Abstract
An intriguing set of neurodegenerative disease are the nine disorders caused by the expansion of a unstable trinucleotide CAG repeat where the repeat is located within the coding of the affected gene, i.e. the polyglutamine (polyQ) diseases. A gain-of-function mechanism for toxicity in polyQ diseases is widely thought to have a major role in pathogenesis. Yet, the specific nature of this gain-of-function is a matter of considerable discussion. The basic issue concerns whether toxicity stems from the native or normal function of the affected protein versus a novel function induced by polyQ expansion. For at least three of the polyQ disease considerable evidence is accumulating that pathology is mediated by a polyQ-induced exaggeration of a native function of the host protein.
Twenty years ago one of the first unstable nucleotide repeat mutations identified was the CAG expansion causing spinal and bulbar muscular atrophy (SBMA/Kennedy disease) [1]. SBMA subsequently proved to be the first member of a group of nine progressive neurodegenerative disorders now known as the polyglutamine (polyQ) diseases [2]. Along with SBMA, the polyQ diseases include Huntington disease, dentatorubral-pallidoluysian atrophy, and spinocerebellar ataxia (SCA) 1, 2, 3, 6, 7 and 17. As a group, the polyQ diseases are one of the more common causes of inherited neurodegeneration.
Like most neurodegenerative diseases, the polyQ disorders are progressive and have the accumulation of mutant protein aggregates as a pathological hallmark. The extent to which these accumulations of mutant protein are pathogenic or protective has evolved over the years and remains under considerable investigation and discussion [3]. While the polyQ stretch alone can be toxic in vivo, for three of the polyQ disorders highlighted here recent studies demonstrate that an expanded polyQ alone is not sufficient to induce neurodegeneration, i.e. residues and regions of the host protein are also critical for pathogenesis. These three disorders provide a strong connection between host protein framework and induction of disease and, thus, argue that the native biochemistry and function are relevant for understanding disease mechanism. Such a link between an affected protein’s normal function and pathogenesis very likely is the basis for the fact that these and other neurodegenerative disorders vary considerably in terms of the region of the CNS they target.
SBMA – The paradigm for linking normal function to pathogenesis
In SBMA the unstable polyQ is located within the androgen receptor (AR) gene located on the X-chromosome [1]. SBMA is a late-onset progressive disorder in which motor neurons in the brainstem and spinal cord in only males degenerate [4]. This latter point, that SBMA is a disease essentially restricted to males, is accentuated by the finding that females homozygous for mutant alleles of the AR have a subclinical presentation of mild symptoms, clearly showing that the disease is influenced by sex-specific factors possibly levels of circulating androgen [5].
Upon generation of SBMA animal models displaying the same male bias in presentation as seen in human patients, it became possible to test directly whether the sex-specific factor important in SBMA was indeed level of circulating androgen. In both a Drosophila and a transgenic mouse model of SBMA, neurodegeneration is linked to the presence of circulating testosterone as is normally present only in males [6,7]. Thus, along with an expanded polyQ tract in the AR, the normal AR ligand is required for manifestation of disease. Nedelsky et al. recently utilized Drosophila genetics to further dissect and characterize the normal interactions and components of the AR/androgen pathway critical for toxicity [8••]. These and earlier results provide a solid platform for exploring the therapeutic potential of hormonal intervention for SBMA.
