Abstract
The E3 ubiquitin ligase MDM2 functions as a crucial negative regulator of the p53 tumor suppressor protein by antagonizing p53 transactivation activity and targeting p53 for degradation. Cellular stress activates p53 by alleviating MDM2-mediated functional inhibition, even though the molecular mechanisms of stress-induced p53 activation still remain poorly understood. Two opposing models have been proposed to describe the functional and structural role in p53 activation of Ser17 phosphorylation in the N-terminal “lid” (residues 1–24) of MDM2. Using the native chemical ligation technique, we synthesized the p53-binding domain (1–109)MDM2 and its Ser17-phosphorylated analog (1–109)MDM2 pS17 as well as (1–109)MDM2 S17D and (25–109)MDM2, and comparatively characterized their interactions with a panel of p53-derived peptide ligands using surface plasmon resonance, fluorescence polarization, and NMR and CD spectroscopic techniques. We found that the lid is partially structured in apo-MDM2 and occludes p53 peptide binding in a ligand size-dependent manner. Binding of (1–109)MDM2 by the (15–29)p53 peptide fully displaces the lid and renders it completely disordered in the peptide-protein complex. Importantly, neither Ser17 phosphorylation nor the phospho-mimetic mutation S17D has any functional impact on p53 peptide binding to MDM2. Although Ser17 phosphorylation or its mutation to Asp contributes marginally to the stability of the lid conformation in apo-MDM2, neither modification stabilizes apo-MDM2 globally or the displaced lid locally. Our findings demonstrate that Ser17 phosphorylation is functionally neutral with respect to p53 binding, suggesting that MDM2 phosphorylation at a single site is unlikely to play a dominant role in stress-induced p53 activation.
Keywords: MDM2, MDMX, p53, phosphorylation, native chemical ligation
Introduction
Protein phosphorylation is an important regulatory mechanism that controls protein structure and function in almost all aspects of cellular life. It is believed that one third of the 30,000 or so human proteins contain covalently bound phosphate, and abnormal phosphorylation is associated with many human diseases1. A significant body of literature exists, describing the intricacy of cellular signaling pathways and networks that are activated or inactivated by protein phosphorylation and de-phosphorylation events2,3. One such extensively studied signaling pathway in biology centers on the tumor suppressor protein p53, a transcription factor of 393 amino acid residues that transactivates in response to cellular stress the expression of many target genes involved in cell cycle arrest, DNA repair, senescence, or apoptosis4. Dubbed the “guardian of the genome” critical for maintaining genetic stability5, p53 constitutes a mainstay of our body’s natural anticancer defense6.
Due to its growth inhibitory and pro-apoptotic activity, p53 is tightly controlled in normal cells by two crucial negative regulators – the E3 ubiquitin ligase MDM2 and its homolog MDMX (also known as MDM4)7–12. Both MDM2 and MDMX contain an N-terminal domain of ~100 amino acid residues that binds with high affinity to the N-terminal transactivation domain of p53, capable of directly blocking p53 transactivation activity13–15. The C-terminal Zn2+-binding RING (really interesting new gene) domain of MDM2 functions as an E3 ubiquitin ligase to facilitate the transfer of E2-conjugated ubiquitin molecules to Lys residues of p53, targeting p53 for proteasomal degradation16–18. Although MDMX lacks E3 ubiquitin ligase activity, growing evidence suggests that MDM2 and MDMX heter-oligomerize via their RING domains to augment p53 degradation10,12,19–21.
Protein posttranslational modifications play important roles in p53 activation and accumulation in the nucleus12. Both p53 and MDM2 are heavily phosphorylated by kinases that are activated in response to stress signals such as DNA damage. However, p53 phosphorylation is not essential for DNA damage-induced p53 stabilization in vivo and only partially contributes to p53 dissociation from its inhibitory complex with MDM212,22. By contrast, MDM2 phosphorylation is functionally multifaceted, exerting a more complex influence on its own activity as well as p53 activation and inhibition12,22. Despite significant progress in understanding stress-induced p53 activation at the cellular level, the precise molecular mechanisms by which p53 escapes from MDM2-mediated functional inactivation remain only partially understood and, in some cases, still controversial.
It has been reported that phosphorylation of Ser17 of MDM2 by the DNA-dependent protein kinase DNA-PK renders MDM2 unable to bind and inhibit p53 in vitro 23. Paradoxically, however, X-ray crystallographic studies of the p53-binding domain of MDM2 in complex with p53 transactivation peptide (residues 15–29) show that Ser17 of MDM2 is situated within a disordered N-terminus of the protein and makes no direct contact with the peptide ligand24. In fact, only residues 25–109 of MDM2 are well defined in the electron density map of its complex structure24,25. A plausible explanation for this paradox first comes from NMR studies of unliganded (16–125)MDM2, which showed that the N-terminal residues 16–24 (TSQIPASEQ) formed a partially helical “lid” occluding the p53-binding site on apo-MDM226. According to this model Ser17 phosphorylation would stabilize intra-molecular electrostatic interactions involving the “lid” peptide and disrupt or prevent the p53-MDM2 interaction, thus leading to p53 activation26. A quantitative dynamics study of (17–125)MDM2 using NMR further indicated that the MDM2 lid, while existing predominantly in the “closed” state in apo-MDM2 (favoring p53 dissociation), could be displaced to become highly disordered in an “open” state by (17–29)p53 peptide, but not by Nutlin-327, a small molecule antagonist of MDM228.
Two most recent reports29,30, however, challenged this model by suggesting that Ser17 phosphorylation “opens” rather than “closes” the p53-binding pocket of MDM2. The supporting evidence for this alternative model stems from biochemical studies of several MDM2 mutants, where the phospho-mimetic mutation S17D was found to increase the stability of the p53-binding domain of MDM2, enhance the p53-MDM2 interaction, and promote MDM2-mediated ubiquitination of p53 in vitro 29,30. These two sharply divided and contradictory models cast uncertainty about the precise physiological role of MDM2 phosphorylation at Ser17 by DNA-PK in p53 activation and stabilization in response to DNA damage. Molecular dynamics simulation studies of (1–119)MDM2 with the lid in ‘open’ and ‘closed’ conformations suggested that the discrepancies in NMR and biochemical data could be partially reconciled by taking into consideration the inherent differences between the phospho-Ser group and the phosphomimetic residue Asp in their ability to interact with partners of an electrostatic nature31. However, definitive experimental evidence that supports either model is still lacking. To resolve this important controversy, we chemically synthesized, via native chemical ligation32,33, (1–109)MDM2 and its Ser17-phosphorylated analog (1–109)MDM2 pS17 as well as (1–109)MDM2 S17D, and comparatively characterized their interactions with a panel of p53-derived peptide ligands.
