Abstract
Sample displacement chromatography (SDC) in reversed-phase and ion-exchange modes was introduced approximately twenty years ago. This method takes advantage of relative binding affinities of components in a sample mixture. During loading, there is a competition among different sample components for the sorption on the surface of the stationary phase. SDC was first used for the preparative purification of proteins. Later, it was demonstrated that this kind of chromatography can also be performed in ion-exchange, affinity and hydrophobic-interaction mode. It has also been shown that SDC can be performed on monoliths and membrane-based supports in both analytical and preparative scale. Recently, SDC in ion-exchange and hydrophobic interaction mode was also employed successfully for the removal of trace proteins from monoclonal antibody preparations and for the enrichment of low abundance proteins from human plasma. In this review, the principals of SDC are introduced, and the potential for separation of proteins and peptides in micro-analytical, analytical and preparative scale is discussed.
1. Introduction
During the last decade proteomics has had an increasing impact on answering key questions and understanding vital functions of biological systems [1]. Proteomics also has an increasingly important role in clinical research, especially in the search for protein biomarkers and targets for new drugs [2, 3]. A number of protein biomarkers have been approved by regulatory agencies and they are now in clinical use [3, 4]. Despite recent optimistic reports [5–7], there have also been very critical statements in the last two years about the application of proteomics to biomarker discovery. These discussions asked whether, compared to existing methods, the progress in proteomics technology has delivered any usable results, or whether a further step in the development of MS-based proteomics, or even a fundamental change of the concept, is necessary before clinically useful results can be expected [6, 8, 9]. Compared to the tremendous number of publications in the scientific literature dealing with the discovery of diagnostic and prognostic biomarkers, only a few that were identified by use of proteomics have been clinically approved [9, 10].
Tissue specimens, cell suspensions, blood plasma and other body fluids are the starting materials that are most frequently used for the identification of new biomarkers [3, 4]. These biological materials are very complex, and they contain thousand of components in a very large dynamic range of concentrations. This range is up to 108–1012 in serum and plasma [11], and up to 105 in cells and tissues [12]. In human plasma, serum albumin (HSA) and immunoglobulins are the most abundant proteins, and they represent over 75% of the weight of all proteins. These two and an additional 20 proteins account for over 99% of overall protein content in this biological fluid. On the other hand, the concentrations of low abundance proteins in plasma range from ng/mL to pg/mL level [2, 4, 11]. Because the minimal range of detection is about 0.1 ng/mL, the large number of trace level proteins require a proper concentration step in order to be identified [2]. Most potential biomarkers and biomarker candidates in plasma and other complex biological mixtures are present in very low levels. Consequently, their detection frequently demands thorough sample preparation. Unfortunately, this step, which can contain dangerous pitfalls, is frequently neglected in MS-based shotgun proteomics. For example, we demonstrated that the unpredicted presence of protease inhibitors in a sample can be a serious obstacle for its tryptic digestion, resulting in a significant reduction of identified proteins [13].
Together with electrophoretic techniques [14], chromatography is the most commonly used method for separation and fractionation of complex biological mixtures [12]. Chromatographic methods are used for both concentration of trace components prior to proteolytic digestion [12, 15], and for fractionation of digested proteins prior to MS analysis [12]. For proteomics analysis of serum or plasma, chromatographic and immunoaffinity chromatographic methods are used for the removal of highly abundant proteins [4, 14]. After their removal, residual proteins can be separated either by SDS-PAGE or 2D electrophoresis [14], or by use of a single chromatographic step, or a combination of different chromatographic and electrophoretic methods [15, 16]. However, the need to improve sample preparation methods still remains [16], and the use of additional fractionation methods such as displacement chromatography for both separation of proteins components prior to proteolytic digestion [17, 18], and digested peptides before prior to mass spectrometric analysis [18–20] has recently been discussed.
In preparative separations of complex mixtures that contain proteins and/or peptides, chromatography is also the method of choice for purification of target components [21, 22]. Ion-exchange (IEC) and different affinity chromatography (AC) methods are most frequently employed for the isolation of biologically active therapeutic proteins [23, 24], but there are additional chromatographic methods, such as hydrophobic-interaction chromatography (HIC) and hydroxyapatite chromatography, that are increasingly being used [25–27]. Reversed-phase chromatography (RPC) has been less frequently used for the separation of therapeutic proteins because of the potential for denaturation and loss of biological activity of target components after elution with organic solvents [28]. On the other hand, RPC is the method of choice for peptide separation [12]. Preparative ion-exchange chromatography, a method widely used for the separation of proteins on industrial scale, can be performed in continuous and step-gradient mode. However, the presence of salts in high concentrations frequently reduces the effective separation factor between the feed solutes [24]. In order to overcome the limitations of preparative gradient chromatography, displacement chromatography was introduced [29]. This method was invented by Tiselius in 1943 [30], used for early separations of amino acids [31], and further developed by Horváth's group in early 1980's [32]. In displacement chromatography, the sample is introduced onto the column, and then displaced by a constant infusion of a displacer solution. The affinity of the displacer for the stationary phase has to be higher than the affinity of any feed components [29]. Displacement chromatography of proteins and peptides is usually performed in ion-exchange mode [29], but hydrophobic interaction mode has also been used [33].
