Abstract
Photochemical ribonucleotide reductases (photoRNRs) have been developed to study the proton-coupled electron transfer (PCET) mechanism of radical transport in class I E. coli ribonucleotide reductase (RNR). The transport of the effective radical occurs along several conserved aromatic residues across two subunits: β2(•Y122→W48→Y356)→ α2(Y731→Y730→C439). The current model for RNR activity suggests that radical transport is strongly controlled by conformational gating. The C-terminal tail peptide (Y- βC19) of β2 is the binding determinant of β2 to α2 and contains the redox active Y356 residue. A photoRNR has been generated synthetically by appending a Re(bpy)(CO)3CN ([Re]) photo-oxidant next to Y356 of the 20-mer peptide. Emission from the [Re] center dramatically increases upon peptide binding, serving as a probe for conformational dynamics and Y356 protonation state. The diffusion coefficient of [Re]-Y-βC19 has been measured (kd1 = 6.1 × 10−7 cm−1 s–1), along with the dissociation rate constant for the [Re]-Y-βC19:α2 complex (7000 s−1 > koff > 400 s−1). Results from detailed time-resolved emission and absorption spectroscopies reveal biexponential kinetics, suggesting a large degree of conformational flexibility in the α2:[Re]-Y-βC19 complex that partitions the N-terminus of the peptide into both bound and solvent-exposed fractions.
Amino acid radicals often serve as cofactors for substrate activation and vehicles for charge transport in enzymes (1). Free amino acid radicals in solution typically have a micro- to milli- second lifetime. Yet Nature has evolved enzymes to manage both proton and electron equivalents so that the oxidative power of amino acid radical intermediates may be harnessed for chemical transformations over disparate timescales (2). Oxidation of amino acids at physiological pH usually involves the loss of both a proton and electron, implicating proton-coupled electron transfer (PCET)φ as the redox mechanism (3-7). In a PCET event, proton tunneling is limited to short distances, usually within hydrogen bonds, whereas the electron, as the lighter particle, may tunnel over tens of ångströms (8-10). Enzymes have evolved to manage the disparate distance requirements of proton and electron tunneling to achieve efficient radical transport and catalysis (11).
Ribonucleotide reductase (RNRs) serves as an exemplar for enzymes that derive their function from amino acid radicals (12). Class I E. coli RNR contributes to DNA replication and repair by catalyzing the reduction of nucleoside diphosphates (NDPs) to deoxynucleoside diphosphates (dNDPs) (1, 13). The enzyme comprises two homodimeric subunits designated α2 and β2, and a complex between the two is required for enzyme activity (14). α2 houses the NDP/NTP substrate/effector binding sites that control the specificity and rate of nucleotide reduction (15). β2 harbors a diferric tyrosyl radical (•Y122) cofactor proposed to initiate nucleotide reduction by oxidizing a cysteine residue (C439) in the active site of α2 (16). Radical transport from •Y122 to C439 occurs over 35 Å (17) via a PCET amino acid radical transport pathway involving conserved amino acid residues β2(•Y122→W48→Y356)→α2(Y731→ Y730→C439) (12, 18).
Photochemical RNRs (photoRNRs) have been designed to probe the intrinsic mechanistic steps attendant to radical transport between the tyrosines of the β2 to α2 subunits (Y356→Y731) and along the α2(Y731→Y730→C439) pathway (19). In this construct, α2 subunits are bound to the 20-mer, C-terminal peptide tail of β2 (Y-βC19) containing a chemically appended photo-oxidant (20-22). Y-βC19 is known to contain both the binding determinant of β2 to α2 (23, 24) and the redox active Y356 residue that facilitates radical transport between the two subunits (25-27). Figure 1 shows the X-ray crystal structure of α2 highlighting the putative residues that comprise the radical transport pathway in this subunit, along with the bound Y-βC19 peptide (18). The N-terminal residues of Y-βC19, including the redox-active “Y356” residue, are not located in the structure. Excitation of the photo-oxidant appended proximal to Y356 on the peptide produces •Y356 (21, 22). When bound to the α2 subunit in the presence of RNR substrate and effector, this photo-triggered radical can be transported into the α2 active site to initiate deoxynucleotide production. PhotoRNRs designed to date have been based on different photo-oxidants of tyrosine including Trp (28), the benzophenone-containing unnatural amino acid (BPA) (21, 29), anthraquinone (Anq) (21), and the inorganic complex ReI(bpy)(CO)3CN ([Re]) (30, 31). Of these various photoRNRs, the [Re]-based RNR affords the highest single-turnover yield (22). In model systems with tyrosine appended to the bpy ligand of [Re], the MLCT excited state of [Re] is only a competent oxidant of the deprotonated, tyrosinate anion (Y−, pH >10) forming the [Re0]-Y• charge separated state (30). Fluorotyrosine unnatural amino acids may be used to lower the pKa of the phenolic proton (29) in order to obtain the tyrosinate form of the amino acid and thus enable photoactivity within the pH window for RNR activity (6 < pH < 8.5) (31). We have directly observed a 3,5-F2Y• radical on a [Re]-3,5-F2Y-βC19 peptide and showed that the peptide bound to the Y731F-α2 protein is competent for initiating RNR turnover at pH 7.5 and 8.2 (22).
Figure 1.
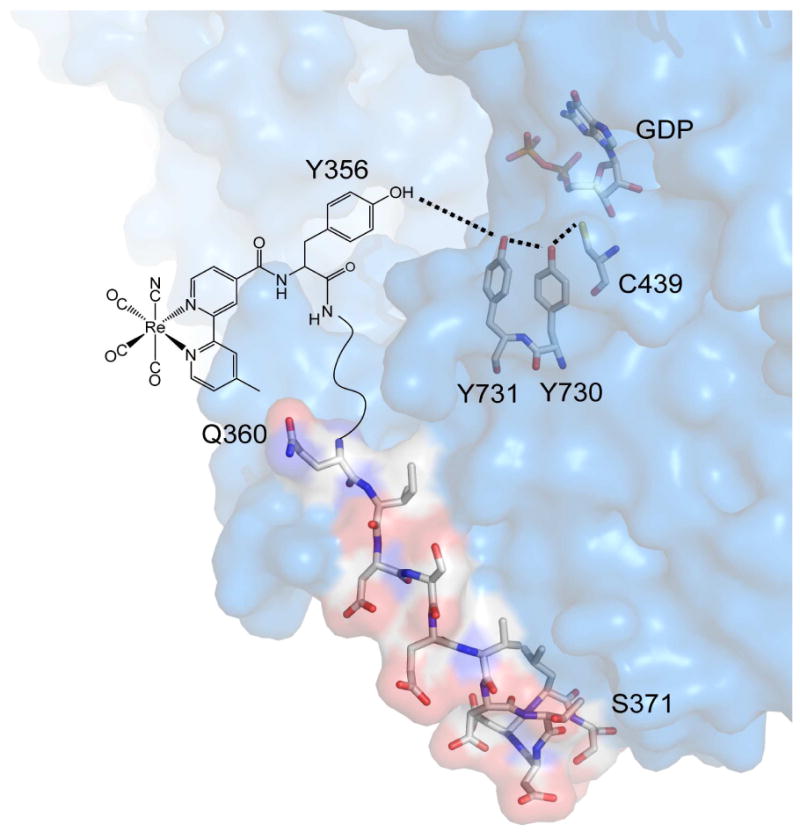
Molecular structure of [Re]-Y-βC19 bound to α2. Peptide and protein coordinates are adapted from the x-ray structure of α2 with Y-βC19 peptide bound (18). The Y731-Y730 and Y730-C439 distances are 3.3 and 3.4 Å, respectively. Y-βC19 residues 359-356 are not located in the structure, thus the Y356-Y731 distance is unknown.
