH 7.8), 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 0.5 μg/ml pepstatin). Protein concentrations were normalized between samples, and for each sample 15 μg of total protein were resolved by SDS-PAGE and analyzed by Western blotting using antibody against the following: CC2D1A (Bethyl Laboratories); PDE4D (FabGennix), phospho-Ser-133-CREB (Cell Signaling); PKA catalytic subunits (PKAcs) (Santa Cruz Biotechnology); PKA regulatory subunits (PKA RegIIα, PKA RII) (Santa Cruz Biotechnology); AKAP95 (Upstate); CREB (Cell Signaling); phospho-Pro-38 (Santa Cruz Biotechnology); phospho-JNK1/2 (Santa Cruz Biotechnology); phospho-ERK1/2 (Santa Cruz Biotechnology); IκB (Santa Cruz Biotechnology); actin (Sigma); tubulin (Santa Cruz Biotechnology); and integrin (Santa Cruz Biotechnology). Following the primary antibodies incubation, membranes were washed with TBST and incubated with secondary antibodies (horseradish peroxidase-linked anti-mouse IgG antibody or a horseradish peroxidase-linked anti-rabbit IgG (Amersham Biosciences)). In the end, the membrane was developed using the ECL detection system (Amersham Biosciences).
Membrane Fractionation
Cells were collected and resuspended in 2.5 volumes of hypotonic swelling buffer, passed through a 25-gauge needle 10 times using a 1-ml syringe, left on ice for 10 min, and then left on ice for 20 min. The nuclear pellet (P1) was centrifuged out at 720 × g (3000 rpm) for 5 min. The supernatant was saved for further processing (see below). The nuclear P1 pellet was washed once by resuspending in 500 μl of fractionation buffer, dispersing the pellet with a pipette, and passing through a 25-gauge needle 10 times, followed by centrifuging at 720 × g for 10 min. The wash buffer supernatant was removed, and the nuclear pellet was resuspended in nuclei buffer (standard lysis buffer with 10% glycerol and 0.1% SDS added). The nuclear pellet was sonicated briefly (3 s) on ice. The supernatant was centrifuged again at 10,000 × g, and the supernatant was placed in a fresh tube. To produce the crude membrane fraction, the supernatant from the first step was centrifuged at 8,000 rpm (100,000 × g) for 1 h at 4 °C. The supernatant contains the cytosol and broken membranes, and the mitochondria are in the pellet. The supernatant was centrifuged in an ultracentrifuge at 40,000 rpm (100,000 × g) for 1 h. The supernatant was considered the cytosol, and the pellet contains crude membranes. The pellet was resuspended in the fractionation buffer (250 mm sucrose, 20 mm HEPES (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, phosphatase inhibitor mixture III) by pipetting using a 25-gauge needle as above and re-centrifuged at 40,000 rpm (100,000 × g) for 45 min. The pellet was resuspended in the same buffer as used for the nuclei. The crude membrane and cytosol fractions were analyzed by Western blots to estimate shifts in the localization of protein pools during signaling.
Immunocytochemistry
Cells cultured on poly-d-lysine-coated coverslips were stimulated with LPS or forskolin, fixed in 4% paraformaldehyde in PBS for 10 min at room temperature, incubated for 10 min on ice with PBS containing 0.3% Triton X-100, and washed again three times with PBS. Thereafter, the coverslips were incubated for 20 min at 37 °C in PBS containing 5% goat serum, and then the corresponding primary antibodies were added and incubated for 20 min at 37 °C (anti-CC2D1A (Bethyl Laboratories), anti-PKAcs (Santa Cruz Biotechnology), anti-MAP2 (Synaptic Systems), and anti-VGlut II (Synaptic Systems)). After that, coverslips were washed with PBS, incubated for 20 min at 37 °C with Texas Red or FITC-conjugated secondary antibody (Chemicon), washed again with PBS, and incubated for 5 min on ice in PBS containing 50 μg/ml DAPI, washed with PBS, and mounted on glass slides. In the end, antibody signals were visualized with a Delta Vision deconvolution microscope, and the images were processed with the SoftWoRx (version 2.5) software package (Applied Precision, Issaquah, WA). For dendrite and synapse formation assays, ImageJ software programs were used to measure the total dendrite length emanating from each neuron and the total number of synapses per neuron. The experiment was carried out three times with similar results. For each experiment, triplicate coverslips were quantified, and the means ± S.E. were reported.
Neuronal Toxicity and Survival Assay
Hippocampus samples from the brains of wild-type and CC2D1A mutant mice were harvested and disrupted. After culturing for 36 h in chemically defined medium (Neurobasal-A media, supplemented with GlutaMAX and B-27, Invitrogen) primary hippocampal neurons were incubated with 30 μm H2O2 for 6 h or with camptothecin (10 μm) for 18 h. After H2O2 incubation, cells were lysed in 200 μl of lysis buffer (0.1× PBS, pH 7.4, 0.4 mm Na2HPO4, 0.15 mm KH2PO4, 13.5 mm NaCl, 0.25 mm KCl, 0.5% Triton X-100, 2 mm MgCl2, and 0.5 g/100 ml acetyl dimethyl ethyl ammonium bromide). Cell lysates (10 μl) were loaded onto a hemocytometer, and the numbers of healthy nuclei were estimated by counting intact nuclei, while excluding those displaying blebbing or disruption of their nuclear membrane. Phase-bright bodies were also excluded from the counts. For each sample, 300 cells were counted, and the count was repeated three times with a new hemocytometer loading. After camptothecin incubations, neurons were fixed with 4% formaldehyde, stained with Hoechst 33258 (0.25 μg/ml), and washed with PBS. The number of healthy nuclei was estimated the same way after H2O2 treatment. The entire experiment was carried out three times with similar results. Triplicate samples were quantified, and the means ± S.E. were reported.
Analysis of Protein Complexes by Mass Spectrometry Immunoprecipitation
Mouse brain tissues were harvested and homogenized in 2.5 volumes of NETN (described above), and cells were collected and lysed by NETEN lysis buffer (100 mm NaCl, 20 mm Tris-Cl (pH 8.0), 0.5 mm EDTA, 0.5% (v/v) Nonidet P-40) for 15 min. Tissues and cell extracts were incubated with anti-CC2D1A (5 μg/ml, Bethyl Laboratories), shaken gently for 4 h at 4 °C, and centrifuged at 60,000 × g for 30 min at 4 °C. Supernatants were incubated with protein A beads (Sigma) and gently shaking for a 1 h at 4 °C. Beads were collected, washed with NETN, and separated by electrophoresis on 4–20% Tris-glycine gels (Invitrogen). Protein bands were cut out of the gel vertically and incubated on a shaker with destaining solution (40% methanol, 50 mm NaHCO3 in water) and then water for 1 day. Gel bands were equilibrated in 50 mm NH4HCO3 and digested by trypsin (1 μl, 25 μg/200 μl) at 37 °C for 4 h. To extract the protein, acetonitrile solution (100% acetonitrile) was added to the digested gel and shaken on high speed for 5 min. The supernatants were dried in a speed vacuum system for 1 h at high temperature. Mass spectrometry was performed by Dr. Sung Yun Jung.
Lipid Binding Assay
To determine the specificity of lipid binding by CC2D1A protein, PIP MicroStrip membranes (Molecular Probes) were used according to the manufacturer's protocol. Briefly, to block nonspecific protein binding, MicroStrips were incubated in 3% fatty acid-free BSA in TBST. Then GST-CC2D1A fusion proteins were eluted with 10 mm glutathione and incubated separately with PIP MicroStrip membranes. Protein-lipid binding was determined by Western blot using commercial mouse monoclonal anti-GST antibodies (Santa Cruz Biotechnology).
