Abstract
MicroRNAs (miRNAs) are a group of small RNAs involved in translation inhibition or mRNA degradation. Due to its large size, Manduca sexta has long been used as a model to study insect physiology and biochemistry. While transcriptome studies have greatly enriched our knowledge on M. sexta structural genes, little is known about posttranscriptional regulation by miRNAs in this lepidopteran species. We constructed four small RNA libraries from embryos, 4th instar feeding larvae, pupae, and adults, obtained 21 million reads of 18-31 nucleotides by Illumina sequencing, and found 163 conserved and 13 novel miRNAs. By searching the M. sexta genome assembly, we identified precursors of 82 conserved miRNAs, 76 of which had mapped reads in one or more of these libraries. After normalization, we compared numbers of miRNA and miRNA-star reads in these libraries and observed abundance changes during development. Interestingly, mse-miR-281-star, mse-miR-31-star, mse-miR-965-star, mse-miR-9a-star, msemiR-9b-star, mse-miR-2a-star, mse-miR-92b-star and mse-miR-279c-star are either more abundant or maintained at similar levels compared to respective mature miRNA strand. Expression profiling of the first set of miRNAs provided insights to their possible involvement in developmental regulation. This study will aid in the annotation of miRNA genes in the genome.
Keywords: metamorphosis, posttranscriptional regulation, Illumina sequencing, small RNA, lepidopteran insect
1. Introduction
Over the last decade, a fundamental role for small RNA-guided post-transcriptional regulation of gene expression has been uncovered. In particular, microRNAs (miRNAs) have attracted attention due to their increasingly appreciated importance in development and disease (Bartel, 2004). MiRNAs are ~22 nt genome-encoded non-coding RNAs processed from hairpin precursors initially by Drosha within the nucleus and then by Dicer in the cytosol (Ambros, 2004; Bartel, 2004). MiRNAs are capable of silencing target gene expression primarily via translational suppression of the target mRNA by guiding the RNA-induced silencing complex (RISC) to the respective target genes. Recognition usually involves the 2-7 nt seed region of miRNA and a downstream complementary area, imperfectly matching 3’-untranslated region of its target. MiRNA-star strand (strand that is complementary to mature miRNA) is usually degraded, but under certain circumstances is used for expression control of target genes (Jagadeeswaran et al., 2010; Kato et al., 2009). MiRNA regulation has been demonstrated in both animals and plants (Huntzinger and Izaurralde, 2011).
Evidence shows that levels of some miRNAs vary temporally and spatially, which provides fine tuning of target gene expression (Chawla and Sokol, 2011). Due to their short life cycles, high reproduction rates, and easy handling, insects are good subjects for miRNA research. Although initial discoveries were made in Drosophila (Elbashir et al., 2001; Lagos-Quintana et al., 2001), computational genome analysis and sequencing small RNA libraries uncovered a large number of miRNAs from a diverse group of insects. These include Drosophila (Lai et al., 2003; Lau et al., 2009; Lu et al., 2008; Ruby et al., 2007; Sandmann and Cohen, 2007; Stark et al., 2007), mosquitoes (Behura et al., 2011; Brunel et al., 2007; Chatterjee and Chaudhuri, 2006; Li et al., 2009; Mendes et al., 2010; Skalsky et al., 2010; Wang et al., 2005), honeybee (Chen et al., 2010; Weaver et al., 2007), silkworm (Cai et al., 2010; Cao et al., 2008; Huang et al., 2010; Jagadeeswaran et al., 2010; Jin et al., 2008; Liu et al., 2011; Tong et al., 2006; Yu et al., 2009; Yu et al., 2008b; Zhang et al., 2009), locust (Jia et al., 2010; Wei et al., 2009), pea aphid (Legeai et al., 2010), neotropical butterfly (Surridge et al., 2011), Blattella germanica (Cristino et al., 2011), and Tribolium castaneum (Singh and Nagaraju, 2008; Yu et al., 2008a).
The tobacco hornworm, Manduca sexta, serves as a biochemical model due to its sheer body size and hemolymph volume at the final larval stadium. It has made significant contributions to our knowledge on hormonal control, neural development, cuticle formation, and other insect physiological processes (Hiruma and Riddiford, 2010). Extensive studies revealed molecular mechanisms underlying its humoral and cellular immune responses against pathogens (Jiang et al., 2011). Recent transcriptome analyses and genome sequencing greatly expanded information on its genetic background and transcriptional regulation (Grosse-Wilde et al., 2011; Pauchet et al., 2010; Zhang et al., 2011; Zou et al., 2008). Sequencing of small RNA component of the transcriptome should help us to understand posttranscriptional regulation of gene expression by miRNAs as well as evolution of species-specific miRNA genes. As a holometabolous insect, M. sexta has also provided insights into developmental control of cell proliferation, differentiation, and apoptosis as well as tissue remodeling (White et al., 1999). Empowered by its genome sequence, we plan to not only study complete metamorphosis at the level of gene regulation, but also use the insights to develop new means of lepidopteran pest control.
In this work, we prepared whole body total RNA from M. sexta embryos, larvae, pupae and adults, constructed and sequenced four small RNA libraries. We identified and predicted conserved and novel miRNAs based on an initial assembly of the genome sequence (http://www.hgsc.bcm.tmc.edu/collaborations/insects/Manduca/Msex_1.0_assembly/). Once located in the genome, fold-back structures of the miRNA precursors were predicted. Clustered miRNAs were sorted out according to their genomic loci. In order to discover development-related miRNAs, we normalized the read numbers and compared abundances of mature miRNAs from the libraries. We also discussed the roles of our first dataset in terms of M. sexta miRNA gene annotation.
2. Materials and methods
2.1. Insect rearing, total RNA extraction, and small RNA library construction
M. sexta eggs were obtained from Carolina Biological Supply (Burlington, NC) and larvae were reared on an artificial diet in a 25°C incubator with a long LD 18:6 photoperiod. Late pupae were placed in a cage with light control and tobacco plants (Nicotiana benthamiana). Newly-laid eggs (0-12 h) were collected for use as embryo sample; three larvae each from days 0, 1, 2, and 3 of 4th instar, three pupae each from days 0, 6, 12, and 18, and three adults each of newly emerged males and females were used as larva, pupa, and adult samples, respectively. Samples were immediately frozen and ground in liquid nitrogen and total RNAs were extracted using TRIZOL Reagent (Life Technologies, Inc.). Separated by 15% denaturing polyacrylamide gel, RNAs in the 15-30 nt range were purified and ligated with the 3’ and 5’ adapters (Jagadeeswaran et al., 2010). They were then reverse transcribed using primer 5’-CAAGCAGAAGACGGCATACGA-3 ’ and proliferated by forward and reverse primers (5’-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA-3 ’ and 5 ’-CAAGCAGAAGACGGCATACGA-3’). PCR products were purified by phenol/chloroform extraction and ethanol precipitation, and shipped to National Center for Genome Resources (Santa Fe, NM) for sequencing.
