Background: TWINKLE is the human mitochondrial DNA helicase associated with heritable neuromuscular diseases.
Results: TWINKLE has NTPase-dependent DNA unwinding activity and NTPase-independent DNA annealing activity. The unwinding activity is enhanced by displaced strand traps.
Conclusion: TWINKLE has more than one ssDNA-binding sites, the one associated with annealing interferes with unwinding in the absence of traps.
Significance: The annealing activity may be involved in recombination-mediated replication initiation.
Keywords: ATPases, Biophysics, DNA-binding Protein, DNA Helicase, DNA Replication, DNA-Protein Interaction, Mitochondrial DNA, Nucleic Acid Enzymology, Protein Expression, Protein Purification
Abstract
TWINKLE is a nucleus-encoded human mitochondrial (mt)DNA helicase. Point mutations in TWINKLE are associated with heritable neuromuscular diseases characterized by deletions in the mtDNA. To understand the biochemical basis of these diseases, it is important to define the roles of TWINKLE in mtDNA metabolism by studying its enzymatic activities. To this end, we purified native TWINKLE from Escherichia coli. The recombinant TWINKLE assembles into hexamers and higher oligomers, and addition of MgUTP stabilizes hexamers over higher oligomers. Probing into the DNA unwinding activity, we discovered that the efficiency of unwinding is greatly enhanced in the presence of a heterologous single strand-binding protein or a single-stranded (ss) DNA that is complementary to the unwound strand. We show that TWINKLE, although a helicase, has an antagonistic activity of annealing two complementary ssDNAs that interferes with unwinding in the absence of gp2.5 or ssDNA trap. Furthermore, only ssDNA and not double-stranded (ds)DNA competitively inhibits the annealing activity, although both DNAs bind with high affinities. This implies that dsDNA binds to a site that is distinct from the ssDNA-binding site that promotes annealing. Fluorescence anisotropy competition binding experiments suggest that TWINKLE has more than one ssDNA-binding sites, and we speculate that a surface-exposed ssDNA-specific site is involved in catalyzing DNA annealing. We propose that the strand annealing activity of TWINKLE may play a role in recombination-mediated replication initiation found in the mitochondria of mammalian brain and heart or in replication fork regression during repair of damaged DNA replication forks.
Introduction
Replication of the human mitochondrial DNA is catalyzed by a minimal replication complex that consists of the helicase (TWINKLE), DNA polymerase (polymerase γA and processivity factor γB), and single strand-binding protein (mtSSB)2 (1). TWINKLE, encoded at 10q24, was discovered in 2001 (2) and was classified as a helicase based on its sequence similarity (46% amino acid sequence similarity and 15% sequence identity) to bacteriophage T7 gene 4 primase-helicase (T7 gp4) (2, 3). It belongs to the superfamily 4 (SF4) class of helicases, which consists of bifunctional primase-helicase proteins that assemble into ring-shaped hexamers and unwind DNA by the strand exclusion model. In this model, the ring-shaped helicase encircles one of the strands of the duplex DNA and uses energy from nucleotide triphosphate (NTP) hydrolysis to translocate unidirectionally along the single-stranded (ss) DNA and unwind the duplex DNA (4). TWINKLE, similar to phage T7 gp4, contains two domains connected by a linker region. The C-terminal domain has five conserved motifs typical to the SF4 helicases (5). The N-terminal domain contains some of the conserved motifs for the primase activity, but it seems to have lost the primase function (5, 6). Deletion studies have indicated that the N-terminal domain of TWINKLE has retained specific binding activity for single strand (ss) DNA (7). Unlike T7 gp4, TWINKLE can assemble into hexamers and heptamers in the absence of any NTP (5, 8).
Since the discovery of TWINKLE, there have been more than 31 dominant mutations and 3 recessive mutations linked to mitochondrial DNA deletions that manifest in genetically inherited diseases such as progressive external ophthalmoplegia, infantile-onset spinocerebellar ataxia, premature aging, and certain types of cancer (9, 10). Most of the mutations lie in the linker region in TWINKLE, and even though some of the mutant proteins have been preliminarily characterized (3, 6, 11–13), the molecular and biochemical basis of these human diseases are not yet well defined. In addition, all the biochemical activities of TWINKLE related to mitochondrial DNA replication have not been comprehended. This is due to the lack of clear understanding of the fundamental mode of action of the helicase, which can be partially attributed to the difficulty in purifying homogeneous and stable protein.
In this paper, we report the purification of stable and near-homogeneous TWINKLE protein from Escherichia coli without contaminating cofactors. We have systematically characterized the oligomerization properties, UTP hydrolysis, DNA binding, and DNA helicase activities to demonstrate that the bacterially expressed TWINKLE protein is comparable with its insect cell expressed counterpart. Importantly, our kinetic studies of DNA unwinding have revealed that TWINKLE possesses a DNA strand annealing activity that competes with the strand separation reaction. We show that efficient dsDNA unwinding by TWINKLE is brought about by trapping of the newly unwound displaced strand. Using competition annealing experiment supported by fluorescence anisotropy competition binding study, we show that although both ssDNA and dsDNA bind TWINKLE tightly, they have distinct binding sites, and ssDNA might have multiple binding sites on TWINKLE apart from the central channel. Taken together, our findings indicate that the role of TWINKLE in mtDNA metabolism might be more complex than presently understood.
EXPERIMENTAL PROCEDURES
Expression and Purification of C-His6-TWINKLE and C-His6-K421A TWINKLE from E. coli
C-His6-TWINKLE clone in pTriEx1 was a kind gift from Prof. Johannes N. Spelbrink (Institute of Medical Technology and Tampere University Hospital, Tampere, Finland). It lacks the mitochondrial targeting sequence (42 amino acids) and contains five extra amino acids (Arg, Pro, Pro, Arg, and Pro) between TWINKLE and His6 with no Tobacco Etch Virus protease cleavage site. The protein was expressed in E. coli Rosetta (DE3) pLacI (Novagen) grown in LB at 37 °C supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) to 0.7 absorbance (A600) and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside (Fermentas) for 4 h at 37 °C. The harvested cell pellet was stored at −80 °C until further use.
The cells were resuspended in buffer A (50 mm Tris-Cl, pH 7.9, 600 mm NaCl, 0.1% Tween 20, 1 mm DTT, 1 mm EDTA, 0.1 mm PMSF, CompleteTM protease inhibitor mixture tablet (Roche Applied Science), 10% sucrose) with 1 mg/ml lysozyme and lysed by five rounds of freeze-thaw in liquid nitrogen and 42 °C water bath followed by sonication. After centrifugation at 60,000 × g for 40 min, the supernatant was treated with 65% (w/v) ammonium sulfate whose pellet was resuspended in buffer A with 20 mm imidazole. The solution was applied to nickel-Sepharose 6 Fast Flow resin (GE Healthcare) equilibrated with buffer A + 20 mm imidazole, washed successively with 25 column volumes of buffer A + 40 mm imidazole, 5 column volumes of buffer A+ 60 mm imidazole followed by elution with a linear gradient of 60 mm to 350 mm imidazole. The peak fractions (∼100–200 mm imidazole) were pooled, diluted with equal volume of buffer B (50 mm Tris-HCl, pH 7.9, 0.1% Tween 20, 10% sucrose), and loaded onto a heparin-Sepharose CL-6B resin (GE Healthcare) equilibrated with buffer B + 300 mm NaCl. After washing with 10 column volumes of buffer B + 300 mm NaCl, the protein was eluted using a linear gradient of 300 mm to 1.5 m NaCl in buffer B. Purity of the peak fractions (600–700 mm NaCl) was analyzed by SDS-PAGE and concentrated using a regenerated cellulose ultrafiltration membrane (cutoff of 30 kDa, Millipore) in an Amicon concentrator. The concentrated fractions were dialyzed against buffer C (50 mm Tris-HCl, pH 7.9, 300 mm NaCl, 50% glycerol), aliquoted, and stored at −80 °C.
DNase contamination of the sample was tested on 72-mer DNA, and nucleic acid contamination was checked by ethidium bromide staining of the sample in a 0.6% agarose gel. The concentration of the final sample was determined spectroscopically by measuring A280 using the extinction coefficient (ϵ) 75,000 m−1 cm−1. T7 gp4A′ was purified from E. coli as described previously (14, 15).
