Abstract
Mapping medical test names into a standardized vocabulary is a prerequisite to sharing test-related data between healthcare entities. One major barrier in this process is the inability to describe tests in sufficient detail to assign the appropriate name in Logical Observation Identifiers, Names, and Codes (LOINC®). Approaches to address mapping of test names with incomplete information have not been well described. We developed a process of "enhancing" local test names by incorporating information required for LOINC mapping into the test names themselves. When using the Regenstrief LOINC Mapping Assistant (RELMA) we found that 73/198 (37%) of "enhanced" test names were successfully mapped to LOINC, compared to 41/191 (21%) of original names (p=0.001). Our approach led to a significantly higher proportion of test names with successful mapping to LOINC, but further efforts are required to achieve more satisfactory results.
Keywords: Standardized terminology mapping, LOINC
1. INTRODUCTION
Representing clinical data consistently across medical centers requires mapping institution-specific information to a standardized terminology. Without this mapping, clinical data cannot be shared, integrated or used in a meaningful way[1–4]. At the University of California, San Diego (UCSD), we are undertaking this mapping process because our medical center is involved in the creation of a community-wide health information exchange that requires sharing clinical data across health care institutions [5–7].
Laboratory and other diagnostic tests are relatively well-defined and smaller in size compared to clinical findings or general intervention and procedure concepts. Therefore, along with medical diagnoses and medications, laboratory and other diagnostic test names often become early targets for standardized terminology encoding. One widely used standard for naming of diagnostic tests is Logical Observation Identifiers Names and Codes (LOINC®)[8]. However, many institutions have their own historical test naming systems. Mapping these test names to LOINC is often challenging [9–15]. LOINC has very specific test names that often incorporate several components of the test, such as component/analyte, specimen type, kind of property, time aspect, type of scale, and type of test method. Local test names may not describe tests to this level of detail, leading to ambiguities in name mapping. A recent study where the authors reviewed the mappings from three different institutions found erroneous mappings in less than 5% of them [15]. However, like any other terminology mapping project, each institution is responsible for establishing the specific processes of conducting LOINC mapping including disambiguating local test names. Few prior works in this domain have described the process of mapping local test names to LOINC in detail.
The overall purpose of this paper is to describe our experience with mapping local test names to LOINC codes, particularly the approach that we took to disambiguate local test names. We hypothesized that addressing ambiguous test names up front by clarifying local name variations and incorporating necessary information on the tests would improve the mapping process, and we report the results of a small-scale comparative evaluation that we conducted to study the effectiveness of this approach.
2. BACKGROUND
2.1. Existing tools for translating local test names to LOINC
LOINC is the publicly available standardized terminology system for diagnostic/laboratory tests and other types of clinical observations maintained by Regenstrief Institute, Inc[8]. Currently LOINC (version 2.34) contains more than 61,200 test and observation concepts. LOINC concept names are defined at a very specific level with six core name parts:(1) component/analyte; (2) property that indicates various kinds of quantities such as mass, substance, number; (3) time aspect of measurement such as random point in time or specific interval; (4) specimen/system; (5) type of scale; and (6) method of performing the test. For example, “Hepatitis C virus RNA (viral load measured as number per volume) in Bone marrow by Probe & target amplification method” is defined in LOINC with the concept code 49370-0 and concept name “Hepatitis C virus RNA:NCnc:Pt:Bonemar:Qn:Probe.amp.tar.” Without knowing these aspects of each test, it is extremely challenging to accurately map a local test name to LOINC.
Regenstrief LOINC Mapping Assistant (RELMA) is a Windows-based tool that provides semi-automated mapping of local test and observation names to LOINC by retrieving potential matches from the LOINC database[16]. RELMA provides an interface in which users can import local terms in delimited or Health Level 7 (HL7) file formats to facilitate the mapping. It also provides lexical cleaning services during the local term import such as correcting spelling errors and replacing abbreviated names with full names. Both LOINC and RELMA are available at no cost from the LOINC website [8].
2.2. Mapping local test and observation names to LOINC
There are a number of articles describing various aspects of mapping local codes to LOINC from content coverage of LOINC [11], to automation of the mapping process [9, 10, 12, 17–19]. The prior works on the automation focused on either cleaning local name variants [9] or facilitating the mapping between LOINC names and local names [10, 12, 17–19]. These works employed dictionary-based approaches to name identification and cleaning. Among the latter studies, Intelligent Mapper (renamed as Lab Auto Mapper in RELMA v5.0), by Vreeman and colleagues, has been integrated into RELMA and is available for public use [17, 18]. Several of these studies concluded that incomplete information about tests was the main reason for the failure of the automated mapping, along with local name variations [9, 12, 17]. However, a detailed description of the corrective process needed to make sufficient information about tests available for the mapping has not been reported.