Despite its importance as a starting material for diagnostic purposes and for discovery of new biomarkers, human plasma is still used as a valuable source for the isolation of therapeutic proteins [34, 35]. Large amounts of intravenous immunoglobulin (IVIG) and human serum albumin preparations originate from human plasma, and thousands of units of clotting factors and inhibitors are still needed in medicine as indispensable therapeutic agents [35–37]. Fractionation of human plasma is a sixty-year old technology, and chromatography is widely used as an integral part of different production processes [34, 37, 38]. However, improvements in yield for production of current therapeutic proteins, and development of fractionation processes for the isolation of new therapeutics are still necessary [34,37].
As mentioned above, chromatography is a crucial method for the separation of proteolytic digests of proteins prior to MS analysis. Chromatography, usually in reversed-phase mode (RPC), is also the basic method for separation of target peptides after synthesis in order to remove trace impurities [39]. However, development of a rapid, facile, reproducible, and cost-effective method for the efficient purification of mixtures containing multiple peptides is still a challenge [40]. Although chromatographic supports and instruments have been further improved, there is still a need for the development of new, complementary methods for the separation of complex mixtures of proteins and peptides in both analytical and preparative scales.
Sample displacement chromatography (SDC) is a method that was introduced by Hodges and Mant [41] for preparative purification of peptides. This method takes advantage of different relative binding affinities of components in a sample mixture. During loading, there is a competition among different sample components for the adsorption on the ligands on the surface of stationary phase. SDC was first introduced for preparative purification of peptides in reversed-phase mode [41, 42]. Later, it was demonstrated that this kind of chromatography can also be performed in ion-exchange [43, 44], affinity [45] and hydrophobic-interaction modes [46]. We also demonstrated that SDC can be performed on small analytical columns for the effective separation of microgram amounts of proteins from human plasma and can be applied as a sample preparation step for subsequent MS analysis of separated proteins [44,46]. Unfortunately, after its introduction over twenty years ago, this method has scarcely been used. In this review, the principals of SDC are introduced, and its potential for fractionation, isolation and purification of proteins and peptides in micro-analytical, analytical, and in preparative scales is discussed.
2. Principles of sample displacement chromatography
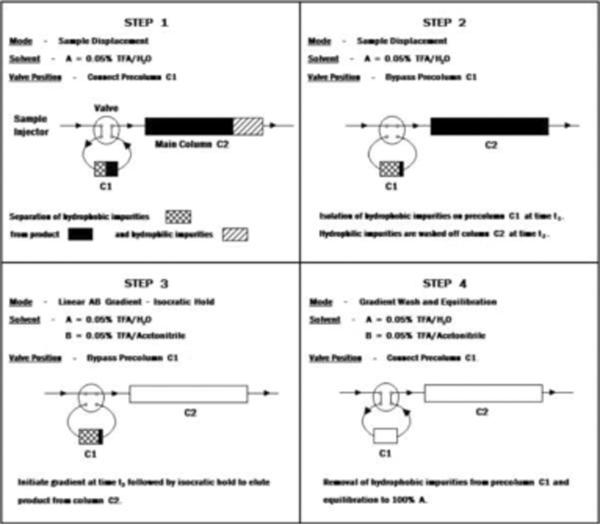
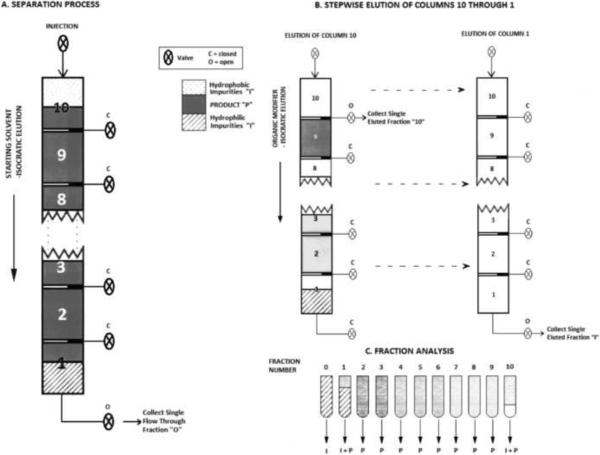
SDC takes advantage of the different affinities of components in a sample mixture for the stationary phase. If the column is optimally loaded with the sample mixture that is dissolved in the starting buffer (solution), there is competition among the sample components for the binding sites (ligands) on the stationary phase. The components with higher affinity for the ligands compete more successfully for the binding to the chromatographic support than the components with lower affinity, and the latter ones can be displaced from the column. The sample displacement can be optimally performed under overloading conditions. Consequently, the main separation occurs during sample loading, and the column capacity is optimally used. Additional fractionation can be achieved in a second step, by use of a linear or step gradient elution of bound components [41]. During early SDC development, Hodges at al. [42] were the first to use the two-column strategy for the separation of peptide mixtures. As shown in Figure 1, one small pre-column was used for retention of very hydrophobic components that have the highest affinity for the chromatographic support (step 1 in Figure 1). At the same time, the less hydrophobic impurities that have the lowest affinity for the support were washed off the main column (step 2 in Figure 1). The product is subsequently eluted from the main column (column 2, see step 3 in Figure 1), along with the more hydrophobic impurities from the pre-column (column 1, see step 4 in Figure 1). This strategy was very quickly extended for use in preparative scale purifications, and a multi-colmn system was developed [40,47]. As shown in Figure 2, ten small column segments have been connected in series, and during loading (step A in Figure 2), sample components (peptides) were distributed throughout the system according to their affinity to the stationary phase (in this case hydrophobicity). In the second step (step 2 in Figure 2), each individual column containing peptides separated according to their binding affinity to the chromatographic support is eluted. The elution can be performed after each column is detached from the system, or automatically by column switching and use of a valve system, integrated in the chromatographic equipment. In optimized SDC, each column represents a pure fraction.