The Walker A mutant of C-His6-TWINKLE was generated by changing the lysine at position 421 to an alanine (K421A mutant) using the QuikChange site-directed mutagenesis XL kit (Stratagene). The presence of the mutation was verified by sequencing the entire gene to ensure that no other secondary mutations apart from K421A were generated.
The mutated clone was transformed into Rosetta BL21 (DE3) placI cells (Novagen). The subsequent steps of cell growth and purification were identical to the purification of wild-type TWINKLE as described above, except that the protein was eluted from the nickel column with the direct addition of 350 mm imidazole instead of a linear gradient, and the heparin-Sepharose column was replaced with a Q-sepharose column (GE Healthcare) for the mutant. The protein was present in the flow-through from the Q column, which was then concentrated using a 30,000 Da molecular weight cutoff membrane in an Amicon concentrator. The protein concentration was determined spectroscopically by measuring A280 using the extinction coefficient (ϵ) 75,000 m−1 cm−1.
Nucleic Acid Substrates
Oligodeoxynucleotides (Table 1) from Integrated DNA Technologies (Coralville, IA) were PAGE-purified, and their concentrations were determined from absorbance at 260 nm. The M13mp18 ssDNA was purchased from United States Biochemical Corp. T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer Life Sciences) were used to radiolabel the DNA, which was subsequently purified by size exclusion chromatography (Bio-Gel P-30 Gel, Bio-Rad). Duplex DNA substrates were generated by mixing complementary strands, heating to 95 °C followed by slow cooling. The 17-bp hairpin loop DNA was made by heating the ssDNA 17hp (Table 1) DNA to 95 °C followed by slow cooling.
TABLE 1.
Oligonucleotide sequences
| Oligonucleotide name | Sequence (5′ to 3′) |
|---|---|
| GC5040tr | 35Ts C TAC GTA GGA GCA TCA CAA GAC TCT CTC GTG ACT CAT CTG GGG |
| GC5040dis | CAG ATG AGT CAC GAG AGA GTC TTG TGA TGC TCC TAC GTA GTT GAA TCT CTT CCA CTA ACC AGC GC |
| M1320tr5′T | ACATGATAAGATACATGGATGAGTTTGGACAAACCACAACGTAAAACGACGGCCAGTGCC |
| M1320tr3′T | GTAAAACGACGGCCAGTGCCCAACACCAAACAGGTTTGAGTAGGTACATAGAATAGTACA |
| Blunt20tr | TATTTATTACTTATATAATA |
| Blunt 20dis | TATTATATAAGTAATAAATA |
| Blunt40tr | CCGGATATAAAACAATTATTTATTACTTATATAATAATAAATAA |
| Blunt 40dis | CCTTATTTATTATTATATAAGTAATAAATAATTGTTTTATATCC |
| Blunt58tr | CCGGATATAAAACAATTATTTATTACTTATATAATAATAAATAAATTATTAATGAATTCC |
| Blunt58dis | CCGGAATTCATTAATAATTTATTTATTATTATATAAGTAATAAATAATTGTTTTATATCC |
| 17hp | GAATTCGCCAGTGTCATGCGTTTCGCATGACACTGGCGAATTC |
| 5′-TMRtr | (TMR-C6)-TAGCGTACTAGCAGCGAAGT |
| M1355tr5′T | ACATGATAAGATACATGGATGAGTTTGGACAAACCACAACGTAAAACGACGGCCAGTGCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATC |
Chemical Cross-linking of TWINKLE
Reaction mixtures contained 50 mm triethanolamine-Cl buffer, pH 8.2, 1 mm EDTA, 5 mm dithiothreitol (DTT), 1.5 m NaCl, 5 μm protein (monomer) with or without 4 mm UTP, 10 mm magnesium acetate, 2 μm DNA (GC5040tr + GC5040dis, see Table 1). The protein was cross-linked at 4 °C for defined times using freshly prepared dimethyl suberimidate (Thermo-Scientific) (DMS, 10 mg/ml) from a 50 mg/ml stock in ice-cold triethanolamine-HCl with pH 8.2 adjusted by 3 n NaOH. The reaction was stopped by the addition of an equal volume of 1 m glycine and analyzed by 4–12% BisTris SDS-PAGE (Invitrogen). The protein bands were detected by Coomassie Brilliant Blue (R-250) staining.
DNA Binding Measured by Fluorescence Anisotropy Titration
Fluorescence anisotropy was measured (16) on a Fluoromax-4 spectrofluorometer (Horiba Scientific, NJ) mounted with Glen-Thomson calcite prism polarizers and a thermoelectrically controlled cell holder. Concentrated TWINKLE was added to 5 nm tetramethylrhodamine (TMR)-labeled ssDNA (5′-TMRtr, see Table 1) or TMR-labeled dsDNA (TMR-labeled 5′-TMRtr annealed to 5′-TMRdis, see Table 1) in a reaction buffer containing 50 mm Tris acetate, pH 7.5, 5 mm magnesium acetate, 1 mm EDTA in the absence of magnesium acetate, 5 mm DTT, 0.01% Tween 20 at 20 °C, and observed anisotropy values (robs) were recorded with excitation at 560 nm and emission at 580 nm (1-s integration time, 4× 8 nm bandpass, at least 10 trials for each point). The change in robs was plotted against TWINKLE hexamer concentrations and fitted to Equation 1 to obtain the equilibrium dissociation constant (Kd),
 |
where rf and rb are the TMR anisotropy on free and protein-bound DNA, respectively
For the competition binding experiment, 6 nm TMR-labeled ds20 complexed with 7 nm hexameric TWINKLE was continuously titrated with nonfluorescent ssDNA (5′-TMRtr). Change of TMR anisotropy was recorded to monitor the replacement until no significant changes in anisotropy by increasing ssDNA concentrations was observed. The fluorescence anisotropy decrease was fit to Equations 2 and 3 to obtain the ssDNA IC50 and Ki,
where robs is the anisotropy value observed at individual ssDNA concentrations; X is the ssDNA concentration, rmin and rmax are the minimum and maximum anisotropy values, respectively; n is the Hill coefficient; IC50 is the ss-DNA concentration, which can replace 50% of dsDNA from protein based on fitting, and x is ssDNA concentration,
where L is the dsDNA concentration; Ki is the dissociation constant for ssDNA, and Kd is the dissociation constant for dsDNA.
NTP Hydrolysis
2 μCi of radioactive UTP was added to reaction mixtures containing 50 mm Tris acetate, pH 7.5, 1 mm edta, twinkle hexamer (100 nm), and indicated amounts of DNA (ss, ds, or M13) and UTP mixed with magnesium acetate (6 mm excess magnesium acetate than total concentrations of EDTA and UTP). Approximately 40 mm NaCl was present from the protein added to the reaction mixture. Reactions were quenched after defined times with 4 n formic acid and analyzed on polyethyleneimine-cellulose TLC (GE Healthcare) developed in 0.4 m potassium phosphate, pH 3.4. Unreacted NTP and product Pi were quantified using a PhosphorImager and ImageQuant (GE Healthcare). The [Pi], after normalizing for the [Pi] at time 0, was plotted against reaction time and fit to a linear equation to obtain the velocities from the slopes. The velocities were plotted against [UTP] to obtain the Km and Vmax from fit to the Michaelis-Menten steady-state equation.