2.3. Laboratory or diagnostic test names at UCSD Medical Center
At the UCSD Medical Center, two sets of laboratory or diagnostic names exist in the electronic medical record system. Physicians order tests from a “Procedure Name” list containing approximately 20,000 entries that physicians use to order a procedure or test. This list includes about 1,600 laboratory or diagnostictest names including ultrasonography and electrocardiography. In addition, this list also contains panel and battery names without specifying the component test names. For example, Liver Function Tests (LFT) is listed as a single test name without further specifying its component test names.
The second set of test names is the “Component Name” list, which shows the basic information about specific tests performed on a single specimen. “Component Names” are used to report test results. For example, component test names of the LFT such as aspartate aminotransferase (AST), alanine aminotransferase (ALT), and lactate dehydrogenase (LDH) are listed in this “Component Name” list. There are 2,064 names in this “Component Name” list.
Neither list provides sufficient information to automatically identify matching test names accurately in LOINC. The “Component Names” list contains more specific test names, but these names are often not associated with critical information such as specimen type and test method. On the other hand, the “Procedure Names” list contains specimen and method information to some extent but often lacks specific component test names of panels or batteries. In addition, the names in the both lists are usually expressed with non-standardized terms including locally created abbreviations.
2.4. Name enhancement approach to LOINC mapping
To assess the feasibility of using the raw test names for LOINC mapping, we first created two sets of 100 test names, one randomly selected from the “Procedure Name” list and the other from the “Component Name” list. We tried to map these lists to LOINC using RELMA without any preprocessing except the brief lexical cleaning services provided by RELMA during the term file importing process. We found this mapping challenging due to the local name variants unrecognized by RELMA and incomplete information about the tests, consistent with the experience reported in the previous studies [9, 12, 17]. We ran another test by adding more information readily available from the test result table such as units, maximum/minimum values, and example result values. However, we found that adding the extra information didn’t improve the mapping results significantly. Specific information on the tests required for conducting LOINC mapping is scattered across different sources making it extremely inconvenient to refer them at once.
Therefore, we performed local test name enhancement by incorporating more information about the test to the name, correcting typos, and replacing local abbreviations with full names. The name enhancement would make test name a bit wordy and unconventional, causing RELMA’s performance on finding potential matches less precise. However, we considered that enhanced test names were still useful as they can provide human coders hints for fine tuning test names and selecting the right matches within the RELMA environment without browsing several different references.
The name enhancement was done manually because (1) the local test name set was relatively small; (2) analyzing the names to come up with rules for automation is time-consuming and automation would need to be validated through human review anyway; (3) we only needed to perform this operation once and (4) idiosyncrasies in local naming convention would make any automated processes hard to generalize to other institutions.
The objective of this paper is to describe the process we used to match our local test names to LOINC codes. We believe the steps in this process will be useful to others facing similar challenges in standardizing local diagnostic or laboratory test names.
3. METHODS AND PROCEDURES
In order to map our institution’s test names to LOINC, we first enhanced the local test names by clarifying naming variations and incorporating detailed information about the test obtained from actual patient records and online reference materials. We first created gold standard mappings to LOINC codes, then we evaluated whether the enhanced test names improved our ability to apply RELMA to map to LOINC codes. The details are described below.
3.1. Identifying active test names for mapping
It is a common view that not every test needs to be mapped to LOINC [11, 13, 14, 20]. Active tests are more likely to be needed for data exchange and integration than rarely used tests thus the former should be prioritized for mapping. To identify these higher-priority tests, we determined the frequencies of the “Component Names” for the test results produced during the past five years. Keeping tests that had been reported more than 1000 times yielded 657 test names, which included laboratory tests (chemistry, hematology, microbiology, etc.), cardiac ultrasonography, and EKG. These 657 test names covered 99.78% of the entire test results reportedfor the same period.