Figure 1.
Strategy for sample displacement chromatography (SDC) in RP mode.
Step 1
More hydrophobic impurities bind to the pre-column. Both the less hydrophobic main product and hydrophilic impurities bind to the main column. The main column is less hydrophobic than the pre-column.
Step 2
The more hydrophobic main product displaces hydrophilic impurities from the main column. The pre-column is switched off.
Step 3
Pure main product is eluted from the column. The pre-column is still switched off.
Step 4
The hydrophobic impurities are eluted from the pre-column. The main column is out of the system.
Adapted from Reference [42] with permission.
Figure 2.
Sample displacement chromatography with a multi-column system containing 10 reversed-phase columns.-Schematic presentation
Step A - sample loading and displacement of less hydrophobic components to the column segments towards detector. More hydrophobic components stay on the columns closest to the inlet.
Step B – Stepwise elution of columns after sample displacement Adapted from Reference [40] with permission.
Veeraragavan et al. [43] applied SDC for separation of proteins in ion-exchange mode. It was demonstrated that in order to achieve optimal separation, the flow rate in SDC has to be reduced to 20% compared to the flow rate in gradient elution separation. This phenomenon was not further discussed, but it seems that the displacement effect is dependent on the flow rate. Consequently, sample displacment is more complete at low flow rate. In these early experiments, two conditions favorable for separation in SDC were met: a) The first peak in the chromatogram is the product. I.e., the product does not bind, or only weakly binds, to the column and can be displaced by stronger binding impurities, or b) The product is the most strongly binding component in the mixture, and it can displace impurities from the column already bound during loading. However, these conditions are rarely met, and a multi column system as shown in Figure 2 has to be used for separation of complex biological mixtures in SDC mode, as demonstrated by Agner [48, 49] and Manseth et al. [50].
We recently “re-discovered” SDC while performing separations of human plasma with different anion- and cation-exchange monolithic supports [44, 46]. The use of membranes and monoliths for SDC can be very favorable because the displacement phenomena by sample components with different affinities for the stationary phase occurs at much lower loading than if bulk, porous chromatographic supports are employed [44, 45, 51]. A similar phenomenon is also observed when non-porous bulk supports are used [51, 52]. However, relatively high backpressure is a serious obstacle when utilizing non-porous supports for SDC separations [51]. The logical conclusion is that in SDC, the sample components can be displaced faster if the interaction between different sample components is not hampered by diffusion into the pores of the bulk support. The fact that the sample displacement effects on membranes and monolithic supports are practically flow rate independent also supports this theory. It has been demonstrated that sample loading on monoliths and membranes can be performed at relatively high flow rates [44, 46]. As mentioned above, in order to achieve optimal sample displacement, Veeraragavan et al. had to reduce the flow rate five-fold when columns packed with porous bulk materials were used for separations of proteins in SDC mode [43]. We also demonstrated that the sample displacement effects on monoliths are independent of column size, and similar results were obtained with either a 340 μL or an 8 mL column (cf. Figure 4) The separation performance is virtually independent on column size, and monoliths are materials suitable for protein separation in microanalytical, analytical and preparative scales.
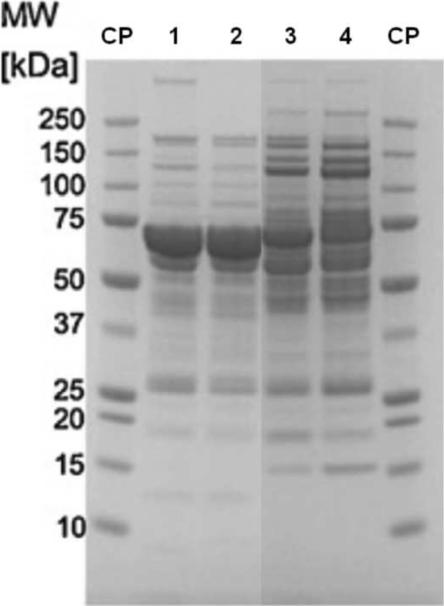
Figure 4.
Sample displacement chromatography on monolithic supports; SDS-PAGE of eluted proteins after different loading of human plasma.
Lanes 1 and 2 – The protein pattern after loading of 15 mg protein/mL support and elution with 20 mM Tris.HCl, pH 7.4 containing 0.5 M NaCl. Lane 1 – CIM DEAE disk, column volume 340 μL; lane 2 – semi-preparative CIM DEAE tube, column volume 8mL. Note – HSA is the major protein that is eluted from both columns under these loading conditions.
Lanes 3 and 4 – The protein pattern after loading of 187.5 mg protein/mL support and elution with 20 mM Tris.HCl, pH 7.4 containing 0.5 M NaCl. Lane 7 – CIM DEAE disk, column volume 340 μL; lane 8 – semi-preparative CIM DEAE tube, column volume 8mL. Note – HSA is almost completely displaced from the column by stronger binding proteins.
The pattern of eluted proteins is very similar and virtually independent of column size (see lanes 1 and 7 for the analytical and lines 2 and 8 for the semi-preparative column).
Adapted from Reference [44] with permission.