Experimental Section
Materials
Boc-amino acids and N-hydroxybenzotriazole (HOBt) were obtained from Peptides International (Louisville, KY); Fmoc-amino acids, Fmoc-Ser(PO(OBzl)OH)-OH and Dawson Dbz AM resin were purchased from Novabiochem (Switzerland); 15N-labeled Fmoc- and Boc-amino acids were from Cambridge Isotope Laboratories, Inc. (Andover, MA); p-Methyl-BHA (MBHA) resin and Boc-Leu-OCH2-PAM resin were purchased from Applied Biosystems (Foster City, CA); Tris-(2-carboxyethyl) phosphine (TCEP), dichloromethane (DCM), N,N-dimethylformamide (DMF), and HPLC grade acetonitrile were obtained from ThermoFisher Scientific (Pittsburgh, PA), and 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluroniumhexafluorophosphate (HBTU) was acquired from Oakwood Products Inc. (West Columbia, SC). Trifluoroacetic acid (TFA) was purchased from Halocarbon (River Edge, NJ) and hydrogen fluoride (HF) was from Matheson Tri-gas (Montgomeryville, PA). Triisopropylsilane (TIS), N,N-diisopropylethylamine (DIEA), thiophenol, and p-cresol were from Sigma-Aldrich (St Louis, MO), and ultrapure guanidine hydrochloride was obtained from ICN Biochemicals (Irvine, CA). BIAcore series S sensor chips CM5 and HBS-EP buffer were purchased from GE Healthcare Bio-Sciences AB (Sweden). Nutlin-3a was obtained from Cayman Chemicals (Ann Arbor, MI).
Peptide synthesis
All peptides were synthesized on appropriate resins on an ABI 433A automated peptide synthesizer using the optimized HBTU activation/DIEA in situ neutralization protocol developed by Kent and colleagues for Boc-chemistry solid phase peptide synthesis (SPPS)34 or an ABI-supplied HBTU/HOBt protocol for Fmoc-chemistry SPPS. After cleavage and deprotection in HF (Boc-chemistry) or in a reagent cocktail containing 95% TFA, 2.5% TIS and 2.5% H2O (Fmoc-chemistry), crude products were precipitated with cold ether and purified to homogeneity by preparative C18 reversed-phase HPLC. The molecular masses were ascertained by electrospray ionization mass spectrometry (ESI-MS).
Native chemical ligation and folding of MDM2 proteins
Native chemical ligation reactions were carried out in 0.1 M phosphate buffer containing 6 M guanidine hydrochloride (GuHCl) and 2% thiophenol, pH 7.4. Removal of Acm in the first ligation product, (36–109)MDM2, was achieved by dissolving the peptide at 1 mg/ml in 50% acetonitrile containing 0.1% TFA, to which 300-fold molar excess of silver acetate was added. The reaction proceeded for 1 h before being quenched by DTT, and the Acm-removed product was purified to homogeneity by preparative C18 RP-HPLC.
Folding of MDM2 proteins was achieved by dissolving the polypeptide in 6 M GuHCl at 1 mg/ml, followed by a 6-fold dilution with phosphate buffered saline (PBS) containing 0.5 mM TCEP, pH 7.4, and an overnight dialysis against the same buffer. Protein solutions were quantified spectroscopically by UV measurements at 280 nm using a molar extinction coefficient of 10430, calculated according to the published algorithm developed by Pace and colleagues35.
Fluorescence polarization (FP) assay
N-terminally acetylated (15–29)p53 was synthesized by Boc-chemistry SPPS and purified to homogeneity by preparative C18 RP-HPLC. Succinimidyl ester-activated carboxyfluorescein (FAM-NHS) was covalently conjugated to N-acetyl-(15–29) via its Lys24 side chain in DMF, and the resultant product N-acetyle-(15–29)p53-FAM HPLC-purified and lyophilized.
The p53-MDM2 binding experiments were performed in 96-well plates (Corning Life Science, Nonbinding Surface or NBS™) on a Tecan Infinite M1000 fluorescence plate reader. Serially diluted MDM2 proteins were prepared in Tris-HCl buffered saline (10 mM Tris, 150 mM NaCl, 1 mM EDTA, pH 7.0) and incubated with 10 nM N-acetyl-(15–29)p53-FAM in a total volume of 100 µl per well. After a 30-min incubation at room temperature, fluorescence polarization was measured at λex = 470 nm and λem = 530 nm. Non-linear regression analyses were performed to give rise to Kd values as previously described36–38 by using the following equation:
where F is measured FP, Fc is FP of the N-acetyl-(15–29)p53-FAM-MDM2 complex, F0 is FP of N-acetyl-(15–29)p53-FAM, [p53] is the final concentration of N-acetyl-(15–29)p53-FAM, and [MDM2] is the total concentration of MDM2 protein.
Surface Plasmon Resonance (SPR)
SPR-based direct and competitive binding assays were carried out at 25 °C on a Biacore T100 instrument, using a CM5 sensor chip to which (15–29)p53 is covalently attached via its N-terminus. The buffer (HBS-EP) was 10 mM HEPES, 150 mM NaCl, 0.005% surfactant P20, pH 7.4. For the competitive binding assay, 50 nM (25–109)MDM2 or 250 nM (1–109)MDM2 proteins (wide-type, S17D or pS17) was incubated at room temperature for 30 min with varying concentrations of inhibitor, and injected at a flow rate of 20 µL/min for 2 min, followed by a 4-min dissociation. The concentration of unbound MDM2 proteins in solution was deduced, based on p53-association RU values, from a calibration curve established by RU measurements of different concentrations of MDM2 proteins injected alone. Nonlinear regression analysis was performed using GraphPad Prism 4.0 to give rise to Kd values using the equation Kd = [peptide][MDM2/]/[complex]. For the direct binding assay, MDM2 proteins prepared in HBS-EP buffer in a 2-fold serial dilution were injected onto the (15–29)p53 peptide-immobilized CM5 sensor chip at a flow rate of 20 µL/min for 2 min, followed by 4 min dissociation. Nonlinear regression analysis was performed using GraphPad Prism 4 to give rise to Kd values according to the equation RU=RUmax•C/(Kd+C).
NMR spectroscopy
NMR spectra were recorded at 20°C on an 800 MHz (800.27 MHz for protons) Bruker Avance-series NMR spectrometer equipped four frequency channels and a 5mm triple-resonance z-axis gradient cryogenic probehead. A one-second relaxation delay was used, and quadrature detection in the indirect dimensions was obtained with states-TPPI phase cycling; initial delays in the indirect dimensions were set to give zero- and first-order phase corrections of 90° and −180°, respectively39,40. Data were processed using the processing program nmrPipe on Mac OS X workstations41. The 1H, 15N-fast HSQC experiment was collected to monitor changes in the backbone 15N and 1H protein resonances42. Typical NMR samples contained synthetic MDM2 protein at 0.12 to 0.2 mM mixed with (15–29)p53 peptide (molar ratio of peptide/protein = 0.5) in PBS containing 0.5 mM TCEP and 10% D2O (v/v).
CD Spectroscopy and Chemical Denaturation of MDM2 proteins
The CD spectra of (1–109)MDM2, (1–109)MDM2 S17D, and (1–109)MDM2 pS17 at 20 µM in 10 mM phosphate buffer, pH 7.2, were obtained at room temperature on a Jasco J-810 spectropolorimeter by using a 1-mm cuvette. GuHCl-induced protein denaturation monitored by CD spectroscopy at 222 nm was carried out at room temperature as previously described43,44. Specifically, an initial 3.0 ml of protein solution prepared at 10 µM in 10 mM phosphate buffer (pH 7.2) was aliquotted into a 10-mm cuvette. An increasing amount of aliquot was withdrawn, followed immediately by addition of an equal volume of denatured protein of the same concentration, prepared in 10 mM phosphate buffer containing 7.2 M GuHCl (pH 7.2). This procedure generated a stepwise increase (0.25 M) in the concentration of GuHCl in the cuvette from 0 to 5 M after 20 withdrawal/addition cycles. The solution in the cuvette was thoroughly mixed before signals at 222 nm were recorded at different GuHCl concentrations. The experimental data were subjected to a six-parameter nonlinear regression analysis by using a published equation that was derived from a two-state protein denaturation model43, yielding the free energy change of unfolding, ΔG° (kcal/mol), at zero GuHCl concentration.