2.2. Sequence analysis and identification of conserved miRNAs of M. sexta
The raw sequence data were analyzed as previously described (Reddy et al., 2009). First, reads with no matches to the proximal 11 nt of the 5’-adaptor were removed. Remaining reads were compared to RepBase (v14, http://www.girinst.org) database and non-coding RNAs from Rfam database (http://www.sanger.ac.uk/Software/Rfam/ftp.shtml) by BLASTN. Mitochondrial RNAs were identified by comparing with M. sexta mitochondrion sequence (gi|165932395|ref| NC_010266.1|, obtained from NCBI Nucleotide database). Reads mapped to the RepBase, Rfam, and mitochondrial databases were removed before further analysis. If reads mapped to silkworm mRNA sequences at SilkDB (http://silkworm.genomics.org.cn/silkdb/), they were considered to be degradation products and, therefore, eliminated. The remaining sequences were aligned to known miRNAs in database miRBase (v16, http://www.miRBase.org/) to obtain the frequencies of conserved miRNAs. Meanwhile, mature miRNA sequences in miRBase were mapped to M. sexta genome. Then, the flanking sequences of matched loci were retrieved to predict their secondary structures using RNAfold (Hofacker, 2003). If they have at least 18 base pairs, only one central loop and low folding energy (≤-18 kCal/mol), the genomic loci were designated to the corresponding miRNAs.
2.3. Identification of novel miRNAs of M. sexta
After mapping of conserved miRNAs, the remaining reads were traced back to the initial assembly of M. sexta genome (http://www.hgsc.bcm.tmc.edu/collaborations/insects/Manduca/Msex_1.0_assembly/). Reads beyond the size range of 18-24 nt were disregarded since sequences of 25-28 nt may represent piRNAs. Unique small RNAs with more than 10 possible genomic loci were removed from further analysis. The flanking regions of the remaining genome-matched sequences were retrieved, and fold-back structures were predicted using the RNAfold program (Hofacker, 2003). We examined the ones that had at least 18 base pairs, only one central loop, and folding energy lower than -18 kCal/mol. We further applied the MIRCHECK program (Jones-Rhoades and Bartel, 2004) to pick up sequences that have ≤ 6 mismatches, ≤2 bulged or asymmetrically unpaired nucleotides, and ≤2 continuous mismatches in the regions of the small RNA reads. They were considered as putative pre-miRNAs. Then, the small RNA sequence, with the highest frequency in all unique sequences mapped to a putative pre-miRNAs, was selected as the mature miRNA of the pre-miRNA. The abundances of the selected mature miRNAs were evaluated using a binomial test by regarding the selected mature miRNA as preferred reads. Then, a self-written program (Zheng et al., unpublished) was used to check the existence of miRNA* of the corresponding mature miRNA, if there were 2 nt overhang(s) at the 3’ end(s) of either the miRNA or miRNA*, or both. The distributions of unique small RNAs on the putative pre-miRNAs were manually examined. Pre-miRNAs without a clear accumulation of reads in the selected mature miRNAs regions, p > 0.05 (binomial test for the selected mature miRNA), were removed from the putative pre-miRNAs because mature miRNAs were expected to be cut out precisely from the pre-miRNAs based on the annotation criteria of miRNAs (Meyers et al., 2008). Small RNAs, if they were unable to match to miRBase but had an accompanying miRNA-star sequence together with predicted fold-back structure (Zuker, 2003) were designated as novel miRNAs. Those without an accompanying miRNA* sequence but with good pre-miRNA fold-back structures were designated as novel miRNA candidates.
3. Results
3.1. Overview of the dataset
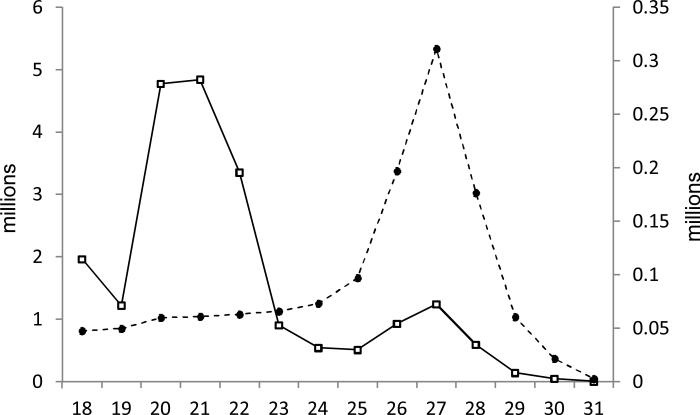
We obtained a dataset of about 21.1 million reads in total: 3,421,611 for embryo, 2,716,576 for larva, 9,284,690 for pupa, and 5,662,414 for adult (Table 1). With all four libraries combined, total read numbers over different lengths show a peak around 20-22 nucleotides (nt) representing typical lengths of miRNAs and a smaller peak at 26-28 nt (Fig. 1). Upon removal of redundant sequences, the curve of unique read numbers exhibits a single peak over 26-28 nt, representing piRNAs. PiRNAs are derived mainly from retrotransposons and other repetitive elements with high sequence diversity (Khurana and Theurkauf, 2010). The peak of unique read distribution over 26-28 nt indicates that the average abundance of piRNA is relatively low, which is common for small RNA libraries (Jagadeeswaran et al., 2010; Lau et al., 2009; Surridge et al., 2011; Wei et al., 2009). For each library, reads mapped to mitochondrial RNAs take up the smallest part, whereas non-coding RNAs (rRNAs, tRNAs, snRNAs, snoRNAs, etc.) are 2.3 times as abundant as the total of mitochondrial RNAs, miRNAs, and mRNAs (Table 1). Reads mapped to non- coding RNAs, mRNAs, and mitochondrial RNAs may be degradation products of their corresponding categories. Due to the lack of complete M. sexta transcript database, we compared the reads to silkworm EST database and found that, although the silkworm and tobacco hornworm belong to the same superfamily (Bomycoidea), only a small portion (around 0.05%) of the four libraries match. Limited by incompleteness of the initial M. sexta genome sequence assembly, we are able to only map 2,087,654 (or 9.9%) of the 21,085,291 reads – a majority of the information remains unexplored in the dataset.
Table 1.