3.2. Creating enhanced test names
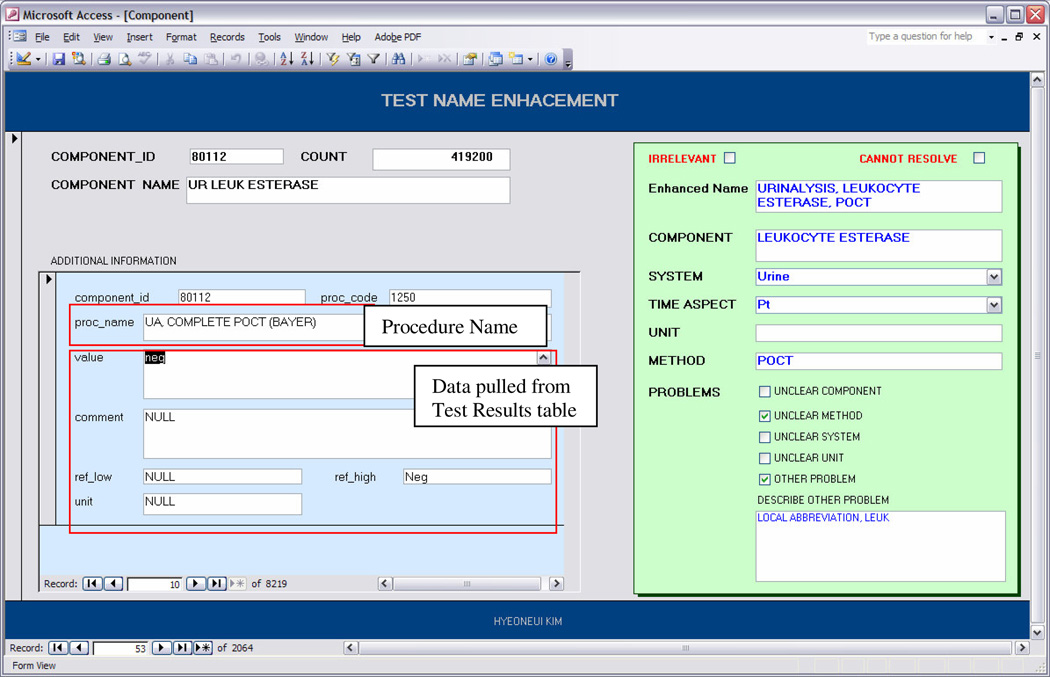
In order to clarify the “Component Names” and to provide sufficient information about the test for mapping, we created enhanced test names that contained the missing name part information. To identify the missing information, wereferred to several different resources, such as (1) the specific test result values and associated comments, (2) reference normal ranges, (3) unit of measure, (4) associated “Procedure Names”, and (5) a website describing some of the procedures performed at our institution. Table 1 shows different types of information required for accurately mapping test names to LOINC and the different information sources accessed to obtain the information. To facilitate reviewing all of the data sources in a single view, we created a graphical user interface tool using Microsoft Access. One of the authors with both a clinical and informatics background (HK) added the additional information found from the difference sources for each test using this tool.
Table 1.
Necessary Information for Test Name Enhancement and Information Source
| Information type | Full Test Name |
Component /Analyte |
Property | Time Aspects |
System | Unit of Measure |
Method |
|---|---|---|---|---|---|---|---|
| Information source | |||||||
| Component Names | X | X | |||||
| Procedure Names | X | X | X | X | |||
| Test Results Values (EMR) | X | X | |||||
| Test Results Comments (EMR) | X | X | X | ||||
| Reference Ranges (EMR) | X | X | X | ||||
| Online reference material | X | X | X | X | X | X | X |
Figure 1 shows a screenshot of the tool’s interface. In the right box, the reviewer specified the information required to identify the correct LOINC match such as various name parts. These name parts are set as optional fields of the local term files that are imported into RELMA. She then created an enhanced name by combining the necessary information. In addition, she recorded several aspects of the name enhancement process, such as type of information unavailable in the original test name (i.e., Component Name) and whether the test name was unsuitable for LOINC mapping. During the name enhancement process, 62 test names were deemed unsuitable for LOINC mapping as they were either referring to properties of a test such as system (e.g., “urine specimen”, “specimen source flu”) and method (e.g., “auto diff”, “gram stain”) information, or workflow related information (e.g., “sent out”, “draw and hold”, “ITL transport”)
Figure 1.
Screenshot of the GUI based Tool for Test Name Enhancement and Test Information Review
3.3. Gold standard LOINC mapping
After excluding the 62 unsuitable names from the 657 most frequently appeared test names, the remaining 595 enhanced test names and their units were imported into RELMA v5.0 as a delimited text file. Additional lexical cleaning was performed as suggested by RELMA. To closely observe the impact of name enhancement, the mapping was done through a term-by-term search and review process without using Lab Auto Mapper, an advanced mapping support that RELMA provides. We kept the search limit in RELMA as follows: (1) selecting terms consistent with local unit, (2) exclude MS terms, (3) include trial LOINC terms, (4) no specific preference on property type, (5) no preference on concept class (i.e., order vs. observation), (6) no restriction on the maximum number of component words, (7) no restriction on lab test types, and (8) no restriction on the availability of methods.
Two members of our team with clinical and informatics experience created the gold standard mappings by independently assigning the enhanced test names to LOINC terms and coming to consensus on disagreements. During this process, these expert reviewers were allowed to refer back to the additional information sources such as the online procedure information pages and the name enhancement tool when it was necessary. The expert reviewers were unable to determine the correct matches for 24 test names because necessary information on those tests was missing. Therefore, gold standard mapping was generated for 571 test names.