Helicase Activity
Reaction mixtures contained 0.4 nm DNA substrate, 25 nm TWINKLE (hexamer), reaction buffer (50 mm Tris acetate, pH 7.5, 0.01% Tween 20, 5 mm dithiothreitol, 1 mm EDTA), indicated amounts of various NTP and magnesium acetate, as well as 40 nm ssDNA trap or 2 μm T7 gp2.5 where indicated. The reactions were incubated at 30 °C for indicated times and stopped with 12 reaction volumes of quenching solution (100 mm EDTA, pH 8.0, 1% SDS, 0.5% bromphenol blue). The amount of dsDNA and unwound ssDNA was quantified from 15% nondenaturing polyacrylamide/TBE gel using PhosphorImager and ImageQuant (GE Healthcare) and fitted using Sigma Plot software. The fraction of ssDNA generated was calculated from Equation 4,
 |
where F is the fraction of unwound substrate, DS and SS are intensities of duplex and unwound substrate bands at a given time, respectively, and DS0 and SS0 are intensities of duplex and unwound substrates at time 0, respectively. The fraction of ssDNA against time was fit to the exponential Equation 5,
where A is the maximum fraction of ssDNA that can be generated enzymatically from the substrates; b is the first-order rate constant of DNA unwinding; t is reaction time. If the reactions did not reach completion in the time scale of measurement, the data were fit to a straight line (Equation 6),
The S.E. were obtained from data fitting in the Sigma Plot software, and the initial rate was determined by multiplying the amplitude with the corresponding rate, and the S.E. for the initial rate was calculated by combining S.E. of both the rate and the amplitude using Equation 7.
 |
DNA Annealing
The DNA annealing reaction was carried out at 30 °C using the indicated amounts of ssDNA and TWINKLE for defined times in reaction buffer containing 50 mm Tris acetate, pH 7.5, 0.01% Tween 20, 5 mm dithiothreitol, 1 mm EDTA with or without 4 mm UTP, and 6 mm free magnesium acetate. Samples were immediately loaded on a nondenaturing polyacrylamide/TBE gel running at 60 V, after quenching with 12× reaction volume of the quenching solution. The amount of annealed DNA and ssDNA was quantified using PhosphorImager and ImageQuant (GE Healthcare) and fit to Equation 4 (by replacing SS0 with DS0 and vice versa) using Sigma Plot software to obtain the rate constant and amplitude of annealing.
Annealing with prebound hairpin DNA (17hp), 20 base dsDNA (5′-TMRtr + 5′-TMRdis without the TAMRA label), or dT100 ssDNA was performed by incubating increasing concentrations of the above DNAs with 10 nm TWINKLE (hexamer) for 15 min at 30 °C followed by addition of 2 nm of the two ssDNAs sequentially complementary to each other with 4 mm UTP and 6 mm free magnesium acetate. Reactions were done at 30 °C for 2 min. Samples were immediately loaded on a 4–20% nondenaturing polyacrylamide gel running at 60 V, after quenching with 5× reaction volumes of the quenching solution.
All protein concentrations mentioned signify hexameric concentration. For the unwinding and annealing assays, the protein was preincubated with DNA in the presence of UTP, and the reaction was started with magnesium acetate. Re-annealing traps for unwinding assay (where mentioned) were added with magnesium acetate. For UTPase assay, the reactions were started by addition of the protein.
RESULTS
Expression and Purification of C-His6-TWINKLE from E. coli
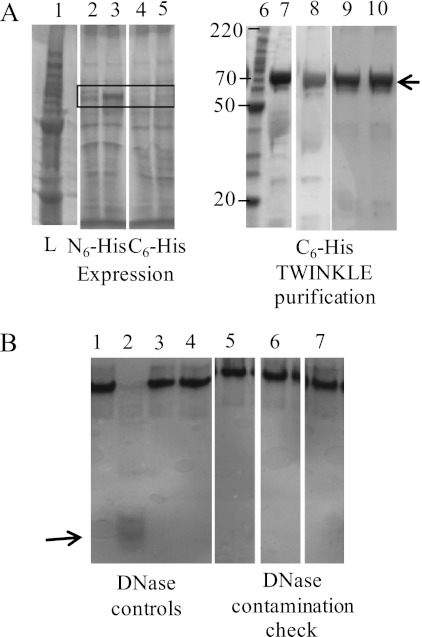
The expression of TWINKLE in E. coli allowed for easy and large scale purification of the homogeneous protein. We estimate that our purification protocol yields 20–30 mg of pure C-His6-TWINKLE per 100 g of cells, which is comparable with the yield using the insect cell system (5). The C-His6-TWINKLE was expressed at low levels in E. coli as visualized after IPTG induction (Fig. 1A, boxed area, lane 5) when compared with the significant amount of expression of the N-His6-TWINKLE (Fig. 1A, boxed area, lane 3). Curiously, the N-His6-TWINKLE protein was insoluble under varied induction temperature conditions, whereas the C-His6-TWINKLE was soluble. TWINKLE resolved by SDS-PAGE was identified by proteolytic digestion followed by nano-LC MS/MS. Using mass spectrometry, we identified a closely associated band as GroEL eluting with TWINKLE from the Ni2+-Sepharose 6 Fast Flow column. A substantial amount of TWINKLE was therefore sacrificed to obtain pure protein with a minimum amount of GroEL (Fig. 1A, lane 7). After chromatography through the heparin-Sepharose column, the protein was ∼90% pure (Fig. 1A, lanes 8 and 9). We observed that 300 mm NaCl and DTT were required for protein solubility and stability, which was further enhanced in the presence of 50% glycerol (Fig. 1A, lane 10). There was no observed DNase contamination in the final sample (Fig. 1B, lanes 5–7). A recent study has reported the purification of TWINKLE from E. coli using a clone in pET21a vector with C-His6 tag preceded by AAALE after the gene sequence. Although the clone was similar to the one we used in this study (pTriEx vector with C-His6 tag preceded by RPPRP after the gene sequence), the purification protocol required the use of Mg2+ and ATP through all the steps to keep TWINKLE soluble (17). However, our protocol provides soluble TWINKLE without added Mg2+ and ATP, and hence the final protein sample is free of MgATP, which allows in vitro studies related to the effect of these cofactors on the biochemical properties of TWINKLE.
FIGURE 1.
A typical profile for the purification of C-His6-TWINKLE. A, representative 4–20% SDS-PAGE shows the purification of C-terminally His-tagged TWINKLE by Ni2+-Sepharose chromatography and heparin-Sepharose chromatography followed by dialysis in storage buffer C (see under “Experimental Procedures”). The purification protocol has been reproduced successfully 11 times. Lane 1, molecular weight ladder (Invitrogen). Lane 2, Pre-IPTG-induced N-His6-TWINKLE lysate. Lane 3, post-IPTG-induced N-His6-TWINKLE lysate. Lane 4, pre-IPTG-induced C-His6-TWINKLE lysate. Lane 5, post- IPTG-induced C-His6-TWINKLE lysate. Lane 6, molecular weight ladder (Invitrogen). Lane 7, Ni2+-Sepharose affinity column eluted fraction. Lane 8, heparin-Sepharose column eluted fraction. Lane 9, heparin-Sepharose column pure fractions combined. Lane 10, sample from lane 9 dialyzed against storage buffer C. The box in lanes 2–5 highlights the induction of TWINKLE by IPTG. The arrow indicates the position of the recombinant protein for lanes 7–10 (75 kDa). B, 20% polyacrylamide/TBE gel confirms the absence of DNase contamination in the purified protein tested from 15 to 60 min at 30 °C on a 72-base DNA. Lane 1, exonuclease III + EDTA. Lane 2, exonuclease III + magnesium acetate. Lane 3, no TWINKLE + EDTA. Lane 4, no TWINKLE + magnesium acetate. Lane 5, reaction time of 15 min. Lane 6, reaction time of 30 min. Lane 7, reaction time of 60 min. Lanes 1 and 2 are the positive controls; lanes 3 and 4 are the negative controls; lanes 5–7 are reactions with TWINKLE, DNA, and magnesium acetate. The arrow indicates the smear for the digested DNA in the presence of ExoIII and Mg2+ in lane 2 (positive control).
E. coli Purified TWINKLE Forms Oligomers Spontaneously but Specifically Forms Hexamers in the Presence of MgUTP
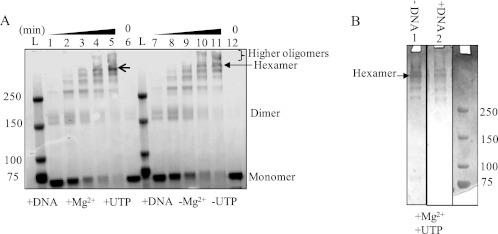
To characterize the oligomeric state of the recombinant TWINKLE purified from E. coli, we performed DMS cross-linking of the protein in the presence and absence of cofactors (Mg2+ and UTP). DMS is a homobifunctional cross-linking reagent that can cross-link surface lysines within 11 Å of each other (18). In the absence of the cross-linking reagent, we observed some dimers (Fig. 2A, lanes 6 and 12) consistent with a previous study where ATPγS was used (8). In the presence of the cross-linking reagent DMS, but in the absence of Mg2+ and UTP, we were able to capture TWINKLE oligomers (Fig. 2A, lanes 7–11). However, when Mg2+ and UTP were present, hexamers accumulated to a greater level as opposed to the higher oligomers (Fig. 2A, lanes 1–5). Our cross-linking results show that Mg2+ and UTP stabilize the hexameric state of TWINKLE. Although dsDNA was present in both experiments, we show that in the presence and absence of the DNA TWINKLE possesses similar oligomeric states (Fig. 2B, lanes 1 and 2). Thus, similar to the protein purified from the insect cells (8), E. coli expressed TWINKLE forms hexamers and higher oligomers in the presence and absence of MgUTP. However, we show that hexamers are stabilized selectively in the presence of MgUTP.