3. Applications of sample displacement chromatography
3. 1. Reversed-phase chromatography
Sample displacement chromatography in reversed-phase mode was developed by Hodges and Mant [41, 42], and has been used almost exclusively by this group for separations of proteins and peptides [40, 53]. The first applications for separations of peptides were published in 1988 [41, 42], and multiple-column reversed phase sample displacement chromatography was introduced in 1991 [47]. Ten years later this group applied SDC to the preparative RP separation of mixtures after multiple peptide synthesis [40] and protein components from a crude sample containing rabbit skeletal troponin [53].
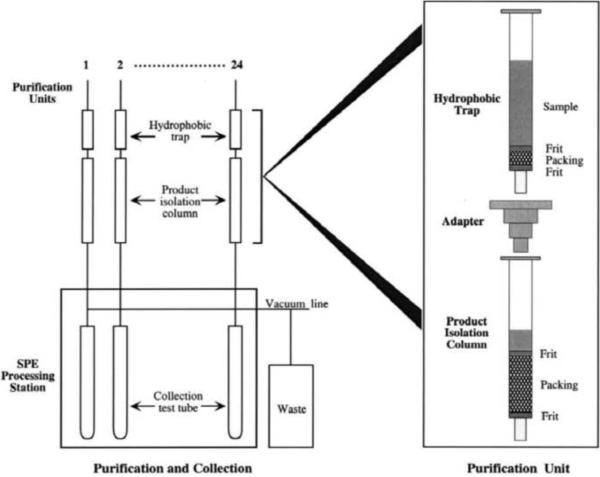
The optimized multiple column system shown in Figure 2 was used for the purification of both peptides and proteins [40, 53]. In Figure 5, a two-column unit developed by Hodges' group for purification of peptides is shown [40]. The first column (pre-column) is packed with a less hydrophobic support and used as a trap for hydrophobic impurities. The peptide of interest is displaced from first column, and binds to the second, more hydrophobic, column. The elution of each column is performed in the second step, after column detachment. Each column can be loaded and eluted by use of low pressure or vacuum, and column construction is designed for use in a robotic system (cf. Figure 5). The most important further development in later years is the above discussed application of different-sized columns, columns with different ligand density, or with ligands that show different affinity for sample components (in RPC and HIC – columns with different hydrophobicity).
Figure 5.
Two-column purification unit for SDC of peptides designed for use in a high-throughput system. Reprinted from Reference [40] with permission.
3.2. Hydrophobic-interaction chromatography
We recently demonstrated that sample displacement can be also used for HIC separation of proteins from human plasma [51]. A three-column system that contains columns packed with chromatographic material with different grades of hydrophobicity (Phenyl, in combination with Butyl and Ether) was used for separation of plasma proteins, and a separation between HSA and low abundance proteins was achieved. More hydrophobic, low-abundance proteins bind to the first two columns and displace HSA, which binds to the third column with a more hydrophobic surface. In each eluted fraction, a significant enrichment of different proteins was achieved. The first two fractions containing more hydrophobic proteins that bind to the first two columns with less hydrophobic surfaces contain some highly concentrated plasma proteins that are used as therapeutics (e.g. clotting factors and inhibitors). In the last fraction, highly enriched HSA was eluted.
In the experiments presented above, concentrated ammonium sulfate was employed as the salt additive in the Buffer A (application buffer). This salt is the most commonly used reagent in HIC of proteins [54]. However, significant numbers of plasma proteins are not soluble in highly concentrated ammonium sulfate, and over 25% of them precipitate in the application buffer (1.7–2.5 M ammonium sulfate). However, it is well known that different salts and salt mixtures can modulate selectivity and capacity of resins used for HIC [54, 55]. By use of sodium chloride as the salt additive to the application buffer, the solubility of plasma proteins significantly increased, and the selectivity of the three-column system also changed. In contrast to ammonium sulfate, only 10–12% of the proteins precipitated when concentrated sodium chloride was added to the application buffer [51].
To the best of our knowledge, this report is the only study performed on HIC in SDC mode. This work has demonstrated that both HIC and SDC in HIC mode may be a welcome complementary method for the separation of proteins from blood plasma on both small and large scales. However, in HIC, like RPC [53], the limited solubility of some proteins under the conditions used for chromatographic separation has to be taken into consideration. Further investigations are needed, especially regarding the choice of column material, use of different salt and salt mixtures, and influence of sample composition on protein separation by SDC in HIC mode.
3.3. Ion-exchange chromatography
In 1991 Veeraragavan et al. [43] introduced SDC in anion exchange (AEX) mode for the fractionation of crude mixtures of ovalbumin or soybean trypsin inhibitor. The separation was optimized in their first experiments with a single AEX column, and two identical columns were later used for preparative purification of these two crude mixtures of model proteins. The use of a multi-column system for optimal separation was also discussed, but no further work was published. The authors also discussed the need for systematic parameter variation in order to maximize throughput, yield and purity of the separated components. However, only two commercial AEX columns (Mono Q) were employed in their experimental work. The possibility to improve the separation by use of a two (or more) column-system with different ligands (e. g. DEAE and Q) and/or different ligand densities combined with optimized dimensions for each column was not explored.
In a study dealing with a protein-purification-parameter screening system performed by the Schlüter group [18, 56], SDC in cation exchange (CEX) mode for the purification of angiotensin-II-generating enzyme from a porcine renal extract was investigated. After optimization by use of small amounts of bulk CE resin in batch-mode in 96-well plates, preparative purification of this enzyme was performed at pH 3.0 containing 500 mM NaCl. [56].
