Abstract
Osteoarthritis (OA) is a degenerative disease of articular cartilage that may be associated with a loss of glycosaminoglycans (GAG). Quantitative sodium MRI is highly specific to GAG content and could be used to assess the biochemical degradation of cartilage in early OA. However, the reproducibility and repeatability of this technique are not well documented. The aim of this study is to test the repeatability and reproducibility of sodium quantification in cartilage in vivo using intra- and inter-day acquisitions at 3T and 7T, with a 3D radial sequence, with and without fluid suppression. Fluid suppression was obtained by adiabatic inversion recovery (IR WURST), and is expected to improve the sensitivity of the method to GAG content. The root mean square of coefficients of variation (CV) are all in the range of 7.5-13.6%. No significant inter-magnet, inter-sequence, intra-day and inter-day differences in the CV were observed. Sodium quantification using IR WURST gave values closer to those reported in the literature for healthy cartilage (220-310 mM) than 3D radial. In conclusion, IR WURST was more accurate in context of sodium measurement, with a reproducibility and repeatability comparable to other compositional MRI techniques of cartilage.
Keywords: Osteoarthritis, Sodium MRI, Reproducibility, Repeatability
Introduction
Osteoarthritis (OA) is the most common form of arthritis in synovial joints and a leading cause of chronic disability, mainly in the elderly population. In 2008, it was estimated that nearly 27 million adults in the United States (11% of the population) had clinical OA and that $185 billion (1.3% of the Gross Domestic Product) are spent annually on medical care due to this condition. It is even predicted that by the year 2030, nearly 67 million adults (25% of the US population) will be affected by OA [1,2]. OA is a degenerative disease of articular cartilage and may be associated with a loss of glycosaminoglycans (GAG), possibly also a change in size and organization of collagen fibers, and increased water content. There is no known cure for OA and present treatments focus mainly on pain management and ultimately, joint replacement.
There are many obstacles to studying OA, including heterogeneity in etiology, variability in progression of disease, and long time periods required to see morphological and structural joint changes. Consequently, we currently lack the ability to predict the course of the disease in individual patients. Given the expense of studying large populations for 3-5 years, the limitation of identifying OA patients at risk for progression, and perhaps more importantly, the lack of available non-invasive early biochemical markers, there has been little progress in the development of potential disease modifying OA drugs (DMOAD’s). Therefore, there is a critical need to identify reliable, objective, non-invasive, and rapid quantitative imaging markers of cartilage compositional changes, which could be translated to the study of the clinical OA population.
Functional magnetic resonance imaging (MRI) techniques are under development to detect biochemical changes in cartilage such as T1ρ mapping [3], T2 mapping [4], delayed Gadolinium-enhanced MRI of cartilage (dGEMRIC) [5], GAG chemical exchange saturation transfer (gagCEST) [6], diffusion MRI [7] and sodium (23Na) MRI [8–10]. All these methods have their own advantages and disadvantages [11,12], but quantitative sodium MRI has been shown to be highly specific to the glycosaminoglycan (GAG) content in cartilage and could therefore be used as a means of detection and assessment of the degree of biochemical degradation of cartilage in very early stages of OA [8–14].
Recent technological developments such as high and ultrahigh magnetic field scanners, novel ultrashort echo time (UTE) pulse sequences, multi-channel radiofrequency (RF) arrays and non-Cartesian reconstruction methods have great potential for improving the performance of multi-nuclear imaging. Although sodium MRI is specific to the GAG content in cartilage, because of sodium’s low tissue concentration, its low nuclear magnetic resonance (NMR) sensitivity and short relaxation times, images must be acquired with specific UTE 3D sequences and long acquisition times (15-20 min in general). Recently, our group has implemented and validated a new sequence with fluid suppression by inversion recovery (IR) which is expected to be more sensitive to sodium/GAG content within cartilage only [15, 16].
The goal of the present study was to test for the first time the reproducibility and repeatability of the sodium quantification in cartilage in vivo by MRI with and without fluid suppression at both 3T and 7T.
From the National Institute of Standards and Technology (NIST, Gaithersburg, MD, USA) guidelines [17] and from the International Union of Pure and Applied Chemistry (IUPAC) Compendium of Chemical Terminology compendium [18], the terms “repeatability” and “reproducibility” can be defined as follows:
Repeatability
Closeness of the agreement between independent results obtained with the same method on identical test material under the same conditions of measurements. It is measured as the “repeatability standard deviation (SD)” or “repeatability coefficient of variation (CV)”. Repeatability conditions include: the same measurement procedure, the same observer/operator, the same measuring instrument used under the same conditions, the same location, repetition over a short period of time.
Reproducibility
Closeness of the agreement between independent results obtained with the same methodon identical test material under different conditions of measurements. It is measured as the “reproducibility standard deviation (SD)” or “reproducibility coefficient of variation (CV)”. Reproducibility conditions include changes in one of these parameters: the principle of measurement, the measurement procedure, the observer/operator, the measuring instrument, the conditions of use of the measuring instrument, the location, time.
Two sequences for sodium imaging were tested in this work: Radial 3D [19] (Abbreviation: R3D) and IR WURST [15] (Abbreviation: IRW), which is the same as Radial 3D but with fluid suppression by inversion recovery (IR) using an adiabatic WURST pulse (Wideband Uniform Rate and Smooth Truncation) [20]. The detailed aims of the present study are as follows:
- To measure the repeatability of sodium quantification:
- at each of 3T and 7T (intra-magnet).
- with each of Radial 3D and IR WURST (intra-sequence).
