Abstract
The derivation of human embryonic stem cells and subsequently human induced pluripotent stem cells (iPSCs) has energized regenerative medicine research and enabled seemingly limitless applications. Although small animal models, such as mouse models, have played an important role in the progression of the field, typically, they are poor representations of the human disease phenotype. As an alternative, large animal models should be explored as a potentially better approach for clinical translation of cellular therapies. However, only fragmented information regarding the derivation, characterization and clinical usefulness of pluripotent large animal cells is currently available. Here, we briefly review the latest advances regarding the derivation and use of large animal iPSCs.
Keywords: induced pluripotent stem cells, large animal iPS, disease modelling, bovine, canine, porcine, primate
The path to induced pluripotency
The isolation of human embryonic stem cells (ESCs) in 1998 stimulated rapid progression of the field of regenerative medicine research [1]. The ability for ESCs to divide endlessly and to differentiate into all body tissues excited researchers and the public alike. However, ESC research also has been plagued by moral and ethical concerns surrounding the use of human embryos [2]. To bypass past such controversies, scientists have derived human ESC equivalents from adult somatic cells. These novel approaches include fusion protocols involving a combination of a pluripotent donor cell or oocyte with a somatic cell [[3],[4],,[5]], methods based on pluripotent cell extracts [[6],[7],,[8]], and somatic cell nuclear transfer (SCNT) [[9],10,,11]. SCNT has been successful in animal cloning, most notably leading to the birth of Dolly the sheep [12]. Due to its need for large numbers of human oocytes, SCNT has been deemed an unethical and unsustainable method of reprogramming by many [13]. Ultimately, it was through the seminal work of Takahashi and Yamanaka that a sophisticated yet simple reprogramming method was invented and implemented 2006. By up-regulating OCT4, SOX2, cMYC and KLF4, Yamanaka and colleagues showed that it is possible to ‘reprogram’ mouse and human somatic cells, effectively inducing pluripotency and leading to the derivation of iPSCs [15,16]. Through reprogramming, researchers have not only avoided the bulk of the moral and ethical problems surrounding pluripotent cell research, but have also opened the door to the possibility of patient-specific regenerative medicine. However, before iPSCs can realize their clinical potential safely, much research is still required. To that end, large animal iPSCs may help usher in a new era of patient-specific cell therapies to the clinic.
Discovery of a pan-species pluripotency network
Although Yamanaka's initial approach to reprogram murine cells in 2006 appeared to successfully result in fully pluripotent cells, the iPSCs produced were not germline competent [14]. Subsequently, Yamanaka's group was able to overcome this hurdle and generate murine iPSCs with germ line competency [15]. Around the same time, Jaenisch's group reported viable chimaeras and germline transmission from murine iPSCs, confirming Yamanaka's initial findings and setting the remarkably fast pace that has since defined the field of iPSC research [17]. Following this major milestone, murine iPSCs have been shown to go beyond chimerism and actually give rise to entire progeny through tetraploid complementation, currently the most stringent test of a cell's pluripotency [18,19,,20]. For obvious ethical reasons, this level of stringency is not an assay of human iPSCs or ESCs. Interestingly, the same four Yamanaka factors that were shown to push mouse somatic cells to a fully pluripotent state can also reprogram human somatic cells [16]. A parallel report from Thomson's group showed that while OCT4 and SOX2 were key factors, cMYC and KLF4 could be replaced by the potentially less oncogenic factors, NANOG and LIN28. This and other studies show that the basic pluripotency network is conserved across species, but may not necessarily be regulated in precisely the same way [21].