Brown et al. [44] applied SDC in AEX and CEX modes as the final steps in the purification of monoclonal antibodies produced by Chinese-hamster ovary cells. In this case, commercially available ion-exchange membrane-based supports were used. After optimization of separation conditions (pH and salt concentration of the application buffer), membranes were overloaded by antibody solution previously purified by protein A affinity chromatography. This intermediate product still contained some trace impurities. Under overloading conditions, these impurities bind to the membrane and displace the product (mAb), which results in consistent removal of impurities such as host cell proteins. This process was independent of the flow-rate and membrane scale.
We applied different AEX and CEX poly-(glycidyl methacrylate) monolithic supports for SDC separation of proteins from cryoprecipitate-depleted human plasma in SDC mode [44, 46]. Under overloading conditions the weakly bound proteins such as HSA in AE and IgG in CEX mode are displaced in the very early stage by stronger interacting proteins, and, as already mentioned above, this phenomenon is not dependent on the size of the monolithic column. We demonstrated that small monolithic columns with a column volume of 100 and 200 μL are ideal supports for high-throughput screening in order to develop new methods for the separation of complex mixtures, especially for the enrichment of strongly-binding low-abundance proteins when SDC in ion-exchange mode is utilized.
Human plasma and serum are the most frequently used biological materials for discovery of new biomarkers [3. 4]. Rapid, reproducible and cost-effective removal of the highly abundant proteins HSA and immunoglobulins still an issue [4, 14]. SDC on ion-exchange monolithic supports offers a very effective and quick method for the removal of high abundance proteins, and concentration of low-abundance ones. The already demonstrated possibility for stacking of two and more different monoliths in one cartridge (“Conjoint chromatography”, see Ref. 57) offers the possibility to perform sample displacement chromatographic separation by optimal use of a combination of ion-exchange monolithic supports with different capacities and affinities to the sample components. A recently developed system containing one or more small monolithic columns mounted in a 96-well plate (Ref. 58, see also Figure 5) can be used for these kinds of separations for sample preparation in proteomic technology.
A big advantage of monoliths and membranes is the relatively simple scale up from small, disk-shaped columns to semi-preparative and preparative radial columns [59], and it has also been shown that displacement effects are almost completely independent of column size (see Figure 4). Lim et al. [60] used anion-exchange chromatography for industrial-scale isolation of inter-alpha-inhibitor proteins (ITIp) for therapeutic use (treatment of sepsis). After optimization of sample load and application buffers (starting pH value and salt concentration), highly pure ITIp concentrate was isolated from a side-fraction that is produced during clotting factor IX (FIX) preparation. The industrial-scale use of membranes for SDC in ion-exchange mode for the purification of antibodies was demonstrated by Brown et al [45].
SDC has been used for years in plasma fractionation for the isolation of vitamin K-dependent clotting factors and inhibitors. However, this purification scheme was developed based on trial-and-error, and it was never properly optimized for both yield and technical performance [37]. Similar to the small-scale strategy presented in Reference [56], undiluted, cryoprecipitate-depleted human plasma was mixed with either weak (DEAE), or strong (quaternary amine) anion exchange resin, usually 2–5 g Sephadex G50/L plasma. After filtration and washing, bound proteins were eluted with a buffer with about neutral pH, containing 0.5–1.0 M NaCl. As shown in Figure 6, this procedure is highly reproducible. Due to the stronger binding of less abundant proteins, HSA is almost completely displaced. For production of F IX, DEAE resin is used, because this resin does not bind clotting factor VII (F VII), and this impurity is removed in first production step (so-called “solid-phase extraction”, see Reference 37). For production of prothrombin complex concentrate (PCC) that contains all four vitamin K-dependent clotting factors (F II, F VII, F IX and F X), and clotting inhibitors protein C, protein S and protein Z, strong anion-exchange resins are used. These resins also bind F VII under the conditions presented above [61].
Figure 6.
The 96-well plates containing small monolithic columns (left), or small micro-columns (right) for high-throughput sample preparation.
The above described applications show that SDC in ion-exchange mode offer further possibilities for improvement of existing processes in plasma fractionation technology, such as the production of vitamin K-dependent clotting factors and inhibitors, or for use in the development of separation processes for the isolation of new physiologically active proteins out of this valuable raw material [34, 37, 60].
3.4. Affinity and pseudo-affinity chromatography
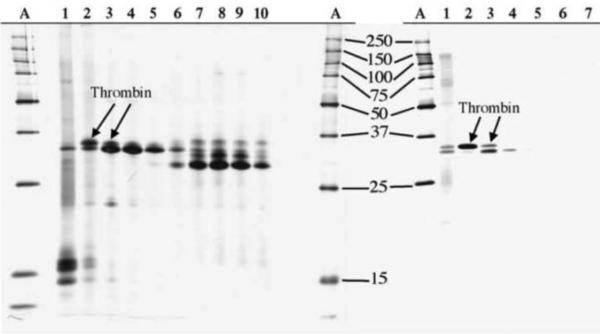
Manseth et al. [50] used heparin-affinity SDC for the separation of thrombin from plasma of Atlantic Salmon. To the best of our knowledge, this report is the only use of sample displacement in affinity chromatography mode. This group demonstrated that a multi-column system containing 10 units can be effectively applied for the isolation of highly pure thrombin. As shown in Figure 7, their approach results in a much higher purity and yield of isolated thrombin than the previously used gradient elution method [62].
Figure 7.
Preparative purification of salmon thrombin by SDC on Heparin Sepharose. SDS-PAGE analysis of eluted fractions (lanes 1–10, lines A – standard proteins).
a – Fractions obtained by stepwise elution from the single column;
b – Fractions eluted from a 10 column system.
