Abstract
The use of tyrosine kinase inhibitors (TKIs) requires the testing for hot spot mutations of the molecular effectors downstream the membrane-bound tyrosine kinases since their wild type status is expected for response to TKI therapy. We report a novel assay that we have called Allele Specific Locked Nucleic Acid quantitative PCR (ASLNAqPCR). The assay uses LNA-modified allele specific primers and LNA-modified beacon probes to increase sensitivity, specificity and to accurately quantify mutations. We designed primers specific for codon 12/13 KRAS mutations and BRAF V600E, and validated the assay with 300 routine samples from a variety of sources, including cytology specimens. All were analyzed by ASLNAqPCR and Sanger sequencing. Discordant cases were pyrosequenced. ASLNAqPCR correctly identified BRAF and KRAS mutations in all discordant cases and all had a mutated/wild type DNA ratio below the analytical sensitivity of the Sanger method. ASLNAqPCR was 100% specific with greater accuracy, positive and negative predictive values compared with Sanger sequencing. The analytical sensitivity of ASLNAqPCR is 0.1%, allowing quantification of mutated DNA in small neoplastic cell clones. ASLNAqPCR can be performed in any laboratory with real-time PCR equipment, is very cost-effective and can easily be adapted to detect hot spot mutations in other oncogenes.
Introduction
Molecular therapy targeting transmembrane receptor tyrosine kinases with a variety of tyrosine kinase inhibitors has become part of the standard treatment for many patients with common forms of cancer. Evidence of both tyrosine kinase activation and lack of activating mutations in the tyrosine kinase downstream effectors is expected as a general precondition for successful patient treatment [1]. Among transmembrane tyrosine kinases the EGF receptor (EGFR) is one of the main therapeutic targets since it is active in both colorectal (CRC) and non-small cell lung cancers (NSCLC). The MAP kinase (MAPK) signaling cascade is a mainstream pathway that modulates many cell functions (e.g. proliferation, differentiation, apoptosis) following the activation of tyrosine kinase receptors like EGFR. KRAS and BRAF are key members of this pathway, constitutively active due to oncogenic mutations in ∼40% of human cancers, with a prevalence of mutation that varies considerably among tumors originating from different tissues [2]. KRAS oncogenic activation, largely due to codon 12–13 mutations, occurs in ∼40% of CRC [3], [4] and in ∼15% of NSCLC [5]. As expected, KRAS mutations have been associated with poor response to anti-EGFR therapy in patients with both CRC [3] and NSCLC [6]. Wild type KRAS is now considered a pre-condition to treat CRC patients with EGFR inhibitors like Cetuximab or Panitumumab [7], [8]. Oncogenic BRAF mutations occur in up to 15% of all human tumors, the vast majority (>90%) being c.1799:T>A substitutions that lead to the replacement of valine with aspartic acid (V600E) causing constitutive BRAF activation [9]. Melanoma (40–60%) [10] and papillary thyroid carcinoma (PTC; 40%–80%) [11] are the tumors with the highest incidence of BRAF mutations. While BRAF mutations are uncommon in NSCLC [12], they occur in ∼10–15% of CRC and are strongly associated with non-Lynch microsatellite unstable tumors and with the CpG island methylator phenotype [13]. Similar to KRAS, BRAF mutation has been correlated with lack of response to EGFR inhibitors in patients with advanced CRC and the impact of BRAF mutations on TKI treatment response is currently being investigated [14]. Furthermore, novel BRAF inhibitor molecules like vemurafenib are proving highly effective to treat patients with BRAF mutated tumors, like melanoma [15].
Additional reasons for molecular testing are the diagnostic or prognostic information that can be obtained by the analysis of tumors with a high prevalence of specific mutations as is the case for KRAS and pancreatic lesions [16] or BRAF and thyroid nodules [11].
The above considerations point to the necessity to test for KRAS and BRAF mutations. In fact, the advent of targeted therapy mandates the analysis of large numbers of tumors and is forcing the integration of molecular data into the routine workflow of cancer patients [1]. This can prove a challenge and underlines the importance of utilizing detection methods that are sensitive, rapid, reproducible and cost-effective.
Sanger sequencing is highly reliable and currently considered the “gold standard” technique for mutation detection [7], [8]. However, when applied to routine diagnostic use suffers from several limitations. Sanger sequencing is low throughput, requires several distinct steps (e.g. PCR, amplicon purification, labelling) each of which is exposed to contamination risk, is relatively dependent on the quality and integrity of DNA, and has a low analytical sensitivity, requiring at least 25% of mutated DNA-corresponding to at least 50% of neoplastic cells with an heterozygous mutated allele. Considering that many routine samples contain large numbers of non-neoplastic reactive/inflammatory cells, dissection of specimens prior to DNA extraction is usually necessary to enrich for neoplastic cells and to avoid false negative results. A variety of more sensitive methods based on different approaches are utilized to overcome the limitations of Sanger sequencing, but many of them can be time-consuming, labor-intensive, expensive or require the use of sophisticated platforms not always affordable by pathology laboratories [17], [18].
We here describe a new assay that we have called Allele Specific Locked Nucleic Acid quantitative PCR (ASLNAqPCR) based on 3′-locked nucleic acid (LNA)-modified primers and the use of a LNA-modified beacon probe. The assay is very cost-effective and not only identifies mutations with high specificity and sensitivity, but unlike other methods gives reliable information about the ratio of mutant and wild-type alleles. We have utilized ASLNAqPCR to identify the most common codon 12 and 13 KRAS mutations and the BRAF V600E mutation, but the test can be easily adapted to detect hot spot mutations in other oncogenes.
Materials and Methods
Ethics Statement
Since KRAS and BRAF mutational analysis is part of proper diagnostic protocols, the need for ethic committee's approval was not necessary for this study, in accordance with medical ethical guidelines of the Azienda Unita' Sanitaria Locale di Bologna (Ufficio Qualita' di Sistema Aziendale, Via Castiglione 29, 40100 Bologna). Accordingly to these guidelines, a comprehensive written informed consent was signed for the surgical treatment that produced the tissue samples and the related diagnostic procedures. All information regarding the human material used in this study was managed using anonymous numerical codes, clinical data were not used and samples were handled in compliance with the Helsinki declaration (http://www.wma.net/en/30publications/10policies/b3/).
Selection of tumor material
Three hundred consecutive tumor samples from the Department of Pathology of the Azienda Unita' Sanitaria Locale di Bologna Ospedale Bellaria-Università di Bologna and corresponding to 281 patients, were analyzed. Of the 300 samples, 220 were primary tumours: 163 from the colon, 29 from the lung, 21 from the pancreas-9 adenocarcinomas and 12 cyst fluid aspirates from pancreatic neoplasms-and 7 from the thyroid. The remaining 80 samples were metastases at various sites from primary tumors of the colon (n = 71) or lung (n = 9) (Table 1 and Table 2). Two hundred and seventy-six samples were obtained from routinely processed formalin-fixed paraffin embedded (FFPE) sections (187 surgical specimens, 89 biopsy samples); 21 were fine needle cytology aspirates from pancreatic and 3 from lung lesions. For FFPE material, Hematoxylin and Eosin (H&E) sections were reviewed to identify paraffin blocks with the highest relative amount of tumor vs. stroma, few infiltrating lymphocytes and little or no tumor necrosis. Six 10 μm thick sections were cut from each block, followed by one H&E control slide. The tumor area selected for the analysis was marked on the control slide to ensure, whenever possible, greater than 70% content of neoplastic cells, in accordance with published guidelines [8]. Tumor material was manually dissected under microscopic guidance from the corresponding 10 μm sections using a sterile blade. Dissected tumor areas ranged from 0.25–1.0 cm2. For cytology preparations the slides with the highest tumor content were selected and material collected after removal of the coverslip. All patient information was handled in accordance with review board approved protocols and in compliance with the Helsinki declaration (http://www.wma.net/en/30publications/10policies/b3/).
