Background: SKI is an oncoprotein with partial understanding of its molecular mechanisms.
Results: Through regulation of Ubc9, SKI can increase MDM2 sumoylation to enhance p53 degradation.
Conclusion: Up-regulation of Ubc9 is a novel biochemical function of SKI and is essential for its transforming activity.
Significance: This is a first example that an oncoprotein can transform cells through regulation of Ubc9, a critical component of cellular sumoylation.
Keywords: Molecular Cell Biology, Oncogene, p53, Sumoylation, Ubiquitin-conjugating Enzyme (Ubc), MDM2, SKI
Abstract
Protooncogene Ski was identified based on its ability to transform avian fibroblasts in vitro. In support of its oncogenic activity, SKI was found to be overexpressed in a variety of human cancers, although the exact molecular mechanism(s) responsible for its oncogenic activity is not fully understood. We found that SKI can negatively regulate p53 by decreasing its level through up-regulation of MDM2 activity, which is mediated by the ability of SKI to enhance sumoylation of MDM2. This stimulation of MDM2 sumoylation is accomplished through a direct interaction of SKI with SUMO-conjugating enzyme E2, Ubc9, resulting in enhanced thioester bond formation and mono-sumoylation of Ubc9. A mutant SKI defective in transformation fails to increase p53 ubiquitination and is unable to increase MDM2 levels and to increase mono-sumoylation of Ubc9, suggesting that the ability of SKI to enhance Ubc9 activity is essential for its transforming function. These results established a detailed molecular mechanism that underlies the ability of SKI to cause cellular transformation while unraveling a novel connection between sumoylation and tumorigenesis, providing potential new therapeutic targets for cancer.
Introduction
Ski was found more than 20 years ago as the only transforming oncogene discovered through in vitro viral replication assay (1). It was shown to be able to transform chicken and quail embryo fibroblasts as evidenced by the overgrowth of virally infected cells in monolayer culture and anchorage-independent colony formation in soft agar, hallmarks of cellular transformation. Consistent with its role as an oncoprotein, SKI was found to be overexpressed in a variety of human cancers, including melanoma (2), leukemia (3), colorectal (4), pancreatic (5), esophageal (6), and gastric (7) cancers. Although there is scarce evidence to suggest that SKI can transform mammalian cells except melanocytes (8), a reduction of SKI through small interfering RNA technology lessens the tumorigenic properties of cancer cells (9). Transgenic mice overexpressing Ski show overgrowth of type II muscle fibers but no enhanced tumor formation (10). Mice lacking the Ski gene result in early postnatal lethality with exencephaly caused by failed closure of the cranial neural tube during neurulation as well as a host of developmental abnormalities (11). Humans diagnosed with a haploid deficiency of SKI due to 1p36 deletion display similar phenotypes as shown in mice with a constitutional lack of Ski gene (12).
The connection of the SKI oncoprotein with the TGFβ signaling pathway was established a decade ago by the finding that SKI can physically interact with Smad proteins, including Smad2, -3, and -4 (13–15). Smad proteins are the central mediators of TGFβ signaling pathways transmitting signals of the activated receptor at the plasma membrane to the nucleus (16). Upon activation of receptor through ligand binding, type I receptor kinase is activated and phosphorylates Smad protein that subsequently oligomerizes with Smad4, and the complex translocates to the nucleus to regulate target gene transcription. SKI regulates the TGFβ signaling at multiple levels; it interacts with the Smad proteins and inhibits the transcriptional activation of target genes, likely by recruiting the nuclear corepressor complex. Indeed, SKI has been found to be a component of the nuclear corepressor complex capable of inhibiting the transcriptional activation of reporter constructs (17). In addition, it has also been found that SKI can interact with the type I TGFβ receptor directly and can lead to repression of the receptor activity (18). Because TGFβ signaling is a major cellular pathway that negatively regulates epithelial cell proliferation, the transforming capability of SKI is at least partially derived from its ability to neutralize the inhibition of cell proliferation by the TGFβ pathway. However, in the initial characterization of Ski oncogene in avian fibroblast cells, TGFβ signaling was thought to be promoting Ski-induced transformation by inhibiting Ski-induced myogenic differentiation (19). Thus, TGFβ signaling cooperates with Ski to transform avian fibroblast cells. This apparent contradiction of the role of TGFβ signaling in Ski-induced cellular transformation has not been fully reconciled.
Small ubiquitin-like modifier (SUMO)3 has been found to be involved in a variety of cellular processes, including intracellular trafficking, stress response, DNA replication, DNA damage repair, as well as transcriptional regulation (20). Four versions of SUMO, 1–4, have been identified. SUMO1–3 is expressed in the precursor forms and requires proteolytic activation to expose the invariant Gly-Gly motif at the C terminus of the mature SUMO for target conjugation. Much like the cellular ubiquitination process, SUMO activation enzyme E1, SUMO-conjugating enzyme E2, and SUMO ligase enzyme E3 are required to transfer the SUMO moiety to target proteins. SUMO E1 activates the SUMO through thioester bond formation between C-terminal Gly of SUMO and the catalytic cysteine residue of E1; the activated SUMO is then transferred to E2 with a similar thioester bond. Although E2 itself can transfer SUMO to target proteins, SUMO ligase E3 through enhancing efficiency and substrate specificity facilitates this process by transferring SUMO from the high energy bond to the ϵ-position of lysine of the target protein. Unlike ubiquitin modification, the main function of sumoylation is not a signal of destruction of the target through proteasomal degradation, instead it plays a role in a variety of cellular processes. SUMO modification in some cases has been shown to antagonize ubiquitination through competitive modification (21–23). Another difference between ubiquitination and sumoylation is their respective core enzymes that carry out the modifications. It appears that there is only one E2 enzyme for sumoylation, although there are many E2s that can fulfill the ubiquitin-conjugating activity. Similar to regulation by ubiquitination, there are also proteases, sentrin-specific proteases, that can hydrolytically remove the SUMO moiety, and thus reverse the modification of sumoylation. These sentrin-specific proteases are also responsible for the activation of SUMO precursor by removing their C-terminal peptides to expose the Gly-Gly motif. Like many biological processes, the core machinery of SUMO modification has also been found to be regulated by other proteins. For example, RSUME and SF2/ASF have been found to enhance SUMO E2 enzyme Ubc9 to regulate target sumoylation (24, 25), although Rhes can enhance cross-sumoylation between E1 and Ubc9 (26).
Several prominent proteins involved in tumorigenesis have been found to be modified by sumoylation. For example, p53 can be modified by sumoylation with either enhancement or reduction of its activity (27). Similarly, MDM2, the main ubiquitin E3 ligase for p53, has been found to be sumoylated (28), and this sumoylation was shown to be associated with increased activity of MDM2 (29). MDM2 can be self-ubiquitinated and degraded. Sumoylation of MDM2 decreases its self-ubiquitination and degradation, whereas de-sumoylation leads to enhanced MDM2 self-ubiquitination. UV damage can decrease MDM2 sumoylation through a SUMO protease SUSP4. MDM2 sumoylation requires its N-terminal domain of amino acids 40–59 (30).