Adapted from Reference [50] with permission.
Heparin affinity chromatography and other affinity and pseudo-affinity chromatographic methods (e.g. Cibachrom-blue chromatography) are frequently applied to the separation of complex biological mixtures. Cibachrom-blue is frequently used as an alternative to mAb for albumin removal from human plasma [57]. Immobilized heparin is frequently applied in plasma fractionation and for isolation of recombinant therapeutic proteins [34, 35, 37, 63], and SDC in affinity mode, as presented by Manseth et al. [50], can be a valuable alternative for further optimization.
4. Conclusions
-
-
Sample displacement chromatography is a very effective method for the separation of complex mixtures containing proteins and peptides.
-
-
SDC can be performed in reversed-phase, hydrophobic interaction, ion-exchange and affinity mode in small as well as preparative scale (see Table 1).
-
-
When applied to the separation of protein mixtures prior to proteolytic digestion as well as for the fractionation of digested peptides prior to MS analysis, SDC in analytical and micro-analytical scale opens new perspectives for sample preparation in proteomics.
Table 1.
Overview of publications dealing with sample displacement chromatography of peptides and proteins.
| Sample | Mode | Scale | Support & number of columns | Reference |
|---|---|---|---|---|
| Peptide mix. | RP | Analytical | Bulk, one and two columns | 41,42 |
| Peptide mix. | RP | Analytical, semiprep. | Bulk, multicolumn system | 47 |
| Proteins | AE | Analytical | Bulk, one column | 43 |
| Proteins (troponin) | RP | Analytical, preparative | Bulk, system containing different columns | 40 |
| Proteins | CE | Analytical | Bulk, screening in 96 vials | 56 |
| Proteins (mAb) | AE&CE | Analytical, preparative. | Membranes, one | 44 |
| Proteins | AE&CE | Analytical, preparative | Monoliths, one | 46,60 |
| Proteins | AE&CE | Analytical | Bulk, multicolumn-system | 45, 48, 49 |
| Proteins | HIC | Analytical | Bulk, multicolumn-system | 45, 48, 49, 51 |
| Proteins | Affinity | Preparative | Bulk, multicolumn system | 50 |
Highlights
Sample displacement chromatography is a method for separation of complex mixtures.
This method can be performed in different chromatographic modes.
SDC in analytical and micro-analytical scale opens new perspectives in proteomics.
In preparative scale, SDC is a method for isolation of low abundance proteins.
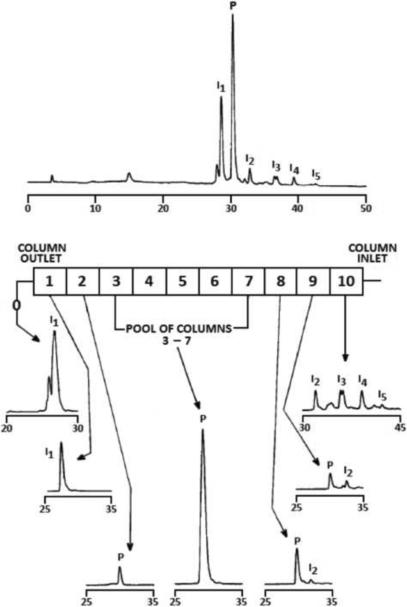
Figure 3.
Chromatograms of the peptide mixture and separated components
Upper part – RPC of the peptide mixture before separation
Lower part – RPC of fractions eluted from individual columns.
Adapted from Reference [40] with permission.
Acknowledgements
This work was supported by National Institutes of Health (NIH), Centers for Biochemical Research Excellence (COBRE), grant No. P20RR017695 and NIH grant No. 1S10RR025623-01 (Dj. Josic), the National Science Foundation (NSF) Experimental Program to Stimulate Competitive Research (EPSCoR), grant No. 1004057 and NIH grant No. 1S10RR020923 (J. Clifton), and the National Science Foundation of Republic of Croatia (Dj. Josic and M. Srajer Gajdosik). M. Srajer Gajdosik was also supported by Fulbright Scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Cravatt BF, Simon GM, Yattes JR., 3rd Nature. 2007;450:991. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- [2].Schiess R, Wollscheid B, Aebersold R. Mol. Oncol. 2009;3:33. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rucevic M, Hixson D, Josic Dj. Electrophoresis. 2011;32:1549. doi: 10.1002/elps.201100212. [DOI] [PubMed] [Google Scholar]