Table 1. KRAS mutations in 300 consecutive samples analyzed by Sanger sequencing and ASLNAqPCR.
| Tissue | KRAS | No amplifiable DNA | ||
| Mutated by SSEQ (%) | Mutated by ASLNA (%) | SSEQ (%) | ASLNA (%) | |
| COLON CRC ( n = 234) | 80/209 (38.3) | 94/221 (44.8) | 25/234 (10.7) | 13/234 (5.6) |
| Primary (n = 163) | 56/146 (38.4) | 69/153 (45.1) | 17/163 (10.4) | 10/163 (6.1) |
| Metastatic (n = 71) | 24/63 (38.1) | 25/68 (36.7) | 8/71 (11.3) | 3/71 (4.2) |
| LUNG NSCLC ( n = 38) | 16/38 (42.1) | 17/37 (45.9) | 0/38 (0) | 1/38 (0) |
| Primary (n = 29) | 9/29 (31.0) | 12/29 (41.4) | 0/29 (0) | 0/29 (0) |
| Metastatic (n = 9) | 7/9 (77.8) | 5/8 (62.5) | 0/9 (0) | 1/9 (11.1) |
| PANCREAS (n = 21) | 7/21 (33.3) | 6/21 (28.6) | 0/21 (0) | 0/21 (0) |
| Carcinoma (n = 9) | 4/9 (44.4) | 4/9 (44.4) | 0/9 (0) | 0/9 (0) |
| Cyst Fluid (n = 12) | 3/12 (25.0) | 2/12 (16.7) | 0/12 (0) | 0/12 (0) |
| THYROID (n = 7) | 0/6 (0) | 0/7 (0) | 1/7 (14.3) | 0/7 (0) |
| PTC-Classic (n = 3) | 0/3 (0) | 0/3 (0) | 0/3 (0) | 0/3 (0) |
| PTC-Others (n = 4) | 0/3 (0) | 0/4 (0) | 1/4 (25) | 0/4 (0) |
SSEQ, Sanger sequencing; ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); CRC, colonic adenocarcinoma; NSCLC, lung adenocarcinoma; PTC, papillary thyroid carcinoma.
Table 2. BRAF mutations analysis in 201 consecutive samples analyzed by Sanger sequencing and ASLNAqPCR.
| Tissue | BRAF | No amplifiable DNA | ||
| Mutated by SSEQ (%) | Mutated by ASLNA (%) | SSEQ (%) | ASLNA (%) | |
| COLON CRC ( n = 159) | 15/153 (9.8) | 19/153 (12.4) | 6/159 (3.8) | 6/159 (3.8) |
| Primary (n = 114) | 13/109 (11.9) | 15/109 (13.8) | 5/114 (4.4) | 5/114 (4.4) |
| Metastatic (n = 45) | 2/44 (4.5) | 4/44 (9.1) | 1/45 (2.2) | 1/45 (2.2) |
| LUNG NSCLC (n = 24) | 0/24 (0) | 0/24 (0) | 0/24 (0) | 0/24 (0) |
| Primary (n = 17) | 0/17 (0) | 0/17 (0) | 0/17 (0) | 0/7 (0) |
| Metastatic (n = 7) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) |
| PANCREAS (n = 11) | 0/10 (0) | 0/10 (0) | 1/11 (9.1) | 1/11 (9.1) |
| Carcinoma (n = 5) | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) |
| Cyst Fluid (n = 6) | 0/5 (0) | 0/5 (0) | 1/6 (16.7) | 1/6 (16.7) |
| THYROID (n = 7) | 0/7 (0) | 1/7 (1.4) | 0/7 (0) | 0/7 (0) |
| PTC-Classic (n = 3) | 0/3 (0) | 1/3 (0) | 0/3 (0) | 0/3 (0) |
| PTC-Others (n = 4) | 0/4 (0) | 0/4 (0) | 0/3 (0) | 0/3 (0) |
SSEQ, Sanger sequencing; ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); CRC, colonic adenocarcinoma; NSCLC, lung adenocarcinoma; PTC, papillary thyroid carcinoma.
Cell line controls
The SW620 (KRAS G12V homozygous, ATCC – American Type Culture Collection, Rockville, MD, USA), CAL62 (KRAS G12R heterozygous), OCUT (BRAF V600E heterozygous), ARO (BRAF V600E heterozygous) and TPC-1 (BRAF wild type, American Type Culture Collection, Rockville, MD, USA) cell lines were used as DNA controls for mutational analysis. The CAL62, OCUT and ARO cell lines have been previously described [19] and were kindly provided by Prof. M. Santoro (University of Naples, Italy). Mutant DNA extracted from the cell lines was spiked in a pool of healthy female donor DNA (DNA Female pool, Cod. G1521, Promega, Madison WI) and serially diluted as 50%, 20%, 10%, 5%, 1%, 0,1%, 0.01% mutant to wild type DNA ratios to determine the analytical sensitivity of both Sanger sequencing and ASLNAqPCR. The minimal amount of input DNA required to obtain reliable mutation detection with the ASLNAqPCR method was determined by serially diluting DNA of the G12V mutated SW620 cell line with normal DNA, as previously described [20].
Primers and molecular beacon probes design
Primers and molecular beacon probes for ASLNAqPCR were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) (Table 3). They identify the seven most common KRAS mutations at codons 12 and 13, present in greater than 95% of tumors with mutated KRAS [3] and the BRAF V600E present in >90% of tumors with a BRAF mutation [9]. Forward ASLNAqPCR mutation-specific primers were modified with LNA nucleotides [21] at the 3′-end terminal of the oligonucleotide sequence (Table 3 and Figure 1). Two internal LNA-modified molecular beacon probes were designed, one for KRAS and one for BRAF real-time analysis (Table 3 and Figure 1). Flanked molecular beacon arms were designed using the OLIGO 6.0 software reaching a temperature between 57°C and 61°C in the stem loop conformation. All primers and probes were tested by MFOLD (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/) to avoid secondary structures. Table 3 also shows the standard set of primers for KRAS [3] and BRAF [22] used for Sanger sequencing.
Table 3. Primers and beacon probes.
| Sanger Sequencing | |||
| Gene | Exon | Forward Primer | Reverse Primer |
| KRAS | 2 | AAGGTGAGTTTGTATTAAAAGGTACTGG | TGGTCCTGCACCAGTAATATGC |
| 3 | TCCAGACTGTGTTTCTCCCTTCTC | AAAACTATAATTACTCCTTAATGTCAGCTT | |
| BRAF | 15 | TCATAATGCTTGCTCTGATAGGA | GGCCAAAAATTTAATCAGTGGA |
bp, base pair. “+” precedes LNA-modified nucleotides.