- To measure the reproducibility of sodium quantification:
- at 3T versus 7T (inter-magnet).
- with Radial 3D versus IR WURST (inter-sequence).
Repeatability and reproducibility will each be assessed in terms of acquisitions on the same day (intra-day) and over different days (inter-day).
Materials and Methods
Acquisition protocol
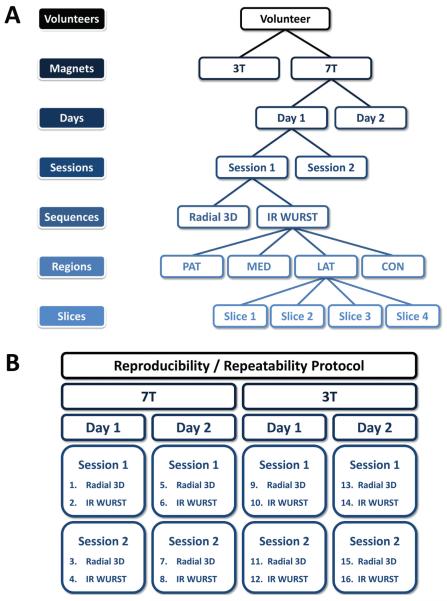
Schematics of the image data acquisition and measurement protocol are presented in Figure 1:
6 asymptomatic volunteers (3 males, 3 females), with average age 37.3±12.7 years were scanned. The study was approved by the institutional review board (IRB) and all volunteers provided informed consent prior to the experiments.
- 2 different magnets were used:
- – 3T: Tim trio (Siemens Medical Solution, Erlangen, Germany), with a transmit/receive sodium RF knee coil (transmit/receive quadrature, birdcage) single-tuned at 32.6 MHz (custom made), of length 21 cm and inner diameter 20 cm.
- – 7T: whole-body (Siemens Medical Solution, Erlangen, Germany), with a transmit/receive sodium RF knee coil (transmit/receive quadrature, birdcage) single-tuned at 78.6 MHz (Rapid MR International, Columbus), of length 27 cm and inner diameter 21 cm.
2 days/magnet: Each volunteer was scanned on each magnet on 2 different days within a week (total: 4times).
2 sessions/day: Each volunteer was scanned during 2 successive acquisition sessions separated by a short pause during which the volunteer was taken out of the center of the magnet and put back in after a brief delay, after repositioning in the center of the magnet.
2 sequences/session: Images were acquired using 2 sequences during each session, radial 3D [19] and IR WURST (radial 3D + Inversion Recovery (IR) for fluid suppression) [15]. The acquisition parameters are given in Table 1. All images were reconstructed using a Non-Uniform Fast Fourier Tranform (NUFFT) algorithm as described in [15] with a Nyquist resolution of 4 mm and interpolated to 100×100×100 voxels (2 mm nominal isotropic resolution). The sequences were written with Sequence-Tree 4.2.2 [21] and compiled within the Siemens environments IDEA VB13 (3T) and VB15 (7T).
Figure 1.
Image data acquisition and measurements protocol for each volunteer. (A) Acquisition protocol tree, (B) The 16 acquisitions obtained on each volunteer. Abbreviations: PAT=Patellar, MED=FT Medial, LAT= FT Lateral, CON=PF Condyle, with PF=Posterior Femoral, FT=Femoro-Tibial.
Table 1.
Acquisition parameters: Number of 3D radial projections (Proj.), echo time (TE), repetition time (TR), flip angle (FA), WURST adiabatic inversion pulse power and duration (WURST), inversion time (TI) and total acquisition time (TA).
| Sequence | Magnet | Proj. | TE (ms) | TR (ms) | FA (°) | WURST (Hz/ms) | TI (ms) | TA (min) |
|---|---|---|---|---|---|---|---|---|
| Radial 3D | 3T | 15,000 | 0.15 | 80 | 90 | - | - | 20 |
| 7T | 10,000 | 0.15 | 100 | 90 | - | - | 17 | |
|
| ||||||||
| IR WURST | 3T | 15,000 | 0.15 | 80 | 90 | 240/10 | 22 | 20 |
| 7T | 10,000 | 0.15 | 140 | 90 | 240/10 | 24 | 25 | |
For each volunteer, all 16 acquired data sets were co-registered using 3-D Voxel Registration in Analyze 10.0 (AnalyzeDirect Inc, Overland Park, KS, USA), which is based on the method of normalized entropy measure [22].
SNR measurements
The standard deviation of the noise (SDnoise) was measured for each volunteer for a large region of interest (ROI) over 4 consecutive slices in the sagittal plane outside of the knee, where no sodium signal is detected. This ROI was generally of the order of 40-50×40-50 voxels and positioned at the center of the slice, where the noise is more homogeneous, in order to avoid reconstruction artifacts from the 3D NUFFT (ringing in the outer part of the images). The mean signal was measured in selected ROIs of 25 voxels in 4 different regions in the cartilage over 4 consecutive slices. The cartilage regions were: patellar (abbreviation: PAT), femoro-tibial medial (MED), femoro-tibial lateral (LAT), and medial posterior femoral condyle (CON). The signal-to-noise ratio (SNR) of each voxel was calculated as the voxel signal divided by SDnoise. Mean value and SD of SNR (SDSNR) were computed as the mean and SD over the 4 consecutive slices (100 voxels) for each region of the cartilage. The coefficient of variation of each SNR measurement (CVSNR) was then calculated as .