With radical generation and competency established at the Y356 position, we have turned our attention toward a thorough characterization of Y356 on the peptide. We now report the utilization of the emission of the [Re]* excited state within the [Re]-Y-βC19:α2 complex to probe conformational dynamics of the N-terminus of the Y-βC19 peptide bound to α2. We show that the [Re]* emission blue-shifts and increases in intensity upon binding to α2. Titration of the 3,5-F2Y phenolic proton in [Re]-3,5-F2Y-βC19:α2 with pH allows for modulation of the [Re]* emission intensity, as the deprotonated 3,5-F2Y− quenches the [Re]* excited state via electron transfer. Plots of the [Re]* emission intensity vs. pH allow for determination of the pKa of 3,5-F2Y on the peptide bound to α2. Stopped-flow and photobleaching experiments provide information on diffusion and binding rate constants between the peptide and the protein. Time-resolved emission spectroscopy reveals multiple populations of the [Re]* excited state. These data, in combination with the X-ray data shown in Figure 1, support a model for a conformationally flexible N-terminus of the Y-βC19 peptide bound to α2.
Materials and Methods
Materials
The peptides [Re]-F-βC19, [Re]-3,5-F2Y-βC19 and [Re]-Y-βC19 were available from a previous study (22), and the synthesis of Ac-Y-βC19 has been described (20). The concentration of each [Re]-peptide stock solution was estimated by UV-vis absorption spectra using the known ε355 = 5300 M−1 cm−1 for the [Re]-3,5-F2Y-OMe dipeptide (31) and the concentration of Ac-Y-βC19 was estimated using ε280 = 1197 M−1 cm−1 for tyrosine (32). E. coli thioredoxin (TR, SA of 40 U/mg), E. coli thioredoxin reductase (TRR, SA of 1800 U/mg) and E. coli β2 (SA of 6800 nmol/min mg) were isolated as previously described (33). E. coli α2 was isolated, purified, and pre-reduced according to an established procedure (21).
Steady-State Emission Spectroscopy
Steady state emission spectra were recorded on an automated Photon Technology International (PTI) QM 4 fluorimeter equipped with a 150-W Xe arc lamp and a Hamamatsu R928 photomultiplier tube. Solutions (200 μL) containing [Re]-peptides and α2 were placed in a 2 × 10 mm quartz micro-cuvette and excited with 315 nm light focused along the 10 mm pathlength. Emission was collected at 90° to the excitation source and passed through a 450 nm long-pass filter prior to entering the detector housing. Emission spectra were recorded as a function of pH for solutions containing 20 μM α2, 5.5 μM [Re]-3,5-F2Y-PC19, 50 mM buffer (either KPi or Tris), 15 mM Mg2+, 3 mM ATP, and 1 mM CDP. Phosphate and Tris buffers were used in the pH range 6.3-7.0 and 7.2-8.9, respectively.
Stopped-Flow Kinetics
Stopped-flow (SF) kinetic experiments were performed on an Applied Photophysics DX.17MV equipped with an emission PMT. Samples were excited with λ = 355 nm and emission detection was limited to λ > 455 nm using a long-pass cut-off filter from Applied Photophysics LTD. The temperature was maintained at 9 °C with a Lauda RE106 circulating water bath. All experiments were performed in assay buffer (50 mM Tris, 15 mM MgSO4, 3 mM ATP, 1 mM CDP, pH = 7.6). Prior to each experiment, the instrument was flushed with copious amounts of water and then with the assay buffer. [Re]-F-βC19 in one syringe was mixed with assay buffer in another syringe in equal volumes to yield a final concentration of 45 μM [Re]-F-βC19. This experiment provided the baseline [Re] emission of the [Re]-F-βC19 peptide. [Re]-F-βC19 and α2 in one syringe were mixed with assay buffer in another syringe in equal volumes to yield a final concentration of 45 μM [Re]-F-βC19 and 1 μM α2. The peptide emits more intensely when bound to the protein (vide infra); hence this experiment provided the maximum emission expected from [Re] when the peptide was bound to α2. Finally, [Re]-F-βC19 and α2 in one syringe were mixed with Ac-Y-βC19 in another syringe in equal volumes to yield a final concentration of 45 μM [Re]-F-βC19 and 1 μM α2, 450 μM Ac-Y-βC19. 2000 evenly spaced data points were collected over 50 ms or 20 s.
Analysis of [Re]-Y-βC19 binding to α2
Binding of the [Re]-Y-βC19 peptide to α2 was monitored by steady state emission spectroscopy. Solutions of 5 μM peptide were titrated with α2 from concentrations of 0 → 65 μM. Emission from [Re] did not significantly change at higher concentrations of α2; thus at 65 μM we concluded that each molecule of α2 was bound to two [Re]-Y-βC19 peptide. The fraction of bound peptide as a function of α concentration, χbound(α), was then calculated from the equation:
| (1) |
where I600nm is the emission intensity at 600 nm. Data were fit to the following simplified binding model:
| (2) |
where P = [Re]-Y-βC19. From this model we can derive an expression for χbound (34):
| (3) |
assuming that KD is the same for binding of the first and second peptide to α2, as previously discussed (23). KD is derived from a plot of χbound vs. [α] and fitting the data to eq. 3.