Absolute read numbers for different RNA categories in the libraries
| embryo | Larva | pupa (# of reads) | adult | combined | |
|---|---|---|---|---|---|
| non-coding RNAs | 73,577 | 34,770 | 58,901 | 16,1398 | 328,646 |
| miRNAs | 69,205 | 22,137 | 30,834 | 64,235 | 186,411 |
| mRNAs | 5,494 | 1,396 | 1,056 | 3,353 | 11,299 |
| mitochondrial RNAs | 1,480 | 860 | 736 | 2,466 | 5,542 |
| subtotal | 149,756 | 59,163 | 91,527 | 231,452 | 531,898 |
| unique small RNAs | 41,582 | 22,061 | 30,774 | 124,873 | 219,290 |
| mapped to genome | 1,085,796 | 181,688 | 181,688 | 547,127 | 2,087,654 |
| total | 3,421,611 | 2,716,576 | 9,284,690 | 5,662,414 | 21,085,291 |
Fig. 1. Size distributions of the numbers and types of reads in four libraries combined.
The solid line shows the total read numbers in reference to the left y-axis for each length, whereas the dashed line shows the unique read numbers to the right y-axis.
3.2. Conserved M. sexta miRNAs
By comparing the library with miRBase, we found 163 conserved miRNAs in M. sexta, 157 of which had mapped reads in at least one small RNA library (Table S1) and the other 6 were predicted from genome sequences (Table 2). The main differences between miRNA and other small RNAs are that miRNA precursors have the low-energy, fold-back structures and that mature miRNAs range from 18 to 24 nt. Probably because of limited read numbers (i.e. low abundances at the chosen stages), we did not find corresponding reads for the 6 predicted genes, which are to be sought after in future M. sexta miRNA studies. For miRNAs whose names do not start with “mse-” (Table S1), we did not find their precursors within the current M. sexta genome assembly and used the same names for all variants. Further identification of corresponding genes (and their variants) in the genome is necessary to account for the gene source (and sequence variations, but for now we consider them all derived from families of miRNA genes). For miRNAs from the let-7, bantam, and miR-iab-4 families, since different variants are well characterized in other organisms, we directly name them as family members without identifying their precursor structures. Based on our dataset, not all conserved miRNAs pair up with the miRNA-star strands. Rapid degradation of miRNA-star is likely responsible for the failure of detection in our deep sequencing data.
Table 2.
Predicted conserved M. sexta miRNAs from genome sequence
| miRNA | mature miRNA sequence | precursor free energy |
|---|---|---|
| mse-miR-1b | UGGGAAGUAAGGAAGCACGGAA | -35.50 |
| mse-miR-450 | GGGAUCAUUUUGCAUCCAU | -22.15 |
| mse-miR-929 | CUCCCUAAUCGAGUCAGGUUGA | -42.30 |
| mse-miR-1926 | AGGAAUUCUAAAGCAAAAA | -32.80 |
| mse-miR-2565 | UGAAAUUUAUUUAUAGGCA | -20.70 |
| mse-miR-3389 | UCGUAGCCGAUGUUCCACAG | -47.80 |
3.3. Novel miRNAs in M. sexta
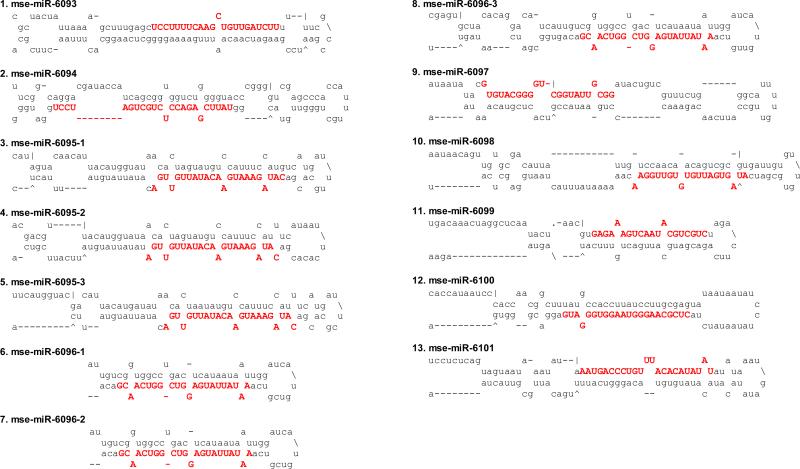
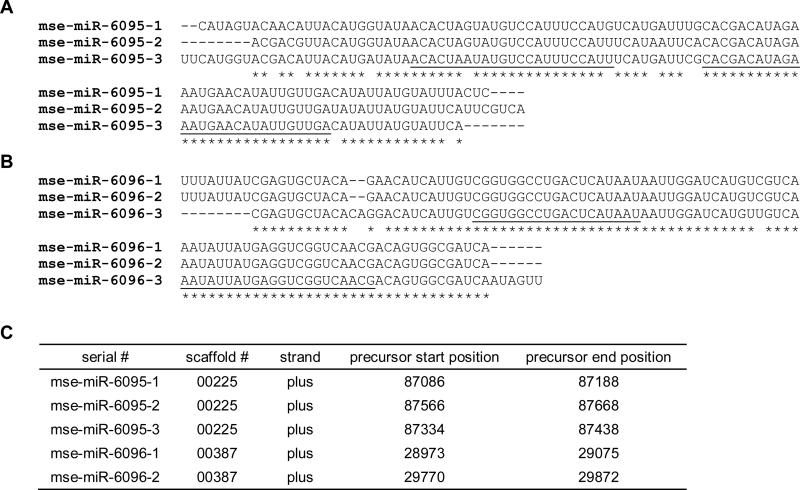
The presence of miRNA-star strand and low-energy, fold-back precursor structure provide solid evidence to annotate miRNAs. If they do not resemble conserved ones in miRBase, we classify them as novel or species-specific (up to now) miRNAs. A total of 13 novel miRNAs were discovered in this study (Table 3) with predicted precursor secondary structures (Fig. 2). If a mature miRNA sequence is mapped to different parts of the genome, these gene loci represent different family members with the same sequence or with a slightly differed sequence by 1 or 2 nt. On the other hand, when several miRNA precursors are located on the same genomic scaffold in the same direction, these genes represent a miRNA cluster. For instance, mse-miR-6095-1, mse-miR-6095-2, and mse-miR-6095-3 have the same mature miRNA sequence and highly similar precursors (Fig. 3A). Their gene loci, located next to each other in the genome, constitute a cluster (Fig. 3C). This cluster probably arose by miRNA gene duplication during evolution. In comparison, mse-miR-6096-1, mse-miR-6096-2 and mse-miR-6096-3 have the identical mature miRNA sequence and similar precursor structures (Fig. 3B), but only the first two are found in the same scaffold (Fig. 3C). Assuming the third gene resides elsewhere in the genome, we tentatively place them in the same family.
Table 3.