PKA Activity Assay
Wild-type and CC2D1A mutant MEF-stimulated cells with LPS, forskolin, or N6-benzoyladenosine-3′,5′-cAMP (Biolog) were collected, lysed in 2.5 volumes of Omnia Cell Extraction Buffer (BIOSOURCE) with added protease and phosphatase inhibitor mixtures 1 and 2 (Sigma) for 30 min on a rotator at 4 °C. Optimal protein concentrations (0.2 μg/ul) of each clarified cell extract (1 μg of total protein) were used for the measurement of PKA activity according to the manufacturer's specifications. The reactions were followed by measuring fluorescence intensity every 30 s for 20 min using a fluorescence microplate reader (excitation 360 nm and emission 485 nm).
RESULTS
Proteomic Analysis of CC2D1A Complexes Identified cAMP Signaling Components as Interacting Proteins
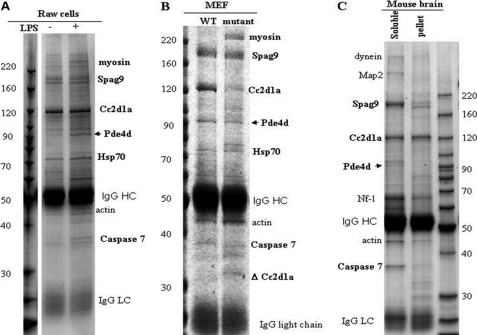
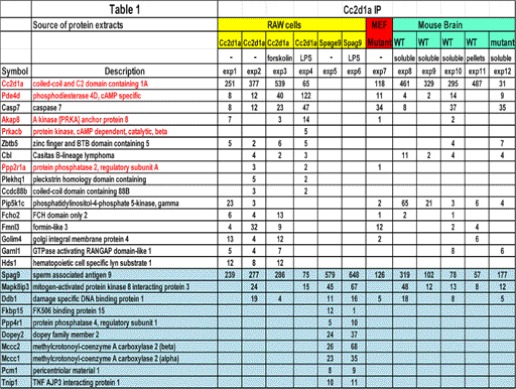
In a proteomic screen of SRC2-interacting proteins, we identified CC2D1A where CC2D1A appeared to associate with SRC2 and TBK1. Therefore, to further investigate this interaction, we immunoprecipitated CC2D1A protein complexes from a RAW macrophage cell line, both before and after stimulation by LPS (a known agonist of Toll-like receptor 4, TLR4) using an anti-CC2D1A antibody for mass spectrometry (MS) analyses (Fig. 1A and Table 1, experiments 1, 2, and 4–6). In addition, we immunoprecipitated the CC2D1A protein complexes from WT and CC2D1A mutant (see below) MEF cells using the anti-CC2D1A antibody for MS analyses (Fig. 1B and Table 1, experiment 7). Because CC2D1A was identified as an NSMR gene, we isolated endogenous murine CC2D1A protein complexes using cytosolic protein extract from day 0 neonatal mouse brain using the same an anti-CC2D1A antibody for MS analyses, to understand CC2D1A function in brain (Fig. 1C and Table 1, experiments 8–12). Surprisingly, in all cases we found that CC2D1A appears to associate with several components of the cAMP/PKA signaling module. As shown (Fig. 1 and Table 1), PDE4D and caspase 7 were consistently identified in multiple experiments in the CC2D1A immunoprecipitation. Also, the identification of PKA catalytic subunit β (PrKacb) in the CC2D1A protein complex suggested a possibility of CC2D1A involvement in cAMP/PKA signaling. This notion is further supported by the immunoprecipitation/MS analysis of the CC2D1A complex from RAW cells stimulated with forskolin (adenylyl cyclase stimulant) (Table 1, experiment 3). Furthermore, CC2D1A immunoprecipitation/MS revealed AKAP8 (AKAP95), Prkacb, and PP2r1a (PP2A regulatory subunit), which can form an AKAP/PKA/PP2A signaling module that is known to be important for PKA activation. These biochemical data suggest a possible role of CC2D1A in the cAMP-PKA signaling pathway.
FIGURE 1.
Proteomic analysis of Cc2d1a complexes identify cAMP signaling components as interacting proteins. Immunoprecipitations of Cc2d1a using an anti-Cc2d1a antibody from cell or tissue extracts under different conditions are shown. A, Raw cells that were cycling and treated with lipopolysaccharide (LPS, 1 μg/ml); B, MEF cells; C, mouse brain extracts. The immunocomplexes were resolved on a 4–20% SDS-polyacrylamide gel and stained with Coomassie Blue. The SDS-PAGE was divided into 12 regions; the bands were digested with trypsin, and the resulting peptides were analyzed with mass spectrometry to identify the proteins.
TABLE 1.
Summary of putative Cc2d1a interacting proteins identified from various experiments
The number of peptides detected is listed in the table, which serves as an estimate of the abundance of the protein in a semi-quantitative way.
Targeted Inactivation of the CC2D1A Gene by Homologous Recombination
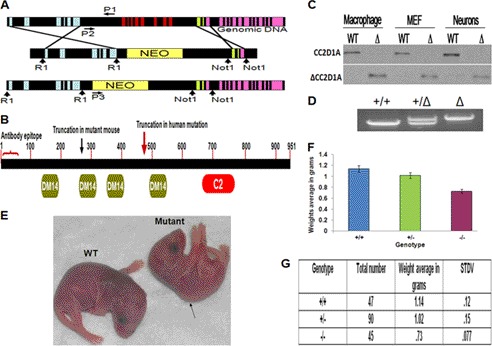
To further explore the potential role of CC2D1A in PKA signaling, we began by gathering genetic evidence in a mouse model. We generated mice with targeted deletion of CC2D1A by homologous recombination (Fig. 2A). Our targeting strategy led to the truncation of most of the CC2D1A protein but left an intact N-terminal fragment (263 amino acids) containing the first DM14 domain (Fig. 2B). We will refer to the mutant mouse as ΔCC2D1A. To detect the truncated protein expressed by the modified CC2D1A locus, different primary cell types (macrophages, MEFs, and neurons) derived from ΔCC2D1A and WT mice were examined by Western blot. On the Western blot, the full-length CC2D1A protein was detected only in the wild-type (WT) cells (∼120 kDa) and not detected in the mutant cells. Because the primary antibodies are directed against the first 15 amino acids of CC2D1A, a truncated form of the protein around 36 kDa could be detected in the mutant cells and not in the WT cells (Fig. 2C) and (Fig. 1B, IP/MS). The levels of the truncated mutant protein were similar to the WT protein. Although CC2D1A+/− heterozygous mice breed and develop normally with no gross differences from the WT mice, the ΔCC2D1A mice weigh significantly less than the WT littermates (Fig. 2, E–G), similar to the PKA−/− mice (33). The ΔCC2D1A mice also displayed a hunched back (Fig. 2E), obvious vascular abnormalities in the brain, and shortness of breath (data not included). In addition, the homozygous ΔCC2D1A mice were born with the expected Mendelian ratio (+/+, +/−, −/−, 1:2:1) but displayed postnatal lethality with most pups dying within 12 h of birth (Fig. 2G). We were unable to carry out electrophysiology and behavior tests because of the postnatal lethality, but we were able to perform biochemical and cellular analyses using the isolated MEF cells, macrophage cells, and neurons from prenatal and postnatal (P0) mutant and WT mice.
FIGURE 2.