The discovery of a pan-species, potentially universal pluripotency network that is catalyzed by a common set of factors led researchers to derive iPSCs from monkeys, rats, pigs, sheep, dogs, cows and, most recently, the endangered white rhinoceros, with reprogramming of other mammalian species likely to be underway [22,23,24,25,26,27,28,29,30,31]. Although rodents have traditionally been the most common research models for studying human genetic diseases and in vivo cell therapies, the derivation of larger animal iPSCs now makes it possible to model autologous cell therapies in animal systems that more closely resemble those of the human body. Several groups have used mouse models to study, ameliorate, and, in some cases, even cure diseases, such as sickle cell anaemia [32], haemophilia [33], diabetes [34], Parkinson's disease [35] and cardiovascular diseases [36]. However, small animal models are limited in their usefulness clinically. For example, while studying heart disease in mice can provide many useful insights, the results are unlikely to be as clinically relevant as those from larger animals (e.g., dogs, pigs and primates), whose lifespan and cardiac physiology are more similar to a human's [37]. See Table for an overview of large animal iPSC derivation.
Table 1.
Large animal iPSC derivation overview
| Date and author | Parental cell source | Feeder layer | Special culture conditions | Differentiation (in vitro) | Differentiation (in vivo) | Markers | Reprogramming factors | |
|---|---|---|---|---|---|---|---|---|
| Canine iPSCs | 2009 Shimada et al. | Canine embryonic fibroblasts | MEFs | 6 ng/ml hbFGF, 1000 U/ml hLIF, 1 mM VPA, 0.5 mM PD0325901, 3 mM CHIR99021, 0.25 mM A83-01 | AFP, FLK1, Beta III-tubulin positive cells | None | AP, Oct3/4 | Retrovirus Canine OKSM |
| 2011 Lee et al. | Canine fibroblasts, Canine adipose stromal cells | MEFs | 5 ng/ml hbFGF, 1000 U/ml hLIF | Embryoid bodies, Endothelial cells | Teratomas | AP, Oct3/4, Sox2, Nanog, TRA- 1-60, and SSEA-4 | Lentivirus Human OKSM | |
| Porcine iPSCs | 2009 Ezashi et al. | Porcine foetal fibroblasts | MEFs | 4 ng/ml hbFGF | Embryoid Bodies | Teratomas | AP, SSEA-1, Oct3/4, Nanog, Sox2 | Lentivirus Human OKSM |
| 2009 Esteban et al | Tibetan miniature pig fibroblast | MEFs | 4 ng/ml hbFGF, mLIF, PD0325901, CHIR99021, 39°C | None | Teratomas | AP, Nanog, Sox2, Klf4, Oct3/4, Rex1, Lin28, SSEA4 | Retrovirus Mouse and Human OKSM | |
| 2009 Wu et al. | Primary ear fibroblast and primary bone marrow cells | MEFs | DMEM/F12 + 20% KSR + DOX | Embryoid bodies | Teratomas | AP, SSEA3, SSEA4, Tra-1-60, Tra-1-81, Oct3/4, Nanog, Sox2, Rex1 and CDH1. | DOX inducible Lentivirus Human OKSM + Nanog, Lin28 | |
| 2010 West et al. | Porcine mesenchymal stem cells | 1. MEFs 2. Matrigel | 1. DMEM + 20% KSR + 10 ng/ml hbFGF 2. mTeSR1 | Embryoid bodies | Chimeric offspring | AP, Oct3/4, Sox2, | Lentivirus Human OKSM + NANOG, LIN28 | |