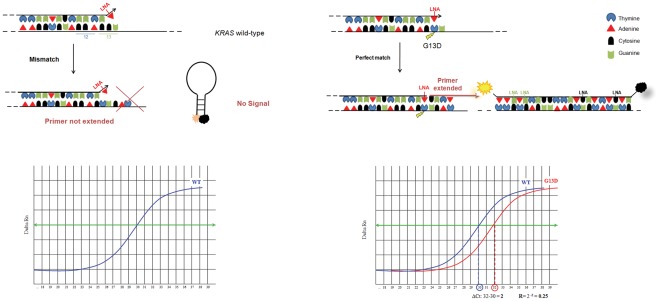
Figure 1. Diagram illustrating ASLNAqPCR.
Left side: a single mismatch of the LNA modified primer does not allow PCR amplification. Right side: in case of a perfect match, the Taq polymerase extends the DNA strand and the amplicon is detected by the internal LNA modified beacon probe.
Mutational Analysis
DNA was extracted from FFPE using the RecoverAll kit (Ambion, Austin TX, U.S.A.), according to the manufacturer's recommendation. DNA from cell lines and FNA samples was extracted using the Gentra Puregene Kit (Qiagen, Hilden, Germany). DNA concentration was measured using the Quant-iT™ dsDNA BR kit (Invitrogen, Carlsbad, CA).
a) Sanger Sequencing
All 300 samples were tested for KRAS, 201 for BRAF. Exon 2 and 3 of KRAS and exon 15 of BRAF were evaluated amplifying fragments of 264 bp, 257 bp, and 223 bp respectively, similar to what previously described [3], [22]. PCR reactions were performed using the FastStartTaq DNA polymerase (Roche Applied Science, Mannheim, Germany) following the instructions of the provider, starting from 30–50 ng for DNA from FFPE and from about 15 ng for cell line DNA. The cycling conditions are shown in Table 4. Sequencing was carried out according to standard procedures using the GenomeLab DTCS Kit (Beckman Coulter, Inc., Fullerton, CA, U.S.A.) and a CEQ2000 XL automatic DNA sequencer (Beckman Coulter, Inc., Fullerton, CA, U.S.A). Strands were screened using forward and reverse primers.
Table 4. PCR conditions for Sanger sequencing and ASLNAqPCR.
| Sanger Sequencing | |||
| Amplicon | Temperature | Time | Cycles |
| BRAF (Ex15) | 95°C | 4′ | 1 |
| 95°C | 30″ | 40 | |
| 53°C | 30″ | 40 | |
| 72°C | 30″ | 40 | |
| 72°C | 10′ | 1 | |
| KRAS (Ex2 and Ex3)a | 95°C | 4′ | 1 |
| 95°C | 30′ | 5 | |
| 63°C–1°C/cycle | 30′ | 5 | |
| 72°C | 30″ | 5 | |
| 95°C | 30″ | 35 | |
| 58°C | 30″ | 35 | |
| 72°C | 30″ | 35 | |
| 72°C | 10′ | 1 | |
Ex, exon; aFor KRAS amplification a touch-down PCR was performed; bPCR for wild type KRAS and all 7 codon 12 and 13 mutations; cPCR for wild type BRAF and BRAF V600E; *Plate reading step.
b) ASLNAqPCR
All 300 samples were tested for KRAS, 201 for BRAF. Fifteen nanograms of DNA purified from fresh cell lines, or 15–50 ng of DNA purified from FFPE, were amplified using the FastStart Universal Probe Master with ROX (Roche Applied Science, Mannheim, Germany) in separate real time reactions for each allele specific primer, but in the same run and following the same cycling conditions shown in Table 4. PCR products were 117 bp for BRAF V600E and 104 to 110 bp for KRAS. Real-time PCR was performed using an ABI SDS 7000TM instrument (Applied Biosystems, Foster City, CA). The relative mutant allele copy number was quantified during the exponential phase of real-time PCR using the ΔCT method [23]. Samples with quantification cycles above 35 for the wild type allele were considered failures and excluded from the study.
c) Pyrosequencing
Twenty-one samples with discrepant results between Sanger sequencing and ASLNAqPCR were tested by pyrosequencing, according to standard procedures using PyroMark Gold Q96 (Qiagen, Gmbh, Hilden Germany) reagents and a PyroMarkTM Q96 ID instrument. Pyrograms outputs were analysed with PyroMarkTM Q96 ID Software (Qiagen, Gmbh, Hilden Germany) using the allele quantification (AQ) mode.
Statistical measures of performance
True positive (TP), false positive (FP), true negative (TN), false negative (FN), test sensitivity (SEN), specificity (SPEC), negative predictive value (NPV), positive predictive value (PPV), accuracy (ACC), false discovery rate (FDR) [24].
Results
Of the 300 consecutive cases, 201 were analyzed for both KRAS and BRAF and 99 only for KRAS. Ten of 300 cases analyzed for KRAS gave no amplifiable products due to excessive DNA degradation with both Sanger sequencing and the ASLNAqPCR technique. Sixteen additional cases gave amplifiable KRAS PCR products by ASLNAqPCR but not by sequencing and four additional cases by sequencing but not by ASLNAqPCR. Seven of 201 cases analyzed for BRAF gave no amplifiable products due to excessive DNA degradation with both Sanger sequencing and ASLNAqPCR.
ASLNAqPCR analytical sensitivity, intra– and inter-assay reproducibility, failure rate
Analytical sensitivity-Sanger sequencing
Analytical sensitivity was tested by serially diluting DNA from the G12V mutated SW620 cell line, the G12R mutated CAL62 cell line, the BRAF V600E mutated ARO cell line, the BRAF V600E mutated OCUT cell line in a pool of healthy female donor DNA. The TPC-1 cell line was used as non-mutated control for the dilution tests. At least 20% of KRAS G12V and KRAS G12R DNA were required to identify the mutations. The BRAF V600E mutation was identified with at least 10% of mutated DNA.
Analytical sensitivity-ASLNAqPCR
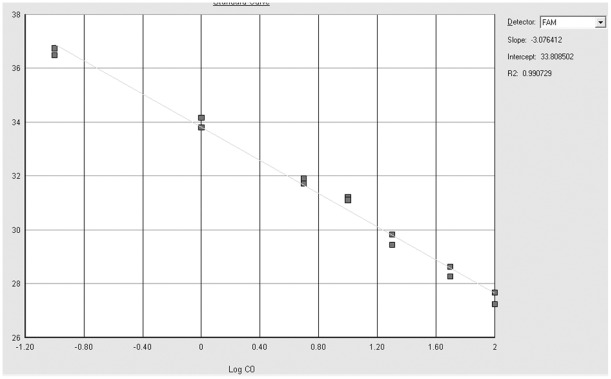
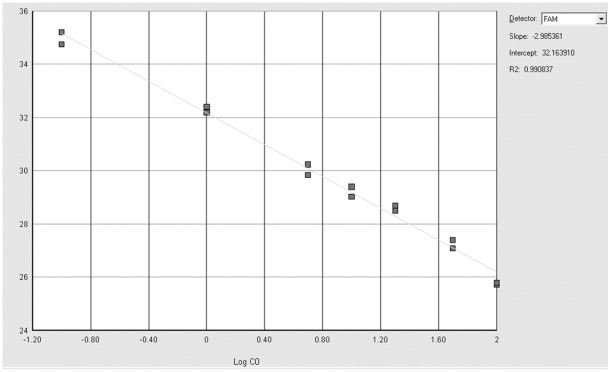
Analytical sensitivity was tested with the same mutated DNA dilutions used for the Sanger sequencing. The KRAS G12V and KRAS G12R mutations were reproducibly detectable at a dilution of 0.1% with a PCR efficiency of 111.3% (slope: −3.0764, R2: 0.9907) [23] (Figure 2). The BRAF V600E mutation was reproducibly detected at a dilution of 0.1% with a PCR efficiency of 116.2% (slope: −2.9854, R2: 0.9908) [23] (Figure 3).
