Abstract
Background
Bt-maize is a transgenic variety of maize expressing the Cry toxin from Bacillus turingiensis. The potential accumulation of the relative effect of the transgenic modification and the cry toxin on the rhizobacterial communities of Bt-maize has been monitored over a period of four years.
Methodology/Principal Findings
The accumulative effects of the cultivation of this transgenic plant have been monitored by means of high throughput DNA pyrosequencing of the bacterial DNA coding for the 16S rRNA hypervariable V6 region from rhizobacterial communities. The obtained sequences were subjected to taxonomic, phylogenetic and taxonomic-independent diversity studies. The results obtained were consistent, indicating that variations detected in the rhizobacterial community structure were possibly due to climatic factors rather than to the presence of the Bt-gene. No variations were observed in the diversity estimates between non-Bt and Bt-maize.
Conclusions/Significance
The cultivation of Bt-maize during the four-year period did not change the maize rhizobacterial communities when compared to those of the non-Bt maize. This is the first study to be conducted with Bt-maize during such a long cultivation period and the first evaluation of rhizobacterial communities to be performed in this transgenic plant using Next Generation Sequencing.
Introduction
Rhizobacterial communities are a key element of soil quality and fertility [1]. The cultivation of genetically modified plants can alter these soil bacterial communities, hence endangering cultivar sustainability. Among these genetically modified maize, the so-called Bt-maize, harbours the Bacillus thuringiensis gene, which encodes the Cry protein, rendering the plant resistant to the attack of corn borer Lepidopterae.
The presence of the Bt protein potentially modifies the composition of root exudates of the transgenic plants and additionally may exerts a direct effect on non-target species of soil microorganisms [2], [3], [4], [5].
Many techniques have been used to analyse soil bacterial communities, including classic approaches based on the cultivation of viable bacteria, metabolic profiling studies and nucleic acid-based methods [6]. Most of the existing studies to date suggest that the cultivation of transgenic Bt-maize plants has minor effects or no effects at all on the rhizobacterial communities. However, these studies have not employed the Next Generation Sequencing (NGS) technique, which, upon being used for the sequencing of the SSU rRNA hypervariable regions, has proven to be very useful for the diversity studies of bacterial communities in many habitats, including soil [7], [8], [9], [10], [11]. Moreover, few studies are available for long-term periods of Bt-maize cultivation. We monitored the potential accumulation of the relative effects of Bt-maize on the rhizobacterial communities when compared to non-Bt maize by using NGS over a four-year period of cultivation. To this end, we have examined the structure of the rhizobacterial communities using a taxonomic approach [12], [13] conducted phylogenetic distances analyses [14], [15], [16], and bacterial diversity estimations. It has recently been reported that differences in diversity estimation may depend on the method used [17], [18], [19], therefore, we employed two commonly used methods to calculate diversity [12], [19], which have proven to be appropriate for the number and length of the sequences obtained [20].
To our knowledge, this is the first study conducted on rhizobacterial communities of Bt-maize during a four-year period using NGS, and the results obtained may be relevant as to a potential renewal of the authorisation to cultivate Bt-maize in the European Union.
Results
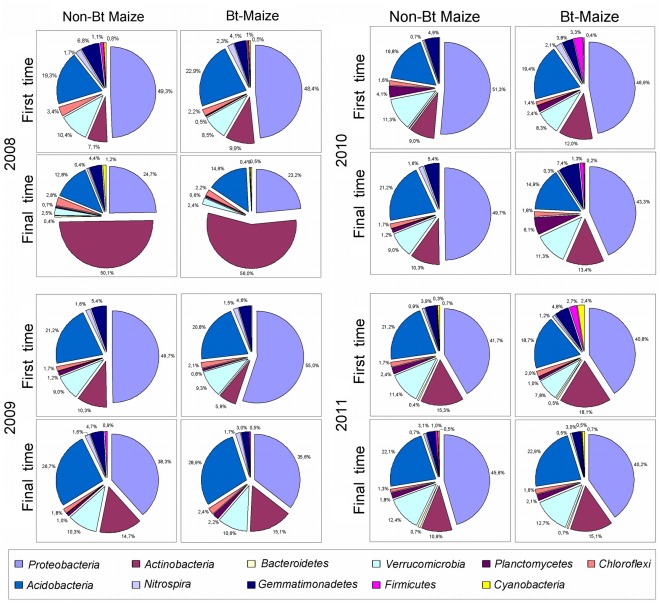
Sequences obtained from the Bt-maize and its non-transgenic counterpart at two different sampling times were compared with the sequences present in the RDP database and grouped using the MEGAN program and NCBI taxonomy to generate the taxonomic trees. Figure 1 shows the taxonomic breakdown as a result of sequencing the hypervariable V6 region of the 16S rDNA genes, and the corresponding taxonomic trees are included in figure S1 as Supplementary Information. The prominent phyla in all the samples were Proteobacteria, Actinobacteria and Acidobacteria.