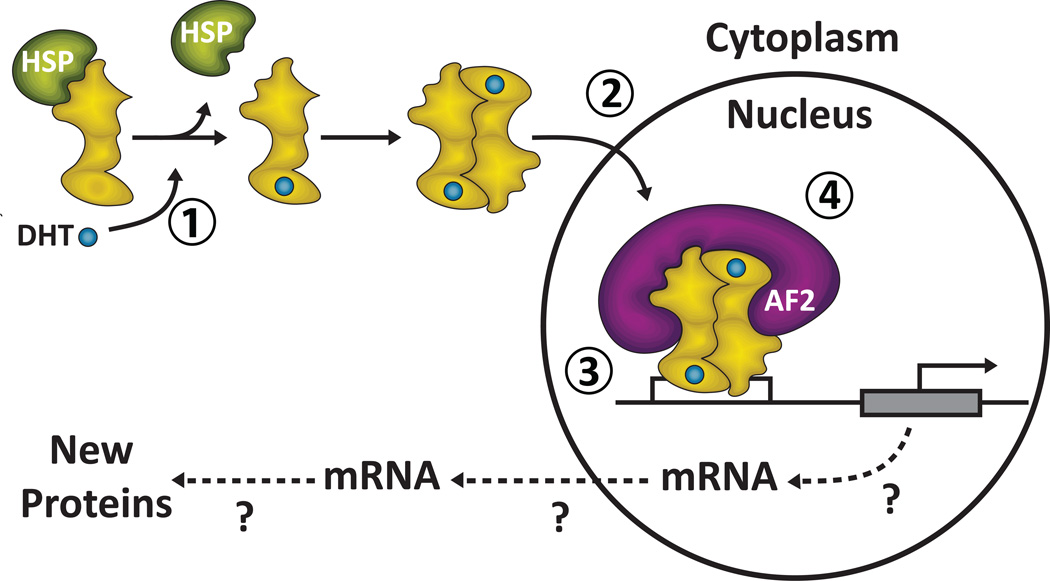
In contrast to many of the polyQ diseases where less is known about the native function and biochemistry of the protein affected, the normal structure/function of AR is well characterized. AR is a nuclear hormone that resides in the cytoplasm in an inactive form. Upon binding androgen, the hormone-receptor complex translocates to the nucleus, binds to DNA at specific recognition sites upstream of its target genes. Expression or repression of these genes occurs through the interaction of two co-regulator regions of AR with other transcription factors. As shown previously, Nedelsky and colleagues found that ligand binding was necessary for expanded polyQ AR to cause degeneration of Drosophila adult photoreceptors and larval motor neurons [8••]. They further showed that upon binding androgen mutant AR had to translocate to the nucleus and bind to DNA to cause degeneration. Lastly, it was found that one of the interaction regions of AR, the AF-2 transactivation domain, is required for toxicity and that several known AF-2 interacting proteins are modifiers of degeneration in a genetic screen. The results of this study show that the mutant AR pathogenic pathway induced by expansion of the polyQ tract overlaps considerably with the normal AR-ligand binding pathway (Figure 1). Nedelsky et al. went on to suggest that disease is induced by the amplification of a yet unidentified aspect of normal AR function involving the AF-2 transduction domain. This idea is supported by their observation that lies overexpressing wild type AR show some signs of disease [8••].
Figure 1. Schematic Depiction of the Minimum Elements of the Native Ligand-Dependent AR Pathway Required for Pathogenesis.
To induce pathogenesis AR with an expanded polyQ must (1) Ligand binds to AR inducing a conformation change that releases the bound chaperone, uncovers the NLS and creates the AF-2 binding surface. Presumably mutant AR forms a dimer prior to (2) AR and bound ligand translocate to the nucleus. (3) AR and bound ligand bind to DNA targets sites. (4) AF-2 interacting co-regulators bind. It remains unclear the extent to which if any downstream events (depicted by dashed lines) such as target gene transcription and subsequent steps contribute to pathogenesis.
SCA – Unbalanced normal function leads to ataxia
Spinocerebellar ataxia type 1 (SCA1) is one of the polyQ disorders where the host protein, Ataxin-1 (ATXN1), is a novel protein whose function was unknown. Subsequent to cloning the SCA1 gene in 1993 [9], studies have identified several functional/structural motifs in ATXN1. These include the AXH domain that folds into an OB-fold that consists of a putative RNA-binding and a second ligand-binding surface [10], a nuclear localization signal (NLS) [11] that overlaps with or is immediately adjacent to a 14-3-3 binding site [12] and a UHM ligand motif (ULM) present in proteins associated with RNA splicing [13•]. Not surprisingly, ATXN1 is reported to interact with RNA, several regulators of transcription, and RNA splicing factors RBM17 and U2AF65 [13•,14••]. Interestingly, mutating the NLS of ATXN1 such that transport of the protein to the nucleus of neurons is dramatically reduced prevents ATXN1 with an expanded polyQ tract from causing disease [11].