We report here that SKI is capable of regulating sumoylation of MDM2 through its ability to interact with and regulate SUMO E2 enzyme Ubc9, resulting in enhanced MDM2 activity and decreased p53 protein. This novel function of SKI is likely a major molecular mechanism for its oncogenic activity as p53 is an important tumor suppressor, and a mechanism that leads to its destruction will greatly contribute to cellular transformation.
EXPERIMENTAL PROCEDURES
Culture of Mammalian Cells
LNCaP was cultured in RPMI 1640 medium; H1299, U2OS, HEK293, 293T, HepG2, and mink lung epithelial cells were cultured in Dulbecco's modified Eagle's medium; HCT116 and HCT116 p53−/− cells were in McCoy's 5A medium, and all were supplemented with 10% fetal calf serum, 2 mm glutamine, penicillin, and streptomycin at 37 °C with 5% CO2.
Nucleic Acids
Chemical siRNA for Ski, PIAS1, and PIAS3 were from IDT Inc. with the following sequences: Ski, sense 5′-rCrCrArGrUrArArGrGrArGrArCrUrUrGrArArArUrUrCrAGA-3′ and antisense 5′-rUrCrUrGrArArUrUrUrCrArArGrUrCrUrCrCrUrUrArCrUrGrGrUrU-3′; PIAS1, sense 5′-rUrGrGrUrUrArUrGrArGrCrCrUrUrArGrArGrUrUrUrCrUGA-3′ and antisense 5′-rUrCrArGrArArArCrUrCrUrArArGrGrCrUrCrArUrArArCrCrArUrU-3′; and PIAS3, sense 5′-rCrArCrUrGrArUrCrArArGrGrArGrArArArUrUrGrArCrUGC-3′ and antisense 5′-rGrCrArGrUrCrArArUrUrUrCrUrCrCrUrUrGrArUrCrArGrUrGrCrC-3′. Dharmafect was used to transfect siRNA into cells. Expression plasmids were introduced into cells with BioT transfection reagent (Bioland Inc.) according to the instruction of the vendor, and empty vector DNA was used to provide equal amount of DNA in each transfection. PCR primers for p53 target gene quantification are as follows: p21, forward 5′-CTGGAGACTCTCAGGGTCGAA-3′ and reverse 5′-GGATTAGGGCTTCCTCTTGGA-3′; Noxa, forward 5′-CTCTTTCCTCCTCGCCACTT-3′ and reverse 5′-CGTGCACCTCCTGAGAAAAC-3′; Tsp1, 5′-GGGAAGAAAATCATGGCTGA-3′ and reverse 5′-GGTCGCACGTTCTAGGAGTC-3′; Mdm2 mRNA (p2 promoter), forward 5′-CGATTGGAGGGTAGACCTGT-3′ and reverse 5′-GGTCTCTTGTTCCGAAGCTG-3′; Mdm2 (last exon), forward 5′-CAGACGGGGACTAGCTTTTG-3′ and reverse 5′-AGGTTGCAGTGAGCCAAGAT-3′; and Gapdh, forward 5′-CATGGGTGTGAACCATGAGA-3′ and reverse 5′-CAGTGATGGCATGGACTGTG-3′. RNA was isolated using Qiagen RNeasy kit, and cDNA was generated using SuperscriptII reverse transcriptase (Invitrogen).
Biochemical and Cell Biological Reagents and Procedures
Anti-SKI, p53, and Ubc9 antibodies were from Santa Cruz Biotechnology. Anti-FLAG M2 antibody was from Sigma (A1205). Anti-HA antibody was from Roche Applied Science. Anti-SUMO1 antibody was from Epitomics. For immunoprecipitation and immunoblotting, cells were collected by scraping or trypsinization and lysed in lysis buffer (20 mm KCl, 150 mm NaCl, 1% IGEPAL, 50 mm Tris-HCl (pH 7.5), 50 mm NaF, 1 mm EGTA, 1 mm DTT, and 1× protease inhibitor mixture (Roche Applied Science) and 10% glycerol). Immunoprecipitations were performed with appropriate antibody and protein A- or G-Sepharose (Upstate Biotechnology). Beads were washed three times in lysis buffer, and immunoprecipitated proteins were separated on SDS-PAGE followed by Western blotting with primary and horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). Immunoreactive proteins were visualized by enhanced chemiluminescence (SuperSignal West Femto, Pierce). CellTiter Glo (Promega Inc.) was used to measure relative cell number according to the manufacturer. For FACS analysis to gauge the sub-G1 DNA content, the cells were collected and fixed in 70% ethanol followed by RNase treatment and 1 μg/ml propidium iodide staining, and the cells were then analyzed by FACS.
MDM2 Sumoylation Assay
Expression plasmid for His6-MDM2 was transfected into 293T cells together with other expression plasmids. 40 h after transfection, cells were collected and lysed in 150 μl of RIPA buffer (regular buffer + 0.1% SDS + 0.5% sodium deoxycholate) followed by centrifugation. The supernatant was mixed with 8 m guanidine hydrochloride to obtain 6 m guanidine hydrochloride with 20 mm sodium phosphate buffer (pH 7.5), 20 mm imidazole, 1 mm DTT. This lysate was centrifuged at room temperature for 10 min, and the supernatant was mixed with 20 μl of nickel-nitrilotriacetic acid beads in 50% bead slurry. After binding for 3 h, the beads were washed three times with urea buffer (8 m urea with 20 mm sodium phosphate buffer (pH 7.5)) followed by incubation with sample buffer at 95 °C for 10 min. The supernatant was then separated by SDS-PAGE followed by immunoblot analysis.