Sodium quantification
All images were acquired with the same calibration tube phantoms that were placed on the knee cap and included in the FOV. Sodium quantitation was then calculated using linear regression in Matlab as follows: ROIs were drawn in 4 calibration phantoms (150, 200, 250 and 300 mM NaCl) and their average signal intensities were corrected for T1, and of the gels as described in [15]. Another ROI was drawn in the noise area and the mean value of the noise was used as a 0 mM sodium concentration phantom. A linear regression curve of these corrected phantom intensities and noise versus sodium concentrations was then calculated and used to extrapolate the sodium 3D maps of the whole sample.
For a better estimation of the sodium concentration [Na+] in cartilage in vivo, after the regression curve calculation from the gel signals but before extrapolation of the images to sodium maps, the images were also corrected for the T1, and of cartilage measured in vivo [23]. Note that in the resulting maps the sodium concentrations in the calibration phantoms were therefore incorrect but the cartilage ones were accurately assigned. As 75% of the volume in cartilage is extracellular and composed of water, and sodium ions are mainly present in this space, the sodium maps were divided by 0.75 in order to obtain the real sodium concentration [24, 25]. Less than 5% of the cartilage volume is composed of cells [26] and the intracellular sodium concentration, estimated around 5-10 mM, can therefore be considered negligible in the present study.
Mean values, SD and coefficients of variation of sodium concentration measurements (CVNa) for each region of the cartilage were calculated as described in the SNR measurements section (same slices, sameROIs).
Statistical analysis
Mixed model analysis of variance (ANOVA) was used to assess and compare sequences in terms of the mean SNR, mean [Na+], CVSNR and CVNa observed for each region of cartilage at each field strength. Mean SNR, mean [Na+], and CVs were used as dependent variables in separate analyses. In each case, the model included sequence (IRW vs. R3D) and imaging session (first vs. second) as fixed classification factors. For all mixed model analyses, statistical dependencies among the observations derived for the same subject were accounted for by assuming data to be symmetrically correlated when acquired from the same subject and independent otherwise. The error variance was allowed to vary across the combinations of sequence and magnet to remove the assumption of variance homogeneity. A Tukey multiple comparison correction was used so that the comparisons for each dependent variable had a familywise type I error probability of 0.05. The root mean square (RMS) of CVSNR and CVNa were calculated for each region of cartilage and each magnet-sequence combination. This RMS CV corresponds therefore to the measure of the uncertainty of the measurements and is used as an indicator of the repeatability of the method. The correlations of SNR with [Na+], and CVSNR with CVNa, were characterized using Pearson correlation coefficients and assessed for statistical significance using mixed model regression.
The CV of the intra- and inter-day component of the overall variance of the mean SNR and sodium concentrations for each magnet-sequence combination, measured in 4 regions in the cartilage were also computed; these CV estimates are denoted as , , and . Each CV is the square root of the variance of the normalized mean SNR or sodium concentration values expressed as a percentage of the overall mean SNR or sodium concentration for the relevant magnet-sequence combination. These CVs correspond therefore to the statistical variation of the results over many measurements (on the same day or over different days, with different magnet-sequence combinations) and is used as an indicator of both the repeatability and reproducibility of the method. Restricted maximum likelihood estimation (REML) of variance components in a random effects model was used to compare the combinations of sequence and magnet with respect to these coefficients of variation intra-day (variation between the 2 acquisitions made on the same day) and inter-day (using as data for each day an average over the acquisitions on that day). In each analysis, the sodium and SNR levels for each magnet-sequence combination were normalized by dividing each value by the overall average level observed for that combination. As a result, the likelihood ratio tests to compare magnet-sequence combinations in terms of the variance of the normalized data were equivalent to comparisons in terms of the CV values on the original scale of measurement. No multiple comparison correction was applied to these analyses since the intent was to establish comparability and the comparisons therefore needed to be conducted with the highest possible statistical power. The correlations for , , and between all pairs of magnet-sequence combinations (3T IRW vs 3T R3D, 3T IRW vs 7T IRW, 3T IRW vs 7T R3D, 3T R3D vs 7T IRW, 3T R3D vs 7T R3D, 7T IRW vs 7T R3D) were characterized using Pearson correlation coefficient and assessed for statistical significance using mixed model regression.
All reported p values are two sided and statistical significance is defined as p<0.05. SAS 9.3 was used for all computations (SAS Institute, Cary, NC).
Results
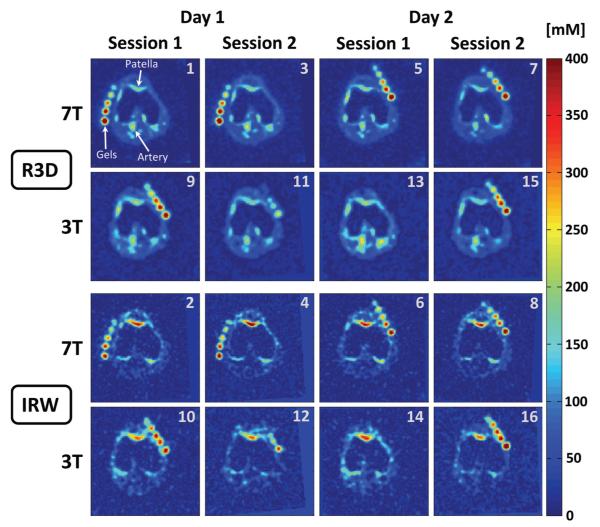
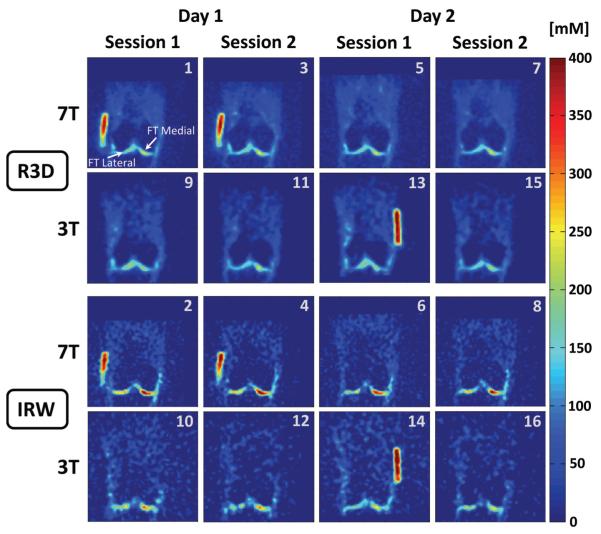
Representative examples of sodium concentration maps are shown in Figure 2 (transverse plane) and Figure 3 (coronal plane). These images were acquired on the same volunteer following the protocol described in Figure 1, and were all co-registered.