Figure 1. Taxonomic breakdown of the more relevant phyla.
The percentages of Proteobacteria, Actinobacteria, Bacteroidetes, Verrucomicrobia, Planctomycetes, Chloroflexi, Acidobacteria, Nitrospira, Gemmatimonadetes, Firmicutes and Cyanobacteria are indicated and do not include the unassigned sequences. Unclassified sequences were not included as they were of no taxonomic use.
A total of 1959 sequences were analysed from the rhizosphere of non-Bt maize at the first sampling time in the first year; 22.6% of these remained unassigned. From the assigned sequences, 49.3% belonged to the Proteobacteria phylum, 19.3% were Acidobacteria, 7.1% Actinobacteria and 23.7% belonged to other taxa. A very similar distribution of taxa (48.4% Proteobacteria, 22.9% Acidobacteria, 9.9% Actinobacteria, and 18.3% other taxa) was found for the 2445 total sequences from the Bt-maize rhizosphere at an equivalent sampling time, where 24.4% of sequences remained unassigned. At the final sampling time, Proteobacteria and Acidobacteria were again prominent among the assigned sequences: 24.7% and 12.8% respectively in the non-Bt maize (2680 total sequences), and 23.2% and 14.6% respectively in the Bt maize (2534 total sequences), while the presence of Actinobacteria markedly increased to 50.1% in the non-Bt maize and 56.0% in Bt maize. 23.6% and 21.2% were unassigned sequences respectively in the non-Bt and Bt-maize.
In the second year, the distribution of taxa in the assigned sequences of the non-Bt and Bt-maize was similar to that of the previous year at the first sampling time; Proteobacteria, Actinobacteria and Acidobacteria were predominant (Fig. 1). At the first sampling time 24.3% and 24.4% of the sequences remained unassigned for the non-Bt and Bt-maize, yielding a total of 2334 and 2777 sequences respectively, while at the second sampling time 22.1% and 22.6% of sequences remained unassigned, with 2177 and 2374 total sequences for the non-Bt and Bt-maize respectively.
The relative presence of the three major phyla was similar in the samples from the third year (Fig. 1), where 22.9% of the sequences were unassigned at the first sampling time for the non-Bt maize (2916 total sequences) and 19.2% for the Bt-maize (3344 total sequences); 21.0% of the sequences were unassigned for the non-Bt maize at the final sampling time, and 22.9% for the Bt-maize, with 4236 and 4523 total sequences respectively.
In the fourth year, an equivalent presence of the predominant phyla was once again found at the first and final sampling time in the non-Bt or Bt-maize, where the relative abundance of Proteobacteria, Actinobacteria and Acidobacteria remained comparatively similar in all cases (Fig. 1). The total number of sequences was 13332 for the non-Bt maize and 24084 for the Bt-maize at the first sampling time, with 15.8% and 16.5% of unassigned sequences respectively. The unassigned sequences were 16.2% for the non-Bt maize and 13.9% for Bt-maize at the final sampling time, with 36314 and 27202 total sequences for the non-Bt and Bt-maize respectively.
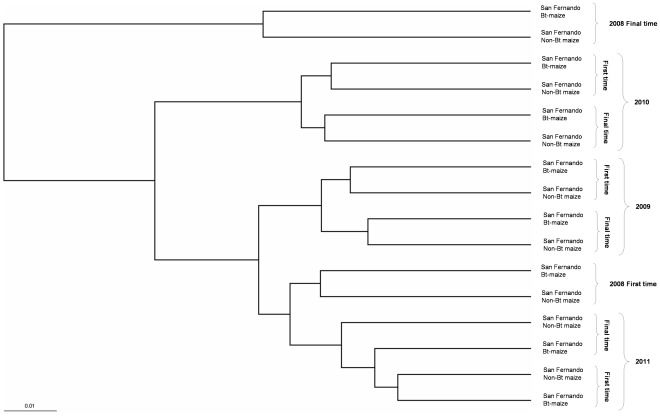
The hierarchical clustering tree of samples based on the UniFrac metric (Fig. 2) shows that the rhizobacterial communities from each particular year are grouped in a separate branch of the tree, except for the year 2008, where samples from the first sampling time were grouped with samples from 2011, and samples from the final sampling time appeared as an independent branch of the tree, as they differ in taxonomic composition (Fig. 1). Samples are also grouped within each year depending on the sampling time. The UniFrac significance test showed no statistical differences between the non-Bt and Bt-maize within each year at any sampling time.