Figure 2. Standard curve titration of ASLNAqPCR for KRAS.
Serial dilution of the KRAS G12V mutated SW620 cell line DNA in wild type DNA. Gray squares correspond to 50%, 20%, 10%, 5%, 1%, 0,1%, 0.01% mutant to wild type DNA ratios (duplicate samples). The titration slope is −3.076, R2 is 0.991 (top right), corresponding to a PCR efficiency of 111.3%.
Figure 3. Standard curve titration of ASLNAqPCR for BRAF.
Serial dilution of the BRAF V600E mutated OCUT cell line DNA in wild type DNA. Gray squares correspond to 50%, 20%, 10%, 5%, 1%, 0,1%, 0.01% mutant to wild type DNA ratios (duplicate samples). The titration slope is −2.985, R2 is 0.991 (top right), corresponding to a PCR efficiency of 116.2%.
Minimal amount of input DNA for ASLNAqPCR at the analytical sensitivity threshold
The amount of a 0.1% dilution of KRAS G12V mutated SW620 cell line DNA and of BRAF V600E mutated ARO cell line DNA spiked with normal DNA was serially decreased to determine the minimal input DNA necessary for mutation detection. A minimal amount of 6.25 ng of DNA from cell lines (equivalent to ∼1000 copies of a diploid human genome) was necessary to detect both mutations. ASLNAqPCR analysis of all clinical samples below the 6.25 ng input DNA threshold was therefore repeated starting with a higher amount of tumor tissue.
ASLNAqPCR intra– and inter-assay reproducibility
Intra-assay reproducibility (i.e. the consistency of results in the same run) has been measured by calculating the Ct (cycle threshold) coefficients of variation of samples run as duplicate in the same plate using serial dilution (50%, 20%, 10%, 5%, 1%, 0,1%) of mutant DNA in wild type DNA for KRAS (CAL62 and SW620 cell lines) and BRAF (ARO and OCUT cell lines). The coefficients of variation for the KRAS mutated DNA ranged between 0.08% and 1.03%. Those for the BRAF mutated DNA ranged between 0.15% and 1.54%. Inter-assay reproducibility (i.e. the consistency of results with the same protocol but in different runs) has been similarly measured by calculating the Ct coefficient of variation of duplicate samples run in different days using the same serial dilutions of mutated DNA mentioned above. The coefficients of variation for the KRAS mutated DNA ranged between 0.91% and 1.62%. Those for the BRAF mutated DNA ranged between 1.12% and 1.73%. Both intra– and inter assay reproducibility results are well within the 10% range considered satisfactory [25].
ASLNAqPCR failure rate
The failure rate was tested by repeating a series of samples with a mutated/wild type ratio thrice the analytical sensitivity threshold, according to published guidelines [26]. Twenty-four samples with a 0.3% dilution of KRAS G12V SW620 cell line DNA and of BRAF V600E ARO cell line DNA were tested. The failure rate was zero, as mutations were consistently detected in all cases.
KRAS and BRAF mutation analysis
Sanger sequencing analysis
Two-hundred-seventy-four samples gave amplifiable DNA and 103 of them (37.6%) showed a KRAS mutation at codon 12, 13 or 61 (Table 1 and Table 5). Out of these, five cases had mutations in exon 3 codon 61 (Q61H or Q61L), not detectable by our ASLNAqPCR method. Fifteen out of the 194 evaluable cases (7.7%) showed the BRAF V600E mutation (Table 2 and Table 5). No BRAF exon 15 mutations other than the V600E were detected. KRAS and BRAF mutations were mutually exclusive on all cases.
Table 5. Frequence of specific KRAS and BRAF mutations cases analyzed by SSEQ and ASLNAqPCR.
| Gene | Mutation | SSEQ (%) | ASLNA (%) |
| KRAS (n = 300) | G12D | 41/274 (14.9) | 48/286 (16.8) |
| G12V | 23/274 (8.4) | 26/286 (9.1) | |
| G13D | 17/274 (6.2) | 19/286 (6.6) | |
| G12C | 8/274 (2.9) | 12/286 (4.2) | |
| G12S | 3/274 (1.1) | 4/286 (1.4) | |
| G12A | 3/274 (1.1) | 3/286 (1.1) | |
| G12R | 2/274 (0.7) | 5/286 (1.7) | |
| G12F | 1/274 (0.4) | NT | |
| Q61H | 3/274 (1.1) | NT | |
| Q61L | 2/274 (0.7) | NT | |
| All mutant cases | 103/274 (37.6) | 117/286 (40.9) | |
| No amplifiable DNA | 26/300 (8.7) | 14/300 (4.7) | |
| BRAF (n = 201) | V600E | 15/194 (7.7) | 20/194 (10.3) |
| No amplifiable DNA | 7/201 (3.5) | 7/201 (3.5) |
SSEQ, Sanger sequencing; ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); NT, not tested, since ASLNAqPCR primers were designed to identify only the seven most common codon 12−13 KRAS mutations.
ASLNAqPCR analysis
Two-hundred-eighty-six samples gave amplifiable DNA and 117 of them (40.9%) showed a KRAS mutation at codon 12, 13 (Table 1 and Table 5), codon 61 mutations were not tested by ASLNAqPCR. Twenty of the 194 evaluable cases (10.3%) showed the BRAF V600E mutation (Table 2 and Table 5). No BRAF exon 15 mutations other than the V600E were tested by ASLNAqPCR. Quantitative real time data always indicated a mutant/wild type ratio equal to or less than 1, consistent with heterozygous mutations. As in the case of Sanger sequencing results, KRAS and BRAF mutations were always mutually exclusive.
The KRAS and BRAF V600E mutation rates detected in our series by both Sanger sequencing and ASLNAqPCR are compatible with the data reported in the literature for colon adenocarcinoma (Table 6) and the other tumors analyzed [3], [4], [5], [11], [16], [27].
Table 6. KRAS mutations in primary colon carcinoma (n = 163) compared with data reported in the literature.
| Mutation | SSEQ (%) | ASLNA (%) | Literature data, % valuesa |
| G12D | 22/146 (15.1) | 27/153 (17.7) | 12.9–15.5 |
| G12V | 14/146 (9.6) | 15/153 (9.8) | 7.7–12.2 |
| G13D | 8/146 (5.5) | 10/153 (6.5) | 5–7.3 |
| G12C | 5/146 (3.4) | 7/153 (4.4) | 2.3−3.6 |
| G12A | 3/146 (2.1) | 3/153 (2.0) | 2.3–2.8 |
| G12S | 3/146 (2.1) | 4/153 (2.6) | 2.6–4.3 |
| G12R | 0/146 (0) | 3/153 (2.0) | 0.3–0.5 |
| G12F | 1/146 (0.7) | NT | 0.2 |
| Q61H | 0/146 (/) | NT | 0.1 |
| Q61L | 0/146 (/) | NT | 0.1 |
| All mutant cases | 56/146 (38.4) | 69/153 (45.1) | 37–42.6 |
| No amplifiable DNA | 17/163 (10.4) | 10/163 (6.1) | ___ |
ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); NT, not tested, since ASLNAqPCR primers were designed to identify only the seven most common codon 12–13 KRAS mutations. a References [Bamford et al., 2004; Karapetis et al., 2008; Neumann et al., 2009].