- [4].Surinova S, Schiess R, Hüttenhein R, Cerciello F, Wollscheid B, Aebersold R. J. Proteome Res. 2011;10:5. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- [5].Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, Clauser KR, Shen D, Lewis GD, Farrell LA, Fifer MA, Sabatine MS, Gerszten RE, Carr SA. Nature Biotechnol. 2011;29:639. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, Sokal I, Yan P, Schoenherr RM, Zhao L, Voytovich UJ, Kelly-Spratt KS, Krasnoselsky A, Gafken PR, Hogan JM, Jones LA, Wang P, Amon L, Chodosh LA, Nelson PS, McIntosh MW, Kemp CJ, Paulovich AG. Nature Biotechnol. 2011;29:625. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Qiu J, Choi G, Li L, Wang H, Potteri SJ, Ferreira-Foca SR, Krasnoselsky AL, Randolph TW, Omenn GS, Edelstein C, Barnett MJ, Thornquist MD, Goodman GE, Brenner DE, Feng Z, Hanash SM. J. Clin. Oncol. 2008;26:5060. doi: 10.1200/JCO.2008.16.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mitchell P. Nature Biotechnol. 2010;28:665. doi: 10.1038/nbt0710-665. [DOI] [PubMed] [Google Scholar]
- [9].Poste G. Nature. 2011;469:156. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- [10].Brower V. Nature. 2011;471:S19. doi: 10.1038/471S19a. [DOI] [PubMed] [Google Scholar]
- [11].Anderson NL, Anderson ND. Mol. Cell. Proteomics. 2002;1:845. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- [12].Nice EC, Rothacker J, Weistock J, Lim L, Catimel B. J. Chromatogr. A. 2007;1168:190. doi: 10.1016/j.chroma.2007.06.015. [DOI] [PubMed] [Google Scholar]
- [13].Clifton J, Huang F, Rucevic M, Cao L, Hixson D, Josic Dj. J. Proteomics. 2011;74:935. doi: 10.1016/j.jprot.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bandow JE. Proteomics. 2010;10:1416. doi: 10.1002/pmic.200900431. [DOI] [PubMed] [Google Scholar]
- [15].Pernemalm M, Ore LM, Lengqvist J, Wilkström P, Lewensohn R, Lehtiö J. J. Proteome Res. 2008;7:2712. doi: 10.1021/pr700821k. [DOI] [PubMed] [Google Scholar]
- [16].Josic Dj., Breen L. Proc. 5th Summer School “Mass Spectrometry in Biotechnology and Medicine”; L 13, Dubrovnik, Croatia. 2011. [Google Scholar]
- [17].Evans ST, Holstein M, Cramer SM. Anal. Chem. 2011;83:4184. doi: 10.1021/ac200486e. [DOI] [PubMed] [Google Scholar]
- [18].Schlüter H. Proc. 5th Summer School “Mass Spectrometry in Biotechnology and Medicine”; L 26, Dubrovnik, Croatia. 2011. [Google Scholar]
- [19].Wilkins JW, Xiang R, Horváth Cs. Anal. Chem. 2002;74:3933. doi: 10.1021/ac025752l. [DOI] [PubMed] [Google Scholar]
- [20].Ahrends R, Lichtner B, Bertsch A, Kohlbacher O, Hildebrand D, Trusch M, Schlüter H. J. Chromatogr. A. 2010;1217:3321. doi: 10.1016/j.chroma.2009.10.028. [DOI] [PubMed] [Google Scholar]
- [21].Jungbauer A. J. Chromatogr. A. 2005;1065:3. doi: 10.1016/j.chroma.2004.08.162. [DOI] [PubMed] [Google Scholar]
- [22].Yang Y, Boysen RI, Harris SJ, Hearn MTW. J. Chromatogr. A. 2009;1216:3767. doi: 10.1016/j.chroma.2009.02.059. [DOI] [PubMed] [Google Scholar]
- [23].Jungbauer A, Kaar W, Schlegl R. Curr. Opin. Biotechnol. 2004;15:487. doi: 10.1016/j.copbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- [24].Gallant SR, Cramer SM. J. Chromatogr. A. 1997;771:9. [Google Scholar]
- [25].Jungbauer A, Hahn R. J. Chromatogr. A. 2008;1184:62. doi: 10.1016/j.chroma.2007.12.087. [DOI] [PubMed] [Google Scholar]
- [26].Müller E, Josic Dj., Schröder T, Moosmann A. J. Chromatogr. A. 2010;1217:4696. doi: 10.1016/j.chroma.2010.05.016. [DOI] [PubMed] [Google Scholar]
- [27].Morrison J, Gagon P, Cramer SM. J. Chromatogr. A. 2010;1217:6484. doi: 10.1016/j.chroma.2010.08.038. [DOI] [PubMed] [Google Scholar]
- [28].Valaya A, Horváth Cs. J. Chromatogr. A. 1998;829:1. doi: 10.1016/s0021-9673(98)00727-4. [DOI] [PubMed] [Google Scholar]
- [29].Kundu A, Barnthouse KA, Cramer SM. Biotechnol. Bioeng. 1997;56:119. doi: 10.1002/(SICI)1097-0290(19971020)56:2<119::AID-BIT1>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- [30].Tiselius A. Ark. Kemi. Mineral. Geol. 1943;16A:1. [Google Scholar]
- [31].Buchanan DC. J. Biol. Chem. 1957;229:211. [PubMed] [Google Scholar]
- [32].Horváth Cs., Nahum A, Frenz J. J. Chromatogr. 1981;218:365. [Google Scholar]
- [33].Shukla AA, Sunasara KM, Rupp RG, Cramer SM. Biotechnol. Bioeng. 2000;68:672. doi: 10.1002/(sici)1097-0290(20000620)68:6<672::aid-bit11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- [34].Burnouf T. Transfusion Med. Rev. 2007;21:101. doi: 10.1016/j.tmrv.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gaso-Sokac D, Kovac S, Clifton J, Josic Dj. Electrophoresis. 2011;32:1104. doi: 10.1002/elps.201000641. [DOI] [PubMed] [Google Scholar]