Figure 2. Fast UniFrac hierarchical clustering tree.
Analysis of the different soil samples was carried out using normalised abundance weights.
When estimating bacterial diversity, we have previously shown that ESPRIT as well as the web-based workflow, RDP pyrosequencing pipeline, produced the more accurate equivalent results as to the size and length of the hypervariable V6 region of the 16S rDNA sequences obtained from the non-Bt and Bt-maize rhizospheres when compared to other methodologies of analysis [20], therefore, we have used both of them in a comparative manner. Table 1 shows a comparison of the OTUs analyses performed using the RDP pyrosequencing pipeline compared to using the ESPRIT package, at three different dissimilarity levels. Species richness was determined using the Chao1 and ACE estimators for the ESPRIT software, and only Chao1 for the RDP software, since it does not use the ACE estimator (Table 1). No significant differences were detected among the bacterial communities from the non-Bt or Bt-maize, or from one sampling time to another or even from one year to another. The number of OTUs estimated when using the ESPRIT package was always slightly lower, as previously reported [19], however the Chao1 estimator was slightly higher when calculated with RDP, probably due to its own implementation of the algorithm. In all cases the results were consistent irrespective of the methodology used.
Table 1. Similarity-based OTUs and species richness estimates at 3%, 5% and 10% dissimilarity levels.
| OTUs | ACE | Chao1 | OTUs | ACE | Chao1 | ||||||||||
| 2008 | First time | non-Bt | RDP | 0.03 | 1148 | NC | 2480±279 | 2010 | First time | non-Bt | RDP | 0.03 | 952 | NC | 2126±266 |
| 0.05 | 846 | NC | 1608±164 | 0.05 | 765 | NC | 1429±172 | ||||||||
| 0.1 | 527 | NC | 670±49 | 0.1 | 465 | NC | 668±77 | ||||||||
| ESPRIT | 0.03 | 1031 | 1842.63 | 1895±194 | ESPRIT | 0.03 | 916 | 2117.53 | 1894±227 | ||||||
| 0.05 | 728 | 1110.54 | 1065±98 | 0.05 | 745 | 1531.48 | 1430±182 | ||||||||
| 0.1 | 316 | 341.97 | 333±14 | 0.1 | 433 | 568.97 | 534±44 | ||||||||
| Bt | RDP | 0.03 | 1135 | NC | 2469±272 | Bt | RDP | 0.03 | 920 | NC | 2199±297 | ||||
| 0.05 | 918 | NC | 1528±173 | 0.05 | 711 | NC | 1289±156 | ||||||||
| 0.1 | 541 | NC | 745±81 | 0.1 | 425 | NC | 592±69 | ||||||||
| ESPRIT | 0.03 | 1035 | 2075 | 2009±215 | ESPRIT | 0.03 | 886 | 2228.54 | 1970±259 | ||||||
| 0.05 | 795 | 1309.08 | 1269±126 | 0.05 | 692 | 1315.88 | 1179±136 | ||||||||
| 0.1 | 447 | 549.7 | 540±42 | 0.1 | 406 | 543.15 | 529±54 | ||||||||
| Final time | non-Bt | RDP | 0.03 | 1053 | NC | 2820±406 | Final time | non-Bt | RDP | 0.03 | 1011 | NC | 2414±311 | ||
| 0.05 | 797 | NC | 1687±211 | 0.05 | 815 | NC | 1595±196 | ||||||||
| 0.1 | 498 | NC | 703±50 | 0.1 | 537 | NC | 807±94 | ||||||||
| ESPRIT | 0.03 | 982 | 1951.57 | 2087±260 | ESPRIT | 0.03 | 1010 | 2508.72 | 2201±260 | ||||||
| 0.05 | 755 | 1247.91 | 1303±153 | 0.05 | 805 | 1556.53 | 1384±146 | ||||||||
| 0.1 | 415 | 502.03 | 506±44 | 0.1 | 469 | 642.18 | 630±65 | ||||||||
| Bt | RDP | 0.03 | 1216 | NC | 2230±258 | Bt | RDP | 0.03 | 999 | NC | 2420±317 | ||||
| 0.05 | 963 | NC | 1474±179 | 0.05 | 819 | NC | 1783±243 | ||||||||
| 0.1 | 568 | NC | 679±71 | 0.1 | 493 | NC | 688±73 | ||||||||
| ESPRIT | 0.03 | 1029 | 1992.92 | 1947±204 | ESPRIT | 0.03 | 1015 | 2679.5 | 2341±293 | ||||||
| 0.05 | 726 | 1095.15 | 1013±84 | 0.05 | 821 | 1692.52 | 1596±196 | ||||||||
| 0.1 | 326 | 352.58 | 342±14 | 0.1 | 485 | 632.34 | 609±51 | ||||||||
| 2009 | First time | non-Bt | RDP | 0.03 | 1307 | NC | 2565±350 | 2011 | First time | non-Bt | RDP | 0.03 | 898 | NC | 2138±296 |