3.4. Testing the effectiveness of conducting LOINC mapping with enhanced test names
In parallel with the gold standard creation, we evaluated the effectiveness of using enhanced test names in LOINC mapping. We first randomly divided the 595 “Component Names” into two sets. The original component names were retained in the first set and replaced with the enhanced names in the second set. We then combined the two sets into one and randomly selected a third of the test names (N=195) for training. One of the authors, who has clinical background and did not participate in gold standard generation, was trained on how to perform LOINC mappings with RELMA using these195 test names. A random subset of 20 of these test names was independently mapped by a one of the expert reviewers to verify adequate reviewer performance.
Our trained reviewer then mapped the remaining test names (N=400) using RELMA. The same RELMA settings used for gold standard generation were applied to this mapping. This set included 204 tests with enhanced names and 196 with the original “Component Names.” The reviewer was blinded to the types of the test names and was not allowed to refer to other resources. However, she was allowed to use the search term manipulation function (i.e., "unselect as a search word" or "add a search word") that RELMA provides. The independent reviewer recorded the total number of potential matches returned by RELMA along with whether she found (1) an exact match to a LOINC code, (2) more than one potential match, (3) no matches, or (4) could not decide whether there was a match. She also recorded the LOINC code when she thought that an exact match was found.
3.5. Analysis
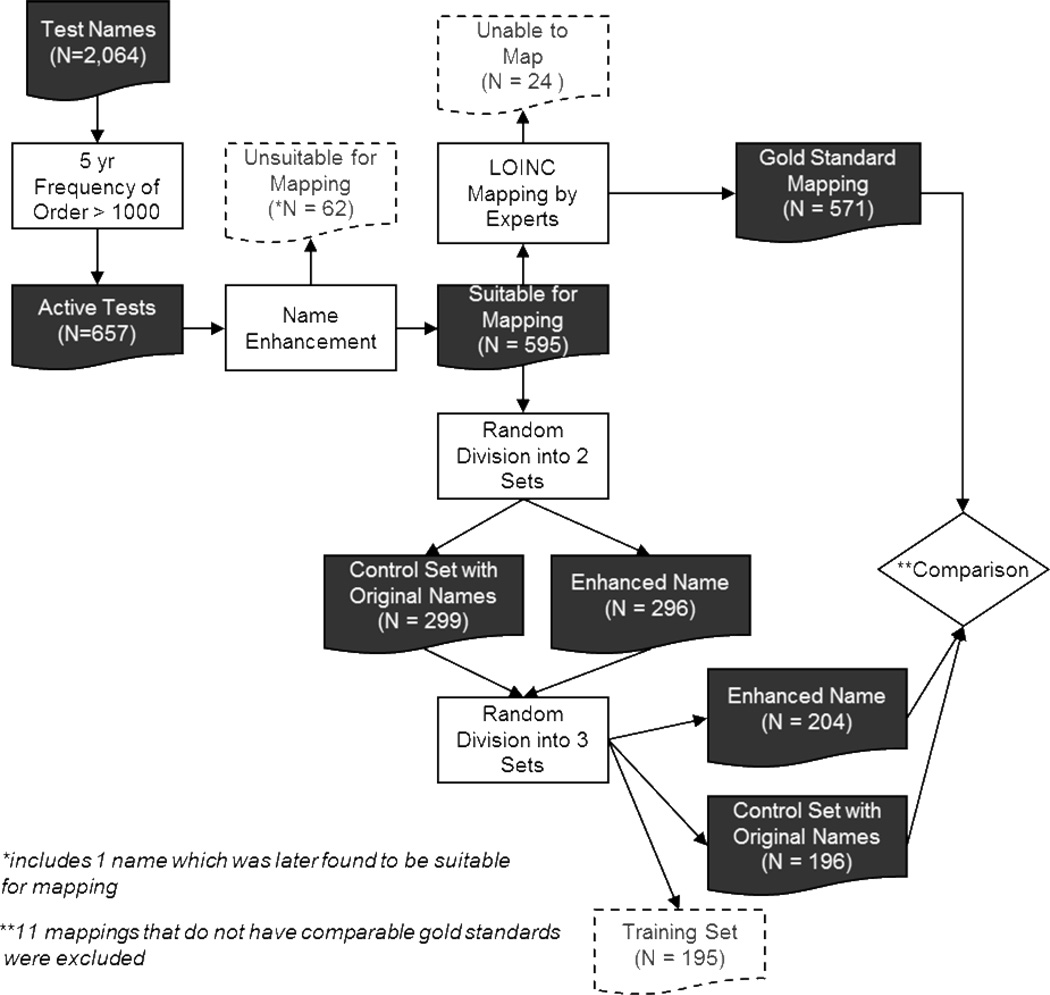
We performed a Wilcoxon Test to compare the number of potential matches returned by RELMA for the cases in which the original “Component Names” were used as search terms against the cases in which the enhanced names were used. We also compared the accuracy of the mapping between the two cases using a Fisher’s Exact Test to compare the proportion of test names mapped to the gold standard LOINC codes. We evaluated the independent mapping results for 389 test names (191 with original “Component Names”, 198 with enhanced names) after excluding 11 test names which fell into the group of 24 test names for which the expert reviewers could not generate any gold standard. All analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC). The overall study process is illustrated in Figure 2.