RESULTS
Overexpression of SKI Leads to Decreased p53
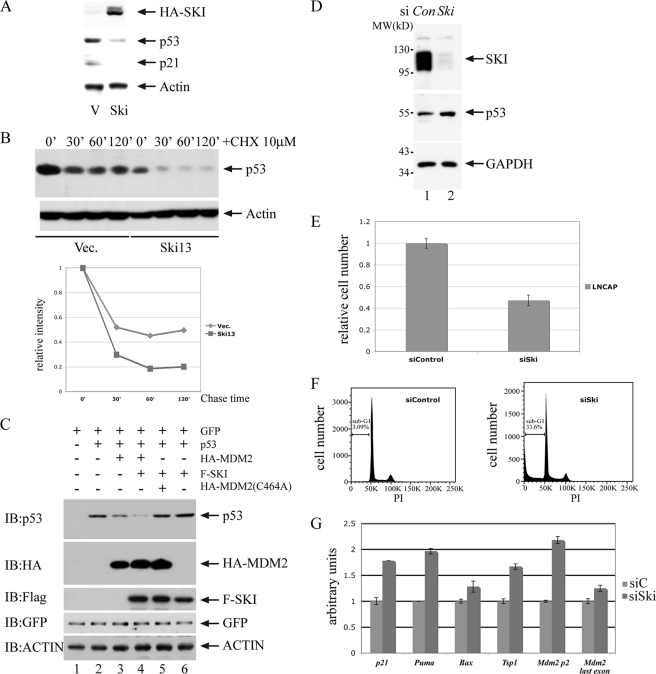
SKI is a transforming oncoprotein for chicken embryo fibroblast cells upon overexpression. Previously, we and others have established that SKI can interact with Smad proteins and negatively regulate TGFβ signal transduction (13–15), and it was widely thought that this inhibition of TGFβ signaling is the underlying mechanism of its transforming activity. Although this mechanistic explanation is plausible, we suspected that other mechanism(s) might also be involved in the oncogenic activity of SKI. Therefore, we examined in cells with overexpression of SKI the abundance of p53 protein, one of the essential tumor suppressor proteins in regulating cell proliferation and differentiation. As shown in Fig. 1A, overexpression of SKI in mink lung epithelial cells leads to a decrease of p53, and this reduction is independent of the status of TGFβ signaling (data not shown). Consistent with this reduction, the protein level of p21waf1, one of the p53 target genes, also decreases. Moreover, this reduced p53 level is likely a result of decreased metabolic stability as its half-life is apparently reduced in cells with overexpression of SKI (Fig. 1B). These findings therefore suggested the possibility that a higher level of SKI results in a reduction of p53, thus resulting in cellular transformation. We further tested whether in a transient overexpression system SKI can also cause a reduction of p53. As shown in Fig. 1C, in human colorectal carcinoma HCT-116 cells with or without endogenous Tp53 (see supplemental Fig. 1), overexpression of SKI by itself cannot decrease p53 levels; however, SKI can enhance the reduction of p53 caused by the wild type MDM2, the main ubiquitin E3 ligase for p53, but not the RING finger mutant of MDM2 (C464A) (31). These results suggest that a higher level of SKI can result in an MDM2-dependent p53 reduction likely through enhanced p53 degradation. To rule out the possibility that SKI can only regulate p53 in an overexpression system, we also examined the impact of SKI depletion on endogenous p53 activity. As shown in Fig. 1D, siRNA-mediated loss-of-function of SKI in the prostate cancer cell line LNCaP results in elevated p53 levels and enhanced apoptosis as determined by cell number enumeration through measurement of ATP levels as well as of sub-G1 DNA content in a fluorescent-activated cell sorter (FACS) analysis (Fig. 1, E and F). Consistent with these findings, we also found that a subset of p53 target genes are up-regulated in response to a reduction of SKI and up-regulation of p53 (Fig. 1G). Taken together, these results strongly suggest that SKI with both physiological and quasi-physiological expressions can negatively regulate p53 through MDM2, and this aspect of SKI is likely a major contributor for its transforming activity.
FIGURE 1.
SKI negatively regulates p53. A, overexpression of SKI leads to a decrease of p53. p53, p21waf1 level was examined with immunoblot analysis in mink lung epithelial cells overexpressing SKI (Ski13) or vector control. B, half-life of p53 was decreased in Ski13 cells. C, SKI enhances decrease of p53 by MDM2 but not by mutant MDM2. Various expression plasmids were introduced into HCT116 cells without Tp53, and the p53 was examined by immunoblot (IB) analysis with anti-p53 antibody. The mutant MDM2 has alanine at amino acid 464 instead of cysteine (C464A). The amount of plasmids was made equal by adding vector DNA, and the transfection efficiency was monitored with GFP expression. MDM2 and SKI expression were monitored with anti-HA and anti-FLAG antibodies. CHX, cycloheximide. D, reduction of SKI results in an increase of p53 level. siRNA for SKI was introduced into LNCaP cells, and the levels of SKI, p53 and GAPDH were examined by immunoblot analysis. E and F, reduction of SKI results in a decrease of cell number and an increase of apoptosis. 48 h after siRNA for SKI was introduced into LNCaP cells, the cell number was measured using CellTiter Glo (E), and the sub-G1 content was measured with FACS analysis (F). G, reduction of SKI results in an increase of several p53 target genes. SYBR quantitative PCR was used to measure the quantity of transcript level of p53 target genes in cells with transfection of control siRNA or siRNA for SKI. The level in control cells was arbitrarily set as 1 followed by correction with relative total RNA level using GAPDH measurements.
SKI Enhances p53 Ubiquitination
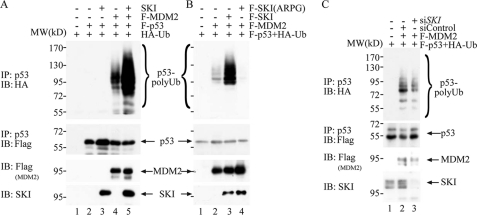
p53 is a major tumor suppressor protein that is inactivated in many human cancers. Nearly half of the human cancers harbor mutations of Tp53, whereas p53 is inactivated in a large proportion of tumors without Tp53 mutation through a multitude of mechanisms. For example, overexpression of MDM2, loss of the p14Arf protein, which is a negative regular of MDM2, and expression of viral oncoproteins, including HPV16 E6, adenovirus E1A, and SV40 large T antigen (32), can all lead to p53 inactivation. Although several ubiquitin E3 ligases have been shown to regulate p53 levels (33), the genetically modified mouse without Mdm2 has unequivocally demonstrated the centrality of Mdm2 in regulating the p53 protein level (34). As described earlier, we found that SKI can enhance the MDM2-mediated p53 decrease (Fig. 1, A and C); therefore, we examined whether SKI can positively regulate MDM2-mediated ubiquitination of p53. As shown in Fig. 2A, although expression of MDM2 alone enhanced the ubiquitination of p53, co-expression of SKI dramatically increased the p53 ubiquitination shown both by the ubiquitin in the p53 immunoprecipitation as well as p53 in the immunoprecipitation of ubiquitin (supplemental Fig. 2). These results suggest that SKI can enhance MDM2-mediated p53 ubiquitination and thus proteasomal degradation, which likely results in a decreased p53 level. It has been shown that a mutant SKI with an insertion of four amino acids, alanine, arginine, proline, and glycine (ARPG) replacing aspartic acid at position 181, inactivated the transforming function of SKI in chicken embryo fibroblast cells (35), and we therefore tested whether the transforming capacity of SKI is correlated with its ability to enhance ubiquitination of p53. As shown in Fig. 2B, lane 4, expression of the SKI mutant ARPG, at a comparable level of wild type SKI protein, did not enhance, if reduced, p53 ubiquitination, suggesting that the transforming ability of SKI is tightly associated with its interaction with MDM2 to result in p53 ubiquitination. In addition, we also tested whether SKI at a physiological level can regulate MDM2-mediated p53 ubiquitination. As shown in Fig. 2C, siRNA-mediated reduction of endogenous SKI protein also reduced p53 ubiquitination conferred upon by MDM2 expression. These results suggest that SKI at both normal and overexpression levels is capable of enhancing MDM2-mediated ubiquitination of p53, and this activity is tightly associated with the transforming function of SKI.
FIGURE 2.