| 0.05 | 1033 | NC | 1661±204 | 0.05 | 691 | NC | 1300±171 | ||||||||
| 0.1 | 608 | NC | 663±85 | 0.1 | 429 | NC | 606±71 | ||||||||
| ESPRIT | 0.03 | 1105 | 2395.43 | 2287±249 | ESPRIT | 0.03 | 847 | 1916.25 | 1774±228 | ||||||
| 0.05 | 865 | 1583.68 | 1602±186 | 0.05 | 680 | 1265.76 | 1204±149 | ||||||||
| 0.1 | 473 | 602.51 | 593±51 | 0.1 | 398 | 523.2 | 517±53 | ||||||||
| Bt | RDP | 0.03 | 1184 | NC | 2454±253 | Bt | RDP | 0.03 | 884 | NC | 2253±332 | ||||
| 0.05 | 910 | NC | 1650±178 | 0.05 | 690 | NC | 1430±208 | ||||||||
| 0.1 | 503 | NC | 654±60 | 0.1 | 407 | NC | 540±58 | ||||||||
| ESPRIT | 0.03 | 1104 | 2315.27 | 2303±255 | ESPRIT | 0.03 | 852 | 1990.86 | 2068±609 | ||||||
| 0.05 | 847 | 1445.27 | 1448±154 | 0.05 | 651 | 1178.58 | 1194±160 | ||||||||
| 0.1 | 470 | 574.01 | 556±39 | 0.1 | 360 | 441.06 | 429±35 | ||||||||
| Final time | non-Bt | RDP | 0.03 | 1252 | NC | 3070±395 | Final time | non-Bt | RDP | 0.03 | 902 | NC | 2068±273 | ||
| 0.05 | 1010 | NC | 1897±241 | 0.05 | 717 | NC | 1318±161 | ||||||||
| 0.1 | 596 | NC | 819±76 | 0.1 | 411 | NC | 512±53 | ||||||||
| ESPRIT | 0.03 | 1063 | 2108.05 | 1899±179 | ESPRIT | 0.03 | 853 | 1961.8 | 1730±213 | ||||||
| 0.05 | 770 | 1266.42 | 1234±128 | 0.05 | 675 | 1235.8 | 1095±119 | ||||||||
| 0.1 | 362 | 397.08 | 387±18 | 0.1 | 370 | 460.93 | 433±32 | ||||||||
| Bt | RDP | 0.03 | 1204 | NC | 2730±301 | Bt | RDP | 0.03 | 868 | NC | 2184±326 | ||||
| 0.05 | 904 | NC | 1636±176 | 0.05 | 665 | NC | 1208±155 | ||||||||
| 0.1 | 539 | NC | 728±71 | 0.1 | 380 | NC | 529±65 | ||||||||
| ESPRIT | 0.03 | 996 | 1808.36 | 1810±186 | ESPRIT | 0.03 | 828 | 1925.67 | 1793±242 | ||||||
| 0.05 | 716 | 1112.28 | 1062±101 | 0.05 | 648 | 1211 | 1153±149 | ||||||||
| 0.1 | 338 | 367.01 | 354±13 | 0.1 | 357 | 456.86 | 444±43 |
The species richness estimates were determined using the RDP pyrosequencing pipeline or the ESPRIT program, as described in Materials and Methods. NC = non computable.
Discussion
In this study the potentially cumulative effect of Bt-maize cultivation on rhizobacterial communities has been monitored over a four-year continuous cultivation of MON810 maize. The effect of Bt-maize is not only exerted by the exudation of the Cry1Ab toxin through the roots [4], as it is constitutively produced within the whole plant and Cry proteins may also confer unexpected properties to the plant, for example, by increasing lignin content [21] or provoking gene instability at or around the insertion site of the Bt coding gene in the plant genome [22]. Furthermore, transgene rearrangements may occur with a potential to produce changes in plant gene expression and phenotype [23]. The fate and effects of insect-resistant toxin in soil ecosystems have been reviewed [2], and the Cry1Ab protein (event MON810) has been detected to remain in soils after four years of Bt-maize cultivation in field conditions, whereas other Cry proteins as Cry3Bb1 (event MON863) were not [24]. Cry proteins are rapidly adsorbed to clay minerals, which render the proteins resistant to biodegradation in soil, thus facilitating a potential longer exposure of non-target organisms to the toxin [2], [25], [26]. The effect of Cry root exudates on different soil non-target organisms (namely earthworms, nematodes and protozoa) has been investigated extensively with apparently little significance or none all, while infective fungal mycorrhizae could colonize Bt maize roots more efficiently than non-Bt ones, and so the persistence of Cry proteins in soil may also be related to the decrease of some microbial activity [2].