FIGURE 2.
Oligomeric state of TWINKLE and effect of cofactors and DNA. A, Coomassie-stained 4–12% BisTris SDS-polyacrylamide gel showing TWINKLE cross-linking with DMS at 4 °C for 1 min (lanes 1 and 7), 8 min (lanes 2 and 8), 15 min (lanes 3 and 9), 30 min (lanes 4 and 10), and 60 min (lanes 5 and 11) in the presence (lanes 1–5) or absence (lanes 7–11) of MgUTP and in the presence of DNA. Lanes 6 and 12 are noncross-linked controls showing both monomers and very small amount of dimers. Lanes marked L are the molecular weight standards. B, Coomassie-stained 4–12% BisTris SDS-PAGE showing the DMS cross-linking of TWINKLE at 4 °C for 60 min in the presence of MgUTP in the presence and absence of DNA.
TWINKLE Binds ssDNA and dsDNA Fragments in the Absence of Cofactors
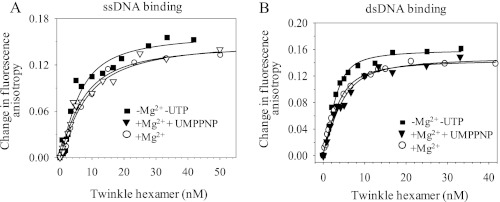
The DNA binding properties of TWINKLE were explored using fluorescence anisotropy titrations, where TMR-labeled ssDNA or dsDNA (attached to the 5′-end) was titrated with increasing TWINKLE concentrations. The titration data clearly show that TWINKLE binds both ssDNA and dsDNA with or without Mg2+ or nucleotides (UTP) (Fig. 3, A and B). The equilibrium measurement provided the Kd of TWINKLE complexed with ssDNA as 3.2 ± 2.8 nm without cofactors, 5.4 ± 2.4 nm in the presence of Mg2+, and 4.4 ± 2.6 nm in the presence of Mg2+ and an unhydrolyzable analog of UTP, UMPPNP (Fig. 3A). TWINKLE binds dsDNA about 2-fold more tightly (Kd is 1.2 ± 0.3 nm without cofactors, 2.1 ± 0.4 nm with Mg2+, and 2.6 ± 1.4 nm with Mg-UMPPNP) (Fig. 3B). This is unlike T7 gp4 helicase, which binds preferentially to ssDNA over dsDNA, and moreover it has a strict requirement of dTTP or dTTP analog to bind ssDNA (19, 20). Thus, our results are consistent with previous reports that indicate that TWINKLE binds to ssDNA (8) and dsDNA (21) without MgUTP, and our equilibrium Kd measurements show that TWINKLE binds to these DNAs very tightly with a preference for binding dsDNA.
FIGURE 3.
Fluorescence anisotropy-based binding of ssDNA and dsDNA to TWINKLE. Changes in anisotropy of TMR-labeled ssDNA (A) and dsDNA (B) substrates in response to the addition of increasing amounts of TWINKLE as described under “Experimental Procedures.” Changes in anisotropy in the presence of Mg2+ (open circles), MgUMPPNP (open triangles) and in the absence of cofactors (filled squares) are shown.
TWINKLE Unwinds dsDNA Using Various NTP Substrates
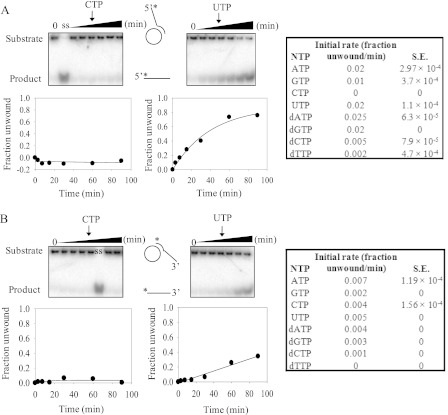
The helicase activity of TWINKLE was measured by monitoring the unwinding of a short radiolabeled ssDNA annealed to M13 ssDNA. Two 60-nucleotide short ssDNAs (M1320tr5′T or M1320tr3′T, see Table 1) were so prepared that they formed a 20-bp duplex region on the M13 ssDNA, whereas the remaining 40-nucleotide region formed either a 5′-tail or a 3′-tail, referred to as the 5′-tailed substrate or the 3′-tailed substrate, respectively. The 20-bp dsDNA length was chosen based on the report that TWINKLE can unwind only short stretches of dsDNA (1). The unwinding reactions were carried out by preincubating 25 nm TWINKLE with the unwinding substrate (0.4 nm) in the presence of various NTPs (4 mm) and initiating the reactions with Mg2+. Control experiments in the absence of TWINKLE or NTP showed no unwinding (supplemental Fig. 1, A and B), indicating that the 20-bp duplex is not spontaneously unwound under our assay conditions and that unwinding is contingent upon NTP hydrolysis-dependent activity of TWINKLE. Additionally, the Walker A mutant K421A of TWINKLE failed to unwind the same DNA with 5′-tail even at higher hexamer concentrations (supplemental Fig. 1C). This demonstrates that the observed NTP-dependent DNA unwinding is due to the activity of TWINKLE and not any other contaminating helicases in the protein sample. Comparison of the initial rates of unwinding (Fig. 4A) of the 5′-tailed substrate revealed most efficient unwinding in the presence of dATP, with slightly lower but comparable rates in the presence of UTP, ATP, and dGTP. The initial rate of unwinding supported by GTP was half of UTP, ATP, and dGTP, whereas dCTP, CTP, and dTTP supported very little unwinding (Fig. 4A and supplemental Fig. 2).
FIGURE 4.
Unwinding kinetics of the 5′- and 3′-tail DNA annealed to M13 ssDNA in the presence of different NTP substrates. The representative 15% acrylamide/TBE gels show the unwinding kinetics of the 5′-tail substrate (A) and 3′-tail substrate (B) at 30 °C in the presence of 25 nm TWINKLE and 4 mm CTP (left panel) and UTP (right panel). Lanes marked ss are the substrate-heated at 95 °C before loading; lanes marked 0 represent time 0. The other lanes in each gel correspond to unwinding reaction times of 3, 7.5, 15, 30, 60, and 90 min. For each NTP, the fraction of DNA unwound at each time point is normalized to the corresponding fraction unwound at time 0, plotted against reaction time, and fitted to Equation 4 for all NTPs except the ones where unwinding was not completed even at 90 min; hence they were fitted to linear Equation 5.
In contrast to the unwinding of the 5′-tailed substrate, the unwinding of the 3′-tailed substrate was not completed and was 3–6 times slower under the same conditions (Fig. 4B). Nonetheless, it was clear that UTP and ATP supported more efficient unwinding and dTTP or CTP did not support any unwinding (Fig. 4B and supplemental Fig. 3). The significantly lower fraction of DNA unwound with the 3′-tailed substrate compared with the 5′-tailed substrate has been recently reported (22). This suggests that TWINKLE favorably binds to the end of the oligonucleotide with the 5′-tail than to the circular ss region of the M13 ssDNA, probably because the free end of the 5′-tail allows threading of the hexameric helicase, as opposed to the circular M13 ssDNA region where the hexamer has to open and assemble around the DNA. This is a strikingly different behavior from T7 gp4 helicase, which preferentially loads on to the long ss region of the M13 ssDNA and therefore unwinds the 3′-tailed substrate more efficiently than the 5′-tailed substrate (22, 23).