Diffusion Coefficients
The diffusion coefficient of [Re]-Y-βC19 (3.65 mM concentration) was determined using Multi-Photon Fluorescence Recovery after Photobleaching (MP-FRaP) (35). The excitation source was a tunable mode-locked Ti:sapphire laser (Mai-Tai Broadband, Spectra-Physics, Mountain View, CA) at 800 nm. The beam was fed into a custom-built multiphoton microscope containing an Axioskop upright microscope (Carl Zeiss, Inc., Thornwood, NY) and Bio-Rad MRC-600 confocal laser scanner (Carl Zeiss, Inc., Thornwood, NY). For each measurement, the laser was focused with a 40× (0.75NA, Zeiss) water-immersion objective to a fixed spot (non-scanning) in the sample at low power (∼5-10 mW at the sample) to monitor fluorescence in the absence of photobleaching. Photons were detected with a photon-counting photomultiplier tube (H7421-40, Hamamatsu, Inc., Bridgewater, NJ) and binned using a multichannel scaler and averaged (SR430, Stanford Research Systems, Sunnyvale, CA). An electro-optic modulator (KD*P Pockels cell, ConOptics, Danbury, CT) and digital delay generator (DG535, Stanford Research Systems, Sunnyvale, CA) were used to rapidly modulate the laser power using custom-built hardware and a user interface developed in LabView (National Instruments, Austin, TX). In a single bleaching pulse, the laser power was increased briefly (<100 μs) to produce a more intense pulse of light (∼40-60 mW at the sample), which caused irreversible photobleaching in the focal volume. The laser power was then returned to its low original value and fluorescence recovery was monitored over several milliseconds. Due to the low concentration of fluorophores within the focal volume, the bleach and recovery process was repeated hundreds of times and fluorescence counts were summed into a single measurement to improve signal to noise. Diffusion coefficients were obtained by fitting a model one-component recovery curve to the change in amplitude of fluorescence over time with non-linear least squares fitting incorporating a trust-region algorithm (35).
The diffusion coefficient of α2 (8 μM concentration) was determined by dynamic light scattering (DLS) performed on a DynaPro Dynamic Light Scatterer. The laser power and temperature were set to 37% and 25 °C, respectively. Each autocorrelation function (ACF) was acquired for 10 s and averaged over 10 measurements. The resulting ACF was fit using Dynamics V6 (Version 6.7.7.9) software from Wyatt Technology Corp. employing a non-negative least-squares fitting algorithm. Hydrodynamic radii were obtained from a mass-weighted size distribution analysis and the reported value is the average of three measurements. The diffusion coefficient of α2, kd2, was calculated with the Einstein-Stokes relationship,
| (4) |
where kB is Boltzmann's constant, T is temperature (25 °C), η (∼1 cP) is the viscosity of the solution, and r is the radius of α2. The radius obtained from DLS experiments was compared to the radius equivalent to a sphere with equal volume to α2,
| (5) |
where d1, d2, and d3 are the diameters of α2 in the x, y, and z directions (36). Eq. (5) is obtained from equating the volume of a sphere with the volume of an ellipsoid and solving for r.
Analysis of [Re]-Y-βC19 Binding to α2
The diffusion limited rate constant, kD, was determined from (37),
| (6) |
where kd1 and kd2 are the diffusion coefficients of the [Re]-Y-βC19 and α2, respectively, and R is the estimated radius of the binding site on α2. The binding site of the [Re]-Y-βC19:α2 complex was estimated from the crystal structure of α2with Ac-Y-19mer (18). Only the 16 residues observed were used and an effective radius (8.97 Å) was calculated from the x, y, and z dimensions of the observed binding site (36.4 Å, 15.4 Å, and 10.3 Å) using eq 5.
Time-Resolved Emission and Transient Absorption
Time resolved emission measurements on the <20 ns timescale were made with the frequency doubled (400 nm) pump light provided by a Ti:sapphire laser system (100 fs pulsewidth) and collected on a Hamamatsu C4334 Streak Scope streak camera (38). Time resolved emission measurements on the >20 ns timescale were made with pump light provided by the third harmonic (355 nm) of an Infinity Nd:YAG laser (Coherent) running at 20 Hz (39). Nanosecond transient absorption (TA) measurements were made with the same laser running at 10 Hz. The sample preparation and experimental setup were identical to that previously described (22).
To produce a transient absorption (TA) spectrum, the series of four spectra were taken: IF (pump on/probe off), I (pump on/probe on), IB (pump off/probe off), and I0 (pump off/probe on). Transient spectra were corrected for fluorescence and background light using these spectra by the calculation: ΔOD = log[(I0 – IB)/(I – IF)]. For experiments involving α2, the spectra reported are an average of 125 of the four-spectra sequences. Sample sizes were typically 200 μL in a 2 × 10 mm cuvette containing a Teflon-coated mini-stirbar. Both the white light and pump beams were focused and overlapped to pass through the 2mm-wide window of the cuvette, providing a total pathlength of 1 cm. To provide optimal beam overlap, the pump beam was reflected off a small mirror in front of the collimating lens for the probe beam after the sample
Results
Photophysics and Photochemistry of [Re]-X-βC19 Peptides
The quenching of the 3[Re]* excited state in the 3[Re]-3,5-F2Y-βC19 peptide can be investigated with steady state emission spectroscopy. Figure 2 plots the emission spectra of the [Re] center as the pH of a solution of [Re]-3,5-F2Y-βC19 peptide is varied from 4.5 to 11.5. Between pH 4.5-5.0 the emission red-shifts and decreases in intensity. We ascribe these spectral changes to an alteration of the peptide conformation, as the carboxylic acid groups of Asp and Glu residues are typically deprotonated in this pH regime. Between pH 5 and 11.5, the emission energy and band shape do not change, however, the overall intensity of the band decreases. This emission quenching is consistent with deprotonation of the 3,5-F2Y phenolic proton, which induces electron transfer from 3,5-F2Y− to the proximate 3[Re]* excited state (31). By titrating the 3[Re]* emission intensity with pH, we obtain a pKa value of 3,5-F2Y when incorporated into the peptide of 7.3 ± 0.1, which is comparable to the pKa = 7.2 of the Ac-3,5-F2Y-NH2 amino acid in solution (29).
Figure 2.
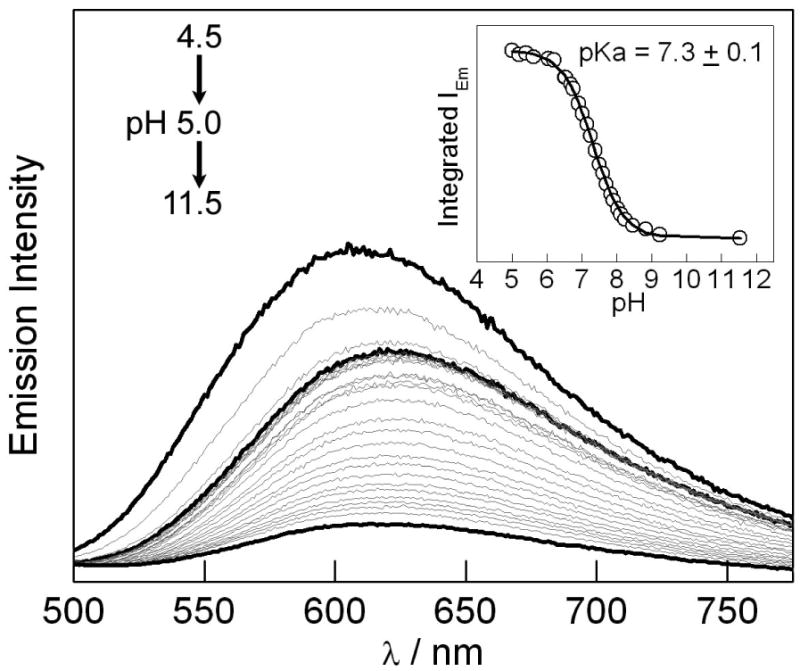
Emission spectra of [Re]-3,5-F2Y-βC19 as a function of pH, which was titrated from 4.5 to 11.5. Black curves correspond to emission at pH 4.5, 5.0 and 11.5. Inset: pH Titration curve of integrated emission intensity from pH 5.0 to 11.5.