| 2010 Montserrat et al. | Pig ear fibroblasts | MEFs or Gelatin only | hES/mES media (1:1) 10 ng/ml hbFGF, 1000 U/ml LIF | Embryoid bodies | Teratomas | Nanog, SSEA-4, TRA-1-60 | Retrovirus pMXs–OKSM–GFP | |
| Sheep iPSCs | 2010 Li et al. | Sheep foetus fibroblast | MEFs | FBS instead of KSR | Embryoid bodies | Teratomas | AP, Oct3/4, Sox2, Nanog, SSEA-4 | DOX inducible lentivirus Mouse OKSM |
| Boving iPSCs | 2011 Sumer et al. | Bovine adult fibroblast | MEFs | 10 ng/ml bovine bFGF, 4 ng/ml hLIF, 39°C | Embryoid bodies | Teratomas | AP, Rex1, Oxt3/4, Sox2, Nanog, c-Myc, Klf4, SSEA1, SSEA4 | Retrovirus OKSM + Nanog |
| Non-human primate iPSCs | 2008 Liu et al. | Adult rhesus macaque ear fibroblast | MEFs | KSR medium | Embryoid bodies | Teratomas | AP, Nanog, Oct3/4, Sox2, SSEA4, TRA-1-60, TRA-1-81 | Retrovirus Monkey OKSM |
| 2010 Zhong et al. | Macaca nemestrina oral fibroblasts | MEFs | 16 ng/ml hbFGF, 0.5 mM PD0325901, 2 mM VPA, and 10 mM Y-27632 | Embryoid bodies Neural precursors, Cardiomyocytes, Hepatocytes | Teratomas | Oct3/4, SSEA3, SSEA4, TRA-1-60, TRA-1-81, Nanog | Retrovirus OKSM | |
| 2011 Deleidi et al. | Cynomolgus macaque skin fibroblasts | MEFs | KSR medium | Neurons | Teratomas | AP, Nanog, Oct3/4, Sox2, Klf4, cMyc, TRA-1-60, TRA-1-81, SSEA4 | Retrovirus hOKSM |
AP: alkaline phosphatase; bFGF: basic fibroblast growth factor; FBS: foetal bovine serum; KSR: knockout serum replacement; LIF: leukaemia inhibitory factor; MEF: mouse embryonic fibroblasts; VPA: valproic acid; MEF: mouse embryonic fibroblast.
Animal iPSCs and disease modelling
The mouse heart beats over twice as fast as the human heart and is therefore under different shear stresses from the human heart and vasculature [37]. Furthermore, the relatively short lifespan of mice (typically <2 years) is often a key limiting factor, and studies have shown that mouse cardiomyocytes are fundamentally different in design, reducing the relevancy of human heart therapies tested on them [38]. In part to address this need for large animal in vivo studies of cardiac disease and injury, we derived canine iPSCs from canine fibroblasts and canine adipose stromal cells (Fig. 1). We then transplanted autologous iPSCs into the same animal and followed fate of transplanted iPSCs using positron emission tomography reporter gene imaging and iron oxide labelling by magnetic resonance imaging [30]. As anticipated, transplanting iPSCs in a large animal model was a significant challenge. However, these cells did demonstrate therapeutic potential while shedding light on the specific hurdles of large animal iPSC transplantation, namely the difficulties involved in in vivo imaging. Undoubtedly, further studies will be required to further optimize both the imaging protocols and iPSC biology to allow successful translation of pluripotent stem cell based therapies to human patients in the future.
Fig 1.