Comparison between Sanger and ASLNAqPCR and pyrosequencing of samples with discordant results
Sanger sequencing and the ASLNAqPCR assay generated discordant results in 22/300 samples for KRAS mutations (7.3%) (Table 7) and in 5/201 samples for BRAF V600E (2.5%) (Table 8). Eighteen discordant KRAS samples and 3 discordant BRAF ones were further analyzed by pyrosequencing. No additional material was available to repeat the test in 4 KRAS and 2 BRAF mutated cases. Among the samples re-tested by pyrosequencing there were two mutated for KRAS Q61H and one for KRAS G12F, not detectable by ASLNAqPCR. Pyrosequencing confirmed all KRAS mutations identified by ASLNAqPCR but not detected by Sanger sequencing (Table 7, Figure 4). Importantly, quantitative real time data showed a mutated/wild type ratio ≤10% –below our KRAS Sanger sequencing analytical sensitivity threshold-for all these samples (Table 7). The same was true for BRAF real time data (Table 8, Figure 5). Review of the pathology material showed variable ratios of tumor to non-neoplastic cells in the areas that were manually dissected for mutational analysis where a mutation was detected by ASLNAqPCR, confirmed by pyrosequencing but not identified by Sanger sequencing. In the majority of samples the discrepant result was simply explained by the low tumor to non-neoplastic cell ratio and the higher analytical sensitivity of ASLNAqPCR (Figure 6, panels A and B). However, in some samples tumor heterogeneity was a contributing factor. ASLNAqPCR quantification of the mutated to wild type allele ratio clearly indicated the presence of tumor cell subclones in 7 of the 16 discrepant KRAS results (cases 1, 8, 9, 10, 11, 12, 16 of Table 7, Figure 6, panels C and D) and in 3 of the 5 discrepant BRAF results (cases 2, 3, 4 of Table 8). In these cases, ASLNAqPCR results and review of the tumor to non-neoplastic cell ratio in the area dissected for DNA extraction were consistent with mutated cells representing <30% of the tumor cell population, assuming that all mutations were heterozygous. In two cases data indicated that mutated cells represented <5% of the tumor (case 1 of Table 7 and case 2 of Table 8, Figure 6, panels C and D).
Table 7. KRAS Pyrosequencing analysis of cases with discordant results between ASLNAqPCR and Sanger sequencing.
| Case number | KRAS-SSEQ | KRAS-ASLNAa | KRAS mutated/wild type (%)b | KRAS Pyrosequencing | Sample | Tumor cells/Non neoplastic cells (%)d |
| 1 | WT | G12D | 1.5 | G12D | CRC, resection | 70 |
| 2 | WT | G12C | 3.0 | G12C | NSCLC, biopsy | 10 |
| 3 | WT | G12D | 8.0 | G12D | CRC, resection | 35 |
| 4 | WT | G12R | 4.0 | G12R | CRC, resection | 15 |
| 5 | WT | G12R | 4.0 | G12R | CRC, resection | 25 |
| 6 | WT | G12S | 8.0 | G12S | CRC, resection | 30 |
| 7 | WT | G12C | 7.0 | G12C | CRC, resection | 45 |
| 8 | WT | G12V | 5.0 | G12V | CRC, resection | 70 |
| 9 | WT | G13D | 7.0 | G13D | CRC, resection | 70 |
| 10 | WT | G12D | 6.0 | G12D | CRC, resection | 45 |
| 11 | WT | G12D | 2.0 | G12D | NSCLC, biopsy | 30 |
| 12 | WT | G13D | 3.0 | G13D | CRC, resection | 35 |
| 13 | WT | G12V | 4.0 | G12V | CRC, resectionc | 10 |
| 14 | WT | G12D | 1.0 | G12D | PC, FNA | <5 |
| 15 | WT | G12C | 3.0 | G12C | NSCLC, biopsy | 5 |
| 16 | WT | G12V | 10.0 | NP | CRC, resection | 80 |
| 17 | G12F | G12C | 20.0 | G12F | CRC, resection | 45 |
| 18 | Q61H | WT | / | Q61H | metNSCLC, LN biopsy | 50 |
| 19 | Q61H | WT | / | Q61H | PC, FNA | 60 |
| 20 | Q61H | WT | / | NP | metCRC, liver biopsy | 70 |
| 21 | Q61L | WT | / | NP | metCRC, lung biopsy | 50 |
| 22 | Q61L | WT | / | NP | PC, FNA | 40 |
SSEQ, Sanger sequencing; ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); WT, wild type; NP, not performed due to lack of additional DNA; CRC, colonic adenocarcinoma; NSCLC, lung adenocarcinoma; met, metastatic; LN, lymph node; FNA, fine needle aspirate; PC, pancreatic carcinoma. aASLNAqPCR primers designed to identify only the seven most common codon 12–13 KRAS mutations, codon 61 KRAS and G12F mutations are not detectable. bReal time ASLNAqPCR quantitative data. cStatus post neoadjuvant chemo– and radiation therapy. dPercentage of the tumor/non neoplastic cells ratio estimated in the area dissected for DNA extraction.
Table 8. BRAF Pyrosequencing analysis of cases with discordant results between ASLNAqPCR and Sanger sequencing.
| Case number | BRAF-SSEQ | BRAF-ASLNAa | BRAF mutated/wild type (%)b | Pyrosequencing | Sample | Tumor cells/Non neoplastic cells (%)c |
| 1 | WT | V600E | 1.25 | V600E | metCRC, LN biopsy | 5 |
| 2 | WT | V600E | 1.25 | V600E | CRC, resection, | 75 |
| 3 | WT | V600E | 1.5 | V600E | CRC, resection | 55 |
| 4 | WT | V600E | 3.0 | NP | PTC, resection | 80 |
| 5 | WT | V600E | 3.0 | NP | CRC, resection | 10 |
SSEQ, Sanger sequencing; ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); WT, wild type; NP, not performed due to lack of additional DNA; CRC, colonic adenocarcinoma; met, metastatic; LN, lymph node; PTC papillary thyroid carcinoma. aASLNAqPCR primers designed to identify only the BRAF V600E mutation. bReal time ASLNAqPCR quantitative data. cPercentage of the tumor/non neoplastic cells ratio estimated in the area dissected for DNA extraction.