The emission of the [Re] excited state on [Re]-(F/Y/3,5-F2Y)-βC19 can be time-resolved to provide insight into the conformational homogeneity of the peptide. The emission decay of [Re]-3,5-F2Y-βC19 at pH 10 fits to a bi-exponential decay function with τ1 = 1.9 ns (66%) and τ2 = 10.7 ns (34%) as does the emission decay of [Re]-Y-βC19 at pH 12 (τ1 = 0.5 ns (86%) and τ2 = 3.0 ns (14%). Both lifetime components for each peptide are faster compared to the emission lifetime of the [Re]-F-βC19 control peptide at all pHs or [Re]-Y-βC19 at pH < 10 (τ = 50 ± 2 ns or 62 ± 2 ns).
Binding of the [Re]-Y-βC19 Peptide to α2
Peptide binding to α2 may be probed by monitoring the emission energy of the 3[Re]* excited state. Figure 3 plots the emission spectra of a 5 μM solution of the [Re]-Y-βC19 peptide as the α2 concentration is increased from 0→65 μM. Upon binding to α2, the emission band blue shifts and increases in intensity. This is consistent with partial desolvation of the [Re] chromophore upon binding to the protein (40, 41). Because there are two binding sites on each α2 subunit (one on each α in the dimer) as shown in the crystal structure (18), the inset of Figure 3 plots the emission maxima as a function of α concentration. The fit of these data to the binding model described by eq 3 yields a dissociation constant, KD, of 9 μM. This value of KD is comparable to the value obtained from competitive inhibition assay (23, 24) measurements of the [Re]-Y-βC19 peptide to α2 (IC50 = 8 μM ≈ KD (22)).
Figure 3.
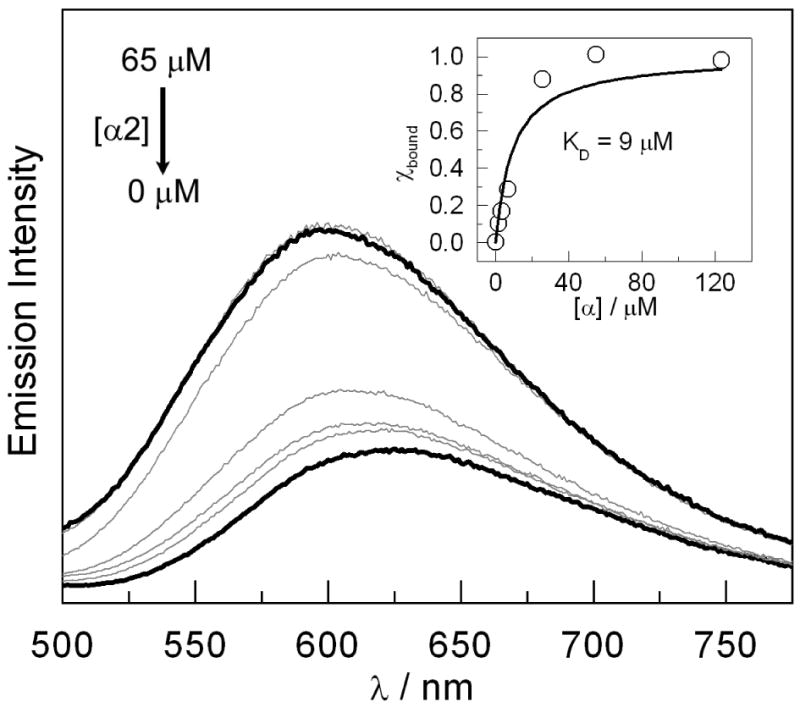
Emission spectra of [Re]-Y-βC19 (5 μM) at pH 7.5 as the α2 concentration is increased from 0 → 65 μM. Inset: Fraction of peptide bound as a function of α concentration (○) with fit (—) to binding model in eq 3.
pKa of 3,5-F2Y356 Bound to α2
The pKa of the 3,5-F2Y on the peptide bound to α2 may be ascertained by monitoring the emission intensity and lifetime of the 3[Re]* excited state. Figure 4 plots the emission intensity of a 5.5 μM solution of [Re]-3,5,-F2Y-βC19 in the presence of 20 μM of α2 as the pH is increased from 6.3 to 9.0. The grey spectra in the figure correspond to the emission of 5 μM [Re]-3,5,-F2Y-βC19 in the absence of α2 at pH 6.3 (lower spectrum) and 9.0 (upper spectrum) and re-emphasize the effect of peptide binding on the [Re] emission band both at pH 6.3 and pH 9.0. As the pH is titrated from 9.0 to 6.3 for the solution containing the bound peptide, the 3[Re]* emission intensity decreases with an apparent pKa of 7.3 ± 0.1. The same pKa was obtained whether the integrated emission or emission maximum (at 600 or 650 nm) was plotted vs. pH. As was observed in Figure 2, deprotonation of the phenol of 3,5-F2Y allows for electron transfer to occur between 3,5-F2Y− and 3[Re]* resulting in a decrease in emission intensity. We thus attribute the emission quenching in Figure 4 to electron transfer between 3[Re]* and 3,5-F2Y− on the peptide bound to α2. Thus the pKa of the bound 3,5-F2Y does not shift significantly from that of the peptide in free solution.
Figure 4.
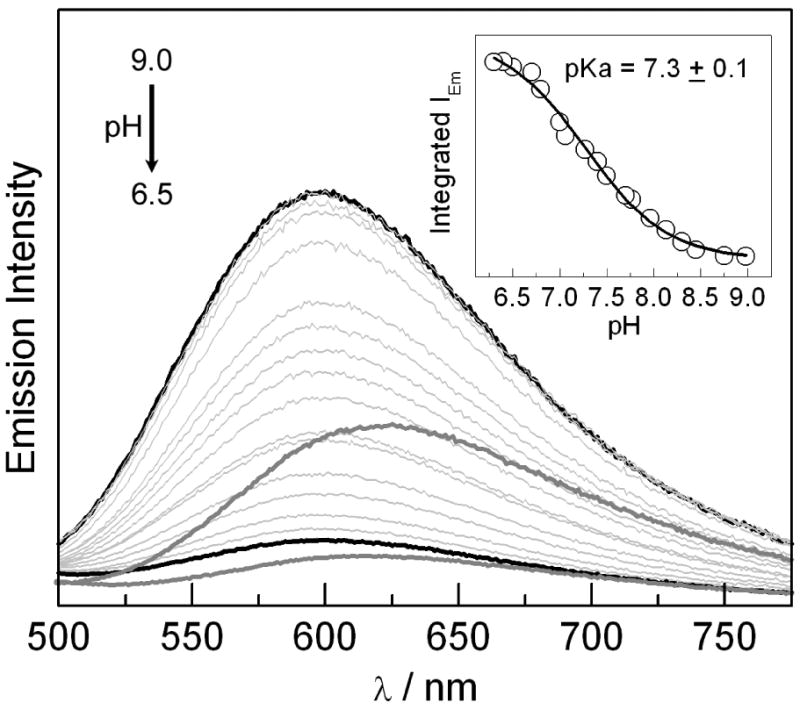
Emission spectra of [Re]-3,5-F2Y-βC19 (5 μM) and α2 (20 μM) as the pH is titrated from 9.0 to 6.3. The solid black curves correspond to the endpoints of the titration at pH 9.0 and pH 6.3. The dark grey traces are spectra recorded at pH 6.3 (lower spectrum) and pH 9.0 (upper spectrum) in the absence of α2. Inset: Integrated emission intensity as a function of pH (○) with fit (—) to monoprotic titration curve.