There has been an increasing concern in recent years with respect to altering the composition of soil microbial communities and the studies performed differ in their objective and methodology. This four-year study was conducted to get a further insight into the possible harmful effect that the continued presence of Bt-toxin may exert on the structure of maize rhizobacterial communities.
In our study we found three predominant phyla in all the rhizospheres: Proteobacteria, Acidobacteria and Actinobacteria. The Proteobacteria are of great importance to global carbon, nitrogen and sulfur cycling [27]; Actinobacteria are an important component of soil communities playing a major role in organic matter turnover in soils, due to their ability to decompose organic materials [28]; Acidobacteria are commonly detected in soils [29] and it has been suggested that members of this phylum are likely to play a relevant role in conducting processes in terrestrial ecosystems [30].
The overall distribution of these three phyla did not change from the non-Bt to the Bt-maize at any sampling time. The reason why the structure of the rhizobacterial community changed at the final sampling time in the first year is still unknown, however, a particular climate change comprising a heavier period of rainfall between the first and final sampling times was registered, when almost three times more rain accumulated than during the same period in subsequent years (http://clima.meteored.com). At this particular sampling time the Actinobacteria became very predominant, which is compensated mostly by a reduction in Proteobacteria, and to a lesser extent in Acidobacteria (Fig. 1). It is not infrequent to see that climate changes alter the structure of rhizobacterial communities, and this has also been reported in maize long-term cultivation studies [31]. It is also remarkable to observe the natural resilience of the rhizobacterial communities, which at the beginning of the second year had recovered completely from the climatic episode.
No consistent statistically significant differences have been reported in the number of different groups of microorganisms, enzyme activities or pH between non-Bt and Bt-maize rhizospheres [2], or in soil improved with Bt-maize biomass [32]. Other short-term experiments using techniques such as Phospholipid Fatty Acid profiles (PLFA) [33], polymerase chain reaction–denaturing gradient gel electrophoresis (PCR–DGGE) [34], automated ribosomal intergenic spacer analysis (ARISA) [35] or genome-wide commercially available DNA microarrays [36] concluded that the effects of transgenic Bt-maize on the bacterial community structure are minimal, and that the growth stage of plant or environmental factors may exert a more noticeable effect on the microbial community. The UniFrac statistical phylogenetic analysis did not show significant differences between the non-Bt maize and Bt- maize rhizospheres at any sampling time, and grouped the samples according to the sampling time, confirming the trends observed in the taxonomic analysis (Fig. 2).
The bacterial diversity analysis showed no differences in species abundance or richness between non-Bt and Bt-maize at any sampling point estimated with the RDP pyrosequencing pipeline or the ESPRIT software (Table 1). Moreover, no differences were found throughout the four-year experiment despite changes in the community structure. Although the two methods used to analyse bacterial diversity showed some minor differences, both of them offered consistent results when estimating the diversity of the bacterial communities. These two methods produced qualitatively equivalent results and have been shown to be an adequate choice, both being equally suitable for assessing the diversity of rhizobacterial communities.
Some studies have suggested that the repeated cultivation of transgenic Bt-plants may lead to the accumulation and persistence of Bt proteins in soil [26], [32], [37]. Taking together the results obtained we can conclude that no effects may be attributed to the transgenic Bt-maize when compared to its respective isogenic counterpart over a four-year period of seasonal cultivation. The differences perceived in the composition of the rhizobacterial communities were most likely due to the fluctuations in climate, which affected the non-Bt and Bt-maize almost equally. Nevertheless, further studies concerning soil microbial communities functioning should contribute to a better understanding of the relationship of the bacterial communities with the plant throughout its development.