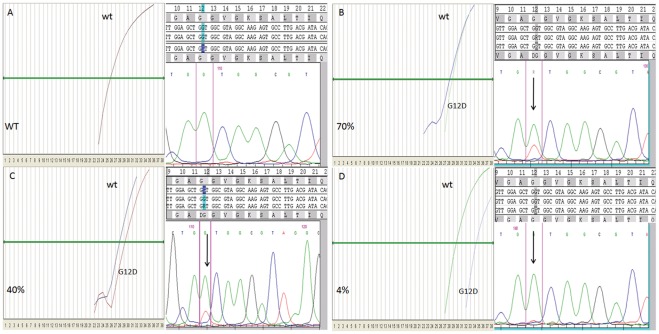
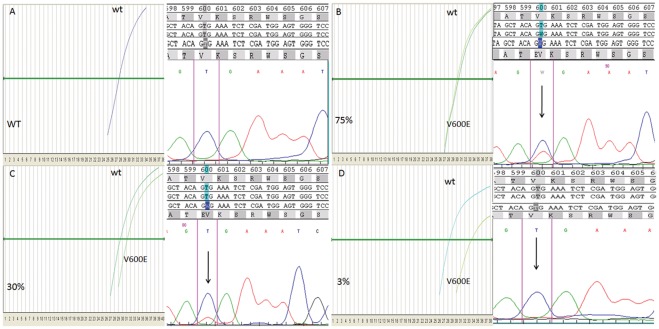
Figure 4. ASLNAqPCR and corresponding Sanger sequencing of four representative tumor samples analyzed for KRAS mutations.
Sample A is wild type, samples B, C and D are KRAS G12D mutated with varying amounts of tumor vs. non neoplastic cells; assuming that KRAS G12D is heterozygous, quantitation of mutated DNA by ASLNAqPCR (ΔCT method) is consistent with 70% of mutated cells in sample B, 40% of mutated cells in sample C, 4% of mutated cells in sample D; in sample D the KRAS G12D mutation is detected only by the ASLNAQPCR due to its high analytical sensitivity.
Figure 5. ASLNAqPCR and corresponding Sanger sequencing of four representative tumor samples analyzed for the BRAF V600E mutation.
Sample A is wild type, samples B, C and D are BRAF V600E mutated with varying amounts of tumor vs. non neoplastic cells; assuming that BRAF V600E is heterozygous, quantitation of mutated DNA by ASLNAqPCR (ΔCT method) is consistent with 75% of mutated cells in sample B, 30% of mutated cells in sample C, 3% of mutated cells in sample D; in sample D the BRAF V600E mutation is detected only by the ASLNAQPCR due to its high analytical sensitivity.
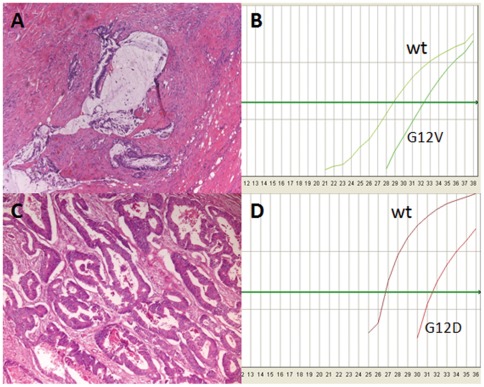
Figure 6. KRAS mutations identified by ASLNAqPCR but not by Sanger sequencing. A.
, Hematoxylin and Eosin (H&E) stained section (X100) of the area of case 13 of Table 7 (rectal adenocarcinoma treated with preoperative chemo– and radiation therapy) dissected for DNA extraction with a tumor vs. non neoplastic cell ratio of ∼10%, below the analytical sensitivity threshold of Sanger sequencing. B, ASLNAqPCR of case 13 of Table 7 shows a KRAS G12V mutation with a mutated/wild type ratio of 4%, corresponding to 8% mutated cells, assuming that the mutation is heterozygous; this is consistent with the mutation being present in the large majority of neoplastic cells. C, H&E stained section (X100) of the area of the colonic adenocarcinoma case 1 of Table 7, dissected for DNA extraction with a tumor vs. non neoplastic cell ratio of ∼70%. D, ASLNAqPCR of case 1 of Table 7 shows a KRAS G12D mutation with a mutated/wild type ratio of 1.5%, corresponding to 3% mutated cells, assuming that the mutation is heterozygous; this is consistent with a small KRAS G12D mutated subclone corresponding to ∼4% of the neoplastic cells.
The only cases mutated by Sanger sequencing and pyrosequencing for which a mutation was not identified by ASLNAqPCR were two cases with KRAS Q61H (cases 18 and 19 of Table 7), not tested by our ASLNAqPCR primers. In one additional case with a G12F (case 17 of Table 7) the Glycine to Phenylalanine mutation was due to a double nucleotide substitution from GGT to TTT on the same KRAS allele, as confirmed by pyrosequencing. The case was identified as mutated by the ASLNAqPCR primer specific for the Glycine to Cysteine mutation (G12C). This is due to the fact that the G12C specific primer correctly recognized the mutated Thymine at the first nucleotide position of the codon. The primer specific for G12V, that recognizes mutated thymines at the second position, was not able to anneal due to the presence, instead of the wild type Guanine, of the mutated Thymine at the beginning of the codon.
Statistical measures of performance
Test sensitivity, specificity and other statistical measures of performance for ASLNAqPCR and Sanger sequencing are shown in Table 9. We considered a result true positive (TP), false positive (FP), true negative (TN) or false negative (FN) as follows. TP were cases that showed the same mutation by both Sanger sequencing and ASLNAqPCR; for cases that gave a discrepant results by the two methods, we considered true positives those with the mutation confirmed by pyrosequencing. FP were cases where a mutation found by one of the two methods (Sanger or ASLNAqPCR) was not confirmed by pyrosequencing. TN were cases that resulted wild type by both Sanger sequencing and ASLNAqPCR; for cases that gave a discrepant results by the two methods, we considered true negatives those that resulted wild type by pyrosequencing. FN were cases where a wild type result by one of the two methods (Sanger or ASLNAqPCR) resulted mutated by pyrosequencing. Test sensitivity (SEN), specificity (SPEC), negative predictive value (NPV), positive predictive value (PPV), accuracy (ACC) and false discovery rate (FDR) were calculated as follows: SEN = TP/(TP+FN); SPEC = TN/(TN+FP) ×100; NPV = TN/(TN+FN) ×100; ACC = (TP+TN)/(TP+FP+TN+FN) ×100; FDR = FP/(FP+VP). Our ASLNAqPCR was designed to detect the seven most common codon 12–13 KRAS mutations and did not detect codon 61 KRAS mutations. For statistical evaluation, all mutations not detected by ASLNAqPCR, including those for which allele specific primers were not designed, were scored as ASLNAqPCR “wild type” results. As shown in Table 9, ASLNAqPCR had 100% specificity, as did Sanger sequencing. The sensitivity of ASLNAqPCR was 95.19%, higher than that of Sanger sequencing (81.37%). Also accuracy and negative predictive value were greater for ASLNAqPCR compared with Sanger sequencing.
Table 9. Statistical measures of performance for Sanger sequencing and ASLNAqPCR.
| Diagnostic Test | Specificity | Sensitivity | PPV | NPV | ACC | FDR |
| SSEQ | 100% | 81.4% | 100% | 90.0% | 94.2% | 0 |
| ASLNA | 100% | 95.2% | 100% | 97.0% | 98.2% | 0 |
SSEQ, Sanger sequencing; ASLNA, allele specific quantitative PCR using 3′-locked nucleic acid modified primers (ASLNAqPCR); PPV, positive predictive value; NPV, negative predictive value; ACC, accuracy; FDR, false discovery rate.
