Abstract
Phosphoinositide lipids were initially discovered as precursors for specific second messengers involved in signal transduction, but have now taken the center stage in controlling many essential processes at virtually every cellular membrane. In particular, phosphoinositides play a critical role in regulating membrane dynamics and vesicular transport. The unique distribution of certain phosphoinositides at specific intracellular membranes makes these molecules uniquely suited to direct organelle-specific trafficking reactions. In this regulatory role, phosphoinositides cooperate specifically with small GTPases from the Arf and Rab families. This review will summarize recent progress in the study of phosphoinositides in membrane trafficking and organellar organization and highlight the particular relevance of these signaling pathways in disease.
1. Introduction
Phosphoinositides are phosphorylated intermediates of phosphatidylinositol (PI) and involved in regulating a plethora of cellular processes. Initially discovered in physiological experiments with brain tissue samples and long thought to be solely precursors of soluble and membrane bound second messengers such as inositol-trisphosphate and diacylglycerol, phosphoinositides have now been recognized to play central roles in regulating a wide array of physiological process at intracellular membranes [1]. Phosphorylation of the phosphatidylinositol headgroup gives rise to seven distinct phosphoinositide species with distinct regulatory functions. Phosphoinositides are minor constituents of phospholipid bilayers and show continuous turnover. Phosphatidylinositol-3,4-bisphosphate (PI(3,4)P2) and phosphatidylinositol-3,4,5-trisphosphate (PI(3,4,5)P3) are short-lived signaling molecules synthesized in response to extracellular stimuli and involved in cell proliferation and survival [2]. The levels of phosphatidylinositol-3-phosphate (PI(3)P), phosphatidylinositol-4-phosphate (PI(4)P), phosphatidylinositol-3,5-bisphosphate (PI(3,5)P2) and phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) can also be regulated by external cues, however, these phosphoinositides show a relatively stable distribution at specific membranes under normal cell growth conditions [1].
Several studies suggest that PI(3)P is concentrated at early endosomes, PI(4)P at Golgi membranes, PI(3,5)P2 at late endocytic compartments and PI(4,5)P2 at the plasma membrane [1]. The compartment-specific distribution of phosphoinositides is coupled to the selective recruitment of effector proteins that bind specifically to individual phosphoinositide species. Recognition of phosphoinositides is mediated by a variety of well-defined modular domains that are present in a subset of cytosolic and membrane proteins [3, 4]. In a growing number of cases it has been found that phosphoinositide-binding proteins additionally interact with activated small GTPases from the Ras protein superfamily. In particular, unique combinations of phosphoinositides and GTP-bound versions of Arf and Rab GTPases, play a major role in a coincidence detection mechanism that ultimately controls the timing and selectivity of effector recruitment to membranes [5]. This review will focus on the function of individual phosphoinositides in controlling vesicular transport and how this regulation interconnects with other dynamic processes at membranes. The particular relevance of these molecules for human disease will be highlighted.