Materials and Methods
Plant Materials and Sampling
Bt-maize MON810, variety DKC6451YG, expressing the Cry1Ab protein (Monsanto Agricultura, Spain) and its isogenic non-Bt line DKC6450 (Monsanto Agricultura, Spain) were grown in experimental maize fields located in San Fernando de Henares, Madrid, Spain (N40° 25′ 14″ W3° 29′ 30″). Current agricultural practises were maintained throughout the four years of cultivation period and crop residues were removed after each vegetative cycle. The surface of each experimental field was 40 m2; both fields were annexed to each other and separated by a four meters wide path. Four extra rows of non-Bt maize surrounded both fields. The non-Bt and Bt-maize plants were harvested at two different growth stages: about 90 days after seeding (first sampling time), when the plants had around 8 leaves, and just before crop harvesting at final growth (final sampling time). The four extra rows were not sampled. Roots and adhered soil measuring approximately 2 mm or less in diameter were separated from the bulk soil by gently shaking the root system. The term “rhizosphere” describes the carefully separated soil adhered to these roots. Given the small size of the maize fields, these were divided into subplots and 3 samples were taken from each subplot at the time of collection. A total of 9 subplots were collected from each maize field at every collection and an equal amount of soil from each of the 27 samples was pooled in all cases. The experiments were performed during development throughout 2008, 2009, 2010 and 2011. The performed studies did not involved human or animal participants and, in this regard, did not required specific permits.
Texture and Chemical Properties of the Soils
The texture of the soil appears to be loamy containing 16.5% clay, 50% silt and 33.5% sand, as determined by Agriquem S.L. (Seville, Spain). Average organic matter (OM) content was 3.03% with a Cationic Exchange Capacity of 4.7 meq/100 g. Total rainfall registered in the experimental field over the four-year study was collected from http://clima.meteored.com.
DNA Extraction, PCR Amplification and Pyrosequencing
Rhizospheres from each of the different collection times were pooled and the soil was subjected to three independent DNA extractions using the PowerMax Soil DNA kits (MO Bio Laboratories Inc., USA) following instructions from the supplier. Soil DNA from each of the three independent extractions was used as template for PCR amplification of the V6 hypervariable region of the 16S rRNA gene. The oligonucleotide design included 454 Life Science’s Titanium A or B sequencing adaptors fused to the 5′ end of primer 967F (5′- CAACGCGAAGAACCTTACC –3′) and 1046R (5′- CGACAGCCATGCANCACCT –3′), where a MID (Multiplex Identifier) was included immediately preceding the V6 specific primer, allowing the samples to be analysed in a single lane of the 454 pyrosequencer first sampling time. A different MID was included for each sample as indicated in Table 2.
Table 2. Multiplex identifiers (MIDs).
| Year | Samplig time | MID Bt-maize | MID Non Bt-maize |
| 2008 | First | MID1 (5′-ACGAGTGCGT-3′) | MID2 (5′- ACGCTCGACA -3′) |
| Final | MID3 (5′- AGACGCACTC -3′) | MID4 (5′- AGCACTGTAG -3′) | |
| 2009 | First | MID5 (5′-ATCAGACACG-3′) | MID6 (5′- CGTGTCTCTA -3′) |
| Final | MID7 (5′- CGTGTCTCTA -3′) | MID8 (5′- CTCGCGTGTC -3′) | |
| 2010 | First | MID1 (5′-ACGAGTGCGT-3′) | MID2 (5′- ACGCTCGACA -3′) |
| Final | MID3 (5′- AGACGCACTC -3′) | MID4 (5′- AGCACTGTAG -3′) | |
| 2011 | First | MID1 (5′-ACGAGTGCGT-3′) | MID2 (5′- ACGCTCGACA -3′) |
| Final | MID3 (5′- AGACGCACTC -3′) | MID4 (5′- AGCACTGTAG -3′) |
The Multiplex identifiers (MIDs) used for pyrosequencing the different samples are shown.
PCR amplification was performed by incubation at 95°C for 5 min, followed by 30 cycles of incubation at 95°C (30 sec), 63°C (45 sec) and 72°C (1 min), with a final extension cycle of 5 min at 72°C. The amplified DNA resulting from the three independent PCR reactions for each DNA template preparation was pooled and cleaned (Illustra GFX PCR DNA purification kit, GE Healthcare), checked with Bioanalyser 2100 (Agilent technologies), quantified with Quant-IT-picogreen (Invitrogen) and used to make the single strands on beads, as required for 454 Titanium pyrosequencing [38]. The obtained sequences were deposited in the NCBI sequence reads archives (accession number SRA009281).
For taxonomical purposes, the 454 reads for the V6 regions of each soil were filtered to eliminate the short sequences that account for 50% of all pyrosequencing errors [17] and then compared with the RDP database version 10 [12] using BLASTN. Files containing the 25 best matches for each of the 454 determined sequences were used as input to generate the corresponding taxonomic trees by means of the MEGAN 2.0 program [13].