Figure 2.
Representative example of all the sodium concentration maps in the transverse plane acquired on one volunteer. All the 16 image data sets were co-registered for each volunteer. The number in the top-right corner of each image corresponds to the acquisition number in the protocol as described in Fig. 1B. Note that in some cases (3T-day 1 and 3T-day 2), the volunteer had to take a break between 2 sessions, therefore the calibrations phantoms (gels) could not be placed exactly at the same place between session 1 and 2 and do not appear at the same position in some slices presented here.
Figure 3.
Representative example of all the sodium concentration maps in the coronal plane acquired on the same volunteer as Fig 2. All the images were co-registered. The number in the top-right corner of each image corresponds to the acquisition number in the protocol as described in Fig.1B. Note that in some cases (3T-day 1 and 3T-day 2), the volunteer had to take a break between 2 sessions, therefore the calibrations phantoms (gels) could not be placed exactly at the same place between session 1 and 2 and do not appear at the same position in some slices presented here.
Mean SNR and sodium concentration
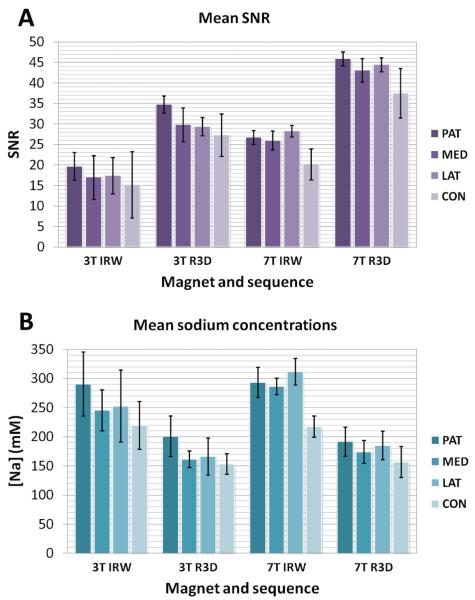
Figure 4 shows the mean SNR measurements (4A) and mean sodium concentrations (4B) over all slices and all volunteers, acquired with both the 3D radial (R3D) and IR WURST (IRW) sequences at 3T and 7T. As the data were acquired with different parameters (TR, number of projections) at 3T and 7T, in order to be able to compare the SNR of the 2 sequences at both fields, the SNR efficiency was calculated as the mean SNR divided by the square root of total acquisition time TA (, in units of min−½). The mean values of SNR, SNR efficiency and sodium concentrations over all cartilage regions are given in Table 2.
Figure 4.
Mean SNR (A) and sodium concentrations (B) for different sequences at different fields measured in 4 regions in the cartilage. Abbreviations: IRW=IR WURST, R3D=Radial 3D, PAT=Patellar, MED=FT Medial, LAT= FT Lateral, CON=PF Condyle, with PF=Posterior Femoral, FT=Femoro-Tibial.
Table 2.
Mean SNR, mean SNR efficiency (SNReff), mean sodium concentrations measured over all cartilage region with each sequence on each magnet.
| Field | Sequence | SNR | SNReff (min−½) | [Na+](mM) |
|---|---|---|---|---|
| 3T | R3D | 30±3 | 6.7±0.7 | 171±21 |
| IRW | 17±2 | 3.8±0.4 | 252±29 | |
|
| ||||
| 7T | R3D | 43±4 | 10.4±1.0 | 177±17 |
| IRW | 25±4 | 5.0±0.8 | 278±41 | |
Coefficients of variation
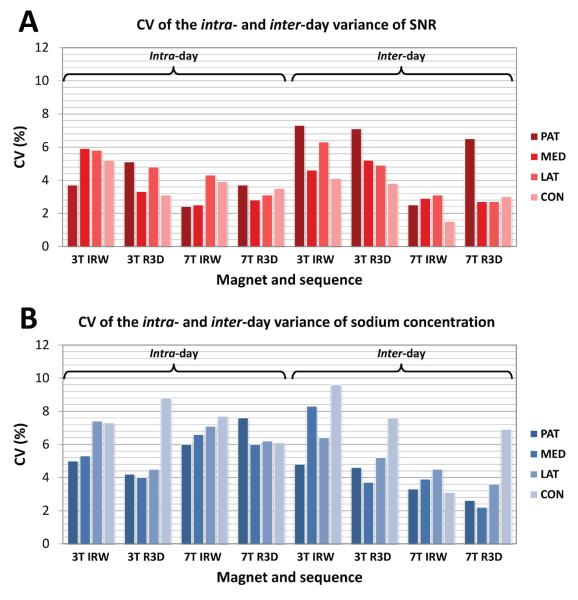
The RMS of CVSNR and CVNa over all cartilage regions are given in Table 3. The intra- and inter-day CV components (, , and ) of SNR and of sodium concentrations are shown in Figures 5A and 5B, respectively.