Figure 2.
Overall Study Process
4. RESULTS
4.1. Name enhancement and gold standard generation
During the name enhancement process, a fully specified name was added for every abbreviation and many typographical errors were corrected. Of these, 260 names required more substantial action during the enhancement such as specifying sample/specimen type, time aspect, method, and component names. Thirty-five of the names required clarification of “Component Names” as they contained only System information or partial Component information. A few examples are “right eye,” “uncorrected,” and “% recovery”. Enhanced names “vision screen, right eye,” “vision screen, without lenses,” and “Thyroglobulin % recovery” were created for these cases. Ninety-five names were augmented with Method concepts and one hundred twenty-three names were added with System concepts. Time Aspect information was added to seven names.
The expert reviewers were able to generate a gold standard LOINC mapping for 571 out of the initially targeted 657 test names. The expert reviewers were unable to map 24 test names because even the enhanced names did not provide sufficient information about the test. The enhanced name creation was an attempt to incorporate as much as possible the necessary information about a test. Completeness of the enhanced name depended on the availability of the necessary information and not every enhanced test name carries complete test information. For example, “strep test, control” was enhanced to “streptococcus screening test, control.” However, this enhanced name did not provide specific method of conducting the screening test or the system (i.e., specimen source) information.
Table 2 shows the results of the mapping that the independent reviewer conducted. With the original “Component Names,” more than half of cases (110/191=58%) were marked as “Cannot Decide on Mapping,” compared with 87/198 (44%) in the enhanced name set. On the other hand, with the enhanced test names, more than half of the cases (113/198=57%) was determined on mapping by the reviewer, either as “match found” or “match doesn’t exist,” compared with 81/191 (42%) in the original “Component Names” set. Overall, 41/191 (21%) of the original “Component Names” were correctly mapped to LOINC, whereas 73/198 (37%) of the enhanced test names (N=198) were correctly mapped (p=0.001).
Table 2.
Mapping Results in Two Different Test Name Sets (numbers represent term counts)
| Component Names | Enhanced Test Names | Row Total | |
|---|---|---|---|
| Cannot Decide on Mapping | 110 | 87 | 205 |
| Cannot perform the mapping | 94 | 76 | 178 |
| More than one possible match | 16 | 11 | 26 |
| Performed Mapping | 81 | 113 | 194 |
| No Matches | 8 | 13 | 22 |
| Mapped to LOINC | 73 | 98 | 172 |
| Accurate Matches | 41 | 73 | 114 |
| Column Total | 191 | 198 | 389 |
1.1. Effectiveness of conducting LOINC mapping with enhanced test names
The average number of potential matches returned by RELMA per test was 144 for the “Component Names”, 27 for the enhanced test names (p < 0.001). RELMA didn’t return any potential matches for two “Component Names” and five enhanced test names. The average number of returns per test was 19 for the accurately mapped test names, 22 for the inaccurately mapped names, and 130 for the unmapped names.
For each category of mapping results, an example case and explanation of failure is given in Table 3.
Table 3.
Example Mapping Cases (mapping was done using the bolded terms.)