An important advance in understanding SCA1 pathogenesis came with the observation that ATXN1 normally forms multimeric complexes with other proteins in the nucleus and that expansion of the polyQ tract shifts the status quo by enhancing certain functional pathways perhaps at the expense of others [14••]. At the center of the imbalance in complex formation induced by mutant ATXN1 are the complexes it forms with the the splicing factor RBM17 (RNA-binding motif protein 17) and the transcription repressor Capicua (Cic). PolyQ expansion in ATXN1 enhances its interaction with RBM17 [14••]. Furthermore, additional evidence indicates that the RBM17/ATXN1 complex is toxic. The biological effect on Cic of ATXN1 polyQ expansion is much more complex. First, in a fly model of SCA1 overexpresison of Cic was protective [15], while in mice partial loss of Cic prolonged life span [16••]. It seems that for some Cic gene targets expanded ATXN1 causes a loss of function while at other targets polyQ-expanded ATXN1 enhances Cic binding inducing a state of hyper-repression. In the mouse, it is postulated that the hyper-repressive effect of expanded polyQ ATXN1 is toxic [16••].
In addition to polyQ expansion, phosphorylation of the Ser residues at position 776 in ATXN1 favors its interaction with RBM17 [14••]. Ser 776 is one of seven endogenous sites of phosphorylation in ATXN1 [17,18]. Phosphorylation of Ser 776 stabilizes ATXN1 and, in the cytoplasm, binding of 14-3-3 blocks dephosphorylation of Ser 776 as well as the transport of ATXN1 to the nucleus [19]. Previous work showed that placing a phosphorylation-resistant Ala at position 776 dramatically decreases the ability of ATXN1 with an expanded polyQ to cause neurodegeneration in vivo [17]. On the other hand a potentially phospho-mimicking Asp at residue 776, a substitution that enhances wild type ATXN1’s interaction with RBM17 [14••], enhances ATXN1 induced pathogenesis such that even ATXN1 with a wild type, unexpanded polyQ becomes pathogenic [20•]. Together these results indicate that phosphorylation of Ser 776 in ATXN1 as well as its subsequent interaction with RBM17 has a critical role in pathogenesis.
Moreover, mild exercise likely involving the Cic pathway in the brainstem improves survival in a mouse model of SCA1 in absence of improving the cerebellar motor dysfunction [16••]. While more vigorous motor activity that engages the cerebellum might restore motor function, it is intriguing to speculate that distinct ATXN1 complexes, i.e. cellular pathways, contribute to disease in different regions of the CNS. Such a hypothesis is supported by the ability of a partial loss of 14-3-3 to improve cerebellar phenotypes and not brainstem phenotypes [21].
HD – Increased ciliogenesis?
Perhaps the more prevalent and widely known polyQ disorder is Huntington disease (HD) where the unstable CAG repeat is located in first exon of the huntingtin (HTT) gene [21]. Clinically, the loss and dysfunction of neurons in the striatum and deep layers of the cortex characterize HD. Symptomatically, HD patients suffer from cognitive, psychiatric, and motor abnormalities. Over the years a number of cellular pathways and proteins have come forward as targets of wild type and mutant Htt [23,24]. One of the first Htt interacting proteins identified is the huntingtin-associated protein 1 (HAP1) [25]. Subsequently, Hap1 was found to interact the p150Glued subunit of dynactin and the pericentriole protein PCM1 [26]. A potential biological relevance of these interactions recently came to light with the observation that Htt/HAP1, impact PCM1’s role in ciliogenesis raising the possibility that HD is at least to some extent a ciliopathy [27••].
Cilia fall into two functional/structural groups; nonmotile primary cilia consisting of nine outer microtubule pairs in a (9 + 0 axoneme) configuration and motile cilia with nine outer microtubule pairs, inner and outer dynein arms, and a pair of central microtubules in a (9 + 2 axoneme) configuration. In the brain, primary cilia are on all neurons and glia. On neurons, primary cilia are located on the cell body or proximal portion of the dendrite where they are believed to have a role in sensing the extracelluar environment and subsequent signal transduction. Motile cilia in the brain are located on ependynmal cells lining the lateral ventricles where they are crucial for maintaining proper flow of the cerebral spinal fluid.