Kinetic Analysis of [Re]* Decay in [Re]-(F/Y)-βC19:α2 Complexes
Time-resolved emission monitoring the decay kinetics from the [Re] excited state ([Re]*) was used to further probe the homogeneity of the peptide:α2 binding interaction. For this experiment, 5 μM peptide and 20 μM α2 (with 3mM ATP and at pH 7.5) were used to ensure that most of the peptide was bound. All experiments were performed in buffered solutions containing 20% glycerol, as this was previously shown to increase the yield of photochemical RNR turnover (22). Figure 5 plots the emission decay trace obtained at 600 nm for 5 μM [Re]-Y-βC19 and 20 μM α2. The same trace was obtained in the presence of 1mM CDP substrate. Under all conditions investigated, the data were best fit with a bi-exponential decay function. One emission decay component was set equal to that of the free peptide while the other component was allowed to vary. We obtained decay components of 60 ns (63%) and 150 ns (37%), which were unaffected by the addition of 1 mM CDP substrate. These data reveal that the conformation of the peptide bound to α2 is heterogeneous, with multiple binding modes. For some binding modes, the [Re] unit may be solvated similar to that in free solution, while for others the [Re] chromophore is partially desolvated resulting in a blue-shifted emission energy with longer emission lifetime.
Figure 5.
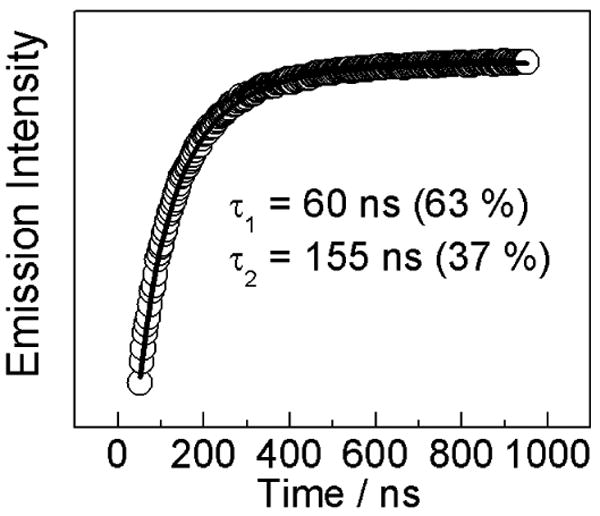
Time resolved emission decay trace recorded at 600 nm for [Re]-Y-βC19 (5 μM) in the presence of α2 (20 μM) and ATP (3 mM).
TA spectroscopy was used to characterize the intermediate species produced upon laser excitation. The [Re]-F-βC19:α2 system, with phenylalanine in place of tyrosine at position 356 on the peptide, was used as a control. Electron transfer photoproducts are not expected when tyrosine is replaced with phenylalanine because its oxidation by the [Re]* excited state is significantly endergonic (30). Figure 6 plots the TA spectra recorded for 100 μM solutions of [Re]-F-βC19 (top) and [Re]-Y-βC19 (bottom) in the presence of 135 μM α2, 1 mM CDP, and 3 mM ATP in 20% glycerol at pH 7.5. The spectra in both panels contain features at 380 and 480 nm and resemble that obtained for the peptides free in solution (Figure S1). These spectra are assigned to the 3[Re]* excited state, as they are identical to that observed in [Re]-Y model compounds (30). For both peptides, the absorbance decay at 480 nm could be fit to a bi-exponential decay function with τ1 = 62 ns (55 %) and τ2 = 150 ns (45 %) for the [Re]-F-R2C19:α2 system and τ1 = 62 ns (62 %) and τ2 = 185 ns (38 %) for [Re]-F-R2C19:α2. These data are consistent with the time-resolved emission results discussed above that suggest the peptide binds to α2 in a heterogeneous fashion.
Figure 6.
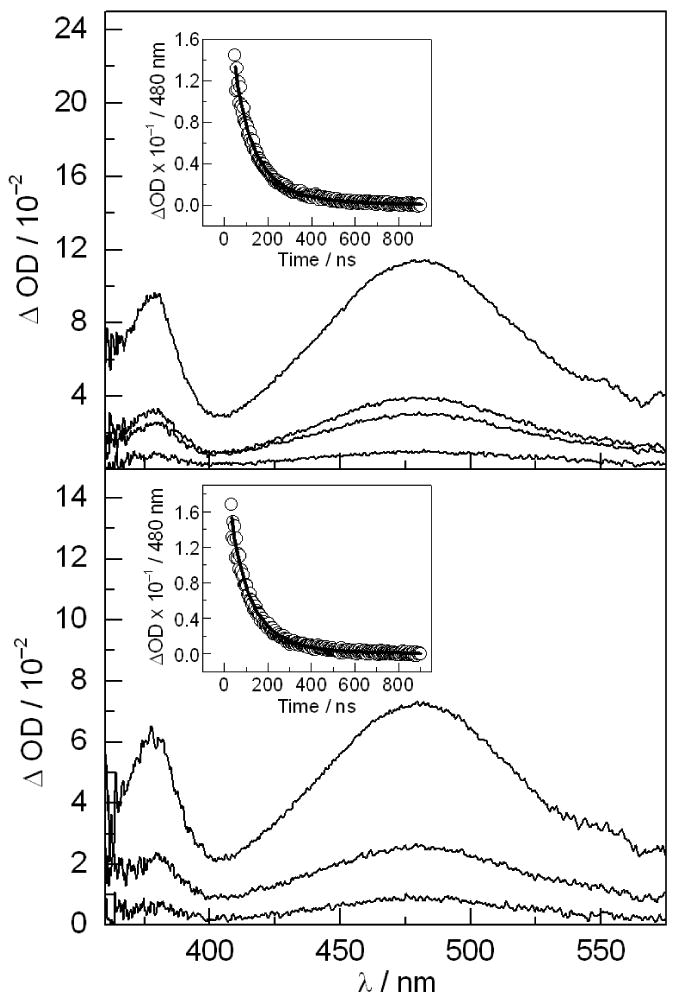
TA spectra recorded for 100 μM solutions of [Re]-F-βC19 (top) and [Re]-Y-βC19 (bottom) in the presence of 135 μM α2, 1 mM CDP, and 3 mM ATP in 20% glycerol at pH 7.5. Spectra are recorded at 65, 115, 215, and 415 ns (top) and at 115, 215, and 415 ns (bottom). Insets: Single wavelength absorbance kinetics data measured at 480 nm.