| Mapping Outcome | Original Test Name |
Enhanced Test Name (with unit of measure information) |
Reasons for Mapping Failure |
|---|---|---|---|
| Correctly mapped |
SIROLOMUS TP/CR RATIO CALC 24 HR |
Sirolimus level, blood 24 hr urine total protein to creatinine ratio |
NA NA |
| Mapped but incorrect |
PCO2, VEN (T) CHLAMIDIA PCR, GENITAL |
VBG, Partial Pressure of Carbon Dioxide (PCO2), Temperature corrected Chlamydia PCR, General Culture, Genital Swab |
(T) was not correctly interpreted. A general component “Chlamydia sp DNA” was selected instead of the correct component “Chlamydia trachomatis DNA.” |
| More than 2 potential matches |
SPEC GRAV, UR AO MEAN GRADIENT |
Specific Gravity – Urine Chemistry Aortic Blood Flow Mean Gradient |
Time Aspect (i.e., Pt vs. 24hr) information was not provided. System information was not provided. |
| Mapper cannot decide whether has a match or not |
PIVKA CARDIAC OUTPUT |
Proteins Induced by Vitamin K Antagonism (PIVKA) 2-D Echocardiogram, Cardiac Output |
The abbreviation was not correctly understood by the mapper. System information was not provided. |
| Incorrectly deemed having no matches |
AVO2 DIFF (VOL%) AORTIC VALVE AREA |
Arterial Mixed Venous Oxygen Difference 2-D Echocardiogram, Aortic Valve Area |
“AVO2” was not modified into a workable search term when RELMA returned 0 matches. A right Component name was not identified. |
5. DISCUSSION
LOINC provided very high level of concept coverage (96%) on the test names used at UCSD Medical Center. However, mapping clinical data to a standardized terminology is are source intensive process. Similar to prior work on LOINC mapping [9, 12, 17], we discovered in this study that clarifying the intended meaning of local test names and making sufficient information about a test available were the two major challenges in mapping local test names to LOINC. In this section we will describe the specific challenges and lessons learned from the name enhancement approach that we used, as well as recommendations for improving efficiency in LOINC mapping.
5.1. Challenges in mapping local test names to LOINC
We encountered various types of challenges during the process of LOINC mapping. The first and largest challenge involved the ambiguous and incomplete local test names. Many typographic errors and locally-developed abbreviations in the local test names made conducting LOINC mapping challenging. In addition, there was no workable single source of test names at our institution, as two sets of test name lists are in use serving slightly different purposes. Neither of these test name lists was sufficiently complete for conducting the mapping. Although combining the two lists would make the mapping process easier, there may be operational factors that require two separate lists.
The lexical cleaning that RELMA provided during the local term import was useful for correcting typographic errors. RELMA also indicated which of the possible matches were commonly used and this often served as an important hint for identifying the most appropriate match given that a commonly used test was very likely the one that we were looking for. Indeed, we found that the right matches we found often fell in the common tests pool. However, considering that not every right match was selected from the common test pool, the common test indication alone is not a reliable source for finding the right match.
The second challenge lies in the fundamental differences in the ways LOINC and UCSD Medical Center create test names. Tests and observations are very precisely defined in LOINC. Fully specified test names are completed in LOINC by pre-coordinating the six name parts. However, some of the fully specified LOINC names are translated into more than one local test name in our institution. For example, “strep culture,” “specimen type” and “gram stain” are defined as separate individual test names in our institution. A fully specified microbiology test name in LOINC requires all the information presented by the three local test names. This difference in representing test names makes it challenging to conduct complete LOINC mapping of the test names in our institution. Precise LOINC mapping becomes possible by post-coordinating the different test names after the necessary information about each test is available. In this case, LOINC mapping needs to be done using test result instances rather than just the test names, as the result instances also contain test properties such as methods and specimen types.
The immediate need of LOINC mapping is to encode test result data stored in the clinical data warehouse in our institution. An approach to this challenge that we are considering involves several steps of: (1) identifying different specimens and methods with which a test can be associated, (2) identifying a LOINC code for each case and establishing mapping between specific specimen and method instances, and (3) assigning the LOINC code to the test result instance during the ETL (Extract, Transfer, and Load) process of the test result data from EMR to the clinical data warehouse. However, as a longer term solution, our institution may need to consider adopting single test names defined at the same level and granularity as LOINC when reporting results.
5.2. Name enhancement approach
As evidenced in our study results, enhancing local test names with information gleaned from the EMR and reference materials facilitated manual LOINC mapping. The major contribution of the enhanced test names was two-fold: (1) providing additional information that human needed to disambiguate among numerous possible LOINC matches and (2) adding extra words to the local term that could be recognized by RELMA and used to automatically retrieve possible matches. Real test result values and related comments from the EMR, along with reference ranges provided useful tips for identifying the correct test. Result values and reference ranges provide specimen and property information (e.g., electrolytes measured with blood vs. urine, numeric values vs. positive/negative values). Comments often provide information on specimen types and methods (e.g., “at bedside” implies point-of-care test).
The number of potential matches returned by RELMA was significantly reduced when enhanced search terms were used. In general, a smaller set of potential matches is considered desirable as it is easier to review and likely to contain fewer false positives. However, we also need to account for the fact that using more search terms can cause false negative such as omission of the correct matches. This is likely to occur especially when the terms used for search are not accurately recognized by RELMA. Therefore, it becomes even more critical that the person who conducts LOINC mapping has sufficient domain knowledge when using enhanced test name. In our experiment, the number of returns increased to more than 11,000 records when the independent reviewer failed to provide sensitive search terms. Ideally, that person should possess sufficient knowledge of tests for effectively manipulating search terms based on the hints provided by the enhanced names to achieve optimal search results. The underlying assumption of this study was that the use of enhanced test names makes the mapping task feasible for non-experts. Our results indicate LOINC mapping is a highly specialized process that requires domain expertise.