Discussion
The therapeutic use of tyrosine kinase receptors inhibitors (TKIs), like Cetuximab or Panitumumab for CRC and Gefitinib or Erlotinib for NSCLC that target EGFR, requires testing of the molecular effectors downstream to the membrane-bound tyrosine kinases and wild type status for these effectors is expected for response to TKIs therapy. Among these, KRAS and BRAF are commonly mutated and the absence of KRAS activating mutations is now a necessary condition to treat CRC patients with Cetuximab or Panitumumab. The need to screen for mutations in a large number of patient samples with rapid turnaround time is a strong motivation to develop methods that are cost-effective, reliable and robust.
Our ASLNAqPCR is a novel allele specific assay with forward mutation-specific primers modified with LNA nucleotides at the 3′-end sequence terminal and an internal LNA-modified beacon probe that detects and quantifies oncogenic mutations with high specificity and sensitivity. Allele specific PCR is ideally suited to detect oncogenic mutations when these are caused by relatively few nucleotide changes at specific hot spots of the gene. However, natural DNA primers in conventional allele specific PCR can miss-anneal the target sequence, particularly when PCR conditions are suboptimal (e.g. due to DNA damaged by formalin fixation or degraded, limiting amounts of the target sequence), thus causing false positive results that may have unwanted consequences for TKI patient treatment.
LNAs are nucleic acid analogs with a 2′-O, 4′-C methylene bridge that “locks” the ribose into a C3′-endo conformation. When LNA-modified nucleotides are incorporated in oligonucleotides the melting DNA heteroduplex temperature (Tm) increases between 1–8°C per LNA-modified nucleotide [21]. Because of the increased Tm, LNA-modified nucleotides have been used for a variety of applications, including in situ hybridization [28], whole genome amplification [29], methylation sensitive PCR [20], germline SNP genotyping [21], as blocker oligonucleotides to suppress wild-type alleles and increase PCR sensitivity. Blocker LNA oligonucleotides have been shown to be particularly useful to detect oncogene mutations with high sensitivity, including KRAS and BRAF [30]. We tested allele specific primers made of unmodified DNA, but with the same base sequence shown in Table 3 for KRAS and BRAF, observing a consistent reduction in PCR specificity compared with the corresponding LNA-modified primers. When performing Allele Specific PCR without LNA modified primers we had false positive results in non-neoplastic samples, including DNA extracted from peripheral blood leukocytes. Specifically, four DNA samples from healthy blood donors and a pool of healthy female donor DNA tested with Allele Specific PCR showed bands compatible with KRAS and BRAF mutations on the agarose gel. The same samples were wild type when tested using ASLNAqPCR with LNA modified primers and probe and after Sanger sequencing (data not shown). In fact, LNA modification has been shown to greatly enhance allelic specificity, while maintaining a high level of sensitivity in comparison with conventional unmodified, natural DNA primers [21].
We have validated ASLNAqPCR analyzing 300 consecutive samples of routinely processed CRC, NSCLC, pancreatic and thyroid tumors, including both primary and metastatic lesions, surgical specimens, biopsy samples and cytology preparations. ASLNAqPCR identified KRAS and BRAF mutations with rates comparable to those reported in the literature. The test was “robust” with excellent intra– and inter-assay reproducibility and with only few routine samples that gave no amplifiable PCR. There were no ASLNAqPCR failures after repeated testing of a limiting ratio of KRAS and BRAF mutated cell line DNA/wild type DNA. ASLNAqPCR was performed in parallel with conventional Sanger sequencing on all cases, results were compared, and discrepant cases analyzed by pyrosequencing to statistically measure the performance of the assay. Our data demonstrate that ASLNAqPCR has 100% specificity and positive predictive value, with higher sensitivity, negative predictive value and accuracy compared with Sanger sequencing. We observed no false positive results, although if qPCR conditions are pushed above 40 cycles false positive results may be expected [21].
The only mutated samples identified by Sanger sequencing and confirmed by pyrosequencying, but not detected by ASLNAqPCR, were two KRAS Q61H mutations. One additional case had a rare double nucleotide substitution at KRAS codon 12 that ASLNAqPCR recognized as mutated but with the wrong aminoacid call. None of these could be identified because our allele specific primers were not designed to cover all possible codon 12 and 13 KRAS mutations, but only the most frequent, and we did not design primers for codon 61 KRAS mutations. A limitation of ASLNAqPCR, common to all hot spot mutation assays, is that it identifies – by definition – only the targeted mutation, while Sanger sequencing can identify all mutations present in the PCR amplicon. Had our study been limited to codon 12 and 13 KRAS mutations, ASLNAqPCR would have been ∼100% sensitive. The way ASLNAqPCR is designed allows for the easy addition of other RAS allele specific primers. In fact, the test can be conveniently adapted to identify hot spot mutations in other genes and we have successfully utilized ASLNAqPCR to identify IDH1-R132H mutation with high specificity and sensitivity in a series of more than 100 gliomas (data not shown).
In all cases where KRAS or BRAF mutations were detected by ASLNAqPCR, but not by Sanger sequencing, this was due to the higher analytical sensitivity of the assay. ASLNAqPCR assay can identify point mutations against a large excess of wild-type allele, in the thousand-fold range. We detected 0.1% KRAS and 0.1% BRAF mutated cell line DNA with high PCR efficiency, even when DNA for mutational analysis was as little as 6.25 ng, the minimal amount of input DNA at the analytical sensitivity threshold of the method. The ASLNAqPCR analytical sensitivity is thus much higher than that of conventional Sanger sequencing (∼25% mutated DNA) and higher than that reported with very sensitive methods such as pyrosequencing (1.25–2.5% mutated DNA) or ARMS PCR with scorpion oligonucleotides (TheraScreen) (1.25% mutated DNA) [31]. ASLNAqPCR is therefore ideally suited to confidently detect mutations in samples were the abundance of inflammatory cells, fibroblasts lymphocytes or other stromal elements results in tumor to non-neoplastic cell ratios that are below the recommended Sanger sequencing threshold of 50% tumor cells – corresponding to 25% DNA with heterozygous mutations [7]. In many of these samples the proportion of tumor cells cannot be effectively enriched by manual dissection. This is a common occurrence when the only material available for testing are metastatic deposits in lymph nodes, samples from patients that have undergone preoperatory chemo– and radiation treatment and fine needle aspiration specimens [30]. On the other hand, when tumor cells are abundant, the use of ASLNAqPCR makes the dissection of tumor material unnecessary, thus obviating the need for a laborious step that is time consuming and increases the potential for sample contamination.
One relevant feature of ASLNAqPCR compared with tests currently utilized to detect oncogenic mutations is that it allows for precise quantification of the mutated allele due to the LNA-modified beacon probe used for real time analysis. This is important when a method with high analytical sensitivity is utilized, since comparison of quantitative mutational data with the proportion of tumor cells in the samples analyzed allows to easily discriminate those where the mutation is widespread from those where the mutation is present only in small neoplastic cell clones. This was clearly the case in a few of our specimens, including both colorectal and lung adenocarcinomas, where quantitative ASLNAqPCR results of KRAS and BRAF analysis showed that mutations were present in a minority of the tumor cells. Although our data do not indicate that this is a particularly common occurrence, the presence of mutated subclones can be an issue for individual cases. Had the DNA from these patients been analyzed with a method that has high analytical sensitivity (e.g. pyrosequencing), but that does not allow precise quantification of the mutated allele, the tumors would have been diagnosed as mutated. This can, at least in the case of KRAS, deny a potentially beneficial treatment to CRC patients whose response to TKI may not be affected by the presence of small mutated clones. On the other hand, Sanger sequencing would have scored the case as negative and failed to predict a possible limited response to TKI. Since tumors are not always made of homogeneous cell populations, their heterogeneity is a relevant concern for therapies that have specific molecular targets. It is currently unclear if and how the presence of small clones with DNA mutations affects the clinical response to molecular inhibitors of oncogenic pathways [32], [33]. The presence and successive selection of mutated clones may indeed explain response failures in some patients [34]. Although the impact of tumor heterogeneity in deciding patient management is a matter of debate, quantitative mutational data may help to clarify the issue, while providing the oncologist with accurate data to manage the patient.