Dissociation Kinetics of [Re]-(F/Y)-βC19:α2
The goal of observing and measuring radical transfer processes from Y356 of modified peptides such as [Re]-Y-βC19 requires that the peptide remain bound to α2 for a sufficiently long time so that radical injection can occur. In other terms, the dissociation rate constant for the [Re]-Y-βC19:α2 complex (koff) must be slower than the PCET rate constant. To this end, we have undertaken experiments to measure the rates of association and dissociation of the peptide:α2 complex.
The data in Figure 3 shows that the emission of [Re]-Y-βC19 increases in the presence of α2 and thus loss of emission intensity could provide a reasonable method to measure the dissociation of the peptide from α2:[Re]-F-βC19 using stopped-flow spectroscopy. The baseline [Re] emission was determined by mixing Re-F-βC19 in one syringe with an equal volume of assay buffer in a second syringe to yield final concentrations of 45 μM. This baseline emission is shown by the black trace in Figure S2. As a second control pre-reduced α2 and Re-F-βC19 in one syringe were then mixed with an equal volume of assay buffer from a second syringe to yield final concentrations of 45 μM and 1 μM, respectively. As expected from Figure 3, the [Re] emission intensity (red trace in Figure S2) is enhanced relative to the baseline owing to the binding of the peptide to α2. To monitor peptide dissociation, a competitive binding experiment was performed in which a complex of Re-F-βC19:α2 is mixed with a 10-fold excess of Ac-Y-βC19. Subsequent to Re-F-βC19 dissociation, the Ac-Y-βC19 outcompetes Re-F-βC19 for rebinding. The Ac-Y-βC19 was chosen for this experiment since it is non-emissive and has a Kd ≈ 20 μM (20). As shown by the green trace in Figure S2, the emission intensity of the [Re] peptide is rapidly decreased upon mixing. This observation is in accordance with the replacement of a Re-F-βC19 by Ac-Y-βC19 on α2. The rapid intensity loss suggests that the peptide dissociation occurs in the dead time of the instrument. This sets a lower limit for the off rate at koff > 400 s−1. Additionally, since Kd = 9 μM, a kon > 4.4 × 10 M−1 s−1 can be calculated.
Diffusion Limited Rate Constants
The upper limit for koff and kon may be ascertained from the determination of the diffusion coefficient for Re-Y-βC19 and α2 and an estimate of the binding site between the two using eq 6. Multiple-photon fluorescence recovery after photobleaching (MP-FRaP) was used to determine the diffusion coefficient of Re-Y-βC19 (kd1). Figure S3 shows the recovery of the emission after an intense pulse of laser light was used to photo-bleach a defined area of the sample. The recovery is attributed to the diffusion of Re-Y-βC19 into the bleached area. A fit of recovery curve yields kd1 = 6.1 × 10−7 cm2 s−1 (35).
The diffusion coefficient of α2 (kd2) was determined using the Einstein-Stokes relationship in eq 4 using the radius found by DLS and that estimated from the crystal structure of α2. The average hydrodynamic radius obtained from DLS from a mass-weighted size distribution analysis was 55.8 Å. The radius of α2 was also estimated as 40.4 Å using eq 5 with d1, d2 and d3 as 100.7 Å, 71.4 Å, and 73.2 Å, respectively. These two radii values yield kd2 for α2 as 3.9 × 10−7 cm2 s−1 and 5.4 × 10−7 cm2 s−1, respectively.
The active site of the α2:[Re]-(Y/F)-βC19 complex was estimated from the crystal structure of Ac-Y-βC19 bound to α2 which has only 16 amino acid residues visible by X-ray crystallography (18). Using eq. 5, the estimated radius of the active site was determined to be 8.97 Å. With eq. 6, the diffusion controlled rate constant for the [Re]-(Y/F)-βC19:α2 complex association (kon) was found to be 6.8 – 7.8 × 10 M−1 s−1. Using Kd = 9 μM, a limiting rate constant for dissociation (koff) was computed to be between 6100 s−1 and 7000 s−1. With these results and the SF kinetic data, the kinetics of association and dissociation for the [Re]-(Y/F)-βC19:α2 complex can limited to 7.8 × 108 M−1 s−1 > kon > 4.4 × 10 M−1 s−1 and 7000 s−1 > koff > 400 s−1.
Discussion
The [Re] complex within the [Re]-(F/Y/3,5-F2Y)-βC19 peptide serves as a probe of both peptide conformation and the pKa of the adjacent Y residue. Excitation of [Re] produces a metal-to-ligand charge transfer (MLCT) excited state, in which a metal d-orbital electron is promoted to an orbital of π* symmetry primarily localized on the bpy ligand resulting in an effective Re I I(bpy•−) electronic configuration. 3[Re (bpy•−)]* serves as a potent excited state oxidant (E°([Re I I(bpy•−)]*/[Re(symbol) (bpy•−)]) = 1.59 V vs. NHE (30)) that can oxidize deprotonated tyrosinate according to the mechanism:
| (7) |
The emission of the photoexcited [Re] center is therefore quenched by the transfer of an electron from the adjacent tyrosine in [Re]-Y-βC19 upon deprotonation of the tyrosine phenol (22). Thus monitoring of the emission band intensity as a function of pH probes the pKa for the tyrosine phenol within the peptide and bound to α2. The solution pKa value of tyrosine is maintained upon incorporation into the peptide and upon binding of the peptide to α2 (pKa(3,5-F2Y) = 7.2, pKa([Re]-3,5-F2Y-βC19) = 7.3, pKa([Re]-3,5-F2Y-βC19:α2) = 7.3). These results show that the photoRNR construct faithfully models RNR in that the pKa of tyrosine at amino acid position 356 is unperturbed relative to the value in solution (42).