The LOINC mapping presented in this study involves two steps of creating enhanced test name and conducting mapping using the enhanced test name. This two-step process turned out to be quite inefficient, as information loss occurs during the transition as implied by the fact that the expert reviewers needed to refer back to the original reference sources when generating gold standard mapping. We suspect that it would be more effective if the same person enhancing the test names also generate the final mapping, because the enhanced test names may not always convey clear meaning to the independent reviewer who conducts mapping solely based on the names.
Nonetheless, the expert reviewers were able to map 96% of the test names by using both the enhanced test names and referring back to the information sources when necessary. This result shows that LOINC provides good concept coverage of the test names eligible for mapping in our institution. Although it is hard to make a direct comparison between the mapping experiences at different institutions, our result seems to be an improvement considering that one study reported that up to 19% of the labs of its institution were not mapped to LOINC due to the ambiguity and/or incompleteness of the local lab names[10].
RELMA provides options to import local test names in the HL7 file format or in the delimited file format. During this importing process actual test results and other information such as units and value ranges can be included. Adding the unit of measure field facilitated the mapping process by reducing the number of false positive matches (i.e., test names with different units of values) returned by RELMA. Also, RELMA automatically identified the correct values for Property and Scale axes based on the unit information. However, the many ambiguities posed in the test name itself, as well as missing other critical information such as methods and system, necessitated the extra step of name enhancement described in this study.
6. CONCLUSION
To address two major challenges in LOINC mapping – idiosyncrasies in local naming conventions and insufficient information about the test – we developed the process of enhancing local test names before conducting the mapping. We used information from real tests recorded in the EMR, along with reference materials to enhance the names. Use of enhanced test names allowed a human to successfully map local names to LOINC codes twice as often as when using the original names. This approach should be generalizable to other institutions.
Although a significantly higher rate of accurate LOINC mapping was achieved with the enhanced test names than with the original local test names, the low success rate (37%) implies that enhanced test names alone do not sufficiently address the identified challenges. As a potential solution to address the limitations with the enhanced name, we consider creating a single test name list that combines “Procedure Name” and “Component Name” lists with additional information such as specimen and methods, where the “Procedure Name” list and the “Component Name” list are hierarchically related. Test orders can be made using the procedure names while the test result reporting can be done using the component names. Also, we will need to form a LOINC mapping task force consisting of a physician, a LOINC expert, and people specialized in laboratory and diagnostic tests to complete the mapping of our institutional test names to LOINC.
In addition, we consider integrating the term mapping interface (i.e., RELMA) with an environment that assembles important test information from various sources and presents it back to the reviewers performing the mapping. The feasibility of this approach will be tested in a future study.
Highlights.
Use of the enhanced test names improves the accuracy of LOINC mapping
Use of the enhanced test names reduces the number of false positive matches in RELMA.
Local test names defined at varying granularity levels cause a mapping challenge.
Acknowledgments
This study was supported in part by the research grants 1U54HL108460-01 (NHLBI/NIH) and R01HS19913-01 (AHRQ). We thank Roberto Rocha MD, PhD, Aziz Boxwala MD, PhD and Lucila Ohno-Machado MD, PhD for providing invaluable input on this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chute CG, Cohn SP, Campbell JR. A framework for comprehensive health terminology systems in the United States: development guidelines, criteria for selection, and public policy implications. ANSI Healthcare Informatics Standards Board Vocabulary Working Group and the Computer-Based Patient Records Institute Working Group on Codes and Structures. J Am Med Inform Assoc. 1998;5(6):503–510. doi: 10.1136/jamia.1998.0050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmadian L, van Engen-Verheul M, Bakhshi-Raiez F, Peek N, Cornet R, de Keizer NF. The role of standardized data and terminological systems in computerized clinical decision support systems: literature review and survey. International journal of medical informatics. 2011;80(2):81–93. doi: 10.1016/j.ijmedinf.2010.11.006. Epub 2010/12/21. [DOI] [PubMed] [Google Scholar]
- 3.Westra BL, Subramanian A, Hart CM, Matney SA, Wilson PS, Huff SM, et al. Achieving "meaningful use" of electronic health records through the integration of the Nursing Management Minimum Data Set. J Nurs Adm. 2010;40(7–8):336–343. doi: 10.1097/NNA.0b013e3181e93994. Epub 2010/07/28. [DOI] [PubMed] [Google Scholar]
- 4.Hamm RA, Knoop SE, Schwarz P, Block AD, Davis WLt. Harmonizing clinical terminologies: driving interoperability in healthcare. Studies in health technology and informatics. 2007;129(Pt 1):660–663. Epub 2007/10/04. [PubMed] [Google Scholar]
- 5.integrating Data for Analysis, Anonymization and Sharing (iDASH) 2011 Mar 15; doi: 10.1136/amiajnl-2011-000538. Available from: http://idash.ucsd.edu. [DOI] [PMC free article] [PubMed]
- 6.Clinical Data Warehouse for Quality Improvement and Resesarch. 2011 Mar 15; Available from: http://dbmi.sdsc.edu/confluence/display/BMI/CDWR.