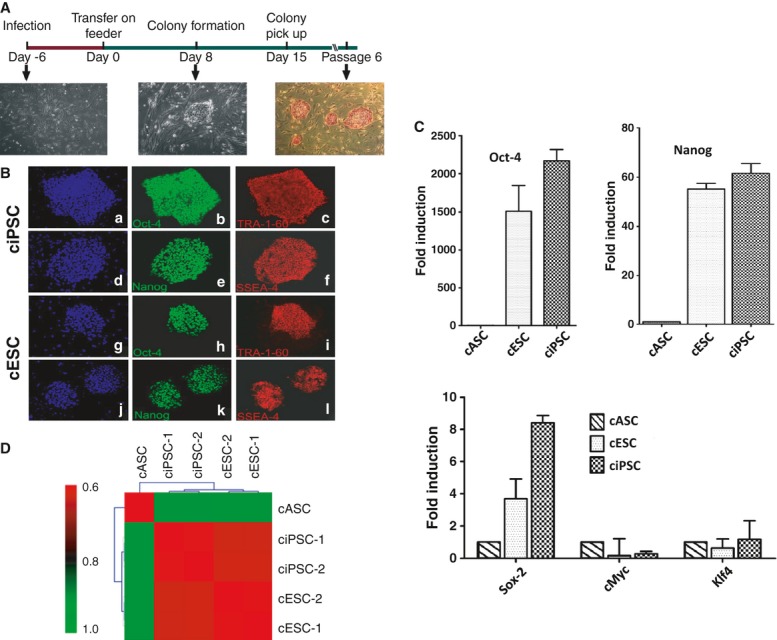
Generation of canine induced pluripotent stem cells (ciPSCs). (A) Schematic diagram of the generation of ciPSCs. ciPSC colonies can be picked out approximately 12–15 days and are alkaline phosphatase-positive. (B) Immunofluorescence staining of pluripotent markers. Similar to cESCs, ciPSCs are positive for pluripotent stem cell markers Oct-4 (b and h), Tra-1-60 (c and i), Nanog (e and k) and SSEA-4 (f and l), with nuclear staining by DAPI (a, d, g and j). (C) Quantitative PCR analysis of expression of pluripotent stem cell markers Oct-4, Nanog, Sox-2, c-Myc and Klf-4. Y-axis value represents fold differences (log2) in expression of select genes. (D) Pearson correlation analysis for gene expression in cASCs, ciPSCs and cESCs using transcripts with SD <0.2 among all samples (17,895 probes, P < 1.0E-15). Reprinted with permission from Lee et al. 2011.
Primates are arguably the best large animal model for comparison with human disease phenotypes. Although both primate ESCs and iPSCs have been previously derived, the use of primates for transplantation experiments remains controversial [39]. In fact, many groups are simply using large animal iPSCs for transplantation in the more traditional mouse model. Zhu et al. reported the generation of insulin-producing pancreatic cells from rhesus monkey iPSCs in 2011, yet only tested their efficacy in a diabetic mouse model 2011. Similarly, Zhong et al. used genetically modified non-human primate iPSCs in a SCID mouse model to show that suicide genes can be conditionally activated to eliminate pluripotent cells in vivo, while leaving the pluripotent state and health of the host unaffected 2011. Although such studies provide clues to how iPSCs might behave following transplantation in humans, transplantation of autologous iPSCs or their derivatives using large animal models would be more insightful by providing better pre-clinical safety data needed to progress towards human clinical trials.
Due to cultural sensitivities, the use of primates and canines as disease research models has generated controversy. The porcine model is less afflicted by the same problems and may prove to be an easier large animal model for pre-clinical iPSC transplantation. Having the appropriate organ size, physiology and lifespan, but with greater ease and availability compared with non-human primates, pigs can serve as a powerful pre-clinical research tool. Porcine iPSCs can be matched to specific pigs, making them essentially ‘patient-specific’, and can be differentiated into specific lineages. These qualities make it possible to use pigs to test transplantation therapies with iPSCs for safety and efficacy before applying the procedures to human patients. In addition to these favourable characteristics of the porcine model, pigs have a long history of medical and therapeutic use such as their role in providing replacement cardiac valves and insulin [42]. Moreover, in terms of pluripotency, reports of porcine ESCs and iPSCs indicate that they may be more similar to human ESCs and iPSCs than their murine counterparts. Both are stereotypically flat with clearly identifiable nuclei and nucleoli and have a high nucleus-to-cytoplasm ratio, comparable colony morphology, dependence on basic fibroblast growth factor (bFGF), and typically express stage specific embryonic antigen 4 (SSEA-4) on their surface [24,25,27,43,44]. Unlike pig ESCs, mouse pluripotent stem cells exhibit a more ‘rounded up’ morphology, require leukaemia inhibitory factor (LIF) to maintain pluripotency, and express SSEA-1 on their surface, an early marker of differentiation on human ESCs and iPSCs [45,46]. These differences are likely tied to the differing pathways that govern pluripotency and self-renewal between mouse and human ESCs. In the mouse, the Janus kinase and signal transducer and activator of transcription (JAK-STAT) pathway, regulated by addition of LIF, and bone morphogenetic proteins, found in serum, are the dominant signalling pathways. By comparison, in human the MEK-ERK pathway is activated by bFGF and transforming growth factor beta signalling is more crucial to maintenance of pluripotency [47]. Although no animal model can fully recapitulate the human disease phenotype, greater effort should be made to ensure that the best models are used for basic research to obtain the most relevant and insightful data while limiting animal use.