Fast UniFrac (http://bmf2.colorado.edu/fastunifrac) [14], [15], [16] was used to perform a hierarchical clustering of the samples based on their phylogenetic distances. The analysis was conducted with the Greengenes core as a reference phylogenetic tree, Jackknife supporting values, and calculated using normalized abundance weights. The Fast UniFrac significance test was used to assess the existence of statistically significant differences among the 16S rDNA sequences from each soil sample, based on their phylogenetic distances.
To assess taxonomic-independent diversity, an equal number of sequences (1959) were randomly selected from each soil, and the selected pools of sequences were procesed with the RDP pyrosequencing pipelines (http://pyro.cme.msu.edu), which build an initial MSA using the Infernal tool [39] and then proceed directly to perform a complete linkage clustering to cluster OTUs (operational taxonomic units), and calculate species richness with the Chao1 estimator. The obtained results were compared with those generated by the ESPRIT software (using the –f parameter), which avoids the initial multiple sequence alignment step by applying an efficient k-tuple based distance filter, subsequently aligning the sequences using the Needleman-Wunsch method and computing pairwise distances using the quickdist algorithm [19]. Hence, a comparison is made of the efficiency of both methods and species diversity estimation. Regarding species richness estimation, confidence intervals were calculated at a 95% confidence level for all Chao1 data.
Supporting Information
Taxonomic trees. Taxonomic trees resulting from pyrosequencing the V6 region of the 16S rDNA extracted from each field at the indicated sampling times are shown. The size of the dots reflects the relative amount of taxa assigned to each particular node.
(TIF)
Acknowledgments
We wish to thank S. Marín for her technical help.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The Spanish Ministry of the Environment and Rural and Marine affairs has commissioned and supported this research (Grant No. EGO22008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, et al. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem. 2008;40:2843–2853. [Google Scholar]
- 2.Icoz I, Stotzky G. Fate and effects of insect-resistant Bt crops in soil ecosystems. Soil Biol Biochem. 2008;40:559–586. [Google Scholar]
- 3.Liu B, Zeng Q, Yan FM, Xu HG, Xu CR. Effects of transgenic plants on soil microorganisms. Plant Soil. 2005;271:1–13. [Google Scholar]
- 4.Saxena D, Stotzky G. Insecticidal toxin from Bacillus thuringiensis is released from roots of transgenic Bt corn in vitro and in situ. FEMS Microbiol Ecol. 2000;33:35–39. doi: 10.1111/j.1574-6941.2000.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 5.Priestley AL, Brownbridge M. Field trials to evaluate effects of Bt-transgenic silage corn expressing the Cry1Ab insecticidal toxin on non-target soil arthropods in northern New England, USA. Transgenic Res. 2009;18:425–43. doi: 10.1007/s11248-008-9234-z. [DOI] [PubMed] [Google Scholar]
- 6.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acosta-Martinez V, Dowb SE, Sun Y, Wester D, Allen V. Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. App Soil Ecol. 2010;45:3–25. [Google Scholar]
- 8.Barriuso J, Marín S, Mellado RP. Effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities: a comparison with pre-emergency applied herbicide consisting of a combination of acetochlor and terbuthylazine. Environ Microbiol. 2010;12:1021–1030. doi: 10.1111/j.1462-2920.2009.02146.x. [DOI] [PubMed] [Google Scholar]
- 9.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, et al. Pyrosequencing enumerates and contrast soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sogin ML, Morison HG, Huber JL, Welch D, Huse SM, et al. Microbial diversity in the deep sea and the unexplored “rare biosphere”. Proc Nat Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barriuso J, Valverde JM, Mellado RP. Effect of the herbicide glyphosate on the culturable fraction of the glyphosate-tolerant maize rhizobacterial communities using two different growth media. Microbes Environ. 2011;26:332–338. doi: 10.1264/jsme2.me11137. [DOI] [PubMed] [Google Scholar]