2. PI(3)P regulates membrane dynamics at early endosomes, phagosomes and autophagosomes
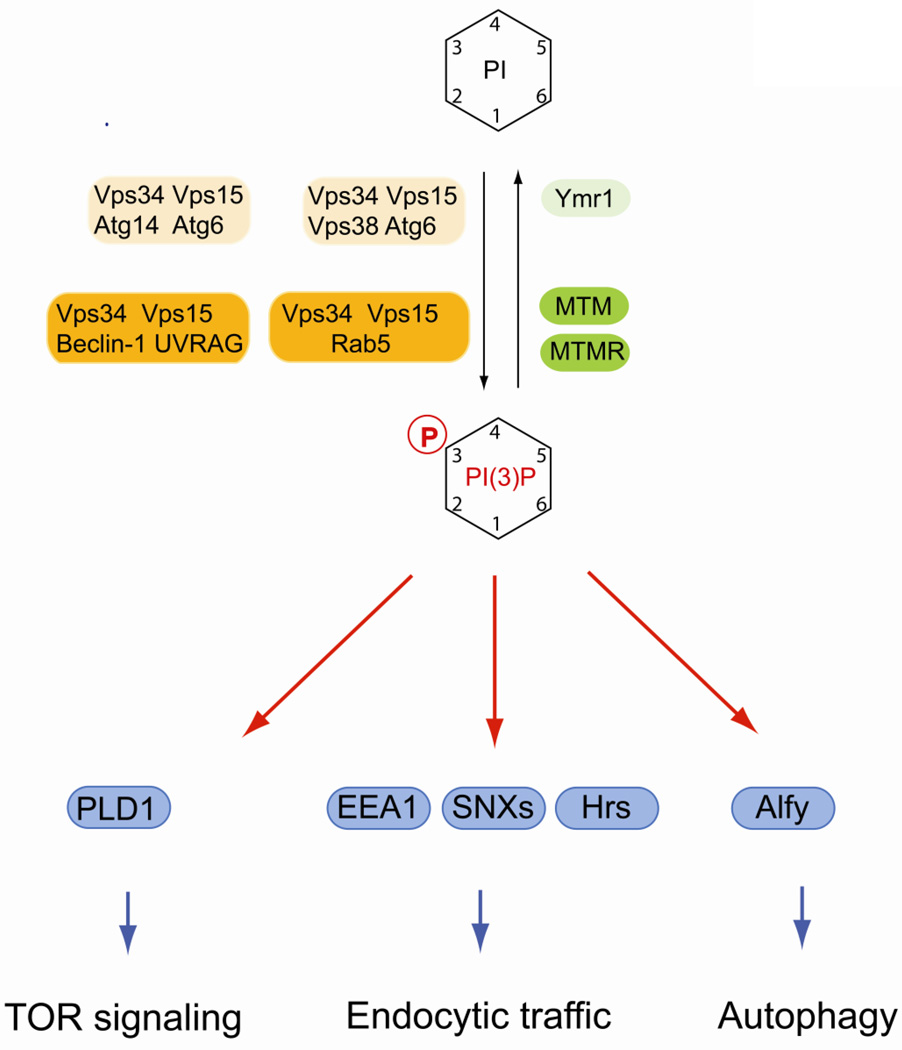
Phosphoinositides phosphorylated at the 3-position of the inositol headgroup were originally discovered as minor components of total phosphoinositides in fibroblasts and brain tumor cells [6, 7]. Subsequently, three families of PI 3-kinases that utilize distinct phospholipid substrates were characterized [2, 8]. Class I PI 3-kinases phosphorylate PI(4,5)P2 and PI(4)P to generate short-lived pools of PI(3,4,5)P3 and PI(3,4)P2 upon stimulation by growth factors or cytokines and are thus directly involved in regulating cell survival and cell growth [2, 8]. Class II isoforms phosphorylate phosphatidylinositol to PI(3)P and like class I enzymes can be activated upon stimulation of tyrosine kinase receptors and G-protein-coupled receptors [9]. A class III PI 3-kinase that generates PI(3)P was first described in yeast and is encoded by the VPS34 gene [10]. Vps34 is an essential component of the vacuolar sorting pathway and is the only PI 3-kinase in yeast. Vps34 interacts with the protein kinase Vps15, which is required for membrane targeting of the lipid kinase complex [11, 12]. The pool of PI(3)P that is generated by the Vps34 complex is largely localized to endosomal compartments and essential for protein sorting to the yeast vacuole but plays an additional important role in autophagy [13] [14]. The mammalian homologue of Vps34 is also involved in endocytic sorting and autophagy.
PI(3)P is critical for membrane recruitment of effector proteins that contain specific lipid-binding regions. Important modular domains that recognize PI(3)P are zinc-finger domains, initially detected in the proteins Fab1, YOTB, Vac1 and EEA1, and termed FYVE domains. In addition, Phox homology (PX) domains bind specifically to PI(3)P [3, 15–17]. Many of the proteins containing these domains play an intimate role in vesicular dynamics in the endosomal system. Many FYVE-domain containing proteins are involved in endocytic trafficking. Prominent examples are early endosomal antigen 1 (EEA1) and Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) (Fig. 1). EEA1 is a Rab5 effector required for endosomal tethering and forms homodimers through a coiled-coiled domain [18–20]. EEA1 also interacts with SNARE proteins such as syntaxin-6 and syntaxin-13 at early endosomes [21, 22]. The FYVE domain of EEA1 exhibits low affinity binding to Rab5-GTP, which enables selective recruitment of EEA1 to endosomes containing both Rab5 and PI(3)P [23, 24]. Higher affinity binding, which is required for efficient tethering and SNARE complex formation, requires a specific N-terminal zinc finger motif in EEA1 [25]. The coinicidence detection of a phosphoinositide and an activated small GTPase is a frequently observed mechanism for controlling timing and specificity of membrane recruitment of effector proteins involved in vesicular traffic. Importantly, Rab5 binds also to Vps34/Vps15 complexes and is required to recruit the PI 3-kinase complex to early endosomal membranes [26, 27]. Thus, Rab5 acts both upstream and in concert with PI(3)P to regulate endosomal membrane dynamics.
Fig. 1.
Regulation of PI(3)P and its role in early endosomal traffic and autophagy. PI(3)P is synthesized by class III PI 3-kinase Vps34 in yeast and mammals. Vps34 forms specific complexes with co-factors that specify its function in either endosmal dynamics or in autophagy. In addition, PI(3)P plays a role in metabolic signaling via PLD1 and mTORC1. Several phosphatases belonging to the myotubularin (MTM) and myotubularin-related protein (MTMR) family can dephosphorylate PI(3)P. Mammalian lipid kinases are marked by orange boxes, lipid phosphatases are in green boxes. Yeast homologs are indicated in lighter shades of the same colors. Effector proteins are in blue boxes.
In contrast to EEA1, Hrs binding to early endosomes occurs independently of Rab5, but is dependent on its FYVE domain [28]. Hrs is involved in the endosomal sorting of ubiquitinated membrane proteins. Hrs contains a clathrin-binding domain and also binds directly to ubiquitin and promotes the endosomal sorting of ubiquitinated membrane proteins [29]. Hrs also associates with phagosomes in a PI(3)P-dependent manner that is required for the maturation of phagosomes and their fusion with late endosomes [30].
Another important group of PI(3)P effectors are PX-domain-containing members of the sorting nexin family (SNX) that function in endosomal membrane dynamics (Fig. 1) [31]. Several SNX proteins contain a Bin, Amphiphysin, Rvs-homology (BAR) domain that triggers dimerization and forms a stable curved structure, which allows these proteins to associate with highly curved membranes and to drive membrane deformation [32]. Not all PX domains of SNX proteins interact with PI(3)P. For example, SNX9 has relatively broad phosphoinositide-binding specificity and is involved clathrin-mediated endocytosis [33].
In both yeast and mammalian cells, class III PI3Ks play an important additional role in autophagy during starvation. In yeast, it was shown that Vps34 forms specific complexes with different sets of cofactors, which specify unique regulatory pathways. Vps34 associates in specific complexes with Vps15, Atg6 and either Vps38 or Atg14 (Fig. 1) [14]. Vps38-containing complexes function in vacuolar protein sorting whereas Atg14-specific complexes are required for autophagy [14]. The mammalian homologue of Atg6 is the protein beclin-1, which also interacts with Bcl2, and plays a prominent role in autophagy [34, 35]. Another binding partner of beclin-1 is the protein encoded by the UV radiation resistance-associated gene (UVRAG) that is involved in regulating hVps34 in autophagy [36]. Mammalian PI(3)P-binding effectors in autophagy include the protein Alfy that functions in the elimination of aggregated proteins by macroautophagy [37, 38]. In addition, the recently discovered FYVE and coiled-coil domain-containing protein 1 (FYCO1) is a novel PI(3)P effector required for microtubule-based transport of autophagic vesicles [39]. Recent evidence suggests that mutations in FYCO1 are linked to congenital cataracts in certain human populations [40].
Recent evidence suggests that human VPS34 has additional functions in metabolic signaling via mammalian target of rapamycin 1C (mTOR1C), which involves PI(3)P-dependent recruitment of phospholipiase D1 (PLD1) to lysosomes in response to amino acids [41–43].
PI(3)P is dephosphorylated by several classes of lipid phosphatases (Fig. 1). Sac1 domain-containing phosphatases show enzymatic activity towards a PI(3)P, PI(4)P and PI(3,5)P2 [44]. In yeast, deletion of the SAC1 gene, which encodes a transmembrane lipid phosphatase and represents the founding member of this protein family, display elevated levels of PI(3)P and PI(4)P [44–46]. Although a physiological role of Sac1 in regulating PI(4)P levels has been clearly established, which will be discussed below, the relevance of this protein in PI(3)P regulation is unclear. In contrast, mammalian myotubularin (MTM) and myotubularin-related (MTMR) lipid phosphatases play established roles in the turnover of PI(3)P and PI(3,5)P2. The MTM/MTMR protein family consists of 15 members, six of which are catalytically inactive [47, 48]. MTM1 localizes to early endosomes and associates directly with the Vps34/Vps15 complex [49]. Whether Vps34/Vps15 and MTM1 are simultaneously active is unclear, but it appears likely that the association of the lipid kinase with its antagonizing phosphatase plays an important role in the tight spatial and temporal control of PI(3)P. MTM2 interacts with Rab7, a late endosomal marker, and probably plays a role in late endosomal dynamics [50]. Mutations in MTMs and MTMRs causes severe neurological diseases in humans such as X-linked myotubular myopathy and Charcot–Marie–Tooth (CMT) disease, characterized by loss of muscle tissue and touch sensation [47, 48]. Complex formation between active and inactive members of the MTM/MTMR family appears to be critical for their proper function. The relevance of these complexes is highlighted by recent findings showing that mutations in both the phosphoinositide 3-phosphatase MTMR2 and in its catalytically inactive binding partner MTM13 cause type 4B CMT [51, 52].
3. PI(3,5)P2 controls late endosomal dynamics
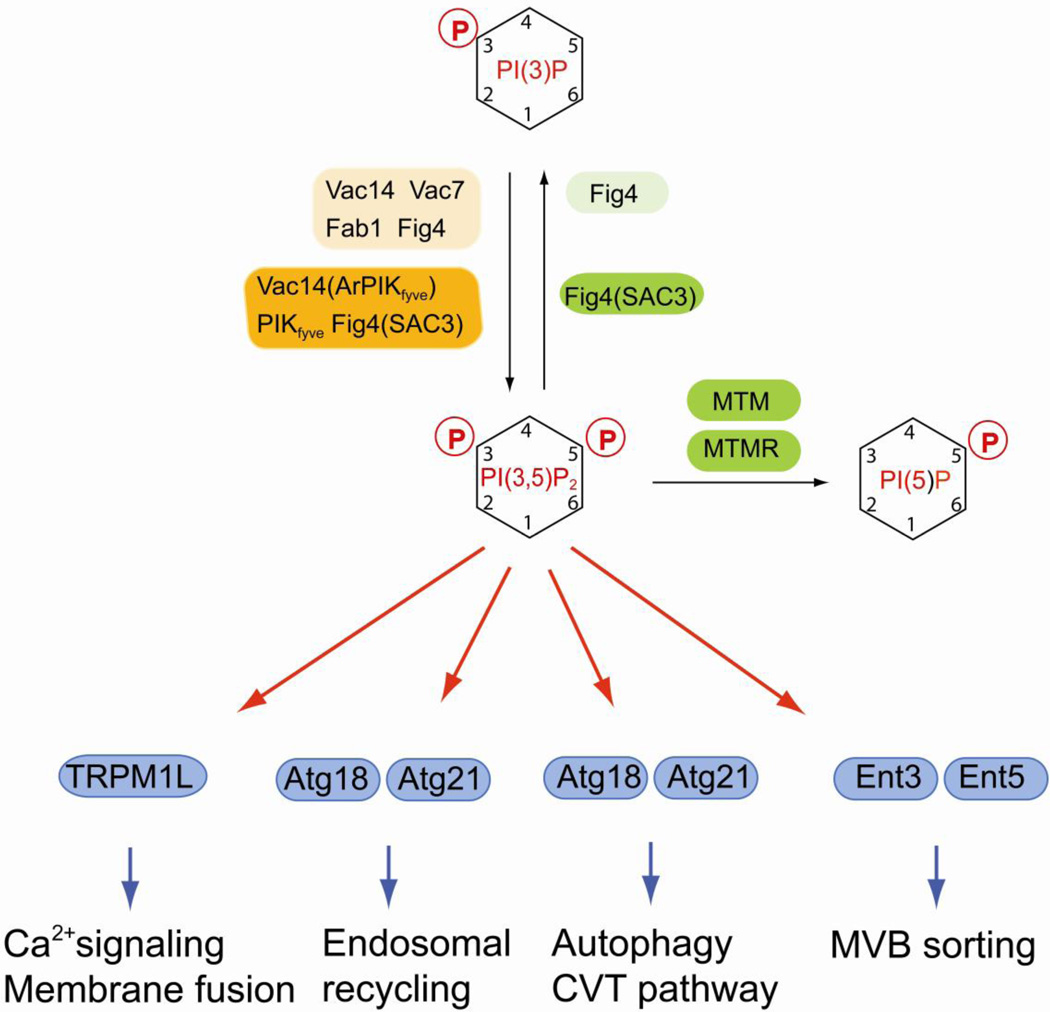
PI(3,5)P2 was first described in yeast, where its synthesis is strongly induced by osmotic stress, but was also discovered as a minor lipid component in non-stimulated mammalian cells [53, 54]. The synthesis of PI(3,5)P2 requires PI(3)P as precursor and therefore depends on a functional Vps34 protein [53]. Studies in yeast identified Fab1 as the kinase responsible for synthesizing PI(3,5)P2 (Fig. 2) [55, 56]. Fab1 is activated by osmotic stress and is involved in regulating vacuolar size and morphology [55, 56]. Fab1 contains a FYVE domain that is important for interaction with PI(3)P and for localization to the endosomal system and the vacuole [56]. The mammalian Fab1 homologue PIKfyve displays similar binding to PI(3)P and is also present in the endosomal/lysosomal system [57, 58]. PI(3,5)P2 appears to play multiple functions in late endosomal dynamics (Fig. 2). The PI(3,5)P2 effector Atg18 (also termed Svp1 and Aut10) localizes to the vacuole and is involved in a retrograde recycling pathway from the vacuole to the Golgi [59]. Atg18 is also required for autophagosome maturation and for the targeting of cytosolic proteases to the vacuole (Cvt pathway) [60]. Atg18 and its paralogue Atg21 are members of a family of novel family of novel beta-propeller proteins with phosphoinositide binding properties and with a few exceptions present in all eukaryotes [61]. Atg18 is thought to regulate membrane recycling and a recent report showed that Atg18 interacts with the myosin adaptor Vac17 [62]. It is therefore possible that Atg18 regulates interaction of membranes with cytoskeletal elements during retrograde transport.
Fig. 2.
Regulation of PI(3,5)P2 and its function in late endosomal dynamics and in autophagy. PI(3,5)P is generated by the FYVE domain-containing lipid kinase Fab1. Synthesis and turnover of PI(3,5)P2 are tightly coupled. Yeast Fab1 is only active when it is in a complex with the scaffold protein Vac14 that also interacts with Vac7 and with the PI(3,5)P2 phosphatase Fig4. A similar complex is also found in mammals containing the lipid kinase PIKfyve, the scaffold protein Vac14 (also termed ArPIKfyve) and the lipid phosphatase Fig4 (also termed Sac3). PI(3,5)P2 plays multiple roles in late endosomal dynamics, autophagy, MVB sorting and Ca2+ mediated membrane fusion. Mammalian lipid kinases or kinase complexes are marked by orange boxes, lipid phosphatases are in green boxes. Alternative names for certain enzymes are in parentheses. Yeast homologs are indicated by lighter shades of the same colors. Effector proteins are in blue boxes.
In yeast, hypertonic shock induces a transient increase in cytosolic Ca2+ that depends on Yvc1p, a vacuolar membrane protein with homology to transient receptor potential (TRP) channels [63, 64]. A recent report now demonstrated that this Ca2+ release requires PI(3,5)P2 production [65]. In addition, the homologous mammalian endolysosome-localized mucolipin transient receptor potential (TRPML) channel is activated by binding to PI(3,5)P2 [65]. These result suggest that TRPML channels are PI(3,5)P2 effectors that control Ca2+-dependent membrane dynamics in response to extracellular stimuli.
Other membrane trafficking regulators that may bind to PI(3,5)P2 are certain members of the epsin family that contain an epsin NH2-terminal homology (ENTH) domain (Fig. 2) [66, 67]. A recent report indicated that Ent3 and Ent5 bind to PI(3,5)P2 in vitro via their ENTH domain and interact with Vps27, a protein that recognizes ubiquitinated cargo [68]. This protein complex is important for ubiquitin-dependent sorting into multivesicular bodies [69]. However, Ent3 and Ent5 also interact with clathrin and the clathrin adaptors Gga2 and AP-1, and are involved in regulating clathrin coat assembly at the Golgi and at endosomes [70] and further studies will be required to determine whether these Ent proteins are physiologically relevant PI(3,5)P2 effectors.
The levels of PI(3,5)P2 are regulated by an unique mechanism (Fig. 2). Fab1 (PIKfyve in mice) forms a protein complex that is essential for regulating its enzymatic activity. One of the interaction partners of Fab1 in this complex is Vac14 (also known as ArPIKfyve in mice), which acts as scaffold protein and also interacts with Atg18 (Fig. 2) [71, 72]. Importantly, Vac14 binds to the PI(3,5)P2-specific phosphatase Fig4 and the presence of Fig4 in the complex is important for efficient PI(3,5)P2 synthesis, thus tightly coupling PI(3,5)P2 synthesis and turnover [71–75].
Deficiencies in PI(3,5)P2 synthesis causes severe neurological phenotypes in mammals. Spontaneous mutations in the Fig4 gene in the “pale tremor” mouse result in a significant reduction in PI(3,5)P2 levels [76]. Similar reduced levels of PI(3,5)P2 are present in mice with mutated Vac14 [77], which highlights the importance of these factors in the enzymatic complex that synthesizes PI(3,5)P2. In both cases, the deficiency in PI(3,5)P2 levels leads to severe neurodegeneration and early lethality. Mutations in FIG4 were also identified in human patients with type 4J CMT, clinically manifested with characteristic motor and sensory neurological defects [76, 78, 79]. Turnover of PI(3,5)P2 is also catalyzed by phosphatases from the myotubularin family, in particular MTM1, MTMR1, MTMR2 and MTMR6 [80]. Different from Fig4 that acts as a 5-phosphatase and generates PI(3)P, myotubularins dephosphorylate PI(3,5)P2 at the 3-position of the inositol headgroup to produce PI(5)P [80]. As mentioned above, mutations in MTMR2 cause type 4B CMT and it is possible that aberrant PI(3,5)P2 levels play a role in this disorder [80].
PI(4)P is a central regulator of Golgi function
PI(4)P is the substrate for PIP 5-kinases and was initially considered solely an intermediate in the biosynthesis of PI(4,5)P2. More recently, many PI(4)P effectors have been identified and it has been established that PI(4)P itself is essential for cell function, which is independent form its role in PI(4,5)P2 signaling (Fig. 3). PI(4)P is highly enriched in the Golgi complex, where it plays a direct role in controlling trafficking and Golgi morphology [81].