Summary of novel M. sexta miRNAs from the four libraries*
| serial # | mature miRNA sequence | miR frequency |
miR* frequency |
||||||
|---|---|---|---|---|---|---|---|---|---|
| embryo | larva | pupa | adult | embryo | larva | pupa | adult | ||
| mse-miR-6093 | UCCUUUUCAAGCUGUUGAUCUU | 322 | 61 | 98 | 237 | 4 | 1 | 0 | 0 |
| mse-miR-6094 | UAUUCGAGACCUCUGCUGAUCCU | 8 | 8 | 6 | 45 | 0 | 0 | 2 | 2 |
| mse-miR-6095-1 | CAUAGAAAUGAACAUAUUGUUGA | 11 | 11 | 8 | 18 | 3 | 1 | 0 | 1 |
| mse-miR-6095-2 | CAUAGAAAUGAACAUAUUGUUGA | 11 | 11 | 8 | 18 | 1 | 0 | 0 | 0 |
| mse-miR-6095-3 | CAUAGAAAUGAACAUAUUGUUGA | 11 | 11 | 8 | 18 | 0 | 0 | 0 | 1 |
| mse-miR-6096** | AAUAUUAUGAGGUCGGUCAACG | 16 | 0 | 2 | 13 | 2 | 0 | 0 | 0 |
| mse-miR-6097 | GUGUACGGGGUCGGUAUUGCGG | 4 | 1 | 0 | 3 | 1 | 1 | 0 | 3 |
| mse-miR-6098 | AUAGUGAUUGUGUGUUGGAA | 2 | 1 | 0 | 5 | 0 | 0 | 0 | 1 |
| mse-miR-6099 | UGAGAAAGUCAAUACGUCGUC | 4 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| mse-miR-6100 | CUCGCAAGGGUAAGGUGGGAUG | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 1 |
| mse-miR-6101 | AAUGACCCUGUUUACACAUAUAU | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Fig. 2. Predicted fold-back precursor structures of novel M. sexta miRNAs.
Mature miRNA sequences are shown in bold capital letters.
Fig. 3. Clustered novel miRNAs in M. sexta.
For sequence alignments, identical residues are marked *. Alignments of fold-back precursor sequences of mse-miR-6095-1, -2, and -3 (A) and mse-miR-6096-1, -2 and -3 (B). (C) Locations of the novel miRNA genes. Note that mse-miR- 6096-3 gene resides on a scaffold different from scaffold 00387.
Due to the absence of predicted miRNA-star strands in our libraries, we did not include other 171 miRNA candidates in Table 3. All these sequences (Table S2) have low-energy, fold-back precursor structures. Further evidence is needed to categorize them as novel miRNAs.
3.4. Development-related fluctuations of miRNA abundances
To examine possible miRNA level variations, we normalized the read numbers of mature miRNAs based on the total read numbers of each library. Here, we focus on 89 miRNAs whose reads are present in at least one of the libraries and whose precursors are identified in the genome (Table 4). In general, most miRNA levels are low in the pupae, a resting stage with low motility and slow metabolism. We divide these miRNAs recovered from the four libraries into five groups. The first group includes 24 miRNAs whose normalized reads in the four libraries are equal to or smaller than 1, such as mse-miR-10b, mse-miR-10c, mse-miR-1000, mse-miR-2765, and mse-miR-6101. Group 2 comprises of 9 miRNAs with similar and low abundances at the chosen stages: mse-miR-33, mse-miR-87, mse-miR-278, mse-miR-745, mse-miR-2768, msemiR-2779, mse-miR-6095-1, mse-miR-6095-2, and mse-miR-6095-3. Group 3 contains 30 miRNAs preferably expressed in embryos, such as mse-miR-8, mse-miR-10a, mse-miR-71, msemiR-2797a, and mse-miR-2797b. The fourth group consists of 16 miRNAs that are more abundant in adult stage, including mse-miR-100, mse-miR-275, mse-miR-277, and mse-miR-989. The remaining 10 miRNAs form the fifth group that is highly expressed in both embryos and adults: mse-miR-1, mse-miR-11, mse-miR-184, mse-miR-276, mse-miR-306, mse-miR-970, mse-miR-2755, mse-miR-2766, mse-bantam, and mse-let-7a.
Table 4.
Developmental profiles of conserved and novel miRNAs of M. sexta*
| miRNA | group | embryo | larva | pupa | adult |
|---|---|---|---|---|---|
| mse-miR-1 | 5 | 2347 | 1883 | 1082 | 2663 |
| mse-miR-2a | 3 | 31 | 10 | 6 | 16 |
| mse-miR-2b | 4 | 4 | 1 | 2 | 7 |
| mse-miR-7 | 3 | 49 | 14 | 3 | 13 |
| mse-miR-8 | 3 | 546 | 260 | 18 | 122 |
| mse-miR-9a | 3 | 16 | 4 | 2 | 6 |
| mse-miR-9b | 3 | 45 | 19 | 6 | 20 |
| mse-miR-10a | 3 | 183 | 54 | 16 | 64 |
| mse-miR-10b | 1 | 1 | 0 | 0 | 0 |
| mse-miR-10c | 1 | 1 | 0 | 0 | 0 |
| mse-miR-11 | 5 | 64 | 23 | 11 | 63 |
| mse-miR-12 | 3 | 27 | 11 | 5 | 18 |
| mse-miR-14 | 4 | 36 | 24 | 14 | 56 |
| mse-miR-31 | 1 | 1 | 0 | 0 | 0 |
| mse-miR-33 | 2 | 2 | 0 | 1 | 2 |
| mse-miR-34 | 4 | 4 | 2 | 4 | 16 |
| mse-miR-71 | 3 | 148 | 42 | 14 | 49 |
| mse-miR-79 | 3 | 30 | 11 | 2 | 5 |
| mse-miR-87 | 2 | 4 | 1 | 2 | 4 |
| mse-miR-92a | 3 | 60 | 14 | 3 | 23 |
| mse-miR-92b | 3 | 29 | 7 | 2 | 7 |
| mse-miR-100 | 4 | 11 | 8 | 7 | 38 |
| mse-miR-124-1 | 1 | 1 | 0 | 0 | 0 |
| mse-miR-133 | 4 | 1 | 1 | 2 | 5 |
| mse-miR-137 | 1 | 0 | 1 | 0 | 0 |
| mse-miR-184 | 5 | 320 | 117 | 68 | 303 |
| mse-miR-190 | 3 | 25 | 11 | 3 | 7 |
| mse-miR-193 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-210 | 1 | 0 | 0 | 0 | 1 |
| mse-miR-252 | 4 | 4 | 3 | 6 | 11 |
| mse-miR-263a | 3 | 567 | 256 | 96 | 325 |
| mse-miR-263b | 4 | 0 | 0 | 0 | 3 |
| mse-miR-275 | 4 | 23 | 10 | 22 | 79 |
| mse-miR-276 | 5 | 253 | 103 | 56 | 200 |
| mse-miR-277 | 4 | 161 | 194 | 201 | 637 |