We measured the UTP hydrolysis Vmax and Km values under similar conditions of unwinding both in the presence and absence of various types of DNA and at varied UTP concentrations. UTP hydrolysis was only marginally stimulated by 25 nm M13mp18 ssDNA or by 1 μm ds fork DNA over no DNA (Table 2). The Km value of UTP in the absence of DNA was 3.4 mm, which is ∼2 times higher than in the presence of M13 ssDNA (1.7 mm). The Km value for ATP in the presence of activated calf thymus DNA has been reported to be 1.4 mm (17), which is similar to what we found for UTP with all DNAs.
TABLE 2.
Determination of Km and Vmax values of TWINKLE with UTP using different DNA substrates
Intrinsic and DNA-stimulated UTP hydrolysis in the presence of 5 nm M13 ssDNA or 200 nm of 78 bases of ssDNA or 78 base-paired dsDNA was measured at varying UTP concentrations (50, 100, 250, and 500 μm and 1, 2, and 4 mm). For determining Km and Vmax values for UTP with each DNA, UTP concentration was plotted against velocity of hydrolysis and data fit to the hyperbolic equation representing Michaelis-Menten steady state.
| DNA | Km | Vmax |
|---|---|---|
| mm | μm/min | |
| ssM13mp18 | 1.7 | 15.6 |
| ssDNA | 1.6 | 12.8 |
| Forked dsDNA | 1.4 | 11.2 |
| No DNA | 3.4 | 16.8 |
Unwinding Activity of TWINKLE Is Facilitated by Trapping of the Displaced Strand
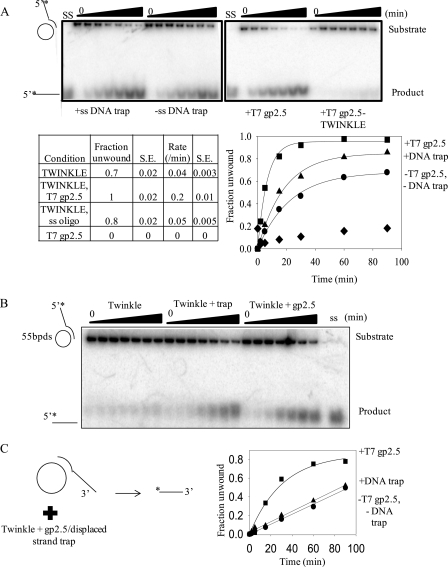
Further analysis of the DNA helicase activity of TWINKLE revealed that the initial unwinding rate of the 5′-tailed substrate was 7-fold faster when we added T7 gp2.5 single strand-binding protein during the addition of Mg2+ (Fig. 5A). It has been shown that mtSSB stimulates unwinding by TWINKLE by almost 8-fold in a helicase assay similar to the M13-based unwinding assay that we have used here (24). Because the heterologous T7 gp2.5 stimulates the helicase activity of TWINKLE, it appears that the stimulation by single strand-binding proteins is not protein-specific. T7 gp2.5 by itself does not unwind dsDNA in the absence of TWINKLE (Fig. 5A). This is unlike E. coli SSB, human replication protein A, and T7 gp32 single strand-binding proteins, which are in many ways similar to mtSSB. These SSB proteins have been shown to capture thermally melted bases to effectively unwind dsDNA (25). One mechanism by which the single strand-binding proteins can increase the helicase activity is by coating the M13mp18 single strand and preventing TWINKLE from getting trapped by the extensive ssDNA region. In this manner, the SSBs can have a nonspecific role in increasing the free TWINKLE concentration for its effective encounter with the 5′-tail of the ssDNA annealed to the circular M13 ssDNA. In addition, the SSB protein can increase the efficiency of DNA unwinding by preventing re-annealing of the newly unwound displaced strand. Consistent with the latter role, we observed increased unwinding when unlabeled M1320tr5′T strand was added at the start of the reaction with the Mg2+(Fig. 5A, left-most panel). The unlabeled M1320tr5′T can anneal to M13 ssDNA and hence acts as a displaced strand trap to prevent re-annealing of the radiolabeled M1320tr5′T back to the M13 ssDNA circle. The displaced strand trap and gp2.5 were essential for unwinding longer DNA tracts of 55 bp by TWINKLE (Fig. 5B). Similarly, TWINKLE could unwind the 3′-tailed M13 partial duplex more efficiently in the presence of gp2.5 and the displaced strand trap than in the absence (Fig. 5C).
FIGURE 5.
Effect of single strand-binding protein and the reannealing DNA trap on the unwinding kinetics by TWINKLE. A, 5′-tailed substrate was unwound in the presence of 25 nm TWINKLE and 4 mm UTP in the presence of 40 nm unlabeled ssDNA displaced strand trap or 2 μm T7 gp2.5. The rates of unwinding and fraction of unwound ssDNA product with the corresponding S.E. under four different conditions tested in the unwinding assay are summarized in the table. A plot of fraction of DNA unwound over time in the presence of T7 gp2.5 trap (square) and ssDNA trap (triangle), in the absence of trap (circle), and in the presence of T7 gp2.5 trap but absence of TWINKLE (diamond) is also shown. The reaction times are 2, 5, 15, 30, 60, and 90 min. B, unwinding of the longer 55-bp M13ds DNA is better in the presence of DNA trap and gp2.5 than in their absence. The unwinding reaction was carried out under the same conditions as the 20-bp M13ds DNA except that the trap DNA is the 40T55-bp in place of the 40T20-bp DNA. C, plot of fraction of 3′-tailed substrate DNA unwound over time in the presence of 2 μm T7 gp2.5 trap (square), 40 nm ssDNA displaced strand trap (triangle), and in the absence of trap (circle).
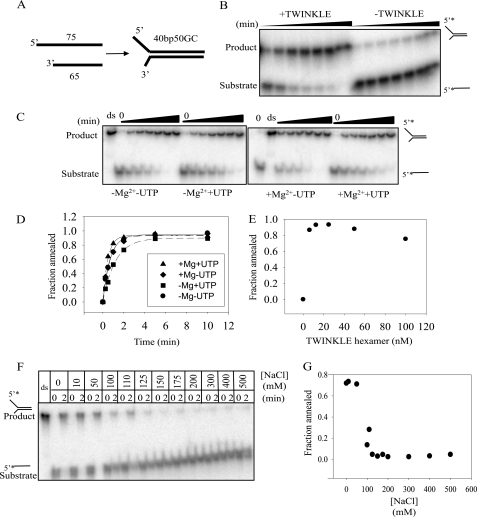
TWINKLE Possesses DNA Strand Annealing Activity
The finding that unwinding is greatly aided by trapping of the newly unwound displaced strand prompted us to test if TWINKLE has the ability to promote annealing of complementary ssDNAs. We tested the annealing of two ssDNA oligonucleotides (GC5040tr and GC5040dis, Table 1) capable of forming a 40-bp duplex region in a fork in the presence and absence of TWINKLE (Fig. 6A). The two ssDNAs at 2 nm concentration formed a duplex within 2 min in the presence of TWINKLE, whereas in the absence of TWINKLE, there was negligible annealing observed (Fig. 6B). T7 gp4 helicase under similar conditions did not exhibit any annealing activity of the partially complementary (GC5040tr and GC5040dis, Table 1) or of the completely complementary DNAs (Blunt 20 or 58tr and to Blunt 20 58dis, see Table 1) (supplemental Fig. 4).
FIGURE 6.
DNA strand annealing activity of TWINKLE. A, schematic representation of two partially complementary ssDNAs annealing to form a 40-bp forked DNA. B, annealing assay was performed by mixing 2 nm ssDNA (GC5040tr + GC5040dis, see Table 1) at 30 °C in the presence and in the absence of 50 nm TWINKLE and 4 mm MgUTP for 1, 5, 10, 15, and 30 min. C, 18% acrylamide/TBE gels show the effect of Mg2+ and UTP on the annealing of the 40-bp forked DNA at 30 °C in the presence of 10 nm TWINKLE and 2 nm DNA for 0,15, 30, 60, 120, 300, and 600 s. D, amount of annealed product formed is normalized to the amount of product at time 0 and plotted against time and fit to Equation 2 to get the rate and amplitude. The initial annealing rate (fraction annealed/min) in the absence of MgUTP is 0.02 ± 0.002, in the presence of magnesium acetate is 0.03 ± 0.004, in the presence of UTP is 0.01 ± 0.001, and in the presence of MgUTP is 0.04 ± 0.003. E, fraction of annealed product in 2 min at varying TWINKLE hexamer concentrations (6, 12.5, 25, 50, and 100 nm). F, 12% acrylamide/TBE gel shows annealing of 2 nm DNA by 10 nm TWINKLE at 30 °C in 2 min at varying NaCl concentrations (0, 10, 50, 100, 110, 125, 150, 175, 200, 300, 400, and 500 mm). The prequenched controls for each salt concentration are shown in the lanes marked 0 under each salt concentration, and the 2-min reaction for each salt concentration is marked as 2. G, amount of annealed product formed is normalized to the product formed at respective 0 and plotted against salt concentration.