Issues with large animal iPSCs
Although human and mouse ESCs and iPSCs are well defined, most other animal models have suffered from a general lack of reliable iPSC and ESC markers. For instance, reports of canine pluripotent cell derivation have shown that the surface marker expression (such as SSEA-1, SSEA-3, SSEA-4, TRA-1-81 and TRA-1-60) varies, despite the common expression of OCT4, SOX2 and NANOG [48,49,,50]. Ugulates, such as pig and cow, also show inconsistencies in surface marker expression [51], and there is a further complication in that porcine and bovine blastocysts show expression of primary pluripotency genes and surface markers, such as OCT4 and SSEA-4, in both the inner cell mass (ICM) and the trophectoderm. Furthermore, as recently reported by[24] the same process which results in pig iPSCs also produces the by-product of trophectoderm-like cells. Like iPSCs, these trophectoderm-like cells can grow seemingly limitlessly in iPSC culture conditions, have high expression of telomerase and a subset of pluripotency genes, making them difficult to distinguish from iPSCs following reprogramming 2011. In addition to problems with characterization, multiple groups have shown that continuous passaging of human ESCs and iPSCs frequently results in chromosomal abnormalities, sometimes even within as few as 20 passages. This last finding suggests that long-term culture of large animal iPSCs may result in similar abnormalities, and therefore should be monitored carefully for culture-induced genetic changes [53,54,,55]. In addition, reports also differ on what surface markers porcine iPSCs may express. Although SSEA-1 is clearly associated with pluripotency in murine cells, it has been shown to be an early marker of differentiation in pluripotent human cells. Interestingly, with ungulates such as pigs and cows, SSEA-1 expression varies. In the bovine blastocyst, SSEA-1 and SSEA-4 are expressed on both the inner cell mass, from which ESCs are derived, as well as the trophectoderm cells. Similarly, pig ESCs have been reported as SSEA-1 positive and SSEA-4 negative 2009; however, another group reported contradictory results of SSEA-4 positive and SSEA-1 negative pig iPSCs [56]. The key may lie in the differences in epiblast development, with different groups reprogramming cells towards different points in development, hence requiring different culture conditions and displaying varying marker profiles.
Conclusions
Despite the rapid progress of the field, iPSCs are difficult to derive from most large animals and there is a general lack of effective reprogramming protocols. Furthermore, more work is needed to develop reliable differentiation protocols capable of becoming different lineages such as neuronal, cardiac, endothelial and hepatic cells. Although no animal study can truly compare with a human study, every effort should be made to ensure that the model system is as close to the human system as possible, particularly when translational medical research is the goal, which is often the case. With this in mind, large animal models should and can play a more significant role in translational research, but they are often overlooked due to difficulty and/or cost. This is especially true with primates because, although they are arguably the most comparable to humans, they typically require special transport, handling and care. Other large animals such as dogs, sheep and pigs offer many of the same benefits, but are simpler in terms of acquisition and care. However, the various differences in pluripotency networks among species remain a hurdle, making maintenance of large animal iPSCs challenging. Furthermore, it has been our experience that imaging and tracking cells following transplantation is quite difficult in large animals, but since this issue is analogous to humans, it will continue to be an important area to focus on in the future. Following further elucidation of the mechanisms of reprogramming, and improvements in iPSC derivation techniques, new methods to simplify and facilitate characterization of iPSC lines will become possible in the future. New technologies, including non-integrating, xeno-free reprogramming strategies and genome-wide epigenetic profiling, will make further progress possible in this exciting field.