Table 3.
Root mean square (RMS) of CVSNR and CVNa as determined for each cartilage region using each sequence on each magnet. Abbreviations: R3D = Radial 3D, IRW = IR WURST (R3D with fluid suppression), PF = Posterior Femoral, FT = Femoro-Tibial.
| RMS of CVSNR | RMS of CVNa | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| R3D | IRW | R3D | IRW | |||||
| 3T | 7T | 3T | 7T | 3T | 7T | 3T | 7T | |
| Patella | 6.87 | 9.85 | 9.52 | 11.74 | 7.48 | 9.97 | 10.51 | 12.85 |
| FT Medial | 9.81 | 12.47 | 10.64 | 12.49 | 11.44 | 12.75 | 12.04 | 13.63 |
| FT Lateral | 8.36 | 9.45 | 11.28 | 12.14 | 9.33 | 9.67 | 12.54 | 13.10 |
| PF Condyle | 6.95 | 9.92 | 8.10 | 8.31 | 8.10 | 10.37 | 9.25 | 9.65 |
| Mean RMS | 8.00±1.39 | 10.42±1.38 | 9.88±1.39 | 11.17±1.93 | 9.09±1.75 | 10.69±1.40 | 11.08±1.50 | 12.31±1.80 |
Figure 5.
Coefficient of variation (CV) of the intra- and inter-day variance of SNR (A) and sodium concentrations (B) for each magnet-sequence combination, measured in 4 regions in the cartilage. Each CV is the square root of the variance expressed as a percentage of the overall mean SNR or sodium concentration for the relevant magnet-sequence combination. Abbreviations: IRW=IR WURST, R3D=Radial 3D, PAT=Patellar, MED=FT Medial, LAT= FT Lateral, CON=PF Condyle, with PF=Posterior Femoral, FT=Femoro-Tibial.
Correlation between SNR and [Na+] and between CVSNR and CVNa
Table 4 and 5 give the Pearson correlation coefficients (r) and corresponding p values calculated for assessing the correlation between mean SNR and mean sodium concentration and between CVSNR and CVNa respectively, as determined for each cartilage region using each sequence on each magnet. The mean correlation coefficient for mean SNR vs. mean [Na+] is 0.74±0.17 for IRW, 0.60±0.22 for R3D, and 0.67±0.20 over all sequences and all magnets (range: 0.22-0.95) with all p<0.05. The mean correlation coefficient for CVSNR versus CVNa is 0.94±0.06 for IRW, 0.95±0.03 for R3D, and 0.94±0.05 over all sequences and all magnets (range: 0.83-1.00) with p<0.0001.
Table 4.
Correlation (r) and corresponding p value for the association between SNR and sodium concentration as determined for each cartilage region using each sequence on each magnet. Abbreviations: R3D = Radial 3D, IRW = IR WURST (R3D with fluid suppression), PF = Posterior Femoral, FT = Femoro-Tibial.
| Correlation between SNR and [Na+]. | |||||
|---|---|---|---|---|---|
| R3D | IRW | ||||
| Cartilage | Magnet | r | p | r | p |
| Patella | 3T | 0.85 | <0.0001 | 0.91 | <0.0001 |
| 7T | 0.84 | <0.0001 | 0.95 | <0.0001 | |
| FT Medial | 3T | 0.56 | 0.0078 | 0.82 | <0.0001 |
| 7T | 0.51 | 0.0023 | 0.70 | 0.0001 | |
| FT Lateral | 3T | 0.42 | 0.0439 | 0.71 | 0.0001 |
| 7T | 0.22 | 0.0470 | 0.39 | 0.0602 | |
| PF Condyle | 3T | 0.64 | 0.0016 | 0.71 | 0.0001 |
| 7T | 0.74 | <0.0001 | 0.73 | <0.0001 | |
| Mean ± SD | 0.60±0.22 | 0.74±0.17 | |||
Table 5.
Correlation (r) and corresponding p value for the association between CVSNR and CVNa as determined for each cartilage region using each sequence on each magnet. Abbreviations: R3D = Radial 3D, IRW = IR WURST (R3D with fluid suppression), PF = Posterior Femoral, FT = Femoro-Tibial.
| Correlation between CVSNR and CVNa. | |||||
|---|---|---|---|---|---|
| R3D | IRW | ||||
| Cartilage | Magnet | r | p | r | p |
| Patella | 3T | 0.98 | <0.0001 | 0.98 | <0.0001 |
| 7T | 0.99 | <0.0001 | 0.99 | <0.0001 | |
| FT Medial | 3T | 0.90 | <0.0001 | 0.90 | <0.0001 |
| 7T | 0.92 | <0.0001 | 0.91 | <0.0001 | |
| FT Lateral | 3T | 0.93 | <0.0001 | 0.99 | <0.0001 |
| 7T | 0.96 | <0.0001 | 1.00 | <0.0001 | |
| PF Condyle | 3T | 0.98 | <0.0001 | 0.89 | <0.0001 |
| 7T | 0.97 | <0.0001 | 0.83 | <0.0001 | |
| Mean ± SD | 0.95±0.03 | 0.94±0.06 | |||
Comparison of magnet-sequence combinations
For SNR, all but one comparisons were highly significant (p<0.0001). Specifically, only the comparison of R3D and IRW at 3T for femoro-tibial medial cartilage failed to achieve significance (p=0.0511, very close to the threshold 0.05 for statistical significance). For sodium concentrations, only the comparisons of R3D versus IRW at each of 3T and 7T for femoral condyle and patellar cartilage failed to achieve significance.