- 7.Improving Health Through Health Information Technology. [Accessed 03/16/2011]; http://healthit.hhs.gov/portal/server.pt?open=512&objID=1805&parentname=CommunityPage&parentid=2&mode=2&cached=true.
- 8.Logical Observation Identifiers Names and Codes (LOINC®) 2011 Feb 15; Available from: http://www.loinc.org.
- 9.Lau LM, Johnson K, Monson K, Lam SH, Huff SM. A method for the automated mapping of laboratory results to LOINC. Proceedings / AMIA Annual Symposium. 2000:472–476. Epub 2000/11/18. [PMC free article] [PubMed] [Google Scholar]
- 10.Khan AN, Russell D, Moore C, Rosario AC, Jr, Griffith SP, Bertolli J. The map to LOINC project. AMIA Annual Symposium proceedings / AMIA Symposium. 2003:890. Epub 2004/01/20. [PMC free article] [PubMed] [Google Scholar]
- 11.Lau LM, Banning PD, Monson K, Knight E, Wilson PS, Shakib SC. Mapping Department of Defense laboratory results to Logical Observation Identifiers Names and Codes (LOINC) AMIA Annual Symposium proceedings / AMIA Symposium. 2005:430–434. Epub 2006/06/17. [PMC free article] [PubMed] [Google Scholar]
- 12.Khan AN, Griffith SP, Moore C, Russell D, Rosario AC, Jr, Bertolli J. Standardizing laboratory data by mapping to LOINC. J Am Med Inform Assoc. 2006;13(3):353–355. doi: 10.1197/jamia.M1935. Epub 2006/02/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter JP, Starmer J, King J, Frisse ME. Mapping laboratory test codes to LOINC for a regional health information exchange. AMIA Annual Symposium proceedings / AMIA Symposium. 2007:1081. Epub 2008/08/13. [PubMed] [Google Scholar]
- 14.Vreeman DJ, Finnell JT, Overhage JM. A rationale for parsimonious laboratory term mapping by frequency. AMIA Annual Symposium proceedings / AMIA Symposium. 2007:771–775. Epub 2008/08/13. [PMC free article] [PubMed] [Google Scholar]
- 15.Lin MC, Vreeman DJ, McDonald CJ, Huff SM. Correctness of Voluntary LOINC Mapping for Laboratory Tests in Three Large Institutions. AMIA Annual Symposium proceedings / AMIA Symposium. 2010;2010:447–451. Epub 2011/02/25. [PMC free article] [PubMed] [Google Scholar]
- 16.RELMA. 2011 Mar 05; Available from: http://loinc.org/relma.
- 17.Vreeman DJ, McDonald CJ. Automated mapping of local radiology terms to LOINC. AMIA Annual Symposium proceedings / AMIA Symposium. 2005:769–773. Epub 2006/06/17. [PMC free article] [PubMed] [Google Scholar]
- 18.Vreeman DJ, McDonald CJ. A comparison of Intelligent Mapper and document similarity scores for mapping local radiology terms to LOINC. AMIA Annual Symposium proceedings / AMIA Symposium. 2006:809–813. Epub 2007/01/24. [PMC free article] [PubMed] [Google Scholar]
- 19.Fiszman M, Shin D, Sneiderman CA, Jin H, Rindflesch TC. A Knowledge Intensive Approach to Mapping Clinical Narrative to LOINC. AMIA Annual Symposium proceedings / AMIA Symposium. 2010;2010:227–231. Epub 2011/02/25. [PMC free article] [PubMed] [Google Scholar]
- 20.Lin MC, Vreeman DJ, McDonald CJ, Huff SM. A Characterization of Local LOINC Mapping for Laboratory Tests in Three Large Institutions. Methods Inf Med. 2010;49(5) doi: 10.3414/ME09-01-0072. Epub 2010/08/21. [DOI] [PMC free article] [PubMed] [Google Scholar]