Results
Total chemical synthesis of (1–109)MDM2 K36C and (1–109)MDM2 K36C/S17D via native chemical ligation
We previously synthesized large quantities of highly pure and correctly folded (25–109)MDM2 for structural and functional studies by ligating H-(25–76)αCOSR (R=CH2CO-Leu-OH) to H-(77–109)-OH25. For the synthesis of (1–109)MDM2, a three-segment ligation strategy was used (Figure 1), where a Lys-to-Cys mutation at position 36 was introduced to enable the second ligation reaction between fragments (1–35) and (36–109). Structural analysis of the p53-binding domain of MDM2 suggested that the K36C mutation would be functionally inconsequential as Lys36 is a surface-exposed, non-contact residue distal to the p53-binding site (Figure 1). For functional verification, however, we also chemically synthesized (25–109)MDM2 K36C using the two-segment ligation strategy as previously described25, and Figure S1 shows the product analyzed by RP-HPLC and ESI-MS. Notably, Lys36 of MDM2 is not involved in any intra-molecular interactions with the negatively charged residue 17 (S17D or pS17) as demonstrated by NMR, mutagenesis, and molecular dynamics simulation studies26,29,31. These studies identify a cluster of C-terminal cationic residues (Lys94, His96, Arg97, Lys98), more than 25 Å apart from Lys36, that potentially form a putative salt bridge with Asp17 or pSer17.
Figure 1.
Amino acid sequence of (1–109)MDM2 and strategy for the synthesis of phosphorylated (1–109)MDM2 at Ser17 using sequential native chemical ligation of three peptide segments in red, green, and blue. Lys36 was mutated to Cys to introduce the second ligation site Leu35-Cys36. As shown in the crystal structure of (25–109)MDM2 in complex with a phage-selected, p53-like peptide ligand (PMI)25 (PDB code: 3EQS), the solvent-exposed side chain of Lys36 is distal to the p53-binding site of MDM2. The K36C mutation is therefore expected to have little impact on the folding and fucntion of MDM2.
For the synthesis of (1–109)MDM2 K36C, the following three peptide fragments were individually assembled on appropriate PAM resins using the HBTU activation/DIEA in situ neutralization protocol developed by Kent and colleagues for Boc-chemistry SPPS34: H-(1–35)αCOSR, H-(36–76)αCOSR, and H-(77–109)-OH. The N-terminal Cys36 of the middle fragment H-(36–76)αCOSR was orthogonally protected by acetamidomethyl (Acm) to prevent an intra-molecular head-to-tail cyclization reaction45,46. The first ligation reaction between H-(36–76)αCOSR and H-(77–109)-OH proceeded to completion overnight, and Acm removal from Cys36 of the ligation product H-(36–109)-OH was achieved quantitatively with AgOAc treatment as described47,48. The subsequent ligation of H-(36–109)-OH to H-(1–35)αCOSR was equally efficient, yielding the full-length product (1–109)MDM2 K36C purified by RP-HPLC to homogeneity and verified by ESI-MS (Figure S2). An identical approach was used for the synthesis of (1–109)MDM2 K36C/S17D, and shown in Figure S3 is the product analyzed by RP-HPLC and ESI-MS. The overall yields of synthesis of these two (1–109)MDM2 proteins are typically 3–5% on a synthetic scale of 0.25 mmol.
Total chemical synthesis of (1–109)MDM2 K36C/pS17 using a combination of Boc- and Fmoc-chemistries coupled with native chemical ligation
Native chemical ligation was originally developed for Boc-chemistry as thioester peptide precursors are stable in TFA but labile in piperidine. Since peptide phosphorylation on solid phase is achievable only with Fmoc-chemistry, total chemical synthesis of phospho-proteins via native chemical ligation necessitates the development of an Fmoc-compatible chemistry for the synthesis of thioester peptides within which the phosphorylation site is located. Considerable effort has been made to develop such chemistries49, among which a robust and efficient approach pioneered by Dawson and colleagues, involving the formation of a C-terminal N-acylurea functionality, holds great promise49. Using the Dawson chemistry, we synthesized (1–35)MDM2 pS17-Nbz (N-acyl-benzimidazolinone) by standard Fmoc-chemistry SPPS. Upon thiolysis (1–35)MDM2 pS17-Nbz yielded a peptide thioester that ligated quantitatively with H-(36–109)-OH. The final product (1–109)MDM2 K36C/pS17 analyzed by RP-HPLC and ESI-MS is shown in Figure 2. The determined molecular mass of 12551.0 Da is within experimental error of the expected value of 12551.6 Da calculated on the basis of the average isotopic compositions of (1–109)MDM2 K36C/pS17. This molecular mass (12551.0 Da) is 80 Da higher than that (12471.1 Da) of (1–109)MDM2 K36C (Figure S2), indicative of the presence of a phosphate group in the synthetic MDM2 protein. Of note, 15N-Val14 and 15N-Ala21 were incorporated in (1–109)MDM2 K36C, (1–109)MDM2 K36C/S17D and (1–109)MDM2 K36C/pS17 to facilitate NMR spectroscopic studies. As will be shown later, all synthetic MDM2 proteins folded correctly and were fully functional.
Figure 2.
(1–109)MDM2 K36C/pS17 characterized by RP-HPLC and ESI-MS. The chromatogram was obtained at 40 °C on a Waters XBridge C18 column (4.6×150 mm, 3.5 µm) running a gradient of 5% to 65% acetonitrile in water, containing 0.1% TFA, at a flow rate of 1 ml/min. The determined molecular mass of 12551.0 Da is with experimental error of the theoretical value of 12551.6 Da calculated on the basis of the average isotopic compositions of (1–109)MDM2 K36C/pS17.
The K36C mutation is functionally neutral as predicted
To evaluate functional ramification of the K36C mutation, we used fluorescence polarization (FP) techniques36–38 to quantify the interaction of (25–109)MDM2 and (25–109)MDM2 K36C with an N-acetyl-(15–29)p53 peptide (SQETFSDLWKLLPEN), to which carboxyfluorescein (FAM) was covalently attached via the side chain of Lys24. Lys24 is a non-contact residue that does not contribute to p53 binding to MDM250. FP measurements are based on the principle that the p53 peptide tumbles significantly faster than its complex with MDM2 in solution, and upon excitation by polarized light, it “scrambles” the polarization of emitted light more efficiently51,52. Thus, MDM2 binding to the fluorescently labeled p53 peptide necessarily causes decreased rotational rates and increased polarization of emitted light. A titration curve is generated from measurements of polarized fluorescence, and the peptide-protein interaction can be quantitatively evaluated using known mathematical models36–38.