SKI enhances MDM2-mediated p53 ubiquitination. A, SKI increases p53 ubiquitination by MDM2. Various plasmids were introduced into H1299 cells, and the amount of plasmids was equalized with empty vector. 40 h after transfection, the cells were treated with 20 μm MG-132 for 4 h, lysed with 2% SDS buffer, and heated at 95 °C for 5 min followed by dilution with regular buffer to 0.1% SDS and centrifugation. The supernatant was immunoprecipitated (IP) with anti-p53 antibody, followed by the immunoblot (IB) analysis with anti-HA antibody. The blot was stripped and probed again with anti-FLAG antibody to gauge the p53 level. A separate portion of the lysate was used to analyze the expression level of MDM2 and SKI with immunoblot analysis. B, wild type but not mutant SKI enhances p53 ubiquitination by MDM2. The experiment was done essentially as in A in H1299 cells. C, endogenous SKI was crucial for MDM2-mediated p53 ubiquitination. The experiment was done essentially as in A except with introduction of siRNA for SKI.
SKI Positively Regulates MDM2 through Sumoylation
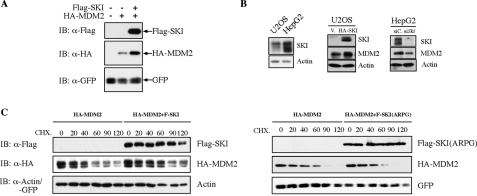
The ability of SKI to enhance MDM2-mediated ubiquitination of p53 suggests several possible modes of action of SKI in the regulation of p53. It is possible that SKI directly regulates p53's propensity of being ubiquitinated by MDM2 followed by proteosomal degradation. It is also possible that SKI can positively enhance the activity of MDM2 through direct physical interaction with MDM2; alternatively, SKI can regulate MDM2 through post-translational modification without direct physical interaction. We established that in an overexpression system, SKI could physically interact with MDM2 (supplemental Fig. 3). However, repeated attempts in a variety of cell types failed to demonstrate this physical interaction at endogenous expression levels. Nevertheless, we can clearly demonstrate that overexpression of SKI increases the abundance of MDM2, consistent with its ability to decrease the p53 level (Fig. 3, A, and middle panel of B). Conversely, a depletion of SKI by siRNA in HepG2 cells can decrease endogenous MDM2 levels (Fig. 3B, right panel), supporting the view that SKI can positively regulate the abundance of MDM2. Moreover, the increased level of MDM2 by SKI is likely a result of increased stability of MDM2 as shown in Fig. 3C, whereas the SKI mutant ARPG fails to increase the stability of MDM2 (Fig. 3C, right panel). These results suggest that SKI can functionally enhance MDM2, likely its protein level or activity, yet it does not do so through high affinity or direct physical interaction with MDM2.
FIGURE 3.
SKI positively regulates MDM2 protein. A, overexpression of SKI leads to an increase of MDM2 levels. 293T cells were transfected with expression plasmids for SKI and/or MDM2, and the exogenous MDM2 protein level was examined by immunoblot (IB) analysis. B, overexpression and reduction of SKI leads to an increase or decrease of endogenous MDM2 protein. Stable expression of SKI in U2OS cells results in increased MDM2 levels compared with the vector cells, although siRNA for SKI in HepG2 cells results in a decrease of MDM2 level. C, SKI increases the metabolic stability of MDM2 protein. 40 h after transfection with expression plasmids for SKI, SKI(ARPG), and MDM2, 293T cells were treated with cycloheximide for the indicated time, and MDM2 protein level was analyzed by immunoblot analysis.
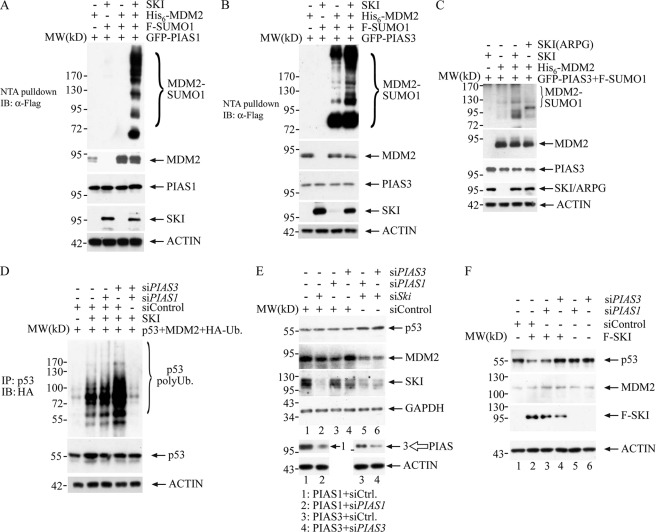
It is known that MDM2 can be broadly regulated by two biochemical mechanisms as follows: one involves post-translational modification, and the other involves proteinaceous regulators that modulate its cellular localization and activity (36–41). For the former, protein kinase B (AKT) can phosphorylate MDM2 and result in its nuclear translocation (42), which is crucial for its ability to degrade p53. In addition, sumoylation of MDM2 has been shown to enhance the MDM2 level and activity (29). Thus, it is possible that SKI can regulate MDM2 activity through these modifications. We tested the latter possibility, and found, as shown in Fig. 4, A and B, that SKI can clearly increase MDM2 sumoylation mediated by SUMO E3 ligases, PIAS1 and PIAS3 (protein inhibitors of activated STAT1 and -3). PIAS1 has been shown to enhance MDM2 sumoylation (28), although its impact on MDM2 activity is not clear. PIAS3 is known to interact with and sumoylate Smad4 protein (43), which has also been shown to interact with SKI (13), thus it is plausible that SKI can alter the substrate specificity of PIAS3 leading to sumoylation of MDM2. In limited cases, protein sumoylation has been found to antagonize ubiquitination thus increasing substrate protein abundance, although hypersumoylation has also been associated with sumoylation-induced ubiquitination with a consequent reduction of target protein abundance (44–46). MDM2 has been shown to be modified by sumoylation, which results in increased MDM2 activity presumably as a result of an increased protein level (29). The ability of SKI to enhance MDM2 sumoylation is thus consistent with its function to enhance MDM2 levels as well as to increase MDM2-mediated p53 ubiquitination. To provide further evidence that SKI can increase MDM2 activity through enhanced sumoylation of MDM2, we examined whether reduction of PIAS1 and/or PIAS3 can result in abrogation of the ability of SKI to up-regulate MDM2 activity. As shown in Fig. 4D, siRNA against PIAS1 or PIAS3 alone does not reduce MDM2-mediated p53 ubiquitination; however, combined reduction of PIAS1 and PIAS3 results in a drastic reduction of p53 ubiquitination by MDM2, suggesting that different PIAS proteins might be able to compensate each other in mediating MDM2 sumoylation and that SKI can communicate with both of them, potentially with different potencies. The potency of the siRNA for PIAS1 and -3 was demonstrated in Fig. 4E, lower panel, where respective siRNA can decrease PIAS1 and -3 protein levels. This result thus suggests that overexpression of SKI positively regulates ubiquitination of p53 by MDM2, likely through the latter's sumoylation by PIAS1 and -3. Consistent with the notion that MDM2 sumoylation by SKI is critical for the transforming ability of SKI, the SKI mutant (ARPG) fails to enhance MDM2 sumoylation as shown in Fig. 4C, providing a mechanistic explanation for the failure of ARPG to enhance MDM2-mediated p53 ubiquitination as demonstrated in Fig. 2B. To examine whether SKI can regulate endogenous p53 through regulation of MDM2 sumoylation, we examined the level of p53 in LNCaP cells in response to reduction of SKI as well as PIAS1 and PIAS3. As shown in Fig. 4E, reduction of SKI, PIAS1, and PIAS3 individually does not lead to a change of p53 protein levels, possibly due to limited