MLCT excitation of d6 polypyridyl complexes is associated with a marked increase in the molecular dipole owing to the intramolecular charge separation. For this reason, the emission energy and intensity are strongly dependent upon polarity of the surrounding environment (43). High polarity solvents tend to lower the energy of the thermally equilibrated excited state, thus resulting in a red shift of the emission. Protic solvents, such as alcohols and water, also strongly quench the excited state lifetime through nonradiative decay mechanisms mediated by vibrational overlap of the excited state with the high frequency O–H stretch of the solvent (40). [Re] attached to the FnY-βC19 peptide therefore serves as an effective probe of peptide conformation and binding to class I RNRs. Removal of water from the solvation sphere of [Re] upon peptide conformational changes or binding events serves to both blue shift the energy of and increase the lifetime for the emission. The monotonic increase of the emission band to higher energy with a concomitant increase in intensity (Figure 3) is consistent with the exclusion of water from the solvation sphere of the [Re] probe upon binding of the peptide to the protein. The shift in the emission profile with addition of the protein therefore provides a direct measure of peptide binding to α2. The similarity of the KD of [Re]-Y-βC19 relative to that measured for Ac-Y-βC19 by competitive inhibition suggests that modification of the peptide with the [Re] moiety does not greatly perturb the interaction of the peptide to the protein. The multiple emission lifetimes observed for [Re]-Y-βC19:α2 in the presence of RNR substrate and effector are consistent with multiple solvation environments of the [Re] chromophore, generally partitioning between protein-bound and solvent exposed. Several other peptides containing photooxidants of different sizes and shapes have been generated, all of which bind to α2 with KDs in the micromolar range (20, 21). In addition, the N-terminal peptide residues (356-360) are not located in the X-ray structure of Ac-Y-βC19:α2 (Figure 1; (18, 44)). Taken together these results are consistent with a flexible peptide N-terminus when bound to α2.
The observation of conformational flexibility within the photoRNR provides a connection to conceptualize the conformational gating of RNR activity at the molecular level. We have shown that the electron and proton are committed to a colinear pathway in α2 (21) for radical transport. The reaction rate for particle transfer is highly dependent on transfer distance because the electron and proton must tunnel. Whereas the electron can transfer over large distances, the proton cannot (9); hence PCET may provide a basis for conformational gating in RNR. The binding of α2 to β2 is known to trigger radical transport from •Y122 to Y356, but only in the presence of RNR substrate and effector (26, 27). Conformational flexibility of the 356-360 region of β2 within the α2:β2 complex would then allow the •Y356 radical to lock into the PCET pathway for radical transport across the subunit interface. Inasmuch as the [Re]-3,5-F2Y-βC19: α2 complex has been shown to turnover upon irradiation (22), hole injection must be faster than the peptide off rate, which has now been determined to be in the range of 7000 s−1 > koff > 400 s−1. The conformational changes in the N-terminal peptide are likely to effect the efficient oxidation of Y731 in α2, making its detection challenging. Studies are now underway to examine how conformational changes of the N-terminal tail affect PCET in the photoRNR.
Supplementary Material
Acknowledgments
We thank Vikash P. Chauhan for his assistance with the MP-FRaP experiments.
Footnotes
These studies were supported by the National Institutes of Health GM29595 (JS) and GM47274 (DGN). D.A.L. is a Jane Coffin Childs Memorial Fund for Medicinal Research Postdoctoral Fellow.
List of Abbreviations: α2, active site-containing subunit of RNR; Anq, anthraquinone;; ATP, adenosine-5′-triphosphate; β2, cofactor-containing subunit of RNR; βC19, 19-residue C-terminal peptide tail of β2; BPA, 4-benzoylphenylalanine; CDP, cytidine-5′-diphosphate; dC, deoxycytidine; DOPA, 3,4-dihydroxyphenylalanine; ET, electron transfer; FPLC, fast protein liquid chromatography; MLCT, metal-to-ligand charge transfer; N3NDP, 2′-azido-nucleotide-5′ diphosphate; PCET, proton-coupled electron transfer; PELDOR, pulsed electron-electron double resonance; photoRNR, photochemical ribonucleotide reductase; [Re]; Re(bpy)(CO)3CN; 3[Re]*, triplet MLCT excited state of [Re]; MP-FRaP, multiple photon-fluorescence recovery after photobleaching; RNR, class I E. coli ribonucleotide reductase; SA, specific activity; SF, stopped-flow; TA, transient absorption; TR, E. coli thioredoxin; TRR, E. coli thioredoxin reductase; Tris, tris(hydroxymethyl) aminomethane; Y•, tyrosyl radical; DLS, dynamic light scattering.
Supporting Information Available. Transient absorption spectra of the [Re]-F-βC19 and [Re]-Y-βC19 peptides in aqueous solution, SF traces, and MP-FRaP trace. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Stubbe J, van der Donk WA. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 2.Stubbe J. Radicals with a controlled lifestyle. Chem Comm. 2003:2511–2513. doi: 10.1039/b307617m. [DOI] [PubMed] [Google Scholar]
- 3.Cukier RI, Nocera DG. Proton-coupled electron transfer. Annu Rev Phys Chem. 1998;49:337–369. doi: 10.1146/annurev.physchem.49.1.337. [DOI] [PubMed] [Google Scholar]
- 4.Mayer JM. Proton-coupled electron transfer: A reaction chemist's view. Annu Rev Phys Chem. 2004;55:363–390. doi: 10.1146/annurev.physchem.55.091602.094446. [DOI] [PubMed] [Google Scholar]
- 5.Hammes-Schiffer S. Theoretical perspectives on proton-coupled electron transfer reactions. Acc Chem Res. 2001;34:273–281. doi: 10.1021/ar9901117. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkiss JM, Rosenthal J, Nocera DG. The relation between hydrogen atom transfer and proton-coupled electron transfer in model systems. In: Hynes JT, Klinman JP, Limbach HH, Schowen RL, editors. Handbook of Hydrogen Transfer Physical and Chemical Aspects of Hydrogen Transfer. Wiley-VCH; Weinheim, Germany: 2006. pp. 503–562. [Google Scholar]
- 7.Chang CJ, Chang MCY, Damrauer NH, Nocera DG. Proton-coupled electron transfer: A unifying mechanism for biological charge transport, amino acid radical initiation and propagation, and bond making/breaking reactions of water and oxygen. Biochim Biophys Acta. 2004;1655:13–28. doi: 10.1016/j.bbabio.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 9.Reece SY, Nocera DG. Proton-coupled electron transfer: The engine that drives radical transport and catalysis in biology. In: Scrutton NS, Alleman R, editors. Quantum Tunneling in Enzyme Catalyzed Reactions. Ch. 15. Royal Society of Chemistry; London: 2009. pp. 351–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray HB, Winkler JR. Electron transfer in proteins. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 11.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: The mechanistic underpinning for radical transport and catalysis in biology. Phil Trans Roy Soc B. 2006;361:1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103:2167–2202. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 13.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Thelander L. Physicochemical characterization of ribonucleoside diphosphate reductase from Escherichia coli. J Biol Chem. 1973;248:4591–4601. [PubMed] [Google Scholar]