As shown in Figure 3, the two synthetic MDM2 proteins differing by only one residue at position 36 bound to N-acetyl-(15–29)p53-FAM with nearly identical affinities (77.2 versus 63.7 nM), confirming that the K36C mutation had no effect on p53-binding activity of MDM2. This finding is entirely consistent with structural studies of MDM2 in complex with peptide ligands25,37,50,53,54 (Figure 1). Given these results, (1–109)MDM2 K36C, (1–109)MDM2 K36C/S17D and (1–109)MDM2 K36C/pS17 are referred to hereafter simply as (1–109)MDM2, (1–109)MDM2 S17D and (1–109)MDM2 pS17, respectively. It is worth pointing out that (1–109)MDM2 proteins can be synthesized via native chemical ligation without resorting to the K36C mutation. An alternative and perhaps more elegant strategy would involve an auxiliary Ala-to-Cys mutation at an appropriate site for ligation followed by desulfurization to revert Cys to Ala after ligation55.
Figure 3.
Binding of N-acetyl-(15–29)p53-FAM to (25–109)MDM2 and (25–109)MDM2 K36C as quantified by fluorescence polarization techniques at room temperature in 10 mM Tris-HCl buffer containing 150 mM NaCl and 1 mM EDTA, pH 7.0. The fluorescently labeled p53 peptide at 10 nM was incubated with varying concentratiuons of MDM2 for 30 min on a 96-well plate before polarization measurements (λex = 470 nm, λem = 530 nm). The Kd values (mean ± SEM, n=3) are from three independent experiments.
The N-terminal lid peptide of MDM2 is detrimental to p53 peptide binding
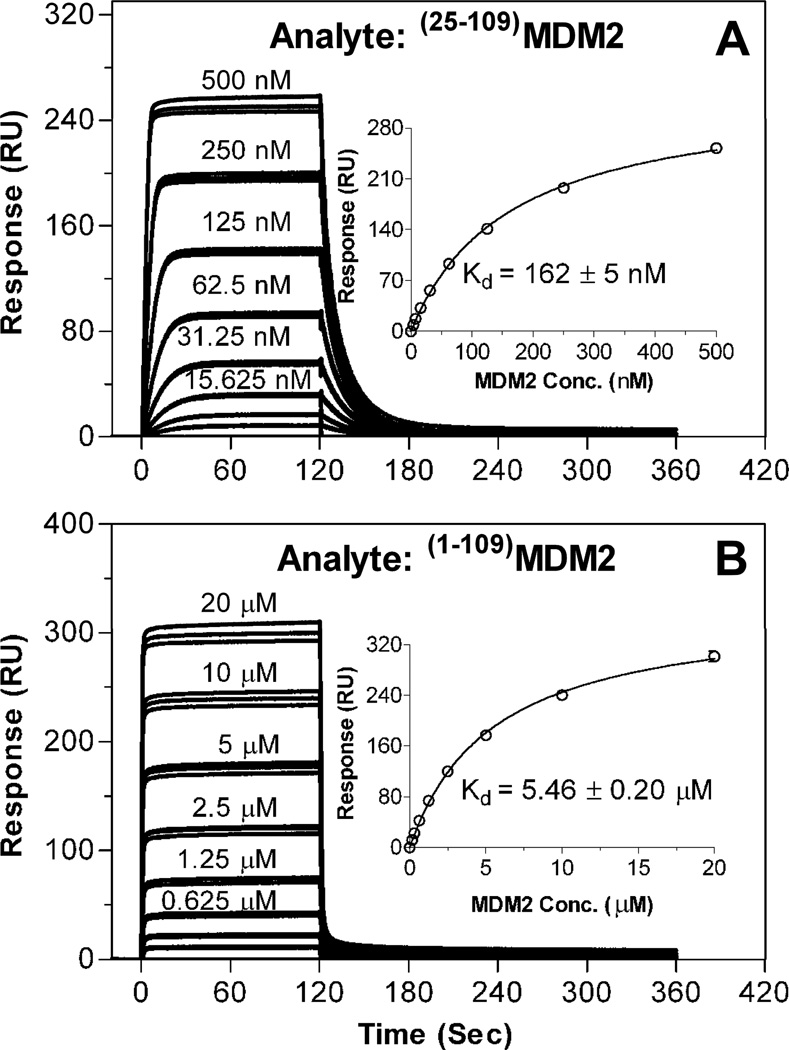
Using the SPR technique, we quantified direct interactions of (25–109)MDM2 and (1–109)MDM2 with (15–29)p53 immobilized as ligand via its N-terminal amino group to a CM5 biosensor chip to capture MDM2 as analyte in solution. Shown in Figure 4 are steady-state binding isotherms of varying concentrations of (25–109)MDM2 and (1–109)MDM2 on 36 response units (RUs) of immobilized (15–29)p53. A non-linear regression analysis yielded a Kd value of 162 nM for (25–109)MDM2, in good agreement with the previously published values of 140 and 144 nM determined by an SPR-based competitive binding assay25 and isothermal titration calorimetry56, respectively. In contrast, (1–109)MDM2 bound to the immobilized p53 peptide at a significantly reduced affinity of 5.5 µM under identical assay conditions, representing a 34-fold reduction in binding affinity attributed by the MDM2 lid.
Figure 4.
Direct binding of varying concentrations of (25–109)MDM2 (A) and (1–109)MDM2 (B) to the (15–29)p53 peptide immobilized on a CM5 sensor chip (36 RU’s) as quantified at 25 °C by SPR techniques in HBS-EP buffer. The steady-state binding isotherms are from a triplicate assay of one representative experiment, and the fitted curves (insets) and Kd values (mean ± SEM, n=2) generated by non-linear regression analyses are from two independent experiments (error bars are too small to be seen).
To better understand the effects of the cis-acting lid peptide on MDM2 function, we performed the SPR-based competitive binding assay25,50,57,58 to measure the binding affinities for (25–109)MDM2 and (1–109)MDM2 of Nutlin-3a and three p53-derived peptides of different lengths, i.e., (19–26)p53 (FSDLWKLL, 8 aa), (17–28)p53 (ETFSDLWKLLPE, 12 aa), and (15–29)p53 (SQETFSDLWKLLPEN, 15aa). The binding curves generated from these in-solution assays are shown in Figure 5, from which the Kd values were derived and tabulated in Table 1. Nutlin-3a, (19–26)p53, (17–28)p53, and (15–29)p53 bound to (25–109)MDM2 at respective affinities of 133 nM, 39.6 µM, 404 nM, and 184 nM, in good agreement with the previously published values determined by the same technique25,50 (Table 1). As expected, the Kd value of Nutlin-3a (133 nM) for (25–109)MDM2 is precisely half that of Nutlin-3 (263 nM)25 for the latter is a racemic mixture of active Nutlin-3a and substantially less active Nutlin-3b28. Importantly, the lid peptide was uniformly deleterious for MDM2 binding by all three p53-derived peptides, registering a decrease in binding affinity by 5.5-fold with (17–28)p53 and as much as 7-fold with (15–29)p53. The 2-fold effect was, however, significantly smaller with (19–26)p53 of 8 amino acid residues – the minimal length required for productive MDM2 binding50. By contrast, the presence of the lid peptide slightly improved the binding affinity of Nutlin-3a for MDM2 from 133 nM to 83 nM. These results suggest that the lid peptide likely imposes steric hindrance to peptide ligands of MMD2 (but not to Nutlin-3a) and weakens their binding in a ligand size-dependent manner, thus consistent with the NMR studies26,27.