To better understand the mechanism of TWINKLE-mediated DNA strand annealing, we first tested the effect of UTP and Mg2+ on its strand annealing activity. TWINKLE-mediated DNA annealing was observed under all conditions, and the yields of the annealed products or the amplitudes of the reaction were comparable, although the initial rate of annealing was almost two times faster in the presence of UTP and Mg2+ than in their absence (Fig. 6, C and D). TWINKLE could anneal the two DNA strands even when UTP was replaced by its nonhydrolysable analog UMPPNP (supplemental Fig. 5).
We also tested the effect of TWINKLE concentration and NaCl concentration on the strand annealing activity. The annealing rate did not show a significant change from 6 to 100 nm TWINKLE hexamer concentration (Fig. 6E). However, the annealing reaction was sensitive to NaCl concentration and showed a drastic decrease in a narrow range between 50 and 125 mm NaCl concentration, and very little annealing was observed beyond 150 mm NaCl (Fig. 6, F and G). The salt dependence is consistent with an active role of TWINKLE in DNA annealing. We expect spontaneous DNA annealing to show an opposite behavior, i.e. base-pairing is more favorable at high concentrations of NaCl as base-stacking interactions are strengthened with an increase in salt concentration (26). It is known that TWINKLE-DNA interactions are salt-sensitive and are lost at 420 mm NaCl (17). The results indicate a role of the DNA-binding site of TWINKLE in the observed DNA strand annealing activity.
We next assessed the dependence of annealing rate and amplitude on the length of the DNA to be annealed. We used 0.2 nm complementary ssDNAs of 20, 40, and 58 bases (Blunt 20, 40, or 58tr and Blunt 20, 40, or 58dis). The short 20-base DNA did not show a clear annealed product but instead a smeary band moving up with increasing reaction times (supplemental Fig. 6). However, both the 40- and 58-base DNAs efficiently annealed in the presence of TWINKLE with similar amplitudes, comparable with the forked DNA (supplemental Fig. 6). Our fluorescence anisotropy-based binding assays above show that a 20-base ssDNA binds tightly to TWINKLE. Therefore, the lower efficiency of the short DNA annealing could be because the DNA-binding site of TWINKLE protects the entire 20-base DNA, and thus most included bases within the binding site of the TWINKLE are not available for base-pairing. The longer 40- or 58-base ssDNAs must have exposed bases that can pair with the complementary ssDNA easily. Interestingly, if T7 gp2.5 was prebound to one of the two strands, strand annealing by TWINKLE was inhibited as expected. The partial inhibition could be because TWINKLE may be able to dislodge T7 gp2.5 (supplemental Fig. 7).
TWINKLE-mediated Strand Annealing Is Inhibited by ssDNA but Not by dsDNA
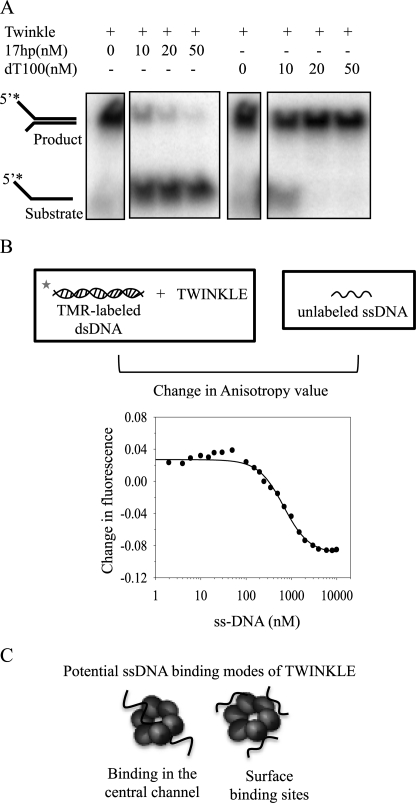
Our DNA binding studies (Fig. 3) showed that ssDNA and dsDNA bind TWINKLE with comparable Kd values. To determine the role of the ssDNA- and dsDNA-binding sites of TWINKLE in strand annealing, we carried out annealing reactions in the presence of increasing amounts of ssDNA or dsDNA competitors (10–50 nm), which were preincubated with TWINKLE before the addition of the annealing substrates and MgUTP. Preincubation of TWINKLE with increasing amounts of ssDNA competitor inhibited the TWINKLE-mediated annealing reaction (Fig. 7A). Interestingly, preincubation of TWINKLE with increasing amounts of the dsDNA competitor, which binds very tightly as shown in Fig. 3A, had no effect on the annealing reaction (Fig. 7A). The distinct effects of ssDNA and dsDNA on the annealing activity of TWINKLE suggest more than one DNA-binding sites on TWINKLE with distinct affinities for ssDNA and dsDNA, and only the ssDNA-specific site appears to promote the strand annealing reaction.
FIGURE 7.
Competition between dsDNA and ssDNA in binding and in TWINKLE-catalyzed strand annealing reaction. A, 12% acrylamide/TBE gels show the annealing of ssDNA to form a 40-bp forked dsDNA in a reaction at 30 °C in the presence of 10 nm TWINKLE, 2 nm DNA, 4 mm UTP, and 6 mm free magnesium acetate conducted for 2 min. In these reactions, TWINKLE was preincubated with increasing concentrations of competitors such as ssDNA (dT100) or dsDNA (17-bp hairpin) for 15 min before adding the ssDNAs to be annealed. B, complex of 6 nm TMR-labeled RD-ds20 and 7 nm TWINKLE was titrated with increasing ssDNAs (5′-TMRtr). The fluorescence anisotropy decrease was fit to Equations 2 and 3 to obtain the ssDNA IC50 of 672 nm and Ki of 174 nm. C, model showing potential ssDNA-binding modes by TWINKLE.
It is proposed that the homologous T7 gp4 hexamer binds ssDNA and dsDNA in its central channel (27), but the N-terminal primase domains are also capable of binding ssDNA (4). It is very likely that TWINKLE similar to T7 gp4 binds ssDNA and dsDNA in the central channel. In addition, as suggested by a previous report (3), the N-terminal domains of TWINKLE may specifically bind ssDNA, and these surface-exposed site(s) may be involved in catalyzing the DNA annealing reaction.
To directly determine whether ssDNA has more than one binding site on TWINKLE, we compared the Kd value of ssDNA (Fig. 3A) on TWINKLE and the Ki value of ssDNA against prebound 20-bp dsDNA on TWINKLE using fluorescence anisotropy-based competition assay. We incubated 7 nm TWINKLE with 6 nm TMR-labeled dsDNA (5′-TMRtr + 5′-TMRdis) and titrated the complex with increasing amounts of unlabeled ssDNA (5′-TMRtr) until no significant change of the TMR anisotropy could be observed. Addition of small amounts of ssDNA led to an increase in anisotropy at first and then a decrease with addition of higher amounts of ssDNA (Fig. 7B). The small increase in anisotropy could be due to ssDNA binding to TWINKLE. The decrease in anisotropy due to unbinding of dsDNA was fit to a competitive binding model, which provided ssDNA Ki of ∼174 nm. This Ki value is much larger than the measured ssDNA Kd of 3–5 nm (Fig. 3A). This result supports two types of ssDNA-binding sites on TWINKLE. We propose that TWINKLE can bind ssDNA both in the central channel and to a surface-exposed site(s) (Fig. 7C).