| mse-miR-278 | 2 | 1 | 1 | 1 | 2 |
| mse-miR-279a | 3 | 371 | 114 | 41 | 137 |
| mse-miR-279b | 3 | 113 | 31 | 13 | 47 |
| mse-miR-279c | 3 | 30 | 8 | 2 | 8 |
| mse-miR-279d | 3 | 89 | 28 | 16 | 40 |
| mse-miR-281 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-282 | 3 | 11 | 11 | 3 | 7 |
| mse-miR-283 | 4 | 3 | 3 | 2 | 6 |
| mse-miR-285 | 4 | 2 | 1 | 4 | 5 |
| mse-miR-306 | 5 | 128 | 57 | 39 | 107 |
| mse-miR-307 | 4 | 7 | 5 | 7 | 15 |
| mse-miR-308 | 3 | 59 | 17 | 6 | 13 |
| mse-miR-316 | 1 | 1 | 0 | 0 | 1 |
| mse-miR-317 | 3 | 47 | 14 | 7 | 21 |
| mse-miR-745 | 2 | 2 | 2 | 2 | 2 |
| mse-miR-750 | 4 | 1 | 1 | 2 | 5 |
| mse-miR-932 | 1 | 1 | 0 | 1 | 1 |
| mse-miR-965 | 1 | 1 | 0 | 0 | 1 |
| mse-miR-970 | 5 | 40 | 18 | 11 | 44 |
| mse-miR-981-1 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-981-2 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-989 | 4 | 33 | 44 | 34 | 144 |
| mse-miR-993 | 3 | 82 | 21 | 4 | 12 |
| mse-miR-998 | 3 | 31 | 7 | 2 | 4 |
| mse-miR-1000 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-2755 | 5 | 242 | 71 | 53 | 177 |
| mse-miR-2763 | 4 | 3 | 0 | 1 | 6 |
| mse-miR-2765 | 1 | 1 | 0 | 1 | 1 |
| mse-miR-2766 | 5 | 234 | 106 | 76 | 298 |
| mse-miR-2767 | 3 | 328 | 96 | 22 | 131 |
| mse-miR-2768 | 2 | 2 | 0 | 1 | 1 |
| mse-miR-2779 | 2 | 3 | 1 | 1 | 3 |
| mse-miR-2788 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-3286 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-3338 | 1 | 0 | 0 | 0 | 0 |
| mse-miR-2796-3 | 1 | 0 | 0 | 0 | 0 |
| mse-bantam | 5 | 42 | 14 | 6 | 29 |
| mse-miR-iab-4-5p | 3 | 8 | 1 | 1 | 4 |
| mse-let-7a | 5 | 152 | 112 | 71 | 204 |
| mse-miR-2797a | 3 | 4469 | 1031 | 83 | 471 |
| mse-miR-2797b | 3 | 854 | 182 | 19 | 78 |
| mse-miR-6093 | 3 | 94 | 22 | 11 | 42 |
| mse-miR-6094 | 4 | 2 | 3 | 1 | 8 |
| mse-miR-6095-1 | 2 | 3 | 4 | 1 | 3 |
| mse-miR-6095-2 | 2 | 3 | 4 | 1 | 3 |
| mse-miR-6095-3 | 2 | 3 | 4 | 1 | 3 |
| mse-miR-6096-1 | 3 | 5 | 0 | 0 | 2 |
| mse-miR-6096-2 | 3 | 5 | 0 | 0 | 2 |
| mse-miR-6096-3 | 3 | 5 | 0 | 0 | 2 |
| mse-miR-6097 | 1 | 1 | 0 | 0 | 1 |
| mse-miR-6098 | 1 | 1 | 0 | 0 | 1 |
| mse-miR-6099 | 1 | 1 | 0 | 0 | 0 |
| mse-miR-6100 | 1 | 0 | 0 | 0 | 1 |
| mse-miR-6101 | 1 | 0 | 0 | 0 | 0 |
All values are normalized for each library as reads per million. If normalized values are below 0.5, they are indicated as zeroes.
3.5. Antisense miRNAs?
Gene transcription usually prefers the sense strand. However, certain miRNA genes also produce antisense miRNAs by convergent transcription, as shown in Drosophila (Tyler et al., 2008) and silkworm (Jagadeeswaran et al., 2010). Since the seed regions of Drosophila miR-iab-4 and miR-iab-4-as are not identical, miR-iab-4-as was predicted to suppress a different set of targets (Stark et al., 2008). In M. sexta, mse-miR-iab-4-as is present in the embryo small RNA library and its star strand exists in embryos, pupae, and adults (Table S1). However, there is no mse-miR-iab-4 in this dataset, which may be present at a low level or in a different stage. mse-miR-1 gene generates sense miRNA (Table S1) and may also yield antisense miRNA. Based on the M. sexta genome sequence, we predict the antisense strand of mse-miR-1 contains a low-energy, fold-back precursor structure (Fig. 4A). Alignment of mse-miR-1 and mse-miR-1b indicated non-identical seed regions (Fig. 4B), implying the targeting of two different gene sets. While mse-miR-1 was highly abundant, there is no read of mse-miR-1b to support our premise. Sequencing of miRNAs from specific tissues or other development stages, together with analysis of 3’-untranslated regions of M. sexta cDNAs, is necessary to verify the hypothesis.
Fig. 4. mse-miR-1b as the antisense miRNA of mse-miR-1?
(A) Fold-back precursor structures of mse-miR-1 and predicted mse-miR-1b. The mature miRNA sequences are shown in bold capital letters. (B) Alignment of the mature mse-miR-1 and mse-miR-1b (predicted). Identical residues are indicated with vertical bars.
3.6. Small RNAs encoded by mse-miR-2779 gene
Deep sequencing of small RNA libraries reveals many types of degradation products (Table 1), which sometimes interfere with miRNA annotation. When reads are randomly mapped to a miRNA precursor, the miRNA appears like a degradation product (Jagadeeswaran et al., 2010). In this study, however, mse-miR-2779 (18 nt) has 37 reads from the four libraries and does represent a genuine miRNA in M. sexta (Table S1). Tracing back to the genome sequence, we find it is a part of the canonical stem-loop structure (Fig. 5A). Interestingly, there are 31 types and 194 total reads of other small RNAs (24-29 nt) mapped to the same precursor (Fig. 5B). These RNA species do not distribute randomly over the mature miRNA sequence region including the seed region.
Fig. 5. mse-miR-2779 precursor encodes many small RNAs of 26-28 nt.