Figure 5.
Quantification of the interactions of (1–109)MDM2 (A) and (25–109)MDM2 (B) with Nutlin-3a, (15–29)p53, (17–28)p53 and (19–26)p53 at 25 °C by an SPR-based competitive binding assay in HBS-EP buffer. (1–109)MDM2 (250 nM) or (25–109)MDM2 (50 nM) was incubated with varying concentrations of test compound, and unbound (free) MDM2 protein quantified by RU measurements on a (15–29)p53-immobilized CM5 sensor chip. Non-linear regression analyses were performed using GraphPad Prism 4 to generate the Kd values (mean ± SEM, n=3, Table 1). Each curve is the mean of three independent measurements with the error bars denoting the SEM.
Table 1.
Equilibrium dissociation constants (Kd, mean ± SEM, nM) for various MDM2 proteins interacting with p53-derived peptide ligands and Nutlin-3a determined by SPR techniques.
| Nutlin-3a | (19–26)p53 | (17–28)p53 | (15–29)p53 | (15–29)p53 pS15 | |
|---|---|---|---|---|---|
| (25–109)MDM2 | 133 ± 9 263 ± 60a,b |
39600 ± 2000 35000 ± 4000c |
404 ± 19 452 ± 11a 442 ± 39c |
184 ± 14 140 ± 5a |
170 ± 12 |
| (1–109)MDM2 | 83.0 ± 6.9 | 88300 ± 3700 | 2250 ± 100 | 1260 ± 70 | 615 ± 32 |
| (1–109)MDM2 pS17 | 108 ± 16 | 95600 ± 6500 | 2630 ± 120 | 1000 ± 30 | 530 ± 56 |
Of note, the deleterious effect of the lid peptide on MDM2 binding by (15–29)p53 was substantially smaller in the SPR-based competitive binding assay (7-fold) than that measured by direct binding (34-fold). A similarly small effect (8-fold) was also observed in the solution-based fluorescence polarization assay (Figure S4). In the SPR-based direct binding assay, the N-terminus of (15–29)p53 was covalently bound to the sensor chip surface via carboxyl moieties on the dextran. Thus, surface immobilization of (15–29)p53 effectively extended its N-terminus, augmenting steric clashes with the lid peptide and exacerbating an already weakened interaction. Given the known effects of the length of a peptide ligand on its interaction with (1–109)MDM2, this disparity (7-fold versus 34-fold) seen in these different binding assays comes as no surprise, and clearly supports the model in that the N-terminal lid peptide of MDM2 “closes” rather than “opens” its p53-binding site to peptide/protein ligands as suggested by the NMR studies26,27.
Neither the S17D mutation nor Ser17 phosphorylation affects the interaction between MDM2 and p53 peptides
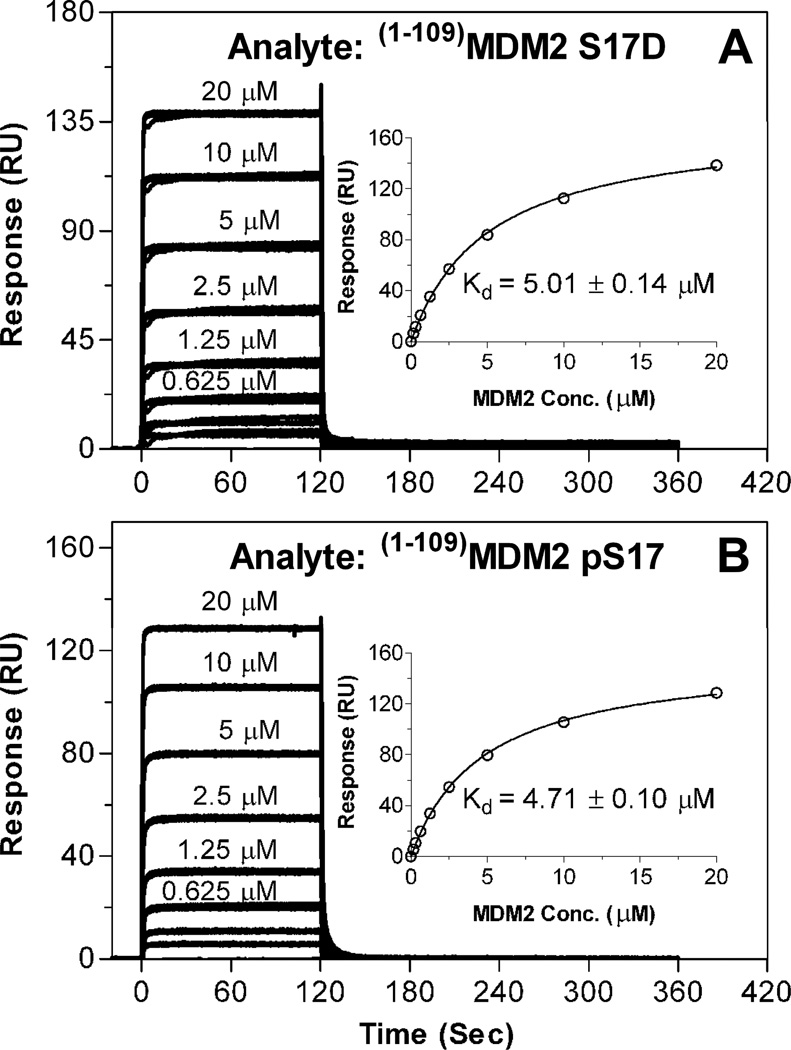
To evaluate functional impact of the S17D mutation and Ser17 phosphorylation on MDM2 binding by the p53 peptides, we first determined the Kd values for (1–109)MDM2 S17D and (1–109)MDM2 pS17 directly interacting with immobilized (15–29)p53 peptide. As shown in Figure 6, (1–109)MDM2 S17D and (1–109)MDM2 pS17 bound to (15–29)p53 at affinities of 5.0 and 4.7 µM, respectively, nearly identical to the Kd value of 5.5 µM determined under the same experimental conditions for (1–109)MDM2 (Figure 4). These results suggest that neither the S17D mutation nor Ser17 phosphorylation is functionally consequential with respect to the (1–109)MDM2-(15–29)p53 interaction.
Figure 6.
Direct binding of varying concentrations of (1–109)MDM2 S17D (A) and (1–109)MDM2 pS17 (B) to immobilized (15–29)p53 (22 RU’s) as quantified at 25 °C by SPR techniques in HBS-EP buffer. The steady-state binding isotherms are from a triplicate assay of one representative experiment, and the fitted curves (insets) and Kd values (mean ± SEM, n=2) are from two independent experiments.