P values to compare combinations of magnet-sequence in terms of intra-day and inter-day CVs are summarized as follows:
For : mean p=0.400±0.277, in the range 0.073-0.968.
For : mean p=0.427±0.263, in the range 0.055-0.967.
For : mean p=0.466±0.276, in the range 0.116-0.949.
For : mean p=0.527±0.201, in the range 0.177-0.852.
Overall, there were no significant inter-magnet or inter-sequence differences in terms of the intra-day and inter-day CV values observed (all p>0.05).
Repeatability and reproducibility of sodium quantification
In summary, the results for the aims of this study are:
- For the repeatability of sodium quantification:
- at 3T (intra-magnet): RMS of CVNa in the range 7.5-12.5%.
- at 7T (intra-magnet): RMS of CVNa in the range 9.7-13.6%.
- with the Radial 3D sequence (intra-sequence): RMS of CVNa in the range 9.7-13.6%.
- with the IR WURST sequence (intra-sequence): RMS of CVNa in the range 7.5-12.8%.
- over different acquisitions on the same day: no significant intra-day difference of the CV (p>0.05).
- over different days of acquisition: no significant inter-day difference of the CV (p>0.05).
- For the reproducibility of sodium quantification:
- at 3T versus 7T: no significant inter-magnet difference of CV (p>0.05) but significant difference for mean [Na+] (p<0.0001).
- with Radial 3D versus IR WURST: no significant inter-sequence difference of the CV (p>0.05) but significant difference for mean [Na+] (p<0.0001).
Discussion
Mean SNR and sodium concentration
For the same sequence, the SNR efficiency is higher at 7T than at 3T: it is 1.55 times higher for R3D and 1.31 times higher for IRW while the field is 2.3 times higher. This shows that although it is better to use the higher field in order to increase the SNR of the images, the technique can be used at 3T without much loss of SNR efficiency.
Mean values of sodium concentrations are of the same order for IRW at 3T and 7T, in the range 220-310 mM, which corresponds to values usually found in the literature for healthy cartilage [9, 27]. The sodium concentrations are also of the same order for R3D at 3T and 7T, in the range 160-200 mM. These latter values underestimate the cartilage [Na+] as R3D does not include fluid suppression, therefore a large partial volume effect due to the presence of synovial fluid (150 mM of [Na+]) and large voxel size leads to a reduction of the average sodium concentration measured in these voxels. Moreover this large reduction in the estimation of sodium concentration is also amplified by uncertainties on T1, and corrections applied during quantification, which have different effects on the quantification from R3D and IRW. In order to understand this issue, measurements of and in vivo with fluid suppression are presently under investigation.
Coefficient of variation
The RMS of CVSNR and CVNa over all magnets and sequence lie in the ranges 6.7-12.5% and 7.5-13.6% respectively.
All the intra- and inter-day CVs of SNR (, ) from different combinations of magnet-sequences, or over all sequences, or over all magnets, and also over all the data, show consistency regardless of the acquisition technique that was used: they all lie in the small range 2.9-5.4 %. The same conclusion canbe drawn for the intra- and inter-day CVs of sodium concentrations (, ) , as all the values lie in the small range 5.2-6.8 %.
For comparison, the ranges of CV for different proton based MRI techniques for assessing OA in cartilage are: Cartilage thickness: 2.1-8.3 % [28,29]; Cartilage volume: 1-7.5 % [28,29]; Delayed Gadolinium-Enhanced MRI of Cartilage (dGEMRIC): 4.7-12.9 % [30]; T2: 3-7 % for global measurements, 6-29 % for regional measurements [31–33]; T1ρ: 7-19 % [33]; Diffusion Tensor Imaging (DTI): Apparent Diffusion Coefficient (ADC) 8.1 %, Fractional Anisotropy (FA) 9.7 % [34].
The repeatability and reproducibility CV of sodium concentrations measured using the proposed quantitative sodium MRI techniques compare therefore favorably with the CV of other proton-based MRI techniques.
Comparison of magnet and sequence combinations
Almost all the SNR values between all combinations of magnet-sequence are significantly different. Only one combination has a p value of 0.0511, which is slightly above the threshold for statistical significance p=0.05. This can probably be attributed to the small number of volunteers and measurements, and can therefore be considered as not relevant for the discussion of the results. This significant difference in SNR is expected as in each pair of magnet-sequence combination compared, one of the combinations included a method that reduced the SNR (3T vs 7T, or IRW vs R3D).
A large majority of the sodium concentrations between all combinations of magnet-sequence are significantly different. A significant difference is actually expected for combinations including different sequences (R3D vs IRW), as we have seen earlier that the R3D sequence underestimates the sodium quantification in cartilage due to partial volume effect and the presence of synovial fluid. On the other hand, no significant difference is expected for combinations including the same sequence but different fields (3T vs 7T). This is what we observe for [Na+] in patellar and posterior femoral condyle, but not for femoro-tibial lateral and medial cartilage. This difference could be either due to different relaxation times at 3T and 7T between the load-bearing cartilage (femoro-tibial) compared to the shear-bearing cartilage (patellar and posterior femoral condyle), systematic bias, lower SNR at 3T and limited spatial resolution. More volunteers are required to isolate this issue.