Although formation of primary and motile cilia differs, the biogenesis of each form of cilia depends on proper trafficking of proteins to the pericentriolar material (PCM). Specifically, if the pericentriolar material protein 1 (PCM1) does not localize to the PCM ciliogenesis is reduced. Keryer et al. made two key observations linking Htt function to ciliogenesis [27••]. First, they found that depletion of Htt in cultured striatal cells using RNAi resulted in a dispersion of PCM1 away from the centrosome and a decease in the number of cells with primary cilia. Moreover, Keryer et al. found that mice lacking Htt in ependymal cells also had an altered distribution of PCM1 and hydrocephalus, a phenotype previously found in mice having a disruption in ciliary function [27]. In contrast to decreased ciliary function with loss of Htt, expression of Htt with an expanded polyQ resulted in hypermorphic cilia and ciliary dysfunction. Again, in both cultured striatal cells and in mice mutant Htt had a similar effect. With mutant Htt, localization of PCM1 was enhanced at the centriole and the length of primary cilia was increased [27••]. Increased localization of PCM1 to the ependymal zone of the lateral ventricle was found in HD patient brain material. Importantly, this altered accumulation of PCM1 directly correlated with severity of pathology. Keryer et al. went on to show that in HD mice CSF flow was altered using an organotypic preparation that included the lateral ventricle. Intriguingly, in the brain of HdhQ111/Q111 mice migration of neuroblasts was altered relatively to the lateral ventricle. In HD mice, alterations in PCM1 and cilia seemed to be an early event with some abnormalities detected as young as 1 month of age.
These latter findings raise the intriguing possibility that in HD migration of developing neurons is abnormal. If indeed HD proves to be a ciliopathy, having a developmental component to pathogenesis would be in line with other ciliopathies [29]. There is suggestive evidence of abnormal neurodevelopment in HD based on HD mutation carriers having a reduced intracranial volume compared to non-carriers prior to onset of disease [30]. In a mouse model of SCA1, data support a link between abnormal development and neurodegeneration in the adult. Expressing a mutant allele of ATXN1 during early postnatal development compromises Purkinje cell development that contributes to the severity of the neurodegeneration in adult mice [31].
Concluding comments
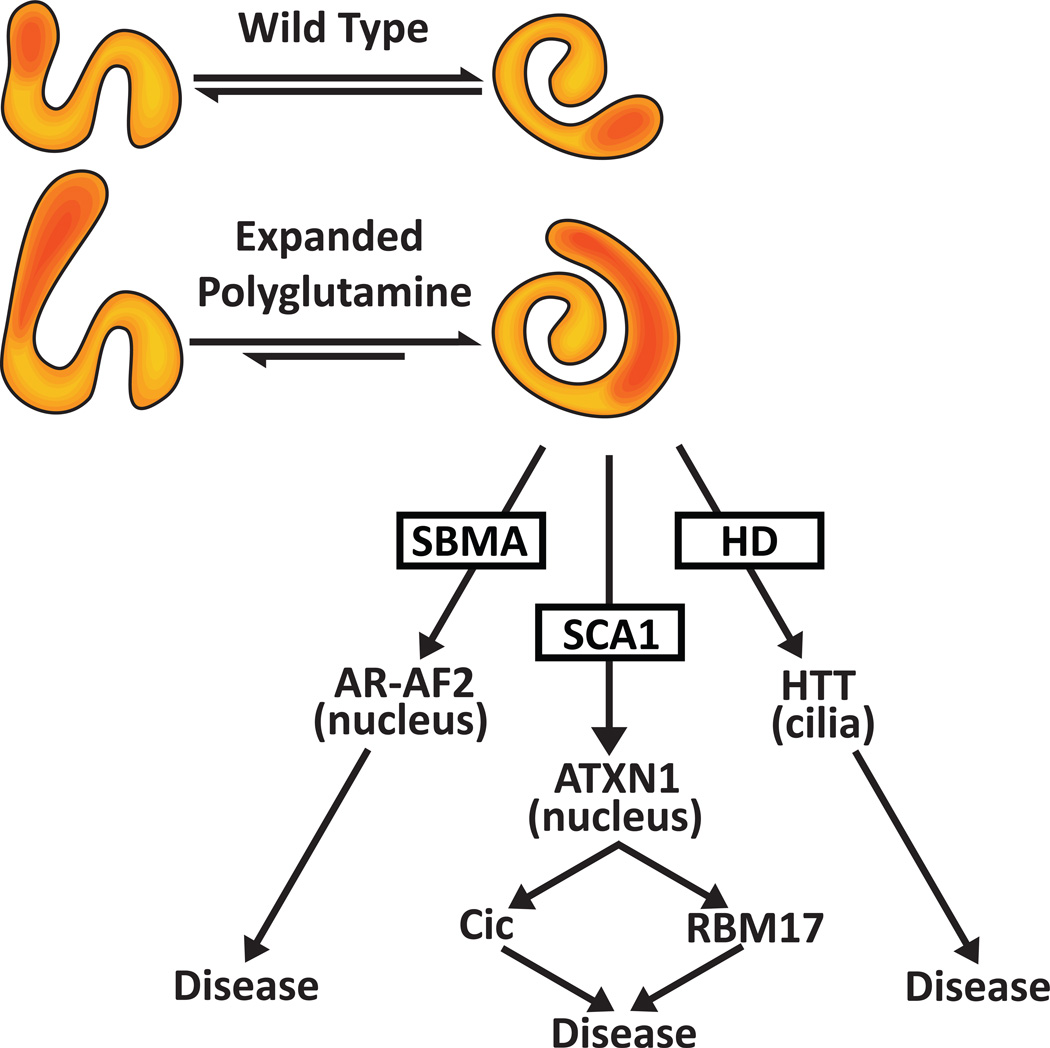
The results for the three polyQ disorders outlined here illustrate the evidence in support of the hypothesis that pathogenesis induced by polyQ expansion is linked to the native function of the host protein, specifically an enhanced level of a normal function (Figure 2). That such a pathogenic mechanism likely extends beyond these three disorders is suggested by the observations that an increase in wild type gene number and presumably function underlies some forms of Alzheimer and Parkinson disease [32,33]. Recently, Kratter and Finkbeiner nicely outlined how this hypothesis might be envisioned at a protein conformation level [34•]. First, likes many proteins, the polyQ host protein normally adopts multiple conformations each associated with a particular function and/or set of