- 12.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acid Res. 2009;37:D141. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC. Methods for comparative metagenomics. BMC Bioinformatics 10(Suppl. 2009;1):S12. doi: 10.1186/1471-2105-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughputphylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozupone C, Hamady M, Knight R. UniFrac – an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–98. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schloss PD. The Effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol. 2010;6(7):e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Cai Y, Liu L, Yu F, Farrell ML, et al. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acid Res. 2009;37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barriuso J, Valverde JR, Mellado RP. Estimation of bacterial diversity using Next Generation Sequencing of 16S rDNA: a comparison of different workflows. BMC Bioinformatics. 2011;12:473. doi: 10.1186/1471-2105-12-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena D, Stotzki G. Bt corn has a higher lignin content than non-Bt corn. Am J Botany. 2001;88:1704–1706. [PubMed] [Google Scholar]
- 22.van Leeuwen W, Ruttnik T, Borst Vrenssen AWM, van der Plas LHW, van der Krol AR. Characterisation of position induced spatial and temporal regulation of transgene promoter activity in plants. J Exp Bot. 2001;52:949–959. doi: 10.1093/jexbot/52.358.949. [DOI] [PubMed] [Google Scholar]
- 23.Windels P, Taverniers I, Depicker A, Van Bockstaele E, De Loose M. Characterisation of the Roundup Ready soybean insert. Eur Food Res Technol. 2001;213:107–112. [Google Scholar]
- 24.Icoz I, Saxena D, Andow DA, Zwahlen C, Stotzky G. Microbial populations and enzyme activities in soil in situ under transgenic corn expressing cry proteins from Bacillus thuringiensis. J Environ Qual. 2008;37:647–62. doi: 10.2134/jeq2007.0352. [DOI] [PubMed] [Google Scholar]
- 25.Koskella J, Stotzky G. Microbial utilization of free and claybound insecticidal toxins from Bacillus thuringiensis and their retention of insecticidal activity after incubation with microbes. App Environ Microbiol. 1997;63:3561–3568. doi: 10.1128/aem.63.9.3561-3568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stotzky G. Persistence and biological activity in soil of the insecticidal proteins from Bacillus thuringiensis, especially from transgenic plants. Plant Soil. 2004;266:77–89. [Google Scholar]
- 27.Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, et al. Introduction to the Proteobacteria. In: Dwarkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds). The Prokaryotes, 3rd edn, vol. 5. Springer: New York, 2006;3–37 [Google Scholar]
- 28.Hodgson DA. Primary metabolism and its control in Streptomycetes a most unusual group of bacteria. Ad Microb Physiol. 2000;42:47–238. doi: 10.1016/s0065-2911(00)42003-5. [DOI] [PubMed] [Google Scholar]
- 29.Kielak AM, Pijl AS, van Veen JA, Kowalchuk GA. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2009;3:378–382. doi: 10.1038/ismej.2008.113. [DOI] [PubMed] [Google Scholar]
- 30.Kielak AM, van Veen JA, Kowalchuk GA. Comparative Analysis of Acidobacterial Genomic Fragments from Terrestrial and Aquatic Metagenomic Libraries, with Emphasis on Acidobacteria Subdivision 6. Appl Environ Microbiol. 2010;76:6769–6777. doi: 10.1128/AEM.00343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barriuso J, Marín S, Mellado RP. Potential accumulative effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities over a three-year cultivation period. PLoS ONE. 2011;6(11):e278558. doi: 10.1371/journal.pone.0027558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxena D, Stotzky G. Bacillus thuringiensis (Bt) toxin released from root exudates and biomass of Bt corn has no apparent effect on earthworms, nematodes, protozoa, bacteria, and fungi in soil. Soil Biol Biochem. 2001;33:1225–1230. [Google Scholar]
- 33.Blackwood CB, Buyer JS. Soil microbial communities associated with Bt and non-Bt corn in three soils. J Environ Qual. 2004;33:832–836. doi: 10.2134/jeq2004.0832. [DOI] [PubMed] [Google Scholar]
- 34.Tan FX, Wang JW, Feng YJ, Chi GL, Kong HL, et al. Bt corn plants and their straw have no apparent impact on soil microbial communities. Plant Soil. 2010;329:349–364. [Google Scholar]
- 35.Brusetti L, Francia P, Bertolini C, Pagliuca A, Borin S, et al. Bacterial communities associated with the rhizosphere of transgenic Bt 176 maize (Zea mays) and its non transgenic counterpart. Plant Soil. 2004;266:11–21. [Google Scholar]
- 36.Val G, Marín S, Mellado RP. A sensitive method to monitor Bacillus subtilis and Streptomyces coelicolor-related bacteria in maize rhizobacterial communities: The use of genome-wide microarrays. Micro Ecol. 2009;58:108–115. doi: 10.1007/s00248-008-9451-2. [DOI] [PubMed] [Google Scholar]
- 37.Muchaonyerwa P, Waladde S, Nyamugafata P, Mpepereki S, Ristori GG. Persistence and impact on microorganisms of Bacillus thuringiensis proteins in some Zimbabwean soils. Plant Soil. 2004;266:41–46. [Google Scholar]
- 38.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1713–1713. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomic trees. Taxonomic trees resulting from pyrosequencing the V6 region of the 16S rDNA extracted from each field at the indicated sampling times are shown. The size of the dots reflects the relative amount of taxa assigned to each particular node.
(TIF)