All the p values for and are above 0.05, showing therefore no significant difference in the variations of the SNR measurements between all magnet-sequence combinations. All the p values for and are above 0.05, showing therefore no significant difference in the variations of the sodium concentration measurements between all magnet-sequence combinations. In other words, the [Na+] measured with sodium MRI show therefore no inter-magnet, no inter-sequence, no inter-day and no intra-day significant difference.
Correlation between SNR and [Na+] and between CVSNR and CVNa
A strong correlation was found between SNR and [Na+] and a very strong correlation was found between CVSNR and CVNa, for all the regions in cartilage, sequences and magnet combinations. It is therefore more suitable to use the highest fields possible for accurate sodium quantification because of their higher SNR.
Conclusion
The repeatability and reproducibility CVs of sodium quantification using sodium MRI at 3T and 7T with and without fluid suppression are in the range 7.5-13.6%, which is comparable to other proton based MRItechniques for assessing OA in cartilage. Sodium quantification in cartilage was more accurate when using an inversion recovery preparation in the sequence (IR WURST), likely due to a reduction of the partial volume effects from synovial fluid, at both 3T and 7T. Future studies will focus on the application of the fluid suppressed sequence at both fields for quantification of sodium and GAG loss in cartilage from OA patients.
Acknowledgments
This research work was supported by US grants NIH 1RO1AR053133, 1RO1AR056260 and 1RO1AR060238.
References
- [1].Lawrence R, Felson D, Helmick C, Arnold L, Choi H, Deyo R, Gabriel S, Hirsch R, Hochberg M, Hunder G, Jordan J, Katz J, Kremers H, Wolfe F. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part II. Arthritis & Rheumatism. 2008;58(1):26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kotlarz H, Gunnarsson C, Fang H, Rizzo J. Insurer and out-of-pocket costs of osteoarthritis in the US: Evidence from national survey data. Arthritis & Rheumatism. 2009;60(12):3546–3553. doi: 10.1002/art.24984. [DOI] [PubMed] [Google Scholar]
- [3].Regatte R, Akella S, Borthakur A, Kneeland J, Reddy R. In Vivo Proton MR Three-dimensional T1ρ Mapping of Human Articular Cartilage: Initial Experience. Radiology. 2003;229(1):269–274. doi: 10.1148/radiol.2291021041. [DOI] [PubMed] [Google Scholar]
- [4].Mosher T, Dardzinski B. Cartilage MRI T2 relaxation time mapping: overview and applications. Seminars in Musculoskeletal Radiology. 2004;volume 8:355–368. doi: 10.1055/s-2004-861764. [DOI] [PubMed] [Google Scholar]
- [5].Bashir A, Gray M, Burstein D. Gd-DTPA2-as a measure of cartilage degradation. Magnetic Resonance in Medicine. 1996;36(5):665–673. doi: 10.1002/mrm.1910360504. [DOI] [PubMed] [Google Scholar]
- [6].Ling W, Regatte R, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proceedings of the National Academy of Sciences. 2008;105(7):2266. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Filidoro L, Dietrich O, Weber J, Rauch E, Oerther T, Wick M, Reiser M, Glaser C. High-resolution diffusion tensor imaging of human patellar cartilage: Feasibility and preliminary findings. Magnetic Resonance in Medicine. 2005;53(5):993–998. doi: 10.1002/mrm.20469. [DOI] [PubMed] [Google Scholar]
- [8].Reddy R, Insko E, Noyszewski E, Dandora R, Kneeland J, Leigh J. Sodium MRI of human articular cartilage in vivo. Magnetic Resonance in Medicine. 1998;39(5):697–701. doi: 10.1002/mrm.1910390505. [DOI] [PubMed] [Google Scholar]
- [9].Shapiro E, Borthakur A, Gougoutas A, Reddy R. 23Na MRI accurately measures fixed charge density in articular cartilage. Magnetic Resonance in Medicine. 2002;47(2):284–291. doi: 10.1002/mrm.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang L, Wu Y, Chang G, Oesingmann N, Schweitzer M, Jerschow A, Regatte R. Rapid isotropic 3D-sodium MRI of the knee joint in vivo at 7T. Journal of Magnetic Resonance Imaging. 2009;30(3):606–614. doi: 10.1002/jmri.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shapiro E, Borthakur A, Dandora R, Kriss A, Leigh J, Reddy R. Sodium visibility and quantitation in intact bovine articular cartilage using high field 23Na MRI and MRS. Journal of Magnetic Resonance. 2000;142(1):24–31. doi: 10.1006/jmre.1999.1932. [DOI] [PubMed] [Google Scholar]
- [12].Lesperance L, Gray M, Burstein D. Determination of fixed charge density in cartilage using nuclear magnetic resonance. Journal of Orthopaedic Research. 1992;10(1):1–13. doi: 10.1002/jor.1100100102. [DOI] [PubMed] [Google Scholar]
- [13].Borthakur A, Shapiro E, Beers J, Kudchodkar S, Kneeland J, Reddy R. Sensitivity of MRI to proteoglycan depletion in cartilage: comparison of sodium and proton MRI. Osteoarthritis and Cartilage. 2000;8(4):288–293. doi: 10.1053/joca.1999.0303. [DOI] [PubMed] [Google Scholar]