Acknowledgments
Due to space limitations, we are unable to include all of the important papers relevant to large animal induced pluripotent stem cell derivation and application; we apologize to those investigators whom we omitted here. This study was supported by the NIH HL089027 and NIH EB009689 (JCW), NIH RC1 HL100490 (MTL) and T32 Training Grant (JRP).
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Munn D. Moral issues of human embryo research. Science. 2001;293:211. doi: 10.1126/science.293.5528.211b. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, et al. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–73. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Tada T. Cell fusion-mediated nuclear reprogramming of somatic cells. Reprod Biomed Online. 2008;16:51–6. doi: 10.1016/s1472-6483(10)60556-1. [DOI] [PubMed] [Google Scholar]
- Flasza M, Shering AF, Smith K, et al. Reprogramming in inter-species embryonal carcinoma-somatic cell hybrids induces expression of pluripotency and differentiation markers. Cloning Stem Cells. 2003;5:339–54. doi: 10.1089/153623003772032844. [DOI] [PubMed] [Google Scholar]
- Hakelien AM, Gaustad KG, Collas P. Modulation of cell fate using nuclear and cytoplasmic extracts. Methods Mol Biol. 2006;325:99–114. doi: 10.1385/1-59745-005-7:99. [DOI] [PubMed] [Google Scholar]
- Collas P, Taranger CK. Epigenetic reprogramming of nuclei using cell extracts. Stem Cell Rev. 2006;2:309–17. doi: 10.1007/BF02698058. [DOI] [PubMed] [Google Scholar]
- Taranger CK, Noer A, Sorensen AL, et al. Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol Biol Cell. 2005;16:5719–35. doi: 10.1091/mbc.E05-06-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu X, Wang H, et al. Human embryos derived by somatic cell nuclear transfer using an alternative enucleation approach. Cloning Stem Cells. 2009;11:39–50. doi: 10.1089/clo.2008.0041. [DOI] [PubMed] [Google Scholar]
- Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc Natl Acad Sci USA. 1952;38:455–63. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–40. [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, et al. Viable offspring derived from foetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Levens ED, DeCherney AH. Human oocyte research: the ethics of donation and donor protection. JAMA. 2008;300:2174–6. doi: 10.1001/jama.2008.601. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, et al. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–8. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–4. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Han X, Han J, Ding F, et al. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 2011;21:1509–12. doi: 10.1038/cr.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Cui C, Chen S, et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–5. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Telugu BP, Alexenko AP, et al. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci USA. 2009;106:10993–8. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Xu J, Yang J, et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634–40. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Trobridge GD, Zhang X, et al. Efficient generation of nonhuman primate induced pluripotent stem cells. Stem Cells Dev. 2011;20:795–807. doi: 10.1089/scd.2010.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West FD, Terlouw SL, Kwon DJ, et al. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211–20. doi: 10.1089/scd.2009.0458. [DOI] [PubMed] [Google Scholar]
- Li Y, Cang M, Lee AS, et al. Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS ONE. 2011;6:e15947. doi: 10.1371/journal.pone.0015947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Nakada A, Hashimoto Y, et al. Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol Reprod Dev. 2010;77:2. doi: 10.1002/mrd.21117. [DOI] [PubMed] [Google Scholar]