- 15.Thelander L, Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 16.Stubbe J, Riggs-Gelasco P. Harnessing free radicals: Formation and function of the tyrosyl radical in ribonucleotide reductase. Tr Biochem Sci. 1998;23:438–443. doi: 10.1016/s0968-0004(98)01296-1. [DOI] [PubMed] [Google Scholar]
- 17.Bennati M, Robblee JH, Mugnaini V, Stubbe J, Freed JH, Borbat P. EPR distance measurements support a model for long-range radical initiation in E. coli ribonucleotide reductase. J Am Chem Soc. 2005;127:15014–15015. doi: 10.1021/ja054991y. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson M, Uhlin U, Ramaswamy S, Ekberg M, Regnström K, Sjöberg BM, Eklund H. Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 19.Reece SY, Nocera DG. Proton-coupled electron transfer in biology: Results from synergistic studies in model and natural systems. Annu Biochem. 2009;78 doi: 10.1146/annurev.biochem.78.080207.092132. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang MCY, Yee CS, Stubbe J, Nocera DG. Turning on ribonucleotide reductase by light-initiated amino acid radical generation. Proc Natl Acad Sci, USA. 2004;101:6882–6887. doi: 10.1073/pnas.0401718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Photoactive peptides for light-initiated tyrosyl radical generation and transport into ribonucleotide reductase. J Am Chem Soc. 2007;129:8500–8509. doi: 10.1021/ja0704434. [DOI] [PubMed] [Google Scholar]
- 22.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Direct observation of a transient tyrosine radical competent for initiating turnover in a photochemical ribonucleotide reductase. J Am Chem Soc. 2007;129:13828–13830. doi: 10.1021/ja074452o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Climent I, Sjöberg BM, Huang CY. Carboxyl-terminal peptides as probes for Escherichia coli ribonucleotide reductase subunit interaction: Kinetic analysis of inhibition studies. Biochemistry. 1991;30:5164–5171. doi: 10.1021/bi00235a008. [DOI] [PubMed] [Google Scholar]
- 24.Climent I, Sjöberg BM, Huang CY. Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry. 1992;31:4801–4807. doi: 10.1021/bi00135a009. [DOI] [PubMed] [Google Scholar]
- 25.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. pH Rate profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli ribonucleotide reductase: Evidence that Y356 is a redox-active amino acid along the radical propagation pathway. J Am Chem Soc. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 26.Seyedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenylalanine in the β2 subunit of E. coli ribonucleotide reductase. J Am Chem Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 27.Seyedsayamdost MR, Stubbe J. Forward and reverse electron transfer with the Y356DOPA-β2 heterodimer of E. coli ribonucleotide reductase. J Am Chem Soc. 2007;129:2226–2227. doi: 10.1021/ja0685607. [DOI] [PubMed] [Google Scholar]
- 28.Reece SY, Stubbe J, Nocera DG. pH dependence of charge transfer between tryptophan and tyrosine in dipeptides. Biochim Biophys Acta. 2005;1706:232–238. doi: 10.1016/j.bbabio.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Seyedsayamdost MR, Reece SY, Nocera DG, Stubbe J. Mono-, di-, tri-, and tetra-substituted fluorotyrosines: New probes for enzymes that use tyrosyl radicals in catalysis. J Am Chem Soc. 2006;128:1569–1579. doi: 10.1021/ja055926r. [DOI] [PubMed] [Google Scholar]
- 30.Reece SY, Nocera DG. Direct tyrosine oxidation using the MLCT excited states of rhenium polypyridyl complexes. J Am Chem Soc. 2005;127:9448–9458. doi: 10.1021/ja0510360. [DOI] [PubMed] [Google Scholar]
- 31.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Electron transfer reactions of fluorotyrosyl radicals. J Am Chem Soc. 2006;128:13654–13655. doi: 10.1021/ja0636688. [DOI] [PubMed] [Google Scholar]
- 32.Fasman GD. Practical Handbook of Biochemistry and Molecular Biology. CRC Press; Boca Raton, FL: 1989. [Google Scholar]
- 33.Salowe S, Bollinger JM, Jr, Ator M, Stubbe J, McCraken J, Peisach J, Samano MC, Robins MJ. Alternative model for mechanism-based inhibition of Escherichia coli ribonucleotide reductase by 2′-azido-2′-deoxyuridine 5′-diphosphate. Biochemistry. 1993;32:12749–12760. doi: 10.1021/bi00210a026. [DOI] [PubMed] [Google Scholar]
- 34.Van Holde KE. Physical Biochemistry. 2nd. Prentice Hall; Englewood Cliffs, NJ: 1985. [Google Scholar]
- 35.Brown EB, Wu ES, Zipfel W, Webb WW. Measurements of molecular diffusion in solution by multiphoton fluorescence photobleaching recovery. Biophys J. 1999;77:2837–2849. doi: 10.1016/S0006-3495(99)77115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mussell RD, Nocera DG. Effect of long-distance electron transfer on chemiluminescence efficiencies. J Am Chem Soc. 1988;110:2764–2772. [Google Scholar]
- 37.Steinfeld JI, Francisco JS, Hase WL. Chemical Kinetics and Dynamics. 2nd. Ch. 4. Prentice Hall; Upper Saddle River, New Jersey: 1999. Reactions in Solution; pp. 124–146. [Google Scholar]
- 38.Damrauer NH, Hodgkiss JM, Rosenthal J, Nocera DG. Observation of proton-coupled electron transfer by transient absorption spectroscopy in a hydrogen-bonded, porphyrin donor-acceptor assembly. J Phys Chem B. 2004;108:6315–6321. doi: 10.1021/jp049296b. [DOI] [PubMed] [Google Scholar]
- 39.Loh ZH, Miller SE, Chang CJ, Carpenter SD, Nocera DG. Excited-state dynamics of cofacial pacman porphyrins. J Phys Chem A. 2002;106:11700–11708. [Google Scholar]
- 40.Chen P, Meyer TJ. Medium effects on charge transfer in metal complexes. Chem Rev. 1998;98:1439–1478. doi: 10.1021/cr941180w. [DOI] [PubMed] [Google Scholar]
- 41.Blanco-Rodriguez AM, Busby M, Gradinăru C, Crane BR, DiBilio AJ, Matousek P, Towrie M, Leigh BS, Richards JH, Vlček A, Gray HB. Excited-state dynamics of structurally characterized [ReI(CO)3(phen)(HisX)]+ (X = 83, 109) Pseudomonas aeruginosa azurins in aqueous solution. J Am Chem Soc. 2006;128:4365–4370. doi: 10.1021/ja057451+. [DOI] [PubMed] [Google Scholar]
- 42.Yee CS, Seyedsayamdost MR, Chang MCY, Nocera DG, Stubbe J. Generation of the R2 subunit of ribonucleotide reductase by intein chemistry: Insertion of 3-nitrotyrosine at residue 356 as a probe of the radical initiation process. Biochemistry. 2003;42:14541–14552. doi: 10.1021/bi0352365. [DOI] [PubMed] [Google Scholar]
- 43.Caspar JV, Meyer TJ. Photochemistry of tris(2,2′-bipyridine)ruthenium (2+) ion (Ru(bpy)32+). Solvent effects. J Am Chem Soc. 1983;105:5583–5590. [Google Scholar]
- 44.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