(A) Predicted stem-loop structure of mse-miR-2779 precursor. (B) Small RNAs encoded by mse-miR-2779 precursor followed by the total read number in the four libraries and sequence length. Precursor sequence and structure are shown below. Mature mse-miR-2779 sequence is shown in bold capital letters in A and underlined in B.
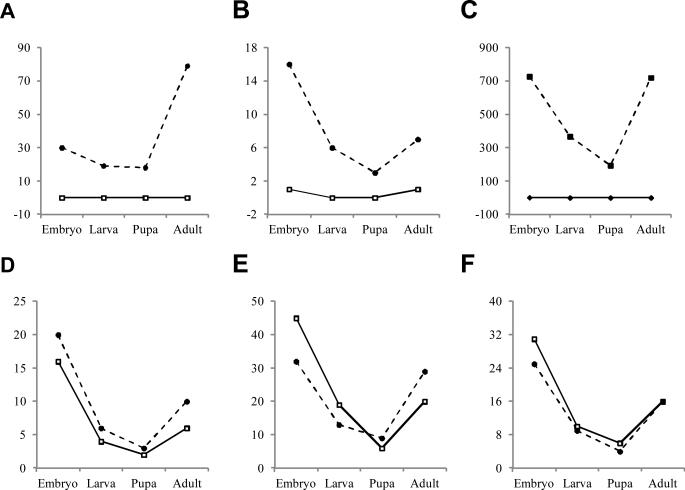
3.7. High abundances of certain miRNA-star strands
Normally, after mature strand is loaded into RISC, its star (*) strand is rapidly degraded or present at a low level. By comparing read numbers (Table S1 and Table 3), we find over 95% of the 170 miRNAs are significantly more abundant in mature strands than in star strands. In sharp contrast, mse-miR-281*, mse-miR-965*, and mse-miR-31* are maintained at much higher levels than respective mature strands in all the developmental stages (Fig. 6, A-C). Both strands of msemiR-9a, mse-miR-9b, and mse-miR-2a are present at similar levels in these stages (Fig. 6, D-F). Their abundance changes relative to the developmental stages remain much the same for both strands. Surprisingly, even though mse-miR-2a and mse-miR-2b belong to the same family, msemiR-2a* was not degraded while mse-miR-2b* was. Similarly, while mse-miR-92a*, mse-miR-279a*, miR-279b* and miR-279d* are mostly degraded, the other family members (mse-miR- 92b and mse-miR-279c) maintained significant levels of the star strands (Table S1).
Fig. 6. Some miRNA-star strands remain at high abundance.
Normalized read numbers for embryo, larva, pupa, and adult are plotted for mse-miR-281 and mse-miR-281* (A), mse-miR- 965 and mse-miR-965* (B), mse-miR-31 and mse-miR-31* (C), mse-miR-9a and mse-miR-9a* (D), mse-miR-9b and mse-miR-9b* (E), as well as mse-miR-2a and mse-miR-2a* (F). Solid lines are for miRNA mature strands and dashed ones for miRNA-star strands.
4. Discussion
4.1. Features of novel miRNA candidates
The 171 miRNA candidates (Table S2), which do not have any star strand read in the libraries, may represent novel miRNAs. Some of them can be traced back to different loci in the genome. Since their precursors can all form low-energy fold-back structures, we predict them as novel miRNAs of the same families. These include: s888171 (2), s60663 (3), s343241 (2), s834258 (3), s324241 (3), s159328 (2), s316155 (2), s496398 (2), s87546 (2), and s1119196 (2), where the numbers in parentheses are gene counts in the same group. If one of them is proven to be miRNA, the other member(s) are likely to be genuine as well (Jagadeeswaran et al., 2010).
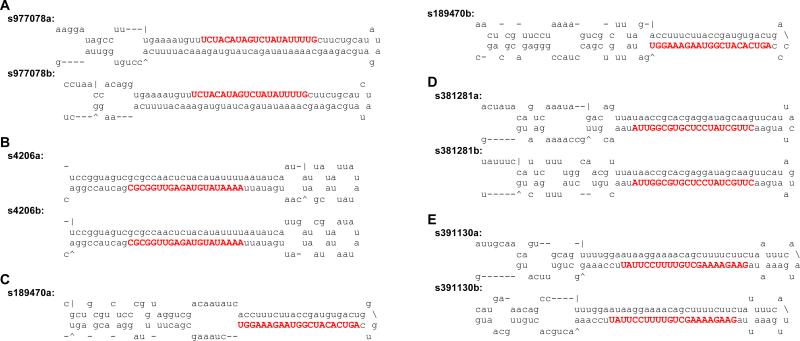
We detected mse-miR-iab-4-as (Table S1) and predicted mse-miR-1b (Fig. 4), because both are highly conserved in animals. For the novel candidates, we identified five possible miRNAs (s977078, s4206, s189470, s381281, and s391130), whose mature strands are identical in sequence to those derived from antisense precursors (Fig. 7). Since the actual read numbers are only 1-2 from the four libraries (Table S2), we are unable to know whether the sense, antisense, or both transcripts were used to generate the mature strand – all the predicted fold-back precursors are low-energy, canonical structures (Fig. 7). Since all the sense and antisense pairs are novel miRNA candidates, M. sexta might serve as a good model to study their transcription and processing.
Fig. 7. Antisense miRNAs of novel miRNA candidates.
These are the predicted fold-back structures of candidate precursors. (A) s977078a and s977078b; (B) s4206a and s4206b; (C) s189470a and s189470b; (D) s381281a and s381281b; (E) s391130a and s391130b. Mature miRNA sequences are shown in bold capital letters.
Based on locations of the putative precursors in the genome assembly, we discovered ten other miRNA clusters. These include s343547-s493484, s343241a-s96924, s1119196as1119196b, s1062476-s595284b, s522556-s487900, s377232-s1048253, s1042305-s449622, and s891216b-s740434 (Table 5). Among them, the s1119196a-s1119196b cluster has the same precursor sequence and, hence, identical mature strand, suggesting the gene pairs arose from recent gene duplications. Future studies on these clustered miRNAs and other (Fig. 3C) may help reveal regulatory mechanisms of miRNA gene expression.
Table 5.