To further verify these findings, we used the SPR-based competitive binding assay for the quantification of the binding affinities of Nutlin-3a, (19–26)p53, (17–28)p53 and (15–29)p53 for (1–109)MDM2 pS17, yielding Kd values of 108 nM, 95.6 µM, 2.63 µM and 1.00 µM, respectively (Figure 7A and Table 1). Pair-wise comparisons with the Kd values determined under identical assay conditions for (1–109)MDM2 confirmed that Ser17 phosphorylation is indeed functionally neutral. Using FP techniques, we also quantified solution interactions of N-acetyl-(15–29)p53-FAM with (1–109)MDM2, (1–109)MDM2 S17D and (1–109)MDM2 pS17. As shown in Figure 7B, the three similar Kd values of 653, 648 and 440 nM obtained for these MDM2 proteins indicate a consistent lack of any meaningful functional role of Ser17 phosphorylation in the interaction between (1–109)MDM2 and (15–29)p53. These somewhat surprising experimental findings are in contrast to the purported importance of Ser17 phosphorylation in MDM2 function as has been suggested by NMR, biochemical, and modeling studies where the phospho-mimetic mutation S17D was used26,27,29–31.
Figure 7.
(A) Quantification of the interaction of (1–109)MDM2 pS17 (250 nM) with varying concentrations of Nutlin-3a, (15–29)p53, (17–28)p53 and (19–26)p53 by the SPR-based competitive binding assay described in the legend of Figure 5. Each curve is the mean of three independent measurements, and the Kd values (mean ± SEM, n=2) are listed in Table 1. (B) Binding of N-acetyl-(15–29)p53-FAM to (1–109)MDM2, (1–109)MDM2 S17D, and (1–109)MDM2 pS17 as quantified by the fluorescence polarization assay described in the legend of Figure 3. Each curve is the mean for a representative assay performed in triplicate, and the Kd values (mean ± SEM, n=3) are from three independent assays.
Structural studies confirm the functional findings
NMR studies of (16–125)MDM2 and (17–125)MDM2 have previously shown that the lid peptide is partially structured in apo-MDM2 with residues 21–24 (Ala-Ser-Glu-Gln) adopting an α-helical conformation26,27, and that, upon peptide ligand binding, the lid peptide becomes displaced and fully disordered27. To facilitate characterization of the p53-MDM2 interaction by NMR spectroscopy, we isotopically labeled Val14 and Ala21 in the lid peptide region of (1–109)MDM2, (1–109)MDM2 S17D and (1–109)MDM2 pS17. The 1H-15N heteronuclear single quantum correlation (HSQC) spectra of the three MDM2 proteins were collected at 20 °C in 10% D2O in the presence of 0.5 equal molar concentration of unlabeled (15–29)p53 (Figure 8 A–C). The two 15N-labled aliphatic residues were unambiguously identified by analyzing the HSQC spectra of a singly labeled 15N-Ala21-(1–24)MDM2 peptide and a doubly labeled 15N-Val14/15N-Ala21-(1–24)MDM2 peptide (Figure 8D). Of note, the 24-residue lid peptide of MDM2, alone, was disordered in aqueous solution as judged by circular dichroism spectroscopy (Figure S5), and no detectable inter-molecular interaction was observed in a FP assay between (25–109)MDM2 at concentrations of up to 60 µM and 100 nM (1–24)MDM2 labeled with FAM at Cys2 (Figure S6).
Figure 8.
NMR spectroscopy. (A) 1H-15N HSQC spectrum of (1–109)MDM2 isotopically labeled at residues Val14 and Ala21 in the presence (red) and absence (black) of p53 peptide. (B) Identically collected 1H-15N HSQC spectrum of S17D mutant (1–109)MDM2 isotopically labeled at residues Val14 and Ala21 in the presence (red) and absence (black) of p53 peptide. (C) HSQC spectrum showing S17 phosphorylated (1–109)MDM2 in the presence (red) and absence (black) of p53 peptide. (D) Overlay of HSQC spectra illustrating the chemical shift assignments of Ala21 and Val14 in (1–24)MDM2.
As shown in Figure 8A, binding of (15–29)p53 to (1–109)MDM2 triggered a significant change in chemical shift to Ala21, but little to Val14. In fact, the cross-peaks of Ala21 and Val14 were substantially more dispersed in the apo protein than those in the peptide-protein complex. These findings suggest that the N-terminal lid peptide in the apo protein (1–109)MDM2 likely adopts a partially ordered conformation of which Ala21 is part but Val14 is not; further, p53 peptide binding to (1–109)MDM2 destabilizes lid conformation involving Ala21. Given that the chemical shifts of 15N-Val14 and 15N-Ala21 of (1–24)MDM2 in its free form (Figure 8D) were nearly identical to the values obtained for (1–109)MDM2 in complex with (15–29)p53, it is obvious that binding of (15–29)p53 to (1–109)MDM2 displaces the partially structured lid peptide, leaving it fully disordered and in no direct contact with the peptide-protein complex. These results are in support of the published NMR studies26,27.
Importantly, the HSQC spectra of all three MDM2 proteins are identical upon binding to (15– 29)p53 (Figure 8A–C), suggesting that the lid peptide is displaced by the ligand and unstructured in the complex irrespective of whether or not24 Ser17 is phosphorylated or mutated to Asp. In the absence of the ligand, the HSQC spectra of the three MDM2 proteins are, again, highly similar, indicating that the S17D mutation or Ser17 phosphorylation does not induce significant conformational changes to the apo protein. A close examination of the HSQC spectra of the three unliganded MDM2 proteins revealed that Ser17 phosphorylation and the S17D mutation caused a modest downfield shift of the 1H signal of Ala21 by 0.1 ppm and 0.05 ppm. We speculate that these changes may reflect a marginal conformational stabilization of the lid peptide in the apo protein afforded by intra-molecular electrostatic interactions involving the negative change(s) introduced at position 17.
Finally, we analyzed the three full-length apo proteins in aqueous buffer using circular dichroism spectroscopy (Figure 9A), and the spectral data revealed the following. First, all three proteins, as expected, adopted a largely α-helical conformation characterized by the double negative peaks at 208 and 222 nm and the single positive peak at 195 nm, consistent with the known structural features of MDM224,25,37. Second, introduction of the negative charge(s) in the lid peptide via the S17D mutation or Ser17 phosphorylation slightly increased protein α-helicity – an outcome presumably attributed by charge-mediated conformational stabilization of the lid peptide, in agreement with the above-mentioned NMR findings. Nevertheless, such stabilization is likely to be local because it clearly did not impact peptide binding to MDM2 in any meaningful way as amply demonstrated by our functional studies. In support of these findings, a chemical denaturation study of the three apo-MDM2 proteins (10 µM each), monitored by CD spectroscopy at 222 nm in response to increasing concentrations of GuHCl43,44, showed that Ser17 phosphorylation had little effect on protein global stability (Figure 9B). In fact, an identical value (4.4 ± 0.1 Kcal/mol) of the free energy change (ΔΔG) associated with protein denaturation was obtained for both (1–109)MDM2 and (1–109)MDM2 pS17.
Figure 9.