reduction of the individual protein by the siRNA; however, a combined reduction of SKI and PIAS1 or PIAS3 results in increased p53 levels, concomitant with a reduction of MDM2 levels. This result supports the notion that SKI and PIAS1 or PIAS3 operate in a linear pathway in that SKI through PIAS1 or PIAS3 can result in increased MDM2 levels through sumoylation, and a combined reduction of both components leads to a reduction of MDM2 sumoylation severe enough to cause a reduction of protein levels with a consequent increase of p53 levels. Similarly, we also found that in human embryonic kidney HEK293 cells, exogenous expression of SKI leads to a reduction of endogenous p53 levels, likely through enhanced MDM2 sumoylation and activity as this reduction of p53 level can be reversed by co-expression of siRNA for PIAS3, but not by PIAS1, suggesting that SKI through PIAS3 can regulate endogenous p53 levels by modulating MDM2 sumoylation. The reason for the failure of PIAS1 to reverse the effect of SKI, despite its collaboration with SKI to enhance MDM2 sumoylation (Fig. 4A) and collaborate with SKI to maintain MDM2 levels (Fig. 4E), remains obscure. We can speculate that additional factors in HEK293 cells might have altered the substrate specificity such that PIAS1 behaves differently when it is in the context of overexpression with SKI as shown in Fig. 4A. These data together strongly suggest that SKI can cause down-regulation of endogenous p53 by up-regulating MDM2 activity through enhanced sumoylation of MDM2 by PIAS1 or PIAS3. It is also clear that because SKI is a modulator of MDM2 through PIAS1 or PIAS3, depending on the particular cellular context, alteration of SKI alone may not be sufficient to modulate the level or activity of MDM2 and ultimately the level of p53.
FIGURE 4.
SKI regulates MDM2 through sumoylation to result in a decrease of endogenous p53. A, SKI can enhance MDM2 sumoylation through PIAS1. 24 h after expression, plasmids were introduced into 293T cells, and cells were lysed in RIPA buffer, and the supernatant was diluted into denaturing buffer. The MDM2 was isolated through nickel-nitrilotriacetic acid (NTA) beads followed by immunoblot (IB) analysis with anti-FLAG antibody to reveal MDM2 sumoylation. The blot was stripped and probed with anti-MDM2 antibody to reveal the MDM2 level. Separate lysate was used to examine PIAS1 and SKI expression levels using anti-PIAS1 and anti-SKI antibody, respectively. B, SKI can enhance MDM2 sumoylation through PIAS3. 40 h after transfection, cells were processed as in A. A portion of the lysate was analyzed to gauge the protein expression through immunoblot with anti-PIAS3 and anti-SKI antibody. C, SKI mutant ARPG fails to increase MDM2 sumoylation. 40 h after transfection, cells were processed as in A. A portion of the lysate was analyzed to gauge the protein expression through immunoblot with anti-PIAS3 and anti-SKI antibody. D, SKI enhances p53 ubiquitination through PIAS1 and -3. H1299 cells were transfected with various combinations of expression plasmids as well as siRNAs. The cells were processed as in Fig. 2. The effectiveness of siRNA for PIAS1 or -3 is shown in E. E, SKI and PIAS1 or -3 are required for maintaining the p53 level in LNCaP cells. Various siRNAs were introduced into LNCaP cells, and 40 h later, cells were lysed, and endogenous p53 as well as other proteins were examined by immunoblot analysis. The effectiveness of siRNA for PIAS1 or -3 was shown by the reduction of PIAS1 or -3 in 293T cells transfected with the expression plasmid alone or with the respective siRNA shown in the lower panel. F, down-regulation of p53 by overexpression of SKI can be reversed by a reduction of PIAS3. Expression plasmids for SKI and siRNAs were introduced into HEK293 cells, and 40 h later, cells were lysed, and endogenous p53 and MDM2 were examined by immunoblot analysis.
SKI Positively Regulates Ubc9 Activity
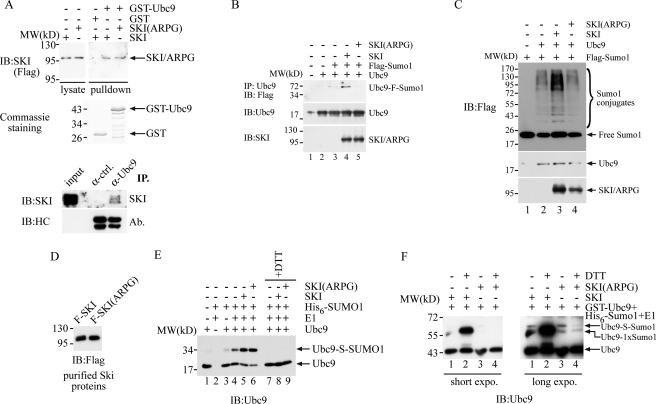
The unique feature of mammalian sumoylation is that there is a single SUMO-conjugating enzyme, Ubc9, that controls the general protein sumoylation. Thus, if a protein like SKI can directly regulate Ubc9, it can broadly regulate substrate sumoylation. We examined this possibility by performing a GST pulldown experiment with GST-Ubc9 and cellular SKI protein. We generated a recombinant GST-Ubc9 fusion protein as well as the control GST protein. As shown in Fig. 5A, when the same amount of these recombinant proteins was incubated with lysates prepared from 293T cells overexpressing SKI or its mutant ARPG protein, SKI as well as the ARPG mutant is capable of interacting with the Ubc9 protein, but not with GST protein, suggesting that SKI can directly interact with Ubc9 protein. This interaction can also be confirmed in vivo with the presence of SKI protein in the immunoprecipitate of anti-Ubc9 antibody as shown in the lower panel of Fig. 5A. Moreover, we found that SKI, but not the mutant ARPG protein, can enhance auto-sumoylation of Ubc9 in a transient expression system (Fig. 5B). We also monitored the in vivo sumoylation profile with expression of Ubc9 in the absence or presence of SKI or its mutant. As shown in Fig. 5C, expression of SKI, but not the ARPG mutant, significantly enhances the detectable protein sumoylation marked by epitope-tagged SUMO1 protein, suggesting that SKI can enhance Ubc9-mediated cellular sumoylation, possibly through direct interaction with Ubc9. This enhancement of Ubc9 is directly linked with its transforming ability as the transformation-defective mutant ARPG fails to do so. To examine the molecular mechanism of this enhanced sumoylation by SKI protein, we reconstituted Ubc9 modification with the SKI protein in vitro. We purified the SKI protein or ARPG mutant by releasing it through FLAG peptide competition from immunoprecipitates of these proteins expressed in 293T cells (Fig. 5D). We then incubated the recombinant Ubc9 protein with SUMO E1 as well as activated SUMO1 protein in the presence of ATP as the energy source. As shown in Fig. 5E, SKI can significantly increase the thioester bond formation of Ubc9 with SUMO1 as the thioester bond between Ubc9 and SUMO1 is sensitive to the reducing agent in the reaction, thus allowing us to monitor their abundance by their disappearance in response to dithiothreitol (DTT). The SUMO1-Ubc9 thioester bond formation serves as an intermediate for Ubc9 before substrate sumoylation, and thus more thioester bond formation will translate into more substrate modification. We also reconstituted this Ubc9 modification using GST-Ubc9 as a substrate. We noticed a higher basal activity of this Ubc9 protein in mediating its SUMO modification (data not shown), presumably due to the artificial dimerization of Ubc9 by the GST moiety. Using