Summary of 16 candidate novel miRNA gene loci
| Serial # | Scaffold # | Strand | Precursor start position | Precursor end position |
|---|---|---|---|---|
| S343547 | 00010 | minus | 1535061 | 1535159 |
| S493484 | 00010 | minus | 1535839 | 1535942 |
| s343241a | 00133 | plus | 389408 | 389508 |
| S96924 | 00133 | plus | 390009 | 390110 |
| s1119196a | 00204 | minus | 38049 | 38148 |
| s1119196b | 00204 | minus | 39063 | 39162 |
| S1062476 | 00241 | minus | 3653 | 3753 |
| s595284b | 00241 | minus | 3844 | 3947 |
| S522556 | 00267 | minus | 298631 | 298729 |
| s487900 | 00267 | minus | 304069 | 304168 |
| s377232 | 00345 | plus | 310355 | 310454 |
| s1048253 | 00345 | plus | 313984 | 314086 |
| s1042305 | 00355 | minus | 373398 | 373499 |
| s449622 | 00355 | minus | 375346 | 375447 |
| s891216b | 00392 | plus | 215730 | 215833 |
| s740434 | 00392 | plus | 217058 | 217162 |
Taken together, the features of novel miRNA candidates should guide our future research and enrich our knowledge on the novel miRNA repertoire of M. sexta.
4.2. Gene expression regulated by miRNA-star strands
After the miRNA strand of a miRNA:miRNA* duplex is loaded onto Argonaute, the star strand is usually degraded. In Drosophila, the first nucleotide in the mature strand is highly conserved and determines correct loading and this is also true for miRNA* (Seitz et al., 2008). The seed regions of miRNAs are well conserved across species so as to regulate similar ranges of target genes. Experimental evidence exists that some miRNA* strands also contain conserved seed regions of 2-7 nt (Guo and Lu, 2010; Okamura et al., 2008; Seitz et al., 2008). Potential regulation of gene expression via miRNA* is assisted by RNA interference pathway (Ghildiyal et al., 2010). Although functional miRNA* strands are usually loaded onto Ago2, Ago1 works as well. Interestingly, mature and star strands from the same duplex sometimes cooperate, but their temporal and spatial expression patterns can be different (Ko et al., 2008; Shen et al., 2010). In our dataset, mse-miR-281*, mse-miR-965*, and six other miR* are abundant and their levels varies in the four stages (Table S1 and Fig. 6). miR-281* and miR-965* are detected at high levels in Bombyx mori (Jagadeeswaran et al., 2010) and, perhaps, other insects in Bomycoidea. mse-miR-9a* and mse-miR-9b* are similar in expression levels to the respective mature strands in all the developmental stages. However, the detailed expression patterns may be worth determining. In vertebrate neural cells and tissues, miR-9:miR-9* is preferably expressed, and their expression patterns change during the developmental course of neural system (Ko et al., 2008; Rosenfeld et al., 2009).
Moreover, miRNA* strands were demonstrated to be associated with hormonal regulation, such as miR-202* with estrogen (Bannister et al., 2011) and miR-488* with androgen receptor (Sikand et al., 2011). To date, functional studies of miRNA* strands have been carried out mostly in vertebrates, and no function is known for these miRNA-star strands in insects. As a model for insect nervous system and hormonal regulation, M. sexta is anticipated to contribute to our knowledge on the regulatory functions of miRNA* strands.
4.3. Development-related functions of M. sexta miRNAs
miRNAs are known to regulate diverse physiological processes in Drosophila (Chawla and Sokol, 2011). Acting at the posttranscriptional level, miRNAs impact various signaling pathways of cell proliferation, differentiation, and apoptosis, which underlie insect development. There is ample evidence that miRNAs affect growth, neural differentiation, wing development and hormonal signaling. Our profiling of M. sexta miRNAs from different developmental stages is the first step toward understanding their specific roles. We found most of the conserved and novel miRNAs are low in pupae. In B. mori, conserved miRNAs are also present at low levels in pupal stage but novel ones are typically higher in pupae (Huang et al., 2010; Jagadeeswaran et al., 2010). Note that these datasets, obtained from total RNA of whole body at limited time points, lack the specificity, depth, and resolution needed to reveal detailed temporal and spatial expression patterns. Furthermore, without 3’-untranslated region database of M. sexta mRNAs, we are unable to predict targets of conserved or novel miRNAs at this time. Nonetheless, we are confident these difficulties for function prediction and testing will soon be overcome through sequencing small RNA libraries of tissues taken from M. sexta under specific physiological conditions and through analysis of extensive RNAseq data in conjunction with the genome sequence.
5. Conclusions
By sequencing the small RNA libraries of embryos, larvae, pupae, and adults, we obtained the first set of M. sexta miRNAs, including 163 conserved and 13 novel ones. Our comparison of their read frequencies in the four stages suggested that M. sexta miRNAs are dynamically regulated throughout the life cycle. Eight miRNA-star strands were maintained at high levels, differentially expressed in these stages, and possibly involved in gene expression control. We detected antisense miRNA (mse-miR-iab-4-as) and predicted mse-miR-1b. Additionally, there were 171 candidate genes encoding novel miRNAs, some forming clusters while others transcribed convergently, perhaps. This dataset serves as a foundation for future miRNA studies in this model species and identification of their targets is expected to greatly enrich our knowledge on the post-transcriptional regulation of gene expression in M. sexta.
Supplementary Material
Acknowledgements
We thank Dr. Ulrich Melcher for his comments on the manuscript. This work was supported by National Institutes of Health Grant GM58634 (to H.J.) and Science and Technology Commission of Shanghai Municipality Grant 10ZR14030000 (to Y.Z.) and a start-up fund from Fudan University (to Y.Z.). This article was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported in part under projects OKLO2450 (to H.J.) and OKLO2611 (to R.S.). We would like to acknowledge Manduca Genome Project for Sequence Assembly 1.0, which is funded by Defense Advanced Research Projects Agency (Gary Blissard, Boyce Thompson Institute) and National Institutes of Health (Michael Kanost, Kansas State University).