Cc2d1a protein structure and targeting strategy for CC2D1A knockouts and phenotypes of the ΔCC2D1A mutant mice. A, schematic of the endogenous CC2D1A gene and the result of homologous recombination with the targeting vector. Disruption resulted in the deletion of exons 7–14 by homologous recombination with the targeting vector. Colored vertical boxes represent exons. Vertical arrows indicate restriction enzyme sites. P1, P2, and P3 are the common, wild-type, and mutant primers, respectively, used for genotyping by PCR. B, schematic of the conserved domains of the Cc2d1a protein, the C2 and the DM14 domains, the truncated form of the Cc2d1a protein expressed in ΔCC2D1A mutant mice, and the truncated form of the Cc2d1a protein expressed in human NSMR patients. The antibody against Cc2d1a recognizes the first 50 amino acids at the N terminus, thereby labeling full-length and all truncation mutants. C, Western blotting shows full-length endogenous Cc2d1a protein (120 kDa) in the wild-type cells (top) and the truncated Cc2d1a protein (lower) expressed only in cells from the mutant mice (Δ)(∼36 kDa). Macrophages, MEF, and hippocampal neurons were cultured from littermates wild-type and mutant (Δ). Newborn (P0) mice were collected, washed with PBS, and lysed in 1× RIPA buffer with protease inhibitors. Fifteen μg of protein was loaded from each lysate. D, PCR genotyping demonstrating the ∼2-kb wild type (+/+), heterozygous (+/−), and ∼8-kb mutant (−/−, Δ) genotype fragments. E, picture of P0 wild-type and mutant mice showing the difference in size between wild-type and mutant. The picture also displays the mutant mouse hunchback phenotype. F, diagram shows the difference in the average weight between wild-type, heterozygous, and mutant mice. (n = 180, p < 0.0001). G, distributions of the genotype and the body weight of P0 mice. STDV refers to means ± S.D.
CC2D1A Is Required for Activation of Protein Kinase A
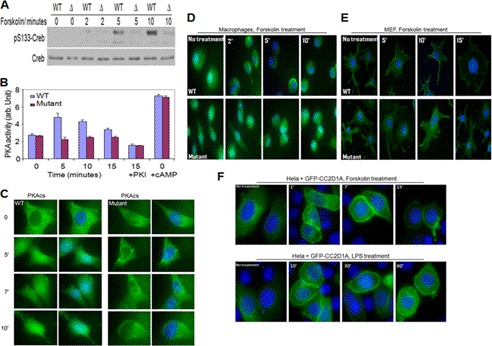
The proteomic data described above suggest that CC2D1A may be involved in the cAMP/PKA pathway, so we investigated CREB phosphorylation at serine 133, which is a downstream event of that pathway. We generated MEF and macrophage cells from CC2D1A WT and mutant E16 (16-day-old embryos), stimulated them with LPS, imidazoquinoline resiquimod R848 (a potent synthetic agonist of the Toll-like receptor 7/8, TLR7/TLR8), and forskolin separately and examined their CREB Ser-133 phosphorylation status by Western blot. As shown in Fig. 3A and supplemental Fig. S1, A and B, the early phosphorylation of the PKA target CREB at Ser-133 appeared to be defective in the CC2D1A mutant cells under all conditions tested. As a positive control experiment, the PKA inhibitor H89 was added to WT mouse MEF cells prior to stimulation with forskolin and LPS, and the phosphorylation status of CREB at Ser-133 was assessed. The kinetics of CREB phosphorylation at Ser-133 in this control was delayed (supplemental Fig. S1, E and F). This is consistent with the published role of PKA in the CREB early activation and strikingly similar manner of CREB phosphorylation observed in the CC2D1A mutant macrophages and MEF cells stimulated with forskolin and LPS. These data suggest that CC2D1A is important for the early CREB phosphorylation at Ser-133 that is mediated by PKA. To gain additional support for this idea, we stimulated WT and mutant MEF cells with agents known to activate the cAMP pathway, including forskolin, estrogen, insulin, and epidermal growth factor (EGF). Each of these treatments displayed similar defects in CREB Ser-133 phosphorylation in the CC2D1A mutant cells (supplemental Fig. S3, A–E). This suggests that CC2D1A is generally required for responses to cAMP signaling. Interestingly, CC2D1A mutant cells did not exhibit any detectable defect in CREB phosphorylation at Ser-133 in response to UV stimulation (supplemental Fig. S3F). A PKA-independent, p38/RK/HOG-1-dependent, p108 serine/threonine protein kinase has been identified as a UV-induced CREB kinase. Thus, the UV results give an indication of the specificity of the involvement of CC2D1A in the cAMP/PKA pathway. Furthermore, because PKA is the primary means by which cAMP exerts its effects on CREB, and given the observed CREB phosphorylation deficit at Ser-133, we turned our attention toward the activation of PKA kinase activity. To assess the activation of PKA under the assay conditions we have been using (see under “Experimental Procedures”), MEF cells were treated with forskolin, and cell lysates were assayed for PKA activity. PKA activity was consistently and significantly lower in the CC2D1A mutant cell lysates, suggesting that CC2D1A is important for PKA activation (Fig. 3B). To confirm this result, several controls were carried out. To ensure that PKA activity was being assayed, N6-benzoyladenosine-3′,5′-cAMP, a potent and selective activator of PKA, stimulated the activity (data not included). Importantly, all of this activity could be inhibited by the specific PKA inhibitory peptide PKI (Fig. 3B). To ensure that the lower PKA activity in the CC2D1A mutant cells was not a consequence of lower PKA catalytic subunit expression in these cells, cAMP was added to untreated sample lysates to measure the maximum amount of PKA catalytic subunit that could have been activated in both WT and CC2D1A mutant cells. The total amount of PKI-inhibitable PKA activity is the same in both WT and mutant cells (Fig. 3B). Thus, the lower PKA activity that we observed in CC2D1A mutant cells is most likely due to unreleased PKA catalytic subunits. This observation suggests that CC2D1A may be a required prerequisite for activating PKA catalytic activity. Finally, based on the notion that CC2D1A is important for PKA catalytic subunit release and activation, and given the fact that PKA catalytic subunits translocate to the nucleus after activation, we followed the PKAcs localization during activation using indirect immunofluorescence of stimulated cells. WT and CC2D1A mutant MEF cells were stimulated with forskolin, fixed with 4% formaldehyde, and stained with antibodies directed against the PKAcs. The translocation of the PKAcs to the nucleus was observed in the WT MEF cells but not detectable in the CC2D1A mutant MEF cells (Fig. 3C). This observation supports the suggestion that CC2D1A is involved in the release of the PKA catalytic subunit allowing its eventual translocation to the nucleus. In contrast, because MAPK pathways, including the p38, JNK, and ERK1/2 protein kinases, are known to be activated by Toll-like receptors and effect CREB phosphorylation, we tested the activation state of these kinases. We prepared primary fetal liver macrophages cells from WT and CC2D1A mutant mouse embryos (E16) and treated them with LPS for different times. After the cells were collected and lysed, we separated their proteins on SDS-polyacrylamide gels to examine these MAPK pathways statuses by Western blot. Each of the protein kinases that we tested (MAPK (p38), ERK1/2, and JNK) appeared to be activated normally (supplemental Fig. S1C). Additionally, because CC2D1A has been reported to positively regulate the NF-κB cascade, using the same macrophages lysates, we tested the degradation of I-κB, the inhibitory subunit of NF-κB. We observed a normal degradation pattern of I-κB, indicating a normal activation state of NF-κB. These results suggest that the CREB phosphorylation defect displayed by the CC2D1A mutant is exclusive to the role of CC2D1A in the cAMP/PKA pathway. Together, these data suggest that CC2D1A is important for PKA-mediated early CREB phosphorylation at Ser-133.
FIGURE 3.
Cc2d1a is required for activation of protein kinase A and subcellular localization of the Cc2d1a protein in response to increases in cAMP. A, Western blot of Ser-133 phosphorylation of CREB in response to forskolin treatment in WT and mutant MEF cells. B, in vitro PKA activity stimulated with forskolin in MEF cells. C, indirect immunofluorescence demonstrating PKAcs translocation into the nucleus in response to forskolin treatment in WT and mutant MEF cells. D and E, immunostaining show the accumulation of Cc2d1a protein toward the cell periphery in both macrophages and MEF cells after stimulation with forskolin. This localization phenotype is not shown in CC2D1A mutant cells neither before nor after forskolin treatment. F, images of HeLa cells show the accumulation of the transfected GFP-Cc2d1a fusion protein toward the cell periphery after stimulation with LPS (1 μg/ml) or forskolin (50 μm). In all cases, cells were fixed and analyzed by immunofluorescence using an anti-Cc2d1a antibody and DAPI to stain the nuclei. Deacon microscopy was used for imaging.