TWINKLE Actively Recruits ssDNA Trap for Complementary Base-pairing with the Displaced Strand during Unwinding
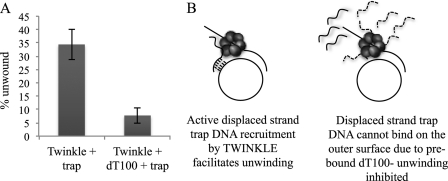
The surface-exposed sites proposed to be involved in the strand annealing reaction are likely to be involved in binding the displaced strand trap during DNA unwinding. If the trap is prebound to TWINKLE, it can effectively base pair with the newly unwound DNA to stimulate the helicase activity of TWINKLE. To test this hypothesis, we designed the following experiments: DNA unwinding of the 55-bp DNA was measured in the presence of the displaced strand trap and in the presence of a noncomplementary dT100 homopolymer together with the displaced strand trap. TWINKLE was mixed with the M13-based forked DNA substrate prior to addition of the DNA traps. We observed that unwinding was efficient in the presence of the displaced strand trap (Fig. 8A) consistent with the above results shown in Fig. 5. Interestingly, unwinding was poor when dT100 was present with the displaced strand trap (Fig. 8A).
FIGURE 8.
TWINKLE actively recruits ssDNA trap for complementary base-pairing with the displaced strand during unwinding. A, unwinding by TWINKLE was measured in the presence of trap with and without prebound dT100. 25 nm TWINKLE hexamer was prebound to 0.4 nm M1355bpds substrate in the presence of 4 mm UTP before addition of 100 nm dT100. The unwinding reaction was initiated with 6 mm free magnesium and 40 nm displaced strand trap and was quenched after 60 min. The percentage unwound was calculated as before and plotted on a bar chart. Mean percentage unwinding ± S.E., n = 3, Twinkle + Trap = 34.4 ± 5.6, and Twinkle + Trap + dT100 = 7.7 ± 2.9. B, model for displaced strand trap recruitment by TWINKLE to facilitate unwinding.
We propose that dT100 (present in excess of the displaced strand trap) binds to the surface-exposed ssDNA-binding sites on TWINKLE and prevents the displaced strand trap from binding (Fig. 8B). Thus, the only way the displaced strand trap can base pair with the newly unwound DNA is after binding from solution, which is an inefficient process at the low concentration of displaced strand trap used in our studies (40 nm). Thus, the results are consistent with the model that TWINKLE binds ssDNA on a surface-exposed site(s), and if the ssDNA happens to be complementary to the unwound DNA, then it stimulates DNA unwinding by trapping the newly unwound DNA strand.
DISCUSSION
In this paper, we report a purification protocol to obtain soluble and stable TWINKLE from E. coli without any co-factor contaminants, and we demonstrate for the first time the DNA strand annealing activity of TWINKLE. Our results also suggest that TWINKLE has distinct duplex and ssDNA-binding sites, and ssDNA can bind to multiple sites outside of the central channel.
A recently reported purification protocol for bacterially expressed TWINKLE used Mg2+ and ATP in the protein preparation buffers to keep the protein soluble, and these cofactors remained through the final steps (17). We show here that TWINKLE can be obtained in soluble form without any cofactor contaminants, which will be important for future structural studies as well as to study the effect of cofactors on the properties of the protein. We observed, as with the insect cell purified TWINKLE, the E. coli expressed protein is more stable at a high salt concentration (>300 mm NaCl). The oligomerization properties and the NTP dependence of the unwinding activity of our protein demonstrate that irrespective of the source of protein, the fundamental biochemical properties of TWINKLE are the same.
In our protein-protein cross-linking study, oligomers were only identifiable at a high salt concentration (1.5 m) possibly because protein aggregation at the lower salt concentrations leads to the formation of multimers that cannot be resolved on a polyacrylamide gel. The protein-protein cross-linking studies revealed that even though TWINKLE forms both hexamers and heptamers, the hexamers are enriched in the presence of MgUTP. The existence of both hexameric and heptameric structures is known among ring-shaped helicases such as the T7 gp4 and Methanothermobacter thermoautotrophicus mini-chromosome maintenance with the existence of double-hexamer or double-heptamer in case of the latter (28, 29). In T7 gp4, heptamers are predominantly present with dTDP, and these species apparently do not bind DNA (29). It has been postulated that the helicase ring opens after the DNA contacts the N-terminal primase domain, and during this process one of the subunits of the heptamer is lost (29–31). Perhaps, in the presence of MgUTP, the preformed TWINKLE heptamer loses one of its subunits to form an open hexameric ring that encircles the DNA.
Although TWINKLE has been demonstrated to unwind short DNA duplexes with a directionality of 5′ to 3′ on the strand it translocates, its helicase activity has not been characterized in detail. In this study, we designed our helicase assays based on the unwinding properties of the T7 gp4, where it is known that on an M13 ssDNA-based assay, the helicase assembles on the M13 ssDNA and moves in the 5′ to 3′ direction to unwind the short duplex with a 3′-tail (23, 32). We, however, observed that TWINKLE does not unwind the 3′-tail DNA substrate very efficiently; instead it unwinds the 5′-tail substrate efficiently. This result indicates that TWINKLE can load more easily on the 5′-tail that has a free end than on the circular ssDNA. This raises an obvious question as to how TWINKLE loads on DNA in the cell, where linear 5′-tail-like structures are not common. Most of the known ring-shaped helicases have an assigned loader that helps them to assemble around the DNA (33). Because TWINKLE can form a ring in the absence of cofactors or DNA, it might need a loader to open the ring and load on circular DNA. Thus, lack of a loader in vitro limits loading of TWINKLE on DNA, leading to slow unwinding rate by TWINKLE as compared with T7 gp4. When this manuscript was in preparation, a study on TWINKLE demonstrated that TWINKLE can load on a closed circular ssDNA molecule to initiate unwinding without the requirement of a specific loader (22). In concurrence with our results, the same study showed that unwinding efficiency with the M13-DNA-based assay was more with the 5′-tail substrate than with the 3′-tail substrate under all the conditions tested. Taken together, this indicates that although TWINKLE can load on a circular ssDNA unaided by a separate loader, the loading efficiency might be significantly higher on an ss tail having a free end than on an ss closed circle. Therefore, we cannot exclude the possibility of the requirement of a loader by TWINKLE during initiation of replication in vivo.
Additionally, we observed that TWINKLE unwinds the DNA more efficiently in the presence of the displaced strand DNA trap or single strand-binding protein. This is observed in some helicases of the RECQ family, e.g. forked dsDNAs are unwound by RECQ5β only in the presence of the human replication protein A and by RECQ4 only in the presence of the displaced strand DNA trap (34, 35). It was shown that both RECQ4 and RECQ5β possess strand annealing activity, which therefore masks their unwinding activity (34, 35). This led us to test and confirm that TWINKLE can anneal two complementary ssDNAs efficiently.
We find that TWINKLE can efficiently anneal two complementary ssDNA with and without Mg2+ and NTP. As of now, the DNA binding properties of the TWINKLE are not well understood. Previous protein-DNA gel-shift assays indicated that TWINKLE binds ssDNA only in the presence of the cofactors (3). However, recent studies by limited proteolysis of TWINKLE have indicated that it binds ssDNA in the absence of cofactors, although its structural conformation differs in their presence (8). Our fluorescence anisotropy-based DNA binding studies show that TWINKLE can bind DNA in the absence of cofactors (Fig. 3). Because TWINKLE can bind ssDNA in the absence of the cofactors, it can catalyze the strand-annealing reaction even without MgUTP. We observed that high salt (>150 mm NaCl) inhibits the strand annealing activity of TWINKLE. At high salt, the protein-DNA interactions are disrupted (26), which leads to a lowering of the strand annealing efficiency. The length of the ssDNA to be annealed also affects the efficiency of strand annealing, and we observed little annealing of the 20-base DNAs. One explanation is that the bound protein occludes a part of the ssDNA; therefore, the longer 40- and 58-base DNAs are annealed with a higher efficiency in the presence of TWINKLE.