protein interactions. Normally the proportion of each confirmation is dynamically regulated by reversible post-translational modifications such as phosphorylation. However, in the presence of an expanded polyQ one confirmation becomes stabilized and, thus, the associated function(s) is accentuated possibly at the expense of others. It is worth noting that a polyQ17 stretch in the exon 1 fragment of HTT was found to assume multiple conformations, indicating that at least a wild type polyQ tract imparts conformational flexibility to a protein [35•]. Regardless, a hope is that as the native functions and pathways for each disease-associated polyQ protein are identified this will lead to additional therapeutic targets.
Figure 2. A Hypothesis in which the Consequence of Expanded PolyQ is to Enhance a Native Function.
Normally the polyQ host exists in a equilibrium of multiple conformations each of which sub serves a native function. Upon expansion of the polyQ (red shaded portion) one native conformation predominates and its associated function is now enhanced in a disease-specific manner.
Acknowledgements
I thank Jillian Frisch for her expert assistance in figure design. The research presented from the author’s was supported by NIH/NINDS grants NS22920-23 and NS45667-09.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Zoghbi HY. Trinucleotide Repeat Disorders. Ann Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Gatxhel JR, Zoghbi HY. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy WR, Alter M, Sung JH. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology. 1968;18:671–680. doi: 10.1212/wnl.18.7.671. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt BJ, Greenber CR, Allingham-Hawkins DJ, Spriggs EL. Expression of X-linked bulbospinal muscular atrophy (Kennedy disease) in two homozygous women. Neurology. 2002;59:770–772. doi: 10.1212/wnl.59.5.770. [DOI] [PubMed] [Google Scholar]
- 6.Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobasyashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 7.Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, Miura M, Tabata T, Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 8. Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. •• This is the most complete and systematic study of the androgen receptor pathway identifying those components from ligand binding to the transactivation domain critical for neurodegeneration in Drosophila model of SBMA.
- 9.Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 10.Chen YW, Allen MD, Veprintsev DB, Lowe J, Bycroft M. The Structure of the AXH Domain of Spinocerebellar Ataxin-1. J. Biol. Chem. 2004;279:3758–3765. doi: 10.1074/jbc.M309817200. [DOI] [PubMed] [Google Scholar]
- 11.Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 12.Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, Aitken A, Skoulakis EM, Orr HT, Botas J, Zoghbi HY. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 13. de Chiara C, Menon RP, Strom M, Gibson TJ, Pastore A. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. 2009 PLoS One. 2009;4:e8372. doi: 10.1371/journal.pone.0008372. • This study identifies and characterizes a UHM ligand motif (ULM) that includes Ser766 of ATXN1. The presence of this motif in ATXN1 provides additional strong evidence for it having role in RNA splicing.