(A) CD spectra of (1–109)MDM2, (1–109)MDM2 S17D and (1–109)MDM2 pS17 collected at room temperature at 20 µM in 10 mM phosphate buffer, pH 7.2. (B) GuHCl-induced two-state denaturation of (1–109)MDM2, (1–109)MDM2 S17D and (1–109)MDM2 pS17, each at 10 µM in 10 mM phosphate buffer, pH 7.2, monitored at room temperature by CD spectroscopy at 222 nm. Non-linear regression analyses yielded the free energy changes (ΔG) associated with protein unfolding of 4.4 ± 0.1 kcal/mol for (1–109)MDM2, 4.8 ± 0.1 kcal/mol for (1–109)MDM2 S17D, and 4.4 ± 0.1 kcal/mol for (1–109)MDM2 pS17.
Discussion
How protein phosphorylation plays a critical regulatory role in stress-induced p53 activation remains poorly understood at the molecular level. This enigma is exemplified by the ongoing debate over the structural and functional importance of MDM2 phosphorylation at Ser17 in the N-ternimal lid peptide region. The controversy is aggravated by the paucity of robust technical means to produce site-specifically phosphorylated MDM2 for biochemical and biophysical studies - a common deficiency in the field of protein phosphorylation. Historically, studying protein phosphorylation at sites of Ser, Thr, or Tyr invariably involves two types of mutations: (1) Ser/Thr-to-Ala and Tyr-to-Phe to prevent phosphorylation, and (2) Ser/Thr-to-Asp/Glu to mimic phosphorylation59. However, since phosphorylated side chains of Ser and Thr differ considerably in chemical structure from the carboxylate group of Asp/Glu, the Ser/Thr-to-Asp/Glu mutation failing to faithfully recapitulate phospho-Ser/Thr functionality is well documented in the literature59. Obviously, site-specifically phosphorylated MDM2 proteins are endowed with a natural advantage over their phospho-mimetic counterparts in unveiling the molecular mechanisms leading to stress-induced p53 activation.
Several powerful technologies enable site-specific incorporation of phospho-amino acids into proteins, including total chemical protein synthesis via native chemical ligation32,33,60, protein semisynthesis via expressed protein ligation59,61,62, and protein in vitro translation via nonsense codon suppression63. In this report we describe the use of native chemical ligation for the synthesis of (1–109)MDM2 and its Ser17-phosphorylated analog (1–109)MDM2 pS17 as well as (1–109)MDM2 S17D. Extensive biochemical and biophysical characterizations of these MDM2 proteins have confirmed some of the published work but also contradicted other aspects of it. For example, we have shown that the N-terminal lid peptide (residues 1–24) adopts a partially structured conformation in (1–109)MDM2, which impairs MDM2 binding by p53-derived peptide ligands in a size-dependent manner, but not by the small antagonist nutlin-3a. Further, (15–29)p53 binding to (1–109)MDM2 fully displaces the lid peptide and renders it completely disordered in the peptide-protein complex. These results largely support, at the structural and functional levels, the “closed” model that the lid peptide occludes the p53-binding site on apo-MDM226,27, but contrast the “open” model that suggests a positive regulatory role of the lid peptide in p53-MDM2 interactions29,30.
In both models, however, Ser17 phosphorylation is hypothesized to be functionally important by formation of a putative salt bridge with some cationic residues on the surface of MDM2 to stabilize the lid peptide albeit opposite functional effects purported26,27,29–31. While our NMR and CD data are in accord with the presence of a pSer17- or Asp17-mediated, lid-stabilizing force of an electrostatic nature in apo-(1–109)MDM2, neither Ser17 phosphorylation nor the phosphomimetic mutation S17D exerts any functional impact on MDM2 binding by p53-derived peptide ligands. In fact, Ser17 phosphorylation or its mutation to Asp does not stabilize apo-MDM2 globally, nor does it stabilize the fully disordered lid peptide of MDM2 upon binding by the (15–29)p53 peptide. These results are consistent with the widely accepted tenet that surface-exposed electrostatic interactions (or salt bridges) contribute little to protein stability64–66. Our work demonstrates that while pSer17 can potentially participate in intra-molecular electrostatic interactions to stabilize the lid peptide in apo-MDM2, the energetic effects are likely localized, transient and weak, as the displacement of the lid peptide in apo-MDM2 by p53-derived peptide ligands is independent of whether Ser17 is phosphorylated or not at both the structural and functional levels.
Our finding that MDM2 phosphorylation at Ser17 is functionally neutral with respect to the p53-MDM2 interaction suggests that protein phosphorylation at a single site is probably not as important as previously thought for p53 activation – a viewpoint that is gaining growing acceptance12,22,67. Our finding also highlights discrepancies that often exist in the literature between biochemical and biological data over the importance of protein phosphorylation for p53 activation. For example, the protein kinases ATM and DNA-PK phosphorylate p53 at Ser15 in vitro and in vivo in response to DNA damage, a process thought to lead p53 activation through impairing the ability of MDM2 to antagonize the tumor suppressor68–70. The highly conserved Ser15 conveniently flanks the transactivation domain of p53 and is in close proximity to MDM2 in the p53-MDM2 complex24. However, biochemical studies show that a p53 peptide phosphorylated at Ser15 is functionally indistinguishable from its un-phosphorylated counterpart towards MDM2 binding67,71. Similarly, we have shown that Ser15 phosphorylation, while improving (15–29)p53 binding to (1–109)MDM2 by a factor of 2, is functionally neutral with the truncated (25–109)MDM2 protein (Table 1 and Figure 10). Importantly, both (1–109)MDM2 and (1–109)MDM2 pS17 display nearly identical binding affinities for (15–29)p53 pS15, indicative of a lack of potential electrostatic repulsion between pS15 of p53 and pS17 of MDM2 in the complex.
Figure 10.
Interactions of (15–29)p53 pS15 with (25–109)MDM2, (1–109)MDM2 and (1–109)MDM2 pS17 as quantified by the SPR-based competitive binding assay (see Figures 5 and 7). Whereas (1–109)MDM2 and (1–109)MDM2 pS17 were used at 250 nM each for the relatively weak binding (A), 50 nM (25–109)MDM2 was used for the strong binding (B). Each curve is the mean of three independent measurements, and the Kd values (mean ± SEM, n=3) are listed in Table 1.
Conclusion
We have found that the partially structured N-terminal lid of apo-MDM2 occludes p53 peptide binding in a ligand size-dependent manner and becomes fully disordered in the peptide-protein complex. Neither Ser17 phosphorylation nor the phospho-mimetic mutation S17D imposes any functional influence on p53 peptide binding to MDM2. As is the case with the studies of p53 phosphorylation at Ser15, our work on MDM2 phosphorylation at Ser17 does not provide a ready explanation for the in vitro finding that DNA-PK phosphorylates Ser17 and renders MDM2 unable to bind p53, leading to p53 activation23. The biochemical data described here also offer no clue as to why the phospho-mimetic mutation S17D is reported to enhance the p53-MDM2 interaction and promote MDM2-mediated p53 degradation in vitro 29,30. The two seemingly contradictory functional observations may be difficult to be reconciled in the absence of any definitive structural information on the interaction between the full-length MDM2 and p53 proteins. It is plausible that Ser17 phosphorylation may “initiate” a series of subsequent phosphorylation events at other sites in MDM2, ultimately resulting in p53 dissociation from the inhibitory complex. Nevertheless, these reported discrepancies clearly demonstrate our still limited understanding of the complexity of protein phosphorylation and of how p53 escapes from MDM2-mediated functional inactivation at the molecular and cellular levels.