The cofactor independent strand annealing activity must depend on TWINKLE bringing together two complementary ssDNAs. Based on our results, a number of mechanistic models can be conceived for the strand annealing activity by TWINKLE. A single ring may bind two ssDNA molecules, and if they are complementary and brought in close proximity by TWINKLE, base-pairing occurs at a faster rate than by simple protein independent collision. Alternatively, two separate rings, each bound to one ssDNA, can closely interact in space bringing the complementary ssDNA together for base-pairing. We have eliminated this second model based on the fact that an increase in protein concentration from 6 to 100 nm does not significantly change the annealing efficiency (Fig. 7E). A third possibility is that TWINKLE may bind ssDNA in such a way that its conformation is conducive for facile base-pairing with a complementary DNA from solution. The ssDNA binding to the N-terminal domain represents an outer surface that may provide such a site for DNA annealing. However, further experiments are needed to test this model.
Strand annealing activity is observed in many helicases (34–37), but its biological role has not been defined precisely. It might be helpful in recombination-mediated replication initiation, which has been proposed to occur in mammalian heart and brain mitochondria (38), or in replication fork regression during the repair of damaged DNA replication forks (36). Therefore, chances are that TWINKLE-mediated strand annealing plays a role in similar processes in mitochondria; however, additional studies with the rest of the components of the replication machinery such as mtSSB and DNA polymerase γAB are needed to establish such roles of the TWINKLE.
Supplementary Material
Acknowledgments
We thank Prof. Johannes. N. Spelbrink (Institute of Medical Technology and Tampere University Hospital, Tampere, Finland) for providing us with the initial clone of TWINKLE and the members of the Patel laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM55310.

This article contains supplemental Figs. 1–7.
- mtSSB
- mitochondrial single strand-binding protein
- ssDNA
- single-stranded DNA
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DMS
- dimethyl suberimidate
- TMR
- tetramethylrhodamine
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- TBE
- Tris/borate/EDTA
- UMPPNP
- uridine-5′-[(β,γ)-imido]triphosphate.
REFERENCES
- 1. Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M. (2004) Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spelbrink J. N., Li F. Y., Tiranti V., Nikali K., Yuan Q. P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., Santoro L., Toscano A., Fabrizi G. M., Somer H., Croxen R., Beeson D., Poulton J., Suomalainen A., Jacobs H. T., Zeviani M., Larsson C. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28, 223–231 [DOI] [PubMed] [Google Scholar]
- 3. Korhonen J. A., Pande V., Holmlund T., Farge G., Pham X. H., Nilsson L., Falkenberg M. (2008) Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J. Mol. Biol. 377, 691–705 [DOI] [PubMed] [Google Scholar]
- 4. Patel S. S., Picha K. M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697 [DOI] [PubMed] [Google Scholar]
- 5. Ziebarth T. D., Farr C. L., Kaguni L. S. (2007) Modular architecture of the hexameric human mitochondrial DNA helicase. J. Mol. Biol. 367, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holmlund T., Farge G., Pande V., Korhonen J., Nilsson L., Falkenberg M. (2009) Structure-function defects of the TWINKLE N-terminal region in progressive external ophthalmoplegia. Biochim. Biophys. Acta 1792, 132–139 [DOI] [PubMed] [Google Scholar]
- 7. Farge G., Pham X. H., Holmlund T., Khorostov I., Falkenberg M. (2007) The accessory subunit B of DNA polymerase γ is required for mitochondrial replisome function. Nucleic Acids Res. 35, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziebarth T. D., Gonzalez-Soltero R., Makowska-Grzyska M. M., Núñez-Ramírez R., Carazo J. M., Kaguni L. S. (2010) Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J. Biol. Chem. 285, 14639–14647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mao C. C., Holt I. J. (2009) Clinical and molecular aspects of diseases of mitochondrial DNA instability. Chang Gung Med. J. 32, 354–369 [PubMed] [Google Scholar]
- 10. Wanrooij S., Falkenberg M. (2010) The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta 1797, 1378–1388 [DOI] [PubMed] [Google Scholar]
- 11. Goffart S., Cooper H. M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J. N. (2009) Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 18, 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsushima Y., Farr C. L., Fan L., Kaguni L. S. (2008) Physiological and biochemical defects in C-terminal mutants of mitochondrial DNA helicase. J. Biol. Chem. 283, 23964–23971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wanrooij S., Goffart S., Pohjoismäki J. L., Yasukawa T., Spelbrink J. N. (2007) Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 35, 3238–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel S. S., Wong I., Johnson K. A. (1991) Presteady-state kinetic analysis of processive DNA replication, including complete characterization of an exonuclease-deficient mutant. Biochemistry 30, 511–525 [DOI] [PubMed] [Google Scholar]
- 15. Patel S. S., Rosenberg A. H., Studier F. W., Johnson K. A. (1992) Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J. Biol. Chem. 267, 15013–15021 [PubMed] [Google Scholar]
- 16. Tang G. Q., Patel S. S. (2006) T7 RNA polymerase-induced bending of promoter DNA is coupled to DNA opening. Biochemistry 45, 4936–4946 [DOI] [PubMed] [Google Scholar]
- 17. Longley M. J., Humble M. M., Sharief F. S., Copeland W. C. (2010) Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis, and helicase activity. J. Biol. Chem. 285, 29690–29702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haller I., Henning U. (1974) Cell envelope and shape of Escherichia coli K12. Cross-linking with dimethyl imidoesters of the whole cell wall. Proc. Natl. Acad. Sci. U.S.A. 71, 2018–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matson S. W., Richardson C. C. (1985) Nucleotide-dependent binding of the gene 4 protein of bacteriophage T7 to single-stranded DNA. J. Biol. Chem. 260, 2281–2287 [PubMed] [Google Scholar]
- 20. Hingorani M. M., Patel S. S. (1993) Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry 32, 12478–12487 [DOI] [PubMed] [Google Scholar]
- 21. Farge G., Holmlund T., Khvorostova J., Rofougaran R., Hofer A., Falkenberg M. (2008) The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 36, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jemt E., Farge G., Bäckström S., Holmlund T., Gustafsson C. M., Falkenberg M. (2011) The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 39, 9238–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matson S. W., Tabor S., Richardson C. C. (1983) The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 258, 14017–14024 [PubMed] [Google Scholar]
- 24. Oliveira M. T., Kaguni L. S. (2010) Functional roles of the N- and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein. PloS ONE 5, e15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delagoutte E., Heneman-Masurel A., Baldacci G. (2011) Single-stranded DNA-binding proteins unwind the newly synthesized double-stranded DNA of model miniforks. Biochemistry 50, 932–944 [DOI] [PubMed] [Google Scholar]
- 26. Yakovchuk P., Protozanova E., Frank-Kamenetskii M. D. (2006) Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 34, 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rasnik I., Jeong Y. J., McKinney S. A., Rajagopal V., Patel S. S., Ha T. (2008) Branch migration enzyme as a Brownian ratchet. EMBO J. 27, 1727–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Costa A., Pape T., van Heel M., Brick P., Patwardhan A., Onesti S. (2006) Structural studies of the archaeal MCM complex in different functional states. J. Struct. Biol. 156, 210–219 [DOI] [PubMed] [Google Scholar]
- 29. Crampton D. J., Ohi M., Qimron U., Walz T., Richardson C. C. (2006) Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J. Mol. Biol. 360, 667–677 [DOI] [PubMed] [Google Scholar]
- 30. Ahnert P., Picha K. M., Patel S. S. (2000) A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 19, 3418–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Picha K. M., Ahnert P., Patel S. S. (2000) DNA binding in the central channel of bacteriophage T7 helicase-primase is a multistep process. Nucleotide hydrolysis is not required. Biochemistry 39, 6401–6409 [DOI] [PubMed] [Google Scholar]
- 32. Picha K. M., Patel S. S. (1998) Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 273, 27315–27319 [DOI] [PubMed] [Google Scholar]
- 33. Remus D., Diffley J. F. (2009) Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21, 771–777 [DOI] [PubMed] [Google Scholar]
- 34. Garcia P. L., Liu Y., Jiricny J., West S. C., Janscak P. (2004) Human RECQ5β, a protein with DNA helicase and strand annealing activities in a single polypeptide. EMBO J. 23, 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu X., Liu Y. (2009) Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 28, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Machwe A., Xiao L., Groden J., Matson S. W., Orren D. K. (2005) RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 280, 23397–23407 [DOI] [PubMed] [Google Scholar]
- 38. Pohjoismäki J. L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P. J., Griffith J. D., Holt I. J., Jacobs H. T. (2009) Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 284, 21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.