CC2D1A Translocates to Plasma Membrane in Response to Ligand Stimulation through Its C Terminus That Binds Phosphatidic Acid (PA) and Phosphatidylserine (PS) in Vitro
The regulated association and dissociation of proteins with the surface of intracellular membranes is essential for a number of cellular functions, such as signal transduction and protein trafficking. Membrane association often involves one or more lipid binding domains within the relevant proteins. Because CC2D1A associates with the cAMP module and has a C2 domain and to further understand the molecular details of CC2D1A-dependent PKA activation, we investigated possible changes in subcellular localization of CC2D1A under different stimulants. We cultured human WT and CC2D1A mutant macrophages and MEF cells on coverslips and treated them with forskolin for different times. After fixing the cells with 4% formaldehyde, we used antibodies directed against the first 15 amino acids of the N-terminal CC2D1A to visualize CC2D1A and reveal its localization in the treated cells. The results are suggestive of an accumulation of CC2D1A toward the cell periphery such that a significant fraction of the signal appears localized at the plasma membrane (Fig. 3, D and E). A similar change in the subcellular localization of CC2D1A was observed in mouse MEF treated with LPS, but macrophage LPS treatment resulted in CC2D1A localization to cortical puncta (supplemental Fig. S2, A and B). These observations suggest that CC2D1A might relocalize to places where the cAMP/PKA module is anchored, in response to the signaling stimulants, to modulate PKA activity. Because the truncated mutant form of the CC2D1A protein includes the first 15 amino acids that are recognized by the CC2D1A antibody we are using, the truncated form of the protein could also be visualized in the mutant cells. Relocalization of the truncated CC2D1A protein was not observed in CC2D1A mutant cells (Fig. 3, D and E, and supplemental Fig. S2, A and B), suggesting that the C2 domain is required for the observed CC2D1A relocalization. Furthermore, we stimulated WT and CC2D1A mutant MEF cells with hormones that are known to activate cAMP pathway, including estrogen, insulin, and EGF, and all of these treatments led to the accumulation of CC2D1A to the cell periphery in the WT cells but not in the CC2D1A mutant cells (supplemental Fig. S3, G–I). Significantly, in agreement with our previous CREB phosphorylation results, CC2D1A did not relocalize in response to stimulating the immortalized MEF cells with UV radiation (supplemental Fig. S3J). These results indicate that CC2D1A is specifically involved in cAMP/PKA signaling. The same phenomenon was observed when a GFP-CC2D1A construct was transfected in HeLa cells, excluding the possibility of antibody cross-reactivity (Fig. 3F). In addition, because CC2D1A responds to stimulation of many pathways that lead to cAMP signaling, this suggests an integral role for CC2D1A in cAMP/PKA signaling where it localizes to the membrane periphery in response to signals that produce cAMP in a variety of cell types.
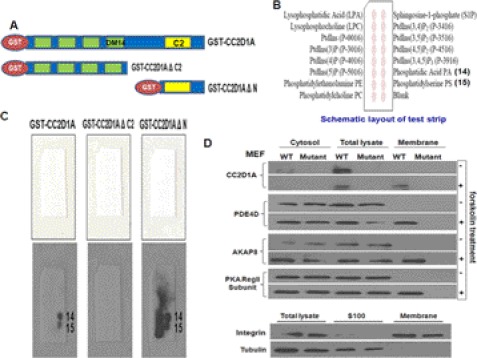
The C2 domains have been characterized as calcium-dependent phospholipid-binding domains, mainly through work on the canonical domain found in PKC (34), but their lipid specificities vary widely, and they can bind without requiring calcium. In addition, based upon our previous results, the CC2D1A C2 domain appears to be required for accumulation of CC2D1A at membranes. Therefore, to determine whether CC2D1A binds phospholipids, we constructed full-length CC2D1A, CC2D1AΔN (the four DM14 domains are truncated), and CC2D1AΔC (the C2 domain is truncated) as N-terminal GST fusion proteins) (Fig. 4A), expressed the proteins in E. coli, and purified them. Using these recombinant proteins, we performed in vitro phospholipid binding assays using PIP Micro Strips membranes from Molecular Probes (Fig. 4B). As shown in Fig. 4C, the results indicated that full-length CC2D1A and CC2D1AΔN (C2 domain intact) bind to phosphatidic acid (PA) and PS, whereas the CC2D1AΔC (DM14 domains intact, C2 domain deleted) did not. These results suggest that the C2 domain mediates binding of CC2D1A to phospholipids and may therefore be potentially responsible for the membrane localization of CC2D1A after cAMP pathway stimulation. Furthermore, PA is the smallest phospholipid that is a major constituent of all cell membranes in the body, and PS is also a major membrane phospholipid, but it is most concentrated in the brain. It's relative abundance in this organ reflects its proven involvement in an assortment of nerve cell functions, including neurotransmitter release and synaptic activity. Thus, it is intriguing that the role of CC2D1A in cognition or neuronal development may depend on its ability to localize to membranes through the binding of PS. Moreover, in agreement with the lipid PIP strip binding assays, the YFP-C2 domain fusion constitutively localizes to vesicle-like structures, and forskolin stimulation does not seem to move the C2 domain to the plasma membrane (supplemental Fig. S2C). Thus, although the C2 domain is essential for phospholipid binding, the N-terminal region of CC2D1A may provide constraints for phospholipid selectivity that regulates translocation of CC2D1A to membranes where high concentrations of PA/PS are transiently produced.
FIGURE 4.
Cc2d1a is required for re-localization of the PKA signaling module to the plasma membrane, and Cc2d1a C2 domain binds phospholipids. A, Western blot showing the presence of Cc2d1a, PDE4D, AKAP8, and PKA regulatory subunit II in wild-type MEF cell membrane fractions only after stimulation and not in CC2D1A mutant MEF cells either before or after stimulation. Crude membranes were purified from WT and CC2D1A mutant MEF cells after stimulation for 5 min. Total cell lysates, crude membrane, and cytosol fractions were analyzed by Western blots to estimate shifts in the localization of proteins. Tubulin and membrane marker proteins were used to show the purity of membrane fractions. B, Western blot showing GST-Cc2d1a and GST-Cc2d1aΔN but not GST-Cc2d1aΔC fusion proteins bind PA and PS. Phospholipid binding assay and Western blots are used to determine the phospholipids types that Cc2d1a C2 domain can bind. Nonspecific protein binding was blocked by incubating the PIP MicroStrip membranes in TBST with 3% fatty acid-free BSA. The membranes were then incubated in 0.5 μg/ml GST-Cc2d1a, of GST-Cc2d1a ΔN, or GST-Cc2d1a ΔC2 separately in TBST plus 3% fatty acid BSA overnight at 4 °C. The membrane strips were then incubated with anti-GST mouse monoclonal antibody and anti-mouse IgG horseradish peroxidase-linked antibody, respectively. In the end, membranes were developed using the ECL detection system. C, schematic diagrams of a PIP MicroStrip membrane showing the types of lipid spotted on the membrane. D, schematic diagram of GST-Cc2d1a, GST-Cc2d1aΔN, and GST-Cc2d1aΔC fusion proteins constructs.
As described above, we found that CC2D1A constitutively binds to PDE4D and associates with the entire cAMP/PKA signaling module after the stimulation of signaling that leads to cAMP accumulation. We also observed that CC2D1A protein accumulated on vesicles or at the plasma membrane, depending on the stimulant used, and that CC2D1A binds PA and PS in vitro. These observations suggest that CC2D1A may be recruiting the PDE4D bound to the PKA module to membranes, using a common lipid-based anchoring mechanism, to facilitate the signal transduction. Based on these data, we investigated PKA module localization to the membrane in response to increases in cAMP concentration. After stimulating WT and CC2D1A mutant MEF cells with forskolin for 5 min, cell fractionation was carried out as described under “Experimental Procedures.” The protein in total cell lysates, as well as crude cytosolic and membrane fractions, were separated on SDS-polyacrylamide gels. We checked the proportion of PKA module components in each of these fractions by Western blot analysis. The results indicated that all detectable CC2D1A, PDE4D, AKAP8, and PKA regulatory subunit II (RII) subunits were present in the crude membrane fraction after stimulation of the cAMP pathway (Fig. 4D). These results suggest the relocalization of the PKA module to the membrane occurs after stimulating the cAMP pathway. Furthermore, segregation of these proteins was not observed in the CC2D1A mutant cells, either before or after stimulation (Fig. 4D). Because the truncated mutant CC2D1A protein does not relocate to the cell membranes, these results support the hypothesis that CC2D1A membrane localization is important for the recruitment of the PKA module to the membrane.
CC2D1A Expression Rescues the CREB Phosphorylation Phenotype in CC2D1A Mutant MEF Cells
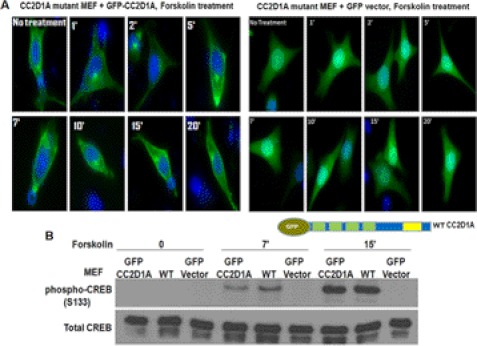
The data presented so far suggest a direct role for CC2D1A in cAMP/PKA signaling. However, there are two alternative explanations for the results with the CC2D1A truncation mutant cells that could challenge those findings. First, the mice that are homozygous for the truncated form of CC2D1A might have suffered from developmental defects that indirectly caused the observed defects in PKA signaling. For example, the levels of some other key regulator of PKA might be reduced or absent in CC2D1A mutants. Second, the truncated CC2D1A protein itself might have a novel function not present in the WT CC2D1A protein that coincidently affects PKA signaling. For example, the truncated CC2D1A might dominantly inhibit the normal function of the WT CC2D1A or its interacting proteins. One way to explore the likelihood of these alternative explanations would be to attempt to rescue some of the phenotypes of the CC2D1A mutant cells by expressing WT CC2D1A protein from a plasmid. Correcting the defects in a cell culture experiment would rule out defective development as the proximal cause of the mutant phenotype. By expressing truncated forms of CC2D1A in WT and heterozygous CC2D1A mutants, one might also begin to test for possible dominant-negative effects of the truncation mutant. To test for a more direct role of CC2D1A in cAMP signaling, we constructed expression plasmids for GFP-CC2D1A fusion protein and transfected the GFP-CC2D1A and GFP control plasmids separately into CC2D1A mutant MEF cells. After 24 h, cells were stimulated with forskolin for different times, collected their lysates, or fixed them with 4% formaldehyde and examined for CREB phosphorylation and GFP-CC2D1A localization. Consistent with the observations of the endogenous WT CC2D1A by immunostaining, the full-length GFP-CC2D1A displayed an accumulation toward the cell periphery, which was not observed in GFP vector control cells (Fig. 5A). These observations suggest that the GFP-CC2D1A plasmid is functional in the CC2D1A mutant MEF cells. Moreover, these results also indicate that the truncated form of CC2D1A in the mutant cells does not act in a dominant-negative fashion with respect to CC2D1A re-localization. This observation also is consistent with the fact that although the heterozygote CC2D1A mice contain the truncated form of CC2D1A, they are alive and healthy as the wild-type mice, indicating that the WT allele is functioning normally in the heterozygous mutants. Interestingly, Ser-133 CREB phosphorylation was restored to the WT level in the CC2D1A mutant cells expressing GFP-CC2D1A from the exogenous plasmid (Fig. 5B). This increased Ser-133 phosphorylation was not detected in CC2D1A mutant cells expressing GFP from the control plasmid. These results show that complementing the CC2D1A mutant MEF cells by expressing wild-type CC2D1A rescues the CREB phosphorylation phenotype, indicating that the CREB phosphorylation defect in the mutant is a direct effect of the absence of the CC2D1A protein.
FIGURE 5.
Cc2d1a expression rescues the CREB phosphorylation phenotype in CC2D1A mutant MEF cells. A, Western blots show that GFP-Cc2d1a expression rescues the early CREB phosphorylation defect in CC2D1A mutant MEF cells to the wild-type level. GFP-Cc2d1a fusion protein was expressed in CC2D1A mutant MEF cells. The transfected mutant cells, along with the wild-type MEF cells, were stimulated with forskolin. GFP vector (control) expression did not rescue the early CREB phosphorylation defect in the CC2D1A mutant cells to the wild-type level, whereas GFP-Cc2d1a expression rescued the phenotype. B, images of CC2D1A mutant MEF cells show the accumulation of the expressed GFP-Cc2d1a fusion protein toward the cell periphery after stimulation with forskolin, whereas GFP protein alone (control) does not.
CC2D1A Is Required for Proper Neuronal Differentiation and Brain Development, and the cAMP/PKA Signaling Pathway Is Compromised in CC2D1A Mutant Neurons
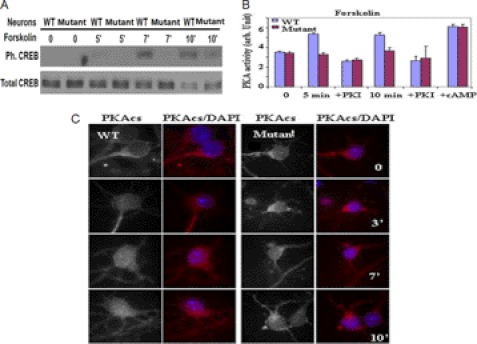
Because the CC2D1A gene was identified as carrying disease-causing mutations in families with severe autosomal recessive NSMR, the study of CC2D1A in the neurons might elucidate the cellular function of CC2D1A and suggest the molecular mechanism by which CC2D1A mutations lead to mental retardation. The postnatal lethality of our ΔCC2D1A mouse prevented us from testing directly the involvement of CC2D1A in NSMR using a mouse model. However, identification of CC2D1A as a positive regulator of the cAMP/PKA pathway in a variety of cell types, including neurons, suggests that an underlying reason for CC2D1A-mediated NSMR is due to defects in cAMP-PKA signaling. Thus, our finding identifies a molecular basis for NSMR with CC2D1A mutations. The cAMP/PKA pathway regulates synaptic plasticity, learning, and memory and thus is an important pathway for mental retardation research (12, 13). As PKA has multiple substrates that play important roles in learning and memory, it remains to be discovered which PKA substrates are involved in the functions of CC2D1A in mental retardation. To investigate the role of CC2D1A in neuronal cells, we started by testing whether CC2D1A alters cAMP signaling in the neuronal system by repeating some of the key experiments that we described above for non-neuronal cell lines. We prepared hippocampal neurons from WT and CC2D1A mutant mice (P0) and cultured them for 14 days. We stimulated the neurons with forskolin for different times, and CREB phosphorylation status at Ser-133 was assessed by Western blot. Consistent with the previous results, we found that CREB early phosphorylation at Ser-133 was defective in the mutant neurons indicating that CC2D1A is important for the early CREB phosphorylation at Ser-133 by PKA in the neurons (Fig. 6A). We also measured PKA activity in WT and CC2D1A mutant neurons. As per Fig. 6B, the PKI-inhibitable kinase activity was much lower in the stimulated CC2D1A mutant neurons compared with WT neurons. We also added cAMP to both WT and CC2D1A mutant untreated lysates to measure the maximum amount of PKA catalytic subunit that could have been activated in either cell type. The total PKA activity was the same in WT and CC2D1A mutant neurons, indicating that the difference in the PKA activity was not due to an unequal amount of activatable PKA catalytic subunit in the cell lines. Consistent with our previous results, this experiment indicates that CC2D1A is important for PKA activation in the neurons. Next, we examined the subcellular re-localization of PKAcs by immunohistochemistry after stimulating cultured WT and CC2D1A mutant hippocampal neurons with forskolin. The translocation of PKAcs to the nucleus was observed in WT neurons but not in CC2D1A mutant neurons, suggesting that CC2D1A is involved in the release of PKA catalytic subunits from the PKA signaling complexes also in neurons (Fig. 6C). We also investigated possible changes in subcellular localization of CC2D1A in hippocampal neurons upon treatment with forskolin. In WT, treatment with forskolin resulted in localization of CC2D1A to the cell periphery (supplemental Fig. S4). These observations suggest that CC2D1A may localize to places where the cAMP/PKA module is anchored in response to the signaling stimulants to modulate PKA activity. Localization of the truncated CC2D1A protein was not observed in CC2D1A mutant neurons, suggesting that the C2 domain may be required for CC2D1A localization also in neurons. These data support that CC2D1A is also involved in regulation of the cAMP/PKA pathway in neurons. Conditional knock-out and transgenic studies have revealed that CREB transcriptional activity is required for neuronal survival, axonal growth (13), and outgrowth of cortical neuron dendrites. Based on the previous data, it is possible that a CREB-regulated transcriptional program is vital for mutations in CC2D1A resulting in NSMR. Because CREB phosphorylation on Ser-133 is defective in CC2D1A mutant neurons, we examined some CREB-dependent functions in CC2D1A mutant neurons.
FIGURE 6.
Cc2d1a regulates PKA activity in the neuronal system. A, Western blot of Ser-133 phosphorylation of CREB in response to forskolin treatment in WT and CC2D1A mutant neurons. B, in vitro PKA activity assay showing the increase in PKA activity only in WT and not in CC2D1A mutant neurons stimulated with forskolin. C, immunostaining demonstrating PKA catalytic subunit translocation into the nucleus in response to forskolin treatment in WT but not CC2D1A mutant neurons.
Survival Rate of Neurons under Oxidative Stress Is Compromised in CC2D1A Mutant Neurons
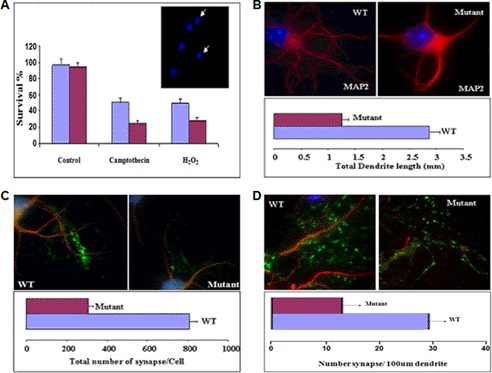
We measured neuronal survival under stress conditions. We cultured neurons for 36 h and then treated them with camptothecin, a DNA topoisomerase I inhibitor, or H2O2 to introduce stress. The concentrations used were aimed to induce 50% cell death in WT neurons. The CC2D1A mutant neurons showed lower survival rates than the WT neurons (Fig. 7A).
FIGURE 7.
Cc2d1a is important for neuronal survival and plasticity. A, immunostaining and dendrite length measurements showing that total dendrite length per cell is lower in CC2D1A mutant neurons compared with WT. Hippocampal neurons were cultured for 14 days, fixed, and stained with antibodies to MAPII. Dendrites were visualized by immunofluorescence microscopy. Fluorescent images were obtained and analyzed to measure total dendrite length per cell (p < 0.001). B, survival assay and immunostaining showing that the survival rate under oxidative stress is compromised in CC2D1A mutant neurons. Hippocampal neurons were cultured for 36 h, and stress was introduced by camptothecin (10 μm) or H2O2 (30 μm) treatment. Then the neurons were fixed and stained with Hoechst or DAPI to score healthy, intact nuclei as an estimate of cell survival. (p < 0.002). Immunostaining and synapse number assay showed that the total synaptic number (C) and the synaptic density (D) is lower in the CC2D1A mutant neurons compared with WT neurons. Hippocampal neurons were cultured for 14 days, fixed, and stained with anti-MAPII and anti-V GlutII antibodies. The number of synapses was counted per cell. Number of synapses per length of dendrite (100 μm) was also calculated. (p < 0.002).
Total Dendrite Length per Cell Is Lower in CC2D1A Mutant Neurons Compared with Wild-type Neurons
Transcriptional regulation carried out by CREB and CREB-binding protein is involved in regulating dendrite growth, and this regulation can be modulated by the level of synaptic firing. However, the mechanism by which CREB-binding protein and CREB do this is not clear (35, 36). To assess the role of CC2D1A in neurons to form dendrites in long term culture, WT and CC2D1A mutant neurons were cultured separately for 14 days, fixed, and stained with antibodies to microtubule-associated protein 2 (MAP2). Fluorescent images were obtained and analyzed to measure total dendrite length per cell. The results show that the CC2D1A mutant neurons produced fewer dendrites and a lower total dendrite length compared with the WT neurons (Fig. 7B). This cellular phenotype is also consistent with our observed CREB phosphorylation defect and the documented role for CREB in dendrite outgrowth.
Total Synaptic Number and Synaptic Density Is Lower in CC2D1A Mutant Neurons Compared with Wild-type Neurons
Synaptic plasticity, the change in the strength of neuronal connections in the brain, is thought to underlie learning and memory. Although how synaptic signals propagate to the nucleus to initiate CREB target gene expression is unclear, well known CREB target genes are implicated in the synaptic plasticity. To further assess the effect of CC2D1A on synapse formation, we cultured primary WT and CC2D1A mutant hippocampal neurons and examined synapse formation. After 14 days of culture, neurons were fixed and stained with anti-MAP2 and anti-V GlutII (a post-synaptic protein) antibodies, and the number of synapses were counted per cell. Consistent with the reduced number of dendrites formed by CC2D1A mutant neurons, the number of synapses that they form per cell is also reduced (Fig. 7C). In addition, the numbers of synapses per length of dendrite were calculated, and the results indicated that this “density” is lower in CC2D1A mutant than WT neurons (Fig. 7D). This indicates that the mutant neurons do not have fewer synapses simply because they have a lower total dendrite length. Given the low survival of CC2D1A mutant neurons under stress, their reduced number and length of dendrites, and their reduced number of synapses per cell, it appears that CC2D1A is important for neuronal survival and plasticity.
DISCUSSION
The cAMP/PKA signaling pathway is critical for many functions during development and cellular homeostasis in adult tissues. Any mutation at any point of the pathway that may cause hypo- or hyperactivity of the pathway can lead to devastating consequences. These mutations can range from fatal mutations that cause death in fetal stages to mutation that can cause different types of human tumorigenesis. Therefore, uncovering the CC2D1A protein as a new component in the cAMP signaling pathway is pivotal and may illuminate another aspect of cAMP signaling specificity. The evidence for the involvement of CC2D1A in cAMP signaling is severalfold. First, it appears that CC2D1A associates with the cAMP module because cAMP module components were isolated from cells and mice brain tissues in complexes with the CC2D1A protein after stimulation of the cAMP signaling transduction pathway. Second, genetic ablation of CC2D1A in the mouse allowed us to demonstrate that CC2D1A is required for PKA activation in vitro, PKAcs translocation to the nucleus, and for the phosphorylation of the transcription factor CREB in primary and immortalized cells as well as in cultured neurons. Because PKA is the main way by which cAMP exerts its effects, the compromised activity of PKA that was detected in the CC2D1A mutant cells suggests the importance of CC2D1A for PKA activation. Also, our controls showed that the amount of PKAcs in both of the WT and mutant cells was equal indicating that the difference in the PKA activity is not due to unequal amount of total PKAcs but may be due to unreleased (in active) catalytic subunits from the regulatory subunits (PKA RII). The previous data are supported by the fact that the translocation of the PKAcs to the nucleus was observed in the WT MEF cells but not in the CC2D1A mutant MEF cells suggesting that CC2D1A may be important for the release of the PKAcs from the regulatory subunit that leads to its activation. Third, the early phosphorylation of CREB at Ser-133 after stimulation of the cAMP pathway that is defective in the CC2D1A mutant cells suggests that CC2D1A is important for CREB phosphorylation by PKA. Although CREB phosphorylation at Ser-133 and PKA activation are significantly compromised in CC2D1A mutant cells, other signaling pathways are unaffected indicating that CC2D1A function is specific to cAMP/PKA activity regulation. Fourth, putting the WT CC2D1A protein back in the CC2D1A mutant cell had rescued the CREB phosphorylation almost to the WT level. Fifth, compartmentalization is an effective and well established mechanism to regulate cAMP activity where AKAP proteins compartmentalize the PKA holoenzyme and other signaling enzymes at precise subcellular sites in close proximity to their physiological substrate(s) and favor specific phosphorylation events. Consistent with the previous knowledge, our data indicate CC2D1A relocalization in the cytosol after cAMP stimulation. CC2D1A relocalization differs depending on the cell and stimulants types. In MEF cells treatment with forskolin resulted in localization of CC2D1A to the cell periphery. A similar change in the cellular localization of CC2D1A was observed in mouse neurons and macrophages treated with forskolin, but macrophages LPS treatment resulted in CC2D1A localization to cortical puncta. These observations suggested that in response to the cAMP elicitors, signaling stimulants, CC2D1A relocalizes to the cAMP pool places where the cAMP/PKA module can be anchored to modulate PKA activity. Additionally, relocalization of the truncated CC2D1A protein was not observed in macrophages and MEF cells derived from the CC2D1A mutants suggesting that the three missing DM14 domains and/or the C2 domain may be required for CC2D1A relocalization. These observations endorse CC2D1A involvement in the cAMP signaling pathway. Moreover, the fractionation results indicated that all of CC2D1A, PDE4D, AKAP8, and PKARII are present in the membrane fraction only after stimulation of the cAMP pathway. Also, the presence of only PDE4D in the CC2D1A mutant cell cytoplasmic fraction before and after stimulation is in agreement with the mass spectrometry results where only PDE4D was identified in CC2D1A mutant protein complex. These results suggest the relocalization of CC2D1A along with the cAMP module to the membrane after stimulating the cAMP pathway. Because our CC2D1A mutant truncated form does not relocate to the cell periphery, the previous fractionation results support our hypothesis that CC2D1A relocalization leads to recruiting the cAMP module to the membrane. Furthermore, CC2D1A appears to bind PA and PS which are highly expressed in the neuronal plasma membrane, in vitro through its C2 domain suggesting that CC2D1A is recruited to the membrane to facilitate signal transduction. In summary of the previous data, we believe that CC2D1A relocalizes after stimulation toward the area where cAMP is made helping to bring the cAMP module close to the cAMP pool and aiding in release of the PKA catalytic subunits from the regulatory subunits. The release of PKA catalytic subunits leads to the PKA activation and translocation to the nucleus, leading to the phosphorylation of CREB and the activation of many gene transcriptions depending on the stimulant type. These results make CC2D1A a new regulator of the cAMP pathway, which opens many doors for research and drug targets for treatments. If that is the case, how is CC2D1A implicated in mental retardation? The recent finding of CC2D1A involvement in NSMR raises the question of the function of CC2D1A in the brain. The cAMP-PKA-CREB signaling pathway plays a diverse role in neuronal functions. It is involved in neuronal survival, processing expression, enhancing neurite outgrowth, alteration of neuronal responses to certain growth factors, overcoming the neurite growth inhibition effects of myelin-associated inhibitory factors, and promoting axonal regeneration in the central nervous system (13, 35). In addition, CREB is among the best studied transcription factors implicated in different neuronal processes such as survival, synaptic plasticity, memory and learning, and drug habituation and addiction (36). Taking this knowledge into consideration, our finding that CC2D1A is a positive regulator of this pathway may provide a molecular explanation for the cause of NSMR in which CC2D1A is mutated. Based upon our results of compromised PKA activity, the CREB phosphorylation defect at Ser-133 along with the reduced translocation of the PKA catalytic subunit to the nucleus in mutant neurons, one may think that CC2D1A plays the same role in the neuronal system. Interestingly, the truncation mutation in humans that is associated with NSMR results from a stop codon at position 480 (485 in the mouse protein), which is similar to the 280-amino acid N-terminal fragment produced in our mutant mouse. However, we have to keep in mind that the mutant mouse that we studied is not identical to the CC2D1A truncation mutation identified in human NSMR. Even if it was the identical mutant protein, there might still be significant differences between mouse and human physiology that could render any of the following observations more or less relevant to the observed defects in cognitive function in NSMR. In agreement with previous knowledge, we have observed several defects in CC2D1A mutant neurons. Because mutant hippocampal neurons display lower survival rate under oxidative stress (37–40), lower synapse number per cell, and lower total dendrite length per cell, which are downstream events of CREB activation, the absence of CC2D1A in neurons may be causing a developmental problem in the brain. If this occurs late in gestation, it would likely lead to the gross neural defects and abnormal brain development, likely leading to deficits in mental function.
Supplementary Material
Acknowledgments
It is a pleasure to thank those who instructed A. A. on how to work with neurons: Hsiao-Tuan Chao and Paymaan Jafar-Nejad from Al Zoghbi Laboratory and Zilai Wang from Huffington Center at Baylor College of Medicine. We also thank Professor Kenneth P Minneman, Dean, Division of Chemical and Life Sciences and Engineering, King Abdullah University of Science and Technology, for guidance and support in preparing this paper.

This article contains supplemental Figs. S1–S4.
- NSMR
- nonsyndromic mental retardation
- CREB
- cAMP-response element-binding protein
- PKAcs
- PKA catalytic subunit
- MEF
- mouse embryonic fibroblast
- PDE
- phosphodiesterase
- PA
- phosphatidic acid
- PS
- phosphatidylserine
- AKAP
- protein kinase A anchoring protein.
REFERENCES
- 1. Basel-Vanagaite L. (2007) Genetics of autosomal recessive nonsyndromic mental retardation. Recent advances. Clin. Genet. 72, 167–174 [DOI] [PubMed] [Google Scholar]
- 2. Ropers H. H. (2006) X-linked mental retardation. Many genes for a complex disorder. Curr. Opin. Genet. Dev. 16, 260–269 [DOI] [PubMed] [Google Scholar]
- 3. Raymond F. L., Tarpey P. (2006) The genetics of mental retardation. Hum. Mol. Genet. 15, R110–R116 [DOI] [PubMed] [Google Scholar]
- 4. Basel-Vanagaite L., Attia R., Yahav M., Ferland R. J., Anteki L., Walsh C. A., Olender T., Straussberg R., Magal N., Taub E., Drasinover V., Alkelai A., Bercovich D., Rechavi G., Simon A. J., Shohat M. (2006) The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive nonsyndromic mental retardation. J. Med. Genet. 43, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kikkawa U., Kishimoto A., Nishizuka Y. (1989) The protein kinase C family. Heterogeneity and its implications. Annu. Rev. Biochem. 58, 31–44 [DOI] [PubMed] [Google Scholar]
- 6. Davletov B. A., Südhof T. C. (1993) A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J. Biol. Chem. 268, 26386–26390 [PubMed] [Google Scholar]
- 7. Sutton R. B., Davletov B. A., Berghuis A. M., Südhof T. C., Sprang S. R. (1995) Structure of the first C2 domain of synaptotagmin I. A novel Ca2+/phospholipid-binding fold. Cell 80, 929–938 [DOI] [PubMed] [Google Scholar]
- 8. Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., Hayashi H., Sugano S. (2003) Large scale identification and characterization of human genes that activate NF-κB and MAPK signaling pathways. Oncogene 22, 3307–3318 [DOI] [PubMed] [Google Scholar]
- 9. Ou X. M., Jafar-Nejad H., Storring J. M., Meng J. H., Lemonde S., Albert P. R. (2000) Novel dual repressor elements for neuronal cell-specific transcription of the rat 5-HT1A receptor gene. J. Biol. Chem. 275, 8161–8168 [DOI] [PubMed] [Google Scholar]
- 10. Ou X. M., Lemonde S., Jafar-Nejad H., Bown C. D., Goto A., Rogaeva A., Albert P. R. (2003) Freud-1. A neuronal calcium-regulated repressor of the 5-HT1A receptor gene. J. Neurosci. 23, 7415–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rogaeva A., Galaraga K., Albert P. R. (2007) The Freud-1/CC2D1A family. Transcriptional regulators implicated in mental retardation. J. Neurosci. Res. 85, 2833–2838 [DOI] [PubMed] [Google Scholar]
- 12. Abel T., Nguyen P. V., Barad M., Deuel T. A., Kandel E. R., Bourtchouladze R. (1997) Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long term memory. Cell 88, 615–626 [DOI] [PubMed] [Google Scholar]
- 13. Nguyen P. V., Woo N. H. (2003) Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog. Neurobiol. 71, 401–437 [DOI] [PubMed] [Google Scholar]
- 14. Taskén K., Skålhegg B. S., Solberg R., Andersson K. B., Taylor S. S., Lea T., Blomhoff H. K., Jahnsen T., Hansson V. (1993) Novel isozymes of cAMP-dependent protein kinase exist in human cells due to formation of RIα-RIβ heterodimeric complexes. J. Biol. Chem. 268, 21276–21283 [PubMed] [Google Scholar]
- 15. Taylor S. S., Buechler J. A., Yonemoto W. (1990) cAMP-dependent protein kinase. Framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 59, 971–1005 [DOI] [PubMed] [Google Scholar]
- 16. Gibbs C. S., Knighton D. R., Sowadski J. M., Taylor S. S., Zoller M. J. (1992) Systematic mutational analysis of cAMP-dependent protein kinase identifies unregulated catalytic subunits and defines regions important for the recognition of the regulatory subunit. J. Biol. Chem. 267, 4806–4814 [PubMed] [Google Scholar]
- 17. Wang L. Y., Salter M. W., MacDonald J. F. (1991) Regulation of kainate receptors by cAMP-dependent protein kinase and phosphatases. Science 253, 1132–1135 [DOI] [PubMed] [Google Scholar]
- 18. Carlezon W. A., Jr., Thome J., Olson V. G., Lane-Ladd S. B., Brodkin E. S., Hiroi N., Duman R. S., Neve R. L., Nestler E. J. (1998) Regulation of cocaine reward by CREB. Science 282, 2272–2275 [DOI] [PubMed] [Google Scholar]
- 19. Gonzalez G. A., Montminy M. R. (1989) Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59, 675–680 [DOI] [PubMed] [Google Scholar]
- 20. Manganiello V. C., Degerman E. (1999) Cyclic nucleotide phosphodiesterases (PDEs). Diverse regulators of cyclic nucleotide signals and inviting molecular targets for novel therapeutic agents. Thromb. Haemost. 82, 407–411 [PubMed] [Google Scholar]
- 21. Maurice D. H., Palmer D., Tilley D. G., Dunkerley H. A., Netherton S. J., Raymond D. R., Elbatarny H. S., Jimmo S. L. (2003) Cyclic nucleotide phosphodiesterase activity, expression, and targeting in cells of the cardiovascular system. Mol. Pharmacol. 64, 533–546 [DOI] [PubMed] [Google Scholar]
- 22. Conti M., Richter W., Mehats C., Livera G., Park J. Y., Jin C. (2003) Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J. Biol. Chem. 278, 5493–5496 [DOI] [PubMed] [Google Scholar]
- 23. Houslay M. D. (2001) PDE4 cAMP-specific phosphodiesterases. Prog. Nucleic Acid Res. Mol. Biol. 69, 249–315 [DOI] [PubMed] [Google Scholar]
- 24. Houslay M. D., Adams D. R. (2003) PDE4 cAMP phosphodiesterases. Modular enzymes that orchestrate signaling cross-talk, desensitization, and compartmentalization. Biochem. J. 370, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barnette M. S., Underwood D. C. (2000) New phosphodiesterase inhibitors as therapeutics for the treatment of chronic lung disease. Curr. Opin. Pulm. Med. 6, 164–169 [DOI] [PubMed] [Google Scholar]
- 26. Méhats C., Jin S. L, Wahlstrom J., Law E., Umetsu D. T., Conti M. (2003) PDE4D plays a critical role in the control of airway smooth muscle contraction. FASEB J. 17, 1831–1841 [DOI] [PubMed] [Google Scholar]
- 27. Spina D. (2003) Phosphodiesterase-4 inhibitors in the treatment of inflammatory lung disease. Drugs 63, 2575–2594 [DOI] [PubMed] [Google Scholar]
- 28. Zhang H. T., Huang Y., Jin S. L., Frith S. A., Suvarna N., Conti M., O'Donnell J. M. (2002) Antidepressant-like profile and reduced sensitivity to rolipram in mice deficient in the PDE4D phosphodiesterase enzyme. Neuropsychopharmacology 27, 587–595 [DOI] [PubMed] [Google Scholar]
- 29. Bauman A. L., Soughayer J., Nguyen B. T., Willoughby D., Carnegie G. K., Wong W., Hoshi N., Langeberg L. K., Cooper D. M., Dessauer C. W., Scott J. D. (2006) Dynamic regulation of cAMP synthesis through anchored PKA-adenylyl cyclase V/VI complexes. Mol. Cell 23, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., Scott J. D. (2005) The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Willoughby D., Wong W., Schaack J., Scott J. D., Cooper D. M. (2006) An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J. 25, 2051–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong W., Scott J. D. (2004) AKAP signaling complexes. Focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 33. Skålhegg B. S., Huang Y., Su T., Idzerda R. L., McKnight G. S., Burton K. A. (2002) Mutation of the Cα subunit of PKA leads to growth retardation and sperm dysfunction. Mol. Endocrinol. 16, 630–639 [DOI] [PubMed] [Google Scholar]
- 34. López-Nicolás R., López-Andreo M. J., Marín-Vicente C., Gómez-Fernández J. C., Corbalán-García S. (2006) Molecular mechanisms of PKCα localization and activation by arachidonic acid. The C2 domain also plays a role. J. Mol. Biol. 357, 1105–1120 [DOI] [PubMed] [Google Scholar]
- 35. Redmond L., Oh S. R., Hicks C., Weinmaster G., Ghosh A. (2000) Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 3, 30–40 [DOI] [PubMed] [Google Scholar]
- 36. Josselyn S. A., Nguyen P. V. (2005) CREB, synapses, and memory disorders. Past progress and future challenges. Curr. Drug Targets CNS Neurol. Disord. 4, 481–497 [DOI] [PubMed] [Google Scholar]
- 37. Green S. P., Cairns B., Rae J., Errett-Baroncini C., Hongo J. A., Erickson R. W., Curnutte J. T. (2001) Induction of gp91-phox, a component of the phagocyte NADPH oxidase, in microglial cells during central nervous system inflammation. J. Cereb. Blood Flow Metab. 21, 374–384 [DOI] [PubMed] [Google Scholar]
- 38. Wyss-Coray T., Mucke L. (2002) Inflammation in neurodegenerative disease. A double-edged sword. Neuron. 35, 419–432 [DOI] [PubMed] [Google Scholar]
- 39. Mattson M. P. (2005) NF-κB in the survival and plasticity of neurons. Neurochem. Res. 30, 883–893 [DOI] [PubMed] [Google Scholar]
- 40. Wilkinson B. L., Landreth G. E. (2006) The microglial NADPH oxidase complex as a source of oxidative stress in Alzheimer disease. J. Neuroinflammation 3, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.