- Lee AS, Xu D, Plews JR, et al. Preclinical derivation and imaging of autologously transplanted canine induced pluripotent stem cells. J Biol Chem. 2011;286:32697–704. doi: 10.1074/jbc.M111.235739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich Ben-Nun I, Montague SC, Houck ML, et al. Induced pluripotent stem cells from highly endangered species. Nat Methods. 2011;8:829–31. doi: 10.1038/nmeth.1706. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Xu D, Alipio Z, Fink LM, et al. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci USA. 2009;106:808–13. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipio Z, Liao W, Roemer EJ, et al. Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. Proc Natl Acad Sci USA. 2010;107:13426–31. doi: 10.1073/pnas.1007884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the foetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci USA. 2008;105:5856–61. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh K, Narsinh KH, Wu JC, et al. Derivation of human induced pluripotent stem cells for cardiovascular disease modelling. Circ Res. 2011;108:1146–56. doi: 10.1161/CIRCRESAHA.111.240374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, Ferrara DE, Sorescu D, et al. Hemodynamic shear stresses in mouse aortas: implications for atherogenesis. Arterioscler Thromb Vasc Biol. 2007;27:346–51. doi: 10.1161/01.ATV.0000253492.45717.46. [DOI] [PubMed] [Google Scholar]
- Lowey S, Lesko LM, Rovner AS, et al. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an alpha- or beta-myosin heavy chain backbone. J Biol Chem. 2008;283:20579–89. doi: 10.1074/jbc.M800554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu F, Yong J, et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–90. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Zhu FF, Zhang PB, Zhang DH, et al. Generation of pancreatic insulin-producing cells from rhesus monkey induced pluripotent stem cells. Diabetologia. 2011;54:2325–36. doi: 10.1007/s00125-011-2246-x. [DOI] [PubMed] [Google Scholar]
- Zhong B, Watts KL, Gori JL, et al. Safeguarding nonhuman primate iPS cells with suicide genes. Mol Ther. 2011;19:1667–75. doi: 10.1038/mt.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall V. Porcine embryonic stem cells: a possible source for cell replacement therapy. Stem Cell Rev. 2008;4:275–82. doi: 10.1007/s12015-008-9040-2. [DOI] [PubMed] [Google Scholar]
- Chen LR, Shiue YL, Bertolini L, et al. Establishment of pluripotent cell lines from porcine preimplantation embryos. Theriogenology. 1999;52:195–212. doi: 10.1016/S0093-691X(99)00122-3. [DOI] [PubMed] [Google Scholar]
- Li M, Zhang D, Hou Y, et al. Isolation and culture of embryonic stem cells from porcine blastocysts. Mol Reprod Dev. 2003;65:429–34. doi: 10.1002/mrd.10301. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–92. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Hayes B, Fagerlie SR, Ramakrishnan A, et al. Derivation, characterization, and in vitro differentiation of canine embryonic stem cells. Stem Cells. 2008;26:465–73. doi: 10.1634/stemcells.2007-0640. [DOI] [PubMed] [Google Scholar]
- Hatoya S, Torii R, Kondo Y, et al. Isolation and characterization of embryonic stem-like cells from canine blastocysts. Mol Reprod Dev. 2006;73:298–305. doi: 10.1002/mrd.20392. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Adler H, Braun J, et al. Canine embryo-derived stem cells–toward clinically relevant animal models for evaluating efficacy and safety of cell therapies. Stem Cells. 2007;25:1850–1. doi: 10.1634/stemcells.2006-0357. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Telugu BP, Ezashi T. Induced pluripotent stem cells from swine (Sus scrofa): why they may prove to be important. Cell Cycle. 2009;8:3078–81. doi: 10.4161/cc.8.19.9589. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Matsuyama H, Telugu BP, et al. Generation of colonies of induced trophoblast cells during standard reprogramming of porcine fibroblasts to induced pluripotent stem cells. Biol Reprod. 2011;85:779–87. doi: 10.1095/biolreprod.111.092809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DE, Harrison NJ, Maltby E, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–15. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Draper JS, Smith K, Gokhale P, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–4. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- Aasen T, Raya A, Barrero MJ, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–84. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Wu Z, Chen J, Ren J, et al. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1:46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]