- [36].Buchacher A, Iberer G. Biotechnol. J. 2006;1:148. doi: 10.1002/biot.200500037. [DOI] [PubMed] [Google Scholar]
- [37].Josic Dj., Hoffer L, Buchacher A. J. Chromatogr. B. 2003;790:183. doi: 10.1016/s1570-0232(03)00082-5. [DOI] [PubMed] [Google Scholar]
- [38].Cohn EJ, Gurd FRN, Surgenor DM, Barnes BA, Brown RK, Derouaux G, Gillespie JM, Kahnt FW, Lever WF, Liu CH, Mittelman D, Mouton RF, Schmid K, Uroma E. J. Am. Chem. Soc. 1950;72:465. [Google Scholar]
- [39].Hanson M, Unger KK, Mant CT, Hodges RS. Trends Anal. Chem. 1996;15:102. [Google Scholar]
- [40].Husband DL, Mant CT, Hodges RS. J. Chromatogr. A. 2000;893:81. doi: 10.1016/s0021-9673(00)00751-2. [DOI] [PubMed] [Google Scholar]
- [41].Lorne Burke TW, Mant CT, Hodges RS. J. Liq. Chromatogr. 1988;11:1229. [Google Scholar]
- [42].Hodges RS, Burke TWL, Mant CT. J. Chromatogr.A. 1988;444:349. doi: 10.1016/s0021-9673(01)94036-1. [DOI] [PubMed] [Google Scholar]
- [43].Veeraragavan K, Bernier A, Braendli E. J. Chromatogr. A. 1991;541:207. [Google Scholar]
- [44].Brgles M, Clifton J, Walsh R, Huang F, Rucevic M, Cao L, Hixson D, Müller E, Josic Dj. J. Chromatogr. A. 2011;1218:2389. doi: 10.1016/j.chroma.2010.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brown A, Bill J, Tully T, Radhamohan A, Dowd C. Biotechnol. Appl. Biochem. 2010;56:59. doi: 10.1042/BA20090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Josic Dj. Presented at Plasma Protein Biotechnology 2011; Paphos, Cyprus, L 506. http://www.bo-conf.com/ppb11/ [Google Scholar]
- [47].Hodges RT, Lorne Burke TW, Mant CT. J. Chromatogr. 1991;548:267. doi: 10.1016/s0021-9673(01)88608-8. [DOI] [PubMed] [Google Scholar]
- [48].Agner E. Method for displacement chromatography. U.S. Patent 6,576,134. 2003
- [49].Agner E. Purification of peptides and oligonucleotides by sample displacement chromatography process and apparatus. U.S. Patent 6,245,238. 2001
- [50].Manseth E, Skjervold PO, Flengsrud R. J. Biochem. Biophys. Methods. 2004;60:39. doi: 10.1016/j.jbbm.2004.04.016. [DOI] [PubMed] [Google Scholar]
- [51].Josic Dj. Proc. 7th HIC/RPC Bioseparation Conference; Estoril, Portugal. 2011. [Google Scholar]
- [52].Jungbauer A. Biotechnol. J. 2011;6:771. doi: 10.1002/biot.201100224. [DOI] [PubMed] [Google Scholar]
- [53].Mant CT, Hodges RS. J. Chromatogr. A. 2002;972:101. doi: 10.1016/s0021-9673(02)01079-8. [DOI] [PubMed] [Google Scholar]
- [54].Xia F, Nagrath D, Garde S, Cramer SM. Biotechnol. Bioeng. 2004;87:354. doi: 10.1002/bit.20120. [DOI] [PubMed] [Google Scholar]
- [55].Senczuk AM, Klinke R, Arakawa T, Vedantham G, Yingzaw Y. Biotechnol. Bioeng. 2009;103:930. doi: 10.1002/bit.22313. [DOI] [PubMed] [Google Scholar]
- [56].Thiemann J, Jankowski J, Rykl J, Kurzawski S, Pohl T, Wittmann-Liebold B, Schlüter H. J. Chromatogr. A. 2004;1043:73. doi: 10.1016/j.chroma.2004.05.074. [DOI] [PubMed] [Google Scholar]
- [57].Cerk-Petric T, Brne P, Gabor B, Govednik L, Barut M, Strancar A, Zupancic Kralj L. J. Pharm. Biomed. Analysis. 2007;43:243. doi: 10.1016/j.jpba.2006.06.019. [DOI] [PubMed] [Google Scholar]
- [58].Pucić M, Knezević A, Vidić J, Adamczyk B, Novokmet M, Polasek O, Gornik O, Supraha-Goreta S, Wormald MR, Redzić I, Campbell H, Wright A, Hastie ND, Wilson JF, Rudan I, Wuhrer M, Rudd PM, Josić Dj., Lauc G. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.010090. mcp.M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Josic Dj., Strancar A. Ind. Eng. Chem. Res. 1999;38:333. [Google Scholar]
- [60].Lim Y-P, Josic Dj., Hixson D. Preparation and composition of inter-alpha inhibitor proteins from humn plasma for therapeutic use. US Patent 7,932,365. 2011 Apr;
- [61].Josic Dj., Hoffer L, Buchacher A, Schwinn H, Frenzel W, Biesert L, Klöcking H-P, Hellstern P, Rokicka-Milewska R, Klukowska A. Thromb. Res. 2000;100:433. doi: 10.1016/s0049-3848(00)00339-x. [DOI] [PubMed] [Google Scholar]
- [62].Manseth E, Skjervpld PO, Fjœra SO, Brosstad FR, Ødegaard, Flengsrud R. J. Food Sci. 2003;68:1648. [Google Scholar]
- [63].Aizawa P, Winge S, Karlsson G. Thromb. Res. 2008;122:560. doi: 10.1016/j.thromres.2007.12.027. [DOI] [PubMed] [Google Scholar]