In addition to quantifying the mutation, ASLNAqPCR has considerable practical advantages over other currently used methods. The assay can be performed in any laboratory with real-time PCR equipment, LNA-modified primers and probes can be easily obtained at low cost and no proprietary reagents, other than those for TaqMan chemistry, are necessary. Once DNA has been extracted, few steps are required for the analysis. All reactions, seven for the KRAS mutations and one for KRAS wild type, one for BRAF V600E and one for BRAF wild type, have been optimized for a single real time run with identical cycling conditions. The entire procedure can be completed in ∼3 hours, including ∼30 minutes operator time to load a 96 well plate, ∼1 hour and 30′ for the real-time run and ∼10′ for data analysis. The short time for the analysis makes it possible to perform several runs in the same day. In addition, since samples are analysed in real-time there is no post-PCR manipulation, avoiding any risk of carry-over contamination.
In summary, we report and validate ASLNAqPCR. The test is rapid, cost-effective, highly sensitive and can accurately quantify oncogenic mutations. It can be proposed as a method of choice to analyze those samples that can not be enriched in neoplastic cell content by tumor dissection prior to DNA extraction. We validated the assay with primers designed to detect the most common KRAS and BRAF mutations in routinely processed samples, but ASLNAqPCR can easily be adapted to detect hot spot mutations in other oncogenes.
Footnotes
Competing Interests: The authors have read the journal's policy and have the following conflicts: LM is a consultant for Alphagenics Biotechnologies S.r.l. Two pending patents related to ASLNAqPCR were filed with the European Patent Office and are currently under review: 1) WO2011104695 (A2) 2011-09-01 for KRAS mutation detection; 2) WO2011104694 (A2) 2011-09-01 for BRAF mutation detection. Alphagenics used a very limited amount of this publicly funded grant to support the authors' work by providing free of charge primers and probes. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This study was supported by: a public grant (1897, 2008) given by the Regione Friuli Venezia Giulia to Alphagenics Diaco Biotechnologies S.r.l., Area Science Park, Basovizza- Building Q1, Trieste Italy (to LM). Alphagenics used a very limited amount of this publicly funded grant to support the authors' work by providing free of charge primers and probes for the initial experimental work that lead to the development of ASLNAqPCR; and an Italian Government MURST grant [grant number 20074zw8la] to GT. DdB is supported with Vanini-Cavagnino grants from the Centro Interdipartimentale di Ricerca sul Cancro G. Prodi, University of Bologna, Bologna, Italy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pao W, Kris MG, Iafrate AJ, Ladanyi M, Janne PA, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15:5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 2.Hoshino R, Chatani Y, Yamori T, Tsuruo T, Oka H, et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999;18:813–822. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 3.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 4.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 8.van Krieken JH, Jung A, Kirchner T, Carneiro F, Seruca R, et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–431. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 9.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 10.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 11.Xing M, Vasko V, Tallini G, Larin A, Wu G, et al. BRAF T1796A transversion mutation in various thyroid neoplasms. J Clin Endocrinol Metab. 2004;89:1365–1368. doi: 10.1210/jc.2003-031488. [DOI] [PubMed] [Google Scholar]
- 12.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29:2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng G, Bell I, Crawley S, Gum J, Terdiman JP, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 14.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 15.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 17.James MR, Dumeni T, Stark MS, Duffy DL, Montgomery GW, et al. Rapid screening of 4000 individuals for germ-line variations in the BRAF gene. Clin Chem. 2006;52:1675–1678. doi: 10.1373/clinchem.2006.070169. [DOI] [PubMed] [Google Scholar]
- 18.Kim IJ, Kang HC, Jang SG, Ahn SA, Yoon HJ, et al. Development and applications of a BRAF oligonucleotide microarray. J Mol Diagn. 2007;9:55–63. doi: 10.2353/jmoldx.2007.060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leboeuf R, Baumgartner JE, Benezra M, Malaguarnera R, Solit D, et al. BRAFV600E mutation is associated with preferential sensitivity to mitogen-activated protein kinase kinase inhibition in thyroid cancer cell lines. J Clin Endocrinol Metab. 2008;93:2194–2201. doi: 10.1210/jc.2007-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morandi L, Franceschi E, de Biase D, Marucci G, Tosoni A, et al. Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference. BMC Cancer. 2010;10:48. doi: 10.1186/1471-2407-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latorra D, Campbell K, Wolter A, Hurley JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- 22.Xing M, Tufano RP, Tufaro AP, Basaria S, Ewertz M, et al. Detection of BRAF mutation on fine needle aspiration biopsy specimens: a new diagnostic tool for papillary thyroid cancer. J Clin Endocrinol Metab. 2004;89:2867–2872. doi: 10.1210/jc.2003-032050. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X-H, Obuchowski NA, McClish DK. Statistical Methods in Diagnostic Medicine: Wiley & Sons. 2011.
- 25.Murray A, Lawrence GP. How should the repeatability of clinical measurements be analysed? An assessment of analysis techniques with data from cardiovascular autonomic function tests. Q J Med. 1993;86:831–836. [PubMed] [Google Scholar]
- 26.Liikanen E. Common technical specification for in vitro diagnostic medical devices. Off J Eur Commun. 2002;L131:17–30. [Google Scholar]
- 27.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Kubota K, Ohashi A, Imachi H, Harada H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl Environ Microbiol. 2006;72:5311–5317. doi: 10.1128/AEM.03039-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Z, Chen Z, Hou X, Li S, Zhu H, et al. Locked nucleic acid pentamers as universal PCR primers for genomic DNA amplification. PLoS One. 2008;3:e3701. doi: 10.1371/journal.pone.0003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2010;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundstrom M, Edlund K, Lindell M, Glimelius B, Birgisson H, et al. KRAS analysis in colorectal carcinoma: analytical aspects of Pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer. 2010;10:660. doi: 10.1186/1471-2407-10-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goranova TE, Ohue M, Shimoharu Y, Kato K. Dynamics of cancer cell subpopulations in primary and metastatic colorectal tumors. Clin Exp Metastasis. 2011;28:427–435. doi: 10.1007/s10585-011-9381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farber L, Efrati E, Elkin H, Peerless Y, Sabo E, et al. Molecular morphometric analysis shows relative intra-tumoural homogeneity for KRAS mutations in colorectal cancer. Virchows Arch. 2011;459:487–493. doi: 10.1007/s00428-011-1158-y. [DOI] [PubMed] [Google Scholar]
- 34.Marchetti A, Milella M, Felicioni L, Cappuzzo F, Irtelli L, et al. Clinical implications of KRAS mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia. 2009;11:1084–1092. doi: 10.1593/neo.09814. [DOI] [PMC free article] [PubMed] [Google Scholar]