Supplementary Material
Acknowledgement
This work was partially supported by the National Institutes of Health Grants AI072732 and AI087423 and the Overseas Scholars Collaborative Research Grant 81128015 (to W.L.) by the National Natural Science Foundation of China.
Footnotes
Supporting information (25–109)MDM2 K36C, (1–109)MDM2 K36C, and (1–109)MDM2 K36C/S17D characterized by RP-HPLC and ESI-MS; interactions of (25–109)MDM2 and/or (1–109)MDM2 with fluorescently labeled (15–29)p53 or (1–24)MDM2 peptides as quantified by fluorescence polarization techniques; circular dichroism spectra of (1–24)MDM2 in solution.
References
- 1.Cohen P. Nat. Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Scott JD. Trends Biochem. Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 5.Lane DP. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 6.Levine AJ, Oren M. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vousden KH, Lane DP. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 8.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Nat. Rev. Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 9.Toledo F, Wahl GM. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 10.Wade M, Wang YV, Wahl GM. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marine J, Dyer M. J. Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 12.Kruse J-P, Gu W. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 14.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 15.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RCA, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 16.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 17.Kubbutat M, Jones S, Vousden K. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 18.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S, Yuan ZM. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vousden K, Prives C. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 21.Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashcroft M, Kubbutat M, Vousden K. Mol. Cell. Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayo LD, Turchi JJ, Berberich SJ. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 24.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 25.Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG, Lu W. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy MA, Gesell JJ, Senior MM, Wyss DF. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1645–1648. doi: 10.1073/pnas.0334477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Showalter SA, Bruschweiler-Li L, Johnson E, Zhang F, Brüschweiler R. J. Am. Chem. Soc. 2008;130:6472–6478. doi: 10.1021/ja800201j. [DOI] [PubMed] [Google Scholar]
- 28.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 29.Worrall EG, Wawrzynow B, Worrall L, Walkinshaw M, Ball KL, Hupp TR. J. Chem. Biol. 2009;2:113–129. doi: 10.1007/s12154-009-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worrall EG, Worrall L, Blackburn E, Walkinshaw M, Hupp TR. J. Mol. Biol. 2010;398:414–428. doi: 10.1016/j.jmb.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 31.Dastidar SG, Raghunathan D, Nicholson J, Hupp TR, Lane DP, Verma CS. Cell Cycle. 2011;10:82–89. doi: 10.4161/cc.10.1.14345. [DOI] [PubMed] [Google Scholar]
- 32.Dawson PE, Kent SBH. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 33.Dawson P, Muir T, Clark-Lewis I, Kent S. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 34.Schnölzer M, Alewwod P, Jones A, Alewood D, Kent SBH. Int. J. Pept. Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 35.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harker EA, Daniels DS, Guarracino DA, Schepartz A. Bioorg. Med. Chem. 2009;17:2038–2046. doi: 10.1016/j.bmc.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czarna A, Popowicz GM, Pecak A, Wolf S, Dubin G, Holak TA. Cell Cycle. 2009;8:1176–1184. doi: 10.4161/cc.8.8.8185. [DOI] [PubMed] [Google Scholar]
- 38.Heyduk T, Lee JC. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1744–1748. doi: 10.1073/pnas.87.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 40.Bax A, Ikura M. J. Biomol. NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6 doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. J. Magn. Reson. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 43.Lu W, Starovasnik M, Dwyer J, Kossiakoff A, Kent S, Lu W. Biochemistry. 2000;39:3575–3584. doi: 10.1021/bi992292q. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z, Alexandratos J, Ericksen B, Lubkowski J, Gallo RC, Lu W. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11587–11592. doi: 10.1073/pnas.0404649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Botos I, Wu Z, Lu W, Wlodawer A. FEBS Lett. 2001;509:90–94. doi: 10.1016/s0014-5793(01)03113-1. [DOI] [PubMed] [Google Scholar]
- 46.Shao Y, Lu W, Kent SBH. Tetrahedron Lett. 1998;39:3911–3914. [Google Scholar]
- 47.Li C, Wu Z, Liu M, Pazgier M, Lu W. Protein Sci. 2008;17:1624–1629. doi: 10.1110/ps.036145.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C, Li X, Lu W. Biopolymers. 2010;94:487–494. doi: 10.1002/bip.21375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blanco-Canosa JB, Dawson PE. Angew. Chem. Int. Ed. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, Pazgier M, Li C, Yuan W, Liu M, Wei G, Lu W-Y, Lu W. J. Mol. Biol. 2010;398:200–213. doi: 10.1016/j.jmb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jameson DM, Ross JA. Chem. Rev. 2010;110:2685–2708. doi: 10.1021/cr900267p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jameson DM, Seifried SE. Methods. 1999;19:222–233. doi: 10.1006/meth.1999.0853. [DOI] [PubMed] [Google Scholar]
- 53.Popowicz GM, Dömling A, Holak TA. Angew. Chem. Int. Ed. 2011;50:2680–2688. doi: 10.1002/anie.201003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M, Li C, Pazgier M, Li C, Mao Y, Lv Y, Gu B, Wei G, Yuan W, Zhan C, Lu W-Y, Lu W. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawson PE. Isr. J. Chem. 2011;51:862–867. [Google Scholar]
- 56.Li C, Pazgier M, Li J, Li C, Liu M, Zou G, Li Z, Chen J, Tarasov SG, Lu W-Y, Lu W. J. Biol. Chem. 2010;285:19572–19581. doi: 10.1074/jbc.M110.116814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu M, Pazgier M, Li C, Yuan W, Li C, Lu W. Angew. Chem. Int. Ed. 2010;49:3649–3652. doi: 10.1002/anie.201000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li C, Pazgier M, Liu M, Lu W-Y, Lu W. Angew. Chem. Int. Ed. 2009;48:8712–8715. doi: 10.1002/anie.200904550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarrant MK, Cole PA. Annu. Rev. Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kent SBH. Chem. Soc. Rev. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 61.Schwarzer D, Cole PA. Curr. Opin. Chem. Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 62.Muir TW, Sondhi D, Cole PA. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noren C, Anthony-Cahill S, Griffith M, Schultz P. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 64.Sali D, Bycroft M, Fersht A. J. Mol. Biol. 1991;220:779–788. doi: 10.1016/0022-2836(91)90117-o. [DOI] [PubMed] [Google Scholar]
- 65.Serrano L, Horovitz A, Avron B, Bycroft M, Fersht A. Biochemistry. 1990;29:9343–9352. doi: 10.1021/bi00492a006. [DOI] [PubMed] [Google Scholar]
- 66.Horovitz A, Serrano L, Avron B, Bycroft M, Fersht A. J. Mol. Biol. 1990;216:1031–1044. doi: 10.1016/S0022-2836(99)80018-7. [DOI] [PubMed] [Google Scholar]
- 67.Lee CW, Ferreon JC, Ferreon ACM, Arai M, Wright PE. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canman C, Lim D, Cimprich K, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M, JD S. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 69.Banin S, Moyal L, Shieh S, Taya Y, Anderson C, Chessa L, Smorodinsky N, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 70.Shieh S-Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 71.Schon O, Friedler A, Bycroft M, Freund SMV, Fersht AR. J. Mol. Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.