GST-Ubc9 as the substrate, as shown in Fig. 5F, SKI but not the ARPG protein can clearly increase Ubc9-mono-sumoylation as evidenced by the DTT-resistant Ubc9-SUMO1 conjugate. It has been reported that sumoylation of Ubc9 can alter substrate sumoylation by Ubc9 (47), and thus an increase of Ubc9 sumoylation likely will result in an enhanced sumoylation of certain substrate proteins. Taken together, we conclude that SKI can directly interact with Ubc9 and enhance its thioester bond formation as well as its own modification with SUMO1, resulting in increased activity of Ubc9. To our knowledge, this is the first demonstration that an oncoprotein can directly enhance Ubc9 activity through up-regulation of its biochemical activity. We also noted that there is a clear difference between wild type SKI and the ARPG mutant in that SKI protein can dramatically increase the mono-sumoylation of Ubc9, whereas ARPG mutant lacks this activity, strongly supporting the notion that the transforming capability of SKI at least partially derives from its up-regulation of Ubc9 activity through enhancing Ubc9 mono-sumoylation.
FIGURE 5.
SKI positively regulates Ubc9 both in vivo and in vitro. A, SKI can directly interact with Ubc9. Approximately equal amounts of GST or GST-Ubc9 protein was mixed with lysate prepared from 293T cells with expression of FLAG-SKI, FLAG-SKI(ARPG), or control transfection. The GST fusion protein was retrieved with glutathione-Sepharose beads and washed three times with lysis buffer. The bead-bound proteins were analyzed by immunoblot (IB) analysis with anti-FLAG antibody to reveal the presence of SKI/ARPG protein in the GST-Ubc9 pulldown. The exposure time for the lysate protein was about 1/10 of that for the pulldown. In the lower panel, LNCaP cells were treated with paraformaldehyde for 10 min before collection, and lysed and immunoprecipitated (IP) with anti-Ubc9 antibody or control antibody. The immunoprecipitates were analyzed together with 10 μg of input lysate with SDS-PAGE followed by immunoblot with anti-SKI antibody or antibody for heavy chain. B, SKI enhances Ubc9 sumoylation in vivo. 293T cells were transfected with various plasmids, and the Ubc9 protein was immunoprecipitated with anti-Ubc9 antibody followed by immunoblot with anti-FLAG antibody to reveal the extent of the Ubc9 sumoylation. C, SKI enhances Ubc9-mediated sumoylation in vivo. 293T cells were transfected with various combinations of expression plasmids, and in vivo sumoylation was determined by immunoblot analysis with anti-FLAG antibody using the total lysate after SDS-PAGE. D, purified SKI and SKI(ARPG) protein. SKI and SKI(ARPG) were first immunoprecipitated with anti-FLAG antibody from 293T cells expressing FLAG-Ski or FLAG-Ski(ARPG), and then the immunoprecipitates were washed and incubated with FLAG peptide. The proteins were then quantitated by immunoblot with anti-FLAG antibody. E, SKI increases thioester bond formation of Ubc9 in vitro. 30 ng of Ubc9 protein was mixed with 60 ng of E1, 250 ng of His6-Sumo1, and purified SKI/ARPG protein in 15 μl at 37 °C for 30 min. The reaction mixture was then separated by SDS-PAGE in the presence or absence of DTT in the sample buffer followed by immunoblot analysis with anti-Ubc9 antibody. F, SKI increases mono-sumoylation of Ubc9 in vitro. 10 ng of GST-Ubc9 protein was mixed with 110 ng of E1, 600 ng of His6-Sumo1 as in E followed by immunoblot analysis with anti-Ubc9 antibody. The designation of Ubc9 conjugated with one SUMO1 molecule is based on its molecular weight increase and resistance to DTT.
DISCUSSION
A multitude of molecular mechanisms has been put forward to explain the oncogenic activity of SKI. The most notable one is its connection with TGFβ signaling by its physical interaction with and inhibition of transcriptional activation by Smad proteins (13–15). It has also been found that SKI is capable of inhibiting transcriptional repression mediated by retinoblastoma tumor suppressor through complex formation (48). Its connection with Sirt1 and SKIP provides additional possibilities that potentially underlie its transforming capability (49, 50). We provided evidence that SKI can interact with the central component of cellular sumoylation machinery, Ubc9, and enhance Ubc9-mediated protein sumoylation. More specifically, SKI can enhance PIAS1- and PIAS3-mediated sumoylation of MDM2 and in this manner increase MDM2 levels and enhance MDM2-mediated p53 ubiquitination and proteasomal degradation. This is the most direct route for SKI to neutralize one of the essential tumor suppressor proteins in the cell and likely the major molecular mechanism that is responsible for its ability to cause cellular transformation.
A lack of transformation of mammalian cells by SKI raises an intriguing question. Overexpression of MDM2 has been shown to readily collaborate with several oncogenes to cause cellular transformation (51), and if SKI is capable of enhancing MDM2, then it should also readily collaborate with other oncogenes to cause transformation. Yet repeated attempts to demonstrate the transforming capability of Ski in mammalian cells yielded inconclusive results at best. Indeed, there are several reports suggesting that SKI is a tumor suppressor rather than an oncoprotein. For example, mouse embryo fibroblasts with a haploid deficiency of Ski grow faster in culture than cells with wild type Ski (52). In light of our evidence that SKI interacts with and enhances the activity of Ubc9, we can now rationalize the transforming ability of SKI in this broad context of cellular sumoylation. In this regard, p53 has also been shown to be sumoylated (53), and SKI can also enhance its sumoylation in mammalian cells through overexpression (supplemental Fig. 4). If sumoylation can compete against ubiquitination, it is conceivable that SKI in this manner can also enhance p53 stability. Thus, SKI can cause opposing effects in regulating p53 activity. On the one hand, more MDM2 sumoylation leads to more protein and activity and thus a reduced p53 level; on the other hand, SKI can directly enhance the p53 level through sumoylation-mediated stabilization or other p53 activities. It is likely that this opposing activity on p53 weakens the potency of SKI in causing cellular transformation and limits its target cells only to avian embryo fibroblast cells. Moreover, it is conceivable that in a certain cellular context, SKI favors the p53 sumoylation, resulting in enhanced tumor suppressor activity, and thus SKI in this fashion acts as a tumor suppressor rather than an oncoprotein. The precise molecular circuitry that dictates the ultimate activity of SKI is not clear; nevertheless, our novel finding will provide a molecular guide to unravel its role in a particular biological context.