- [14].Borthakur A, Mellon E, Niyogi S, Witschey W, Kneeland J, Reddy R. Sodium and T1ρ MRI for molecular and diagnostic imaging of articular cartilage. NMR in Biomedicine. 2006;19(7):781–821. doi: 10.1002/nbm.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Madelin G, Lee J, Inati S, Jerschow A, Regatte R. Sodium Inversion Recovery MRI of the Knee Joint In Vivo at 7T. Journal of Magnetic Resonance. 2010;207:42–52. doi: 10.1016/j.jmr.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Madelin G, Jerschow A, Regatte R. Sodium MRI with fluid suppression: will it improve early detection of osteoarthritis? Imaging in Medicine. 2011;3(1):1–4. [Google Scholar]
- [17].Taylor B. Guidelines for Evaluating and Expressing the Uncertainty of NIST Measurement Results. DIANE Publishing; 2009. [Google Scholar]
- [18].McNaught A, Wilkinson A. IUPAC compendium of chemical terminology. volume 2. Blackwell Scientific Publications; 1997. [Google Scholar]
- [19].Nielles-Vallespin S, Weber M, Bock M, Bongers A, Speier P, Combs S, Wohrle J, Lehmann-Horn F, Essig M, Schad L. 3D radial projection technique with ultrashort echo times for sodium MRI: clinical applications in human brain and skeletal muscle. Magnetic Resonance in Medicine. 2006;57(1):74–81. doi: 10.1002/mrm.21104. [DOI] [PubMed] [Google Scholar]
- [20].Kupce E, Freeman R. Adiabatic pulses for wide-band inversion and broad-band pulse. Journal of Magnetic Resonance A. 1995;115:273–276. [Google Scholar]
- [21].Magland J, Wehrli F. Pulse sequence programming in a dynamic visual environment. Proceedings of the 14th Annual Meeting of ISMRM; Seattle, WA, USA. 2006. [Google Scholar]
- [22].Studholme C, Hawkes D, Hill D. Normalized entropy measure for multimodality image alignment. Proceedings of SPIE.1998. p. 132. [Google Scholar]
- [23].Madelin G, Jerschow A, Regatte R. Sodium relaxation times in the knee joint in vivo at 7T. NMR in Biomedicine. 2011 doi: 10.1002/nbm.1768. XX(XX):XX–XX. In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shapiro E, Borthakur A, Kaufman J, Leigh J, Reddy R. Water distribution patterns inside bovine articular cartilage as visualized by1H magnetic resonance imaging. Osteoarthritis and Cartilage. 2001;9(6):533–538. doi: 10.1053/joca.2001.0428. [DOI] [PubMed] [Google Scholar]
- [25].Borthakur A, Shapiro E, Akella S, Gougoutas A, Kneeland J, Reddy R. Quantifying Sodium in the Human Wrist in Vivo by Using MR Imaging1. Radiology. 2002;224(2):598. doi: 10.1148/radiol.2242011039. [DOI] [PubMed] [Google Scholar]
- [26].Kuettner K. Biochemistry of articular cartilage in health and disease. Clinical Biochemistry. 1992;25(3):155–163. doi: 10.1016/0009-9120(92)90224-g. [DOI] [PubMed] [Google Scholar]
- [27].Wheaton A, Borthakur A, Shapiro E, Regatte R, Akella S, Kneeland J, Reddy R. Proteoglycan Loss in Human Knee Cartilage: Quantitation with Sodium MR Imaging - Feasibility Study. Radiology. 2004;231(3):900. doi: 10.1148/radiol.2313030521. [DOI] [PubMed] [Google Scholar]
- [28].Koo S, Gold G, Andriacchi T. Considerations in measuring cartilage thickness using MRI: factors influencing reproducibility and accuracy. Osteoarthritis and cartilage. 2005;13(9):782–789. doi: 10.1016/j.joca.2005.04.013. [DOI] [PubMed] [Google Scholar]
- [29].Glaser C, Burgkart R, Kutschera A, Englmeier K, Reiser M, Eckstein F. Femoro-tibial cartilage metrics from coronal MR image data: Technique, test–retest reproducibility, and findings in osteoarthritis. Magnetic Resonance in Medicine. 2003;50(6):1229–1236. doi: 10.1002/mrm.10648. [DOI] [PubMed] [Google Scholar]
- [30].Multanen J, Rauvala E, Lammentausta E, Ojala R, Kiviranta I, Häkkinen A, Nieminen M, Heinonen A. Reproducibility of imaging human knee cartilage by delayed gadolinium-enhanced MRI of cartilage (dGEMRIC) at 1.5 Tesla. Osteoarthritis and Cartilage. 2009;17(5):559–564. doi: 10.1016/j.joca.2008.12.001. [DOI] [PubMed] [Google Scholar]
- [31].Glaser C, Mendlik T, Dinges J, Weber J, Stahl R, Trumm C, Reiser M. Global and regional reproducibility of T2 relaxation time measurements in human patellar cartilage. Magnetic Resonance in Medicine. 2006;56(3):527–534. doi: 10.1002/mrm.21005. [DOI] [PubMed] [Google Scholar]
- [32].Raya J, Horng A, Dietrich O, Weber J, Dinges J, Mützel E, Reiser M, Glaser C. Voxel-based reproducibility of T2 relaxation time in patellar cartilage at 1.5 T with a new validated 3D rigid registration algorithm. Magnetic Resonance Materials in Physics, Biology and Medicine. 2009;22(4):229–239. doi: 10.1007/s10334-009-0168-0. [DOI] [PubMed] [Google Scholar]
- [33].Mosher T, Zhang Z, Reddy R, Boudhar S, Milestone B, Morrison W, Kwoh C, Eckstein F, Witschey W, Borthakur A. Knee Articular Cartilage Damage in Osteoarthritis: Analysis of MR Image Biomarker Reproducibility in ACRIN-PA 4001 Multicenter Trial. Radiology. 2011;258(3):832. doi: 10.1148/radiol.10101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Raya J. 2011. Personal communication.