- 14. Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–719. doi: 10.1038/nature06731. •• This study presents two major advances regarding SCA1 and the polyQ field in general. It demonstrates how expansion can simultaneously convey a gain of function to one pathway while imparting a loss of function to another. In addition, it reveals that a single amino acid substitution is able to “convert” wild type ATXN1 into a form having a biochemical property characteristic of ATXN1 with an expanded polyQ.
- 15.Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun E, Duvick LA, Orr HT, Botas J, Zoghbi HY. Mutant ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 16. Fryer JD, Yu P, Kang H, Mandel-Brehm C, Carter A, Crespo-Barreto J, Gao Y, Flora A, Shaw C, Orr H, Zoghbi HY. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334:690–693. doi: 10.1126/science.1212673. •• In this study it is found that modest exercise extends life span in a SCA1 mouse. This effect is linked to increased levels of EGF and a decrease in Cic.
- 17.Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY, Clark HB, Orr HT. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 18.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai S, O’Callaghan B, Zoghbi HY, Orr HT. 14-3-3 binding to ataxin-1(ATXN1) regulates its dephosphorylation at S776 and transport to the nucleus. J Biol Chem. 2011;286:34606–34616. doi: 10.1074/jbc.M111.238527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J, Giesler GJ, Zoghbi HY, Orr HT. SCA1-like disease in mice expressing wild type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. • A follow up to the study by Lim at al [14••] showing that the same amino acid replacement in ATXN1, Ser 776 to Asp 776, that promotes its binding with RBM17 also enhances pathogenesis in mice.
- 21.Jafar-Nejad P, Ward CS, Richman R, Orr HT, Zoghbi HY. Regional rescue of spinocerebellar ataxia type 1 phenotypes by 14-3-3{epsilon} haploinsufficiency in mice underscores complex pathogenicity in neurodegeneration. Proc Natl Acad Sci U S A. 2001;108:2142–2147. doi: 10.1073/pnas.1018748108. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.HD Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 23.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;9:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 24.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 25.Li X-J, Li S-H, Sharp AH, Nucifora FC, Schilling G, Lanahan A, Worley P, Snyder SH, Ross CA. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 26.Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur ELF, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 27. Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, Smith K, Cordelières FP, Spassky N, Ferrante RJ, Dragatsis I, Saudou F. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121:4372–4382. doi: 10.1172/JCI57552. •• This is a comprehensive study on the role of Htt in ciliogenesis and ciliary function. An intriguing aspect of the findings is that loss of Htt resulted in hypomorphic ciliary phenotypes while Htt with an expanded polyQ yielded hypermorphic phenotypes.
- 28.Ibanez-Tallion I, Gorokhova S, Heintz N. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum Mol Genet. 2002;11:715–721. doi: 10.1093/hmg/11.6.715. [DOI] [PubMed] [Google Scholar]
- 29.Millen KJ, Gleeson JG. Cerebellar development and disease. Curr Opin Neurobiol. 2008;18:12–19. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nopoulos PC, Aylward EC, Ross CA, Mills JA, Langbehn DR, Johnson HJ, Magnotta VA, Pierson RK, Beglinger LJ, Nance MA, Berker RA, Paulsen JS, PRED Investigators and Coordinators of the Huntington Study Group Smaller intracranial volume in prodromal Huntington disease evidence for abnormal neurodevelopment. Brain. 2011;134:137–142. doi: 10.1093/brain/awq280. • A titillating study in which magnetic resonance imaging of a large cohort of HD at risk individuals suggesting that HD mutation carriers show signs that mutant Htt contributes to abnormal brain development.
- 31.Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, Lysholm A, Burright E, Zoghbi HY, Clark HB, Andresen JM, Orr HT. RORa-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 32.Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 33.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. (2003) [DOI] [PubMed] [Google Scholar]
- 34. Kratter IH, Finkbeiner S. PolyQ diseases too many Qs, too much function. Neuron. 2010;67:897–899. doi: 10.1016/j.neuron.2010.09.012. • In a Preview written in conjunction with refs. 8 and 19, these authors present a simple but elegant model for how polyQ expansion influences host protein structure and function.
- 35. Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. • The first and a tour de force protein crystallographic study showing that the exon 1 encoded Htt fragment containing a wild type polyQ adopts multiple structures indicating considerable conformational flexibility.