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bannister SC, Smith CA, Roeszler KN, Doran TJ, Sinclair AH, Tizard ML. Manipulation of estrogen synthesis alters MIR202* expression in embryonic chicken gonads. Biol Reprod. 2011;85:22–30. doi: 10.1095/biolreprod.110.088476. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behura SK, Haugen M, Flannery E, Sarro J, Tessier CR, Severson DW, Duman-Scheel M. Comparative genomic analysis of Drosophila melanogaster and vector mosquito developmental genes. PLoS One. 2011;6:e21504. doi: 10.1371/journal.pone.0021504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel C, Winter F, Edaye S, Huttenhofer A. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu X, Zhou Q, Yu C, Hu H, Liu J, Lin H, Yang J, Zhang B, Cui P, Hu S, Yu J. Novel microRNAs in silkworm (Bombyx mori). Funct Integr Genomics. 2010;10:405–415. doi: 10.1007/s10142-010-0162-7. [DOI] [PubMed] [Google Scholar]
- Cao J, Tong C, Wu X, Lv J, Yang Z, Jin Y. Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. Insect Biochem Mol Biol. 2008;38:1066–1071. doi: 10.1016/j.ibmb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Chatterjee R, Chaudhuri K. An approach for the identification of microRNA with an application to Anopheles gambiae. Acta Biochim Pol. 2006;53:303–309. [PubMed] [Google Scholar]
- Chawla G, Sokol NS. MicroRNAs in Drosophila development. Int Rev Cell Mol Biol. 2011;286:1–65. doi: 10.1016/B978-0-12-385859-7.00001-X. [DOI] [PubMed] [Google Scholar]
- Chen X, Yu X, Cai Y, Zheng H, Yu D, Liu G, Zhou Q, Hu S, Hu F. Next-generation small RNA sequencing for microRNAs profiling in the honey bee Apis mellifera. Insect Mol Biol. 2010;19:799–805. doi: 10.1111/j.1365-2583.2010.01039.x. [DOI] [PubMed] [Google Scholar]
- Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X. Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae). PLoS One. 2011;6:e19350. doi: 10.1371/journal.pone.0019350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci U S A. 2011;108:7449–7454. doi: 10.1073/pnas.1017963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K, Riddiford LM. Developmental expression of mRNAs for epidermal and fat body proteins and hormonally regulated transcription factors in the tobacco hornworm, Manduca sexta. J Insect Physiol. 2010;56:1390–1395. doi: 10.1016/j.jinsphys.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zou Q, Tang SM, Wang LG, Shen XJ. Computational identification and characteristics of novel microRNAs from the silkworm (Bombyx mori L.). Mol Biol Rep. 2010;37:3171–3176. doi: 10.1007/s11033-009-9897-4. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese EL, Soulages JL, Zhang W, Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Lin K, Liang J, Yu L, Li F. Discovering conserved insect microRNAs from expressed sequence tags. J Insect Physiol. 2010;56:1763–1769. doi: 10.1016/j.jinsphys.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2011;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YF, He PA, Nie ZM, Chen JQ, Chen J, Lv ZB, Sheng Q, Zhou SP, Gao XL, Kong LY, Wu XF, Zhang YZ. Identification and characteristics of microRNAs from Bombyx mori. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MH, Kim S, Hwang do W, Ko HY, Kim YH, Lee DS. Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 2008;275:2605–2616. doi: 10.1111/j.1742-4658.2008.06408.x. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legeai F, Rizk G, Walsh T, Edwards O, Gordon K, Lavenier D, Leterme N, Mereau A, Nicolas J, Tagu D, Jaubert-Possamai S. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genomics. 2010;11:281. doi: 10.1186/1471-2164-11-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mead EA, Liang S, Tu Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics. 2009;10:581. doi: 10.1186/1471-2164-10-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang L, Li Q, Zhao P, Duan J, Cheng D, Xiang Z, Xia Q. MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori). BMC Genomics. 2011;12:284. doi: 10.1186/1471-2164-12-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Vasconcelos AT, Sagot MF. Combination of measures distinguishes pre-miRNAs from other stem-loops in the genome of the newly sequenced Anopheles darlingi. BMC Genomics. 2010;11:529. doi: 10.1186/1471-2164-11-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauchet Y, Wilkinson P, Vogel H, Nelson DR, Reynolds SE, Heckel DG, ffrench-Constant RH. Pyrosequencing the Manduca sexta larval midgut transcriptome: messages for digestion, detoxification and defence. Insect Mol Biol. 2010;19:61–75. doi: 10.1111/j.1365-2583.2009.00936.x. [DOI] [PubMed] [Google Scholar]
- Reddy AM, Zheng Y, Jagadeeswaran G, Macmil SL, Graham WB, Roe BA, Desilva U, Zhang W, Sunkar R. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genomics. 2009;10:65. doi: 10.1186/1471-2164-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, Sitbon E, Yanai GL, Elyakim E, Cholakh H, Gibori H, Spector Y, Bentwich Z, Barshack I. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–383. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Cohen SM. Identification of novel Drosophila melanogaster microRNAs. PLoS One. 2007;2:e1265. doi: 10.1371/journal.pone.0001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5' precision of both microRNAs and their miRNA* strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Zhou HB, Huang XF, Cui HJ, Luo XB, Tang YJ, Chen SL, Wu L. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- Sikand K, Slaibi JE, Singh R, Slane SD, Shukla GC. miR 488* inhibits androgen receptor expression in prostate carcinoma cells. Int J Cancer. 2011;129:810–819. doi: 10.1002/ijc.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Nagaraju J. In silico prediction and characterization of microRNAs from red flour beetle (Tribolium castaneum). Insect Mol Biol. 2008;17:427–436. doi: 10.1111/j.1365-2583.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surridge AK, Lopez-Gomollon S, Moxon S, Maroja LS, Rathjen T, Nadeau NJ, Dalmay T, Jiggins CD. Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene. BMC Genomics. 2011;12:62. doi: 10.1186/1471-2164-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong CZ, Jin YF, Zhang YZ. Computational prediction of microRNA genes in silkworm genome. J Zhejiang Univ Sci B. 2006;7:806–816. doi: 10.1631/jzus.2006.B0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- Weaver DB, Anzola JM, Evans JD, Reid JG, Reese JT, Childs KL, Zdobnov EM, Samanta MP, Miller J, Elsik CG. Computational and transcriptional evidence for microRNAs in the honey bee genome. Genome Biol. 2007;8:R97. doi: 10.1186/gb-2007-8-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen S, Yang P, Ma Z, Kang L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009;10:R6. doi: 10.1186/gb-2009-10-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- Yu J, Luo QB, Zhou Q, Yu XM, Lin HB, Hu SN. Genome-wide mapping of conserved microRNAs and their host transcripts in Tribolium castaneum. J Genet Genomics. 2008a;35:349–355. doi: 10.1016/S1673-8527(08)60051-X. [DOI] [PubMed] [Google Scholar]
- Yu X, Zhou Q, Cai Y, Luo Q, Lin H, Hu S, Yu J. A discovery of novel microRNAs in the silkworm (Bombyx mori) genome. Genomics. 2009;94:438–444. doi: 10.1016/j.ygeno.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Yu XM, Zhou Q, Li SC, Luo QB, Cai YM, Lin WC, Chen H, Yang Y, Hu SN, Yu J. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS One. 2008b;3 doi: 10.1371/journal.pone.0002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gunaratna RT, Zhang X, Najar F, Wang Y, Roe B, Jiang H. Pyrosequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta, a lepidopteran model insect. Insect Biochem Mol Biol. 2011;41:733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Ge X, Jiang J, Li M, Jia S, Yang X, Kan Y, Miao X, Zhao G, Li F, Huang Y. Insect-specific microRNA involved in the development of the silkworm Bombyx mori. PLoS One. 2009;4:e4677. doi: 10.1371/journal.pone.0004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem Mol Biol. 2008;38:677–682. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.