In contrast to cellular ubiquitination machinery, there are a limited number of core components mediating protein sumoylation. There are so far one SUMO-activating enzyme E1 and one conjugating enzyme E2, and a handful of SUMO E3s, including PIAS1–4, Polycomb protein Pc2, and RanGAP1. A recent report of the TRIM protein family likely will expand the SUMO E3 family (54). As with many cellular enzymes, there will be positive and negative regulators for their functions. RSUME and ASF2 have been shown to positively regulate Ubc9 activity, although with distinct molecular mechanisms. RSUME acts as an enhancer for Ubc9 activity leading to increased SUMO-Ubc9 thioester bond formation (24), although ASF2 does not lead to enhanced thioester bond formation while augmenting substrate sumoylation by Ubc9 (25). In this regard, SKI is shown to both enhance Ubc9-SUMO1 thioester bond formation as well as sumoylation of Ubc9 itself. Because it is known that Ubc9 sumoylation can alter substrate sumoylation, it is conceivable that SKI can indirectly alter substrate sumoylation in a more dramatic manner by also enhancing thioester bond formation thus providing more SUMO donor. In this regard, SKI can also be viewed as a SUMO E3 for Ubc9.
We have provided evidence that oncoprotein SKI can interact with Ubc9 protein, and up-regulate MDM2 activity through enhanced sumoylation with a consequent down-regulation of the p53 level. This activity of SKI likely is a major determinant of its transforming ability as it directly links to the central tumor suppressor, p53, in the cell. This finding suggests that in tumors where SKI is overexpressed but the p53 protein is intact, SKI may contribute to tumor initiation and progression through down-regulation of p53 via increased sumoylation. At the same time, a reduction of cellular sumoylation cascade might provide therapeutic benefits in causing inactivation of MDM2 protein with a reactivation of p53.
Supplementary Material
Acknowledgments
We thank Dr. Moshe Oren for sending expression plasmids for wild type MDM2; Dr. Xinhua Feng for expression plasmid of FLAG-Sumo1; Dr. Steven B. McMahon for FLAG-p53; and Dr. Bert Vogelstein for sending HCT-116 with or without Tp53.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA093848 (to Y. S.).

This article contains supplemental Figs. 1–4.
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Li Y., Turck C. M., Teumer J. K., Stavnezer E. (1986) Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene. J. Virol. 57, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medrano E. E. (2003) Repression of TGF-β signaling by the oncogenic protein SKI in human melanomas. Consequences for proliferation, survival, and metastasis. Oncogene 22, 3123–3129 [DOI] [PubMed] [Google Scholar]
- 3. Ritter M., Kattmann D., Teichler S., Hartmann O., Samuelsson M. K., Burchert A., Bach J. P., Kim T. D., Berwanger B., Thiede C., Jäger R., Ehninger G., Schäfer H., Ueki N., Hayman M. J., Eilers M., Neubauer A. (2006) Inhibition of retinoic acid receptor signaling by Ski in acute myeloid leukemia. Leukemia 20, 437–443 [DOI] [PubMed] [Google Scholar]
- 4. Buess M., Terracciano L., Reuter J., Ballabeni P., Boulay J. L., Laffer U., Metzger U., Herrmann R., Rochlitz C. (2004) Amplification of SKI is a prognostic marker in early colorectal cancer. Neoplasia 6, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heider T. R., Lyman S., Schoonhoven R., Behrns K. E. (2007) Ski promotes tumor growth through abrogation of transforming growth factor-β signaling in pancreatic cancer. Ann. Surg. 246, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fukuchi M., Nakajima M., Fukai Y., Miyazaki T., Masuda N., Sohda M., Manda R., Tsukada K., Kato H., Kuwano H. (2004) Increased expression of c-Ski as a co-repressor in transforming growth factor-β signaling correlates with progression of esophageal squamous cell carcinoma. Int. J. Cancer 108, 818–824 [DOI] [PubMed] [Google Scholar]
- 7. Takahata M., Inoue Y., Tsuda H., Imoto I., Koinuma D., Hayashi M., Ichikura T., Yamori T., Nagasaki K., Yoshida M., Matsuoka M., Morishita K., Yuki K., Hanyu A., Miyazawa K., Inazawa J., Miyazono K., Imamura T. (2009) SKI and MEL1 cooperate to inhibit transforming growth factor-β signal in gastric cancer cells. J. Biol. Chem. 284, 3334–3344 [DOI] [PubMed] [Google Scholar]
- 8. Barkas A. (1986) Ph.D. dissertation New York University [Google Scholar]
- 9. Chen D., Lin Q., Box N., Roop D., Ishii S., Matsuzaki K., Fan T., Hornyak T. J., Reed J. A., Stavnezer E., Timchenko N. A., Medrano E. E. (2009) SKI knockdown inhibits human melanoma tumor growth in vivo. Pigment Cell Melanoma Res. 22, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sutrave P., Kelly A. M., Hughes S. H. (1990) Ski can cause selective growth of skeletal muscle in transgenic mice. Genes Dev. 4, 1462–1472 [DOI] [PubMed] [Google Scholar]
- 11. Berk M., Desai S. Y., Heyman H. C., Colmenares C. (1997) Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 11, 2029–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colmenares C., Heilstedt H. A., Shaffer L. G., Schwartz S., Berk M., Murray J. C., Stavnezer E. (2002) Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski−/− mice. Nat. Genet. 30, 106–109 [DOI] [PubMed] [Google Scholar]
- 13. Luo K., Stroschein S. L., Wang W., Chen D., Martens E., Zhou S., Zhou Q. (1999) The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes Dev. 13, 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y., Liu X., Eaton E. N., Lane W. S., Lodish H. F., Weinberg R. A. (1999) Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol. Cell 4, 499–509 [DOI] [PubMed] [Google Scholar]
- 15. Xu W., Angelis K., Danielpour D., Haddad M. M., Bischof O., Campisi J., Stavnezer E., Medrano E. E. (2000) Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type β transforming growth factor. Proc. Natl. Acad. Sci. U.S.A. 97, 5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kretzschmar M., Massagué J. (1998) SMADs. Mediators and regulators of TGF-β signaling. Curr. Opin. Genet. Dev. 8, 103–111 [DOI] [PubMed] [Google Scholar]
- 17. Nomura T., Khan M. M., Kaul S. C., Dong H. D., Wadhwa R., Colmenares C., Kohno I., Ishii S. (1999) Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferrand N., Atfi A., Prunier C. (2010) The oncoprotein c-Ski functions as a direct antagonist of the transforming growth factor-β type I receptor. Cancer Res. 70, 8457–8466 [DOI] [PubMed] [Google Scholar]
- 19. Colmenares C., Stavnezer E. (1989) The ski oncogene induces muscle differentiation in quail embryo cells. Cell 59, 293–303 [DOI] [PubMed] [Google Scholar]
- 20. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation. A decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 21. Bae S. H., Jeong J. W., Park J. A., Kim S. H., Bae M. K., Choi S. J., Kim K. W. (2004) Sumoylation increases HIF-1α stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 324, 394–400 [DOI] [PubMed] [Google Scholar]
- 22. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 23. Sun Y., Perera J., Rubin B. P., Huang J. (2011) SYT-SSX1 (Synovial Sarcoma Translocated) regulates PIASy to cause overexpression of NCOA3. J. Biol. Chem. 286, 18623–18632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carbia-Nagashima A., Gerez J., Perez-Castro C., Paez-Pereda M., Silberstein S., Stalla G. K., Holsboer F., Arzt E. (2007) RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1α during hypoxia. Cell 131, 309–323 [DOI] [PubMed] [Google Scholar]
- 25. Pelisch F., Gerez J., Druker J., Schor I. E., Muñoz M. J., Risso G., Petrillo E., Westman B. J., Lamond A. I., Arzt E., Srebrow A. (2010) The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc. Natl. Acad. Sci. U.S.A. 107, 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subramaniam S., Mealer R. G., Sixt K. M., Barrow R. K., Usiello A., Snyder S. H. (2010) Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 285, 20428–20432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hock A., Vousden K. H. (2010) Regulation of the p53 pathway by ubiquitin and related proteins. Int. J. Biochem. Cell Biol. 42, 1618–1621 [DOI] [PubMed] [Google Scholar]
- 28. Miyauchi Y., Yogosawa S., Honda R., Nishida T., Yasuda H. (2002) Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277, 50131–50136 [DOI] [PubMed] [Google Scholar]
- 29. Lee M. H., Lee S. W., Lee E. J., Choi S. J., Chung S. S., Lee J. I., Cho J. M., Seol J. H., Baek S. H., Kim K. I., Chiba T., Tanaka K., Bang O. S., Chung C. H. (2006) SUMO-specific protease SUSP4 positively regulates p53 by promoting Mdm2 self-ubiquitination. Nat. Cell Biol. 8, 1424–1431 [DOI] [PubMed] [Google Scholar]
- 30. Buschmann T., Lerner D., Lee C. G., Ronai Z. (2001) The Mdm-2 amino terminus is required for Mdm2 binding and SUMO-1 conjugation by the E2 SUMO-1 conjugating enzyme Ubc9. J. Biol. Chem. 276, 40389–40395 [DOI] [PubMed] [Google Scholar]
- 31. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 32. Hollstein M., Hainaut P. (2010) Massively regulated genes. the example of Tp53. J. Pathol. 220, 164–173 [DOI] [PubMed] [Google Scholar]
- 33. Brooks C. L., Gu W. (2006) p53 ubiquitination. Mdm2 and beyond. Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toledo F., Wahl G. M. (2006) Regulating the p53 pathway. In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 35. Colmenares C., Teumer J. K., Stavnezer E. (1991) Transformation-defective v-ski induces MyoD and myogenin expression but not myotube formation. Mol. Cell. Biol. 11, 1167–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sui G., Affar el B., Shi Y., Brignone C., Wall N. R., Yin P., Donohoe M., Luke M. P., Calvo D., Grossman S. R., Shi Y. (2004) Yin Yang 1 is a negative regulator of p53. Cell 117, 859–872 [DOI] [PubMed] [Google Scholar]
- 37. Linares L. K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. (2003) HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. U.S.A. 100, 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu C., Miloslavskaya I., Demontis S., Maestro R., Galaktionov K. (2004) Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature 432, 640–645 [DOI] [PubMed] [Google Scholar]
- 39. Aylon Y., Michael D., Shmueli A., Yabuta N., Nojima H., Oren M. (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 20, 2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pomerantz J., Schreiber-Agus N., Liégeois N. J., Silverman A., Alland L., Chin L., Potes J., Chen K., Orlow I., Lee H. W., Cordon-Cardo C., DePinho R. A. (1998) The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92, 713–723 [DOI] [PubMed] [Google Scholar]
- 41. Dai M. S., Shi D., Jin Y., Sun X. X., Zhang Y., Grossman S. R., Lu H. (2006) Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J. Biol. Chem. 281, 24304–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mayo L. D., Donner D. B. (2001) A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Long J., Wang G., Matsuura I., He D., Liu F. (2004) Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3). Proc. Natl. Acad. Sci. U.S.A. 101, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 45. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 46. Zhang X. W., Yan X. J., Zhou Z. R., Yang F. F., Wu Z. Y., Sun H. B., Liang W. X., Song A. X., Lallemand-Breitenbach V., Jeanne M., Zhang Q. Y., Yang H. Y., Huang Q. H., Zhou G. B., Tong J. H., Zhang Y., Wu J. H., Hu H. Y., de Thé H., Chen S. J., Chen Z. (2010) Arsenic trioxide controls the fate of the PML-RARα oncoprotein by directly binding PML. Science 328, 240–243 [DOI] [PubMed] [Google Scholar]
- 47. Knipscheer P., Flotho A., Klug H., Olsen J. V., van Dijk W. J., Fish A., Johnson E. S., Mann M., Sixma T. K., Pichler A. (2008) Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell 31, 371–382 [DOI] [PubMed] [Google Scholar]
- 48. Tokitou F., Nomura T., Khan M. M., Kaul S. C., Wadhwa R., Yasukawa T., Kohno I., Ishii S. (1999) Viral ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion. J. Biol. Chem. 274, 4485–4488 [DOI] [PubMed] [Google Scholar]
- 49. Inoue Y., Iemura S., Natsume T., Miyazawa K., Imamura T. (2011) Suppression of p53 activity through the cooperative action of Ski and histone deacetylase SIRT1. J. Biol. Chem. 286, 6311–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dahl R., Wani B., Hayman M. J. (1998) The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene 16, 1579–1586 [DOI] [PubMed] [Google Scholar]
- 51. Akagi T. (2004) Oncogenic transformation of human cells. Shortcomings of rodent model systems. Trends Mol. Med. 10, 542–548 [DOI] [PubMed] [Google Scholar]
- 52. Shinagawa T., Nomura T., Colmenares C., Ohira M., Nakagawara A., Ishii S. (2001) Increased susceptibility to tumorigenesis of ski-deficient heterozygous mice. Oncogene 20, 8100–8108 [DOI] [PubMed] [Google Scholar]
- 53. Carter S., Vousden K. H. (2008) p53-Ubl fusions as models of ubiquitination, sumoylation, and neddylation of p53. Cell Cycle 7, 2519–2528 [DOI] [PubMed] [Google Scholar]
- 54. Chu Y., Yang X. (2011) SUMO E3 ligase activity of TRIM proteins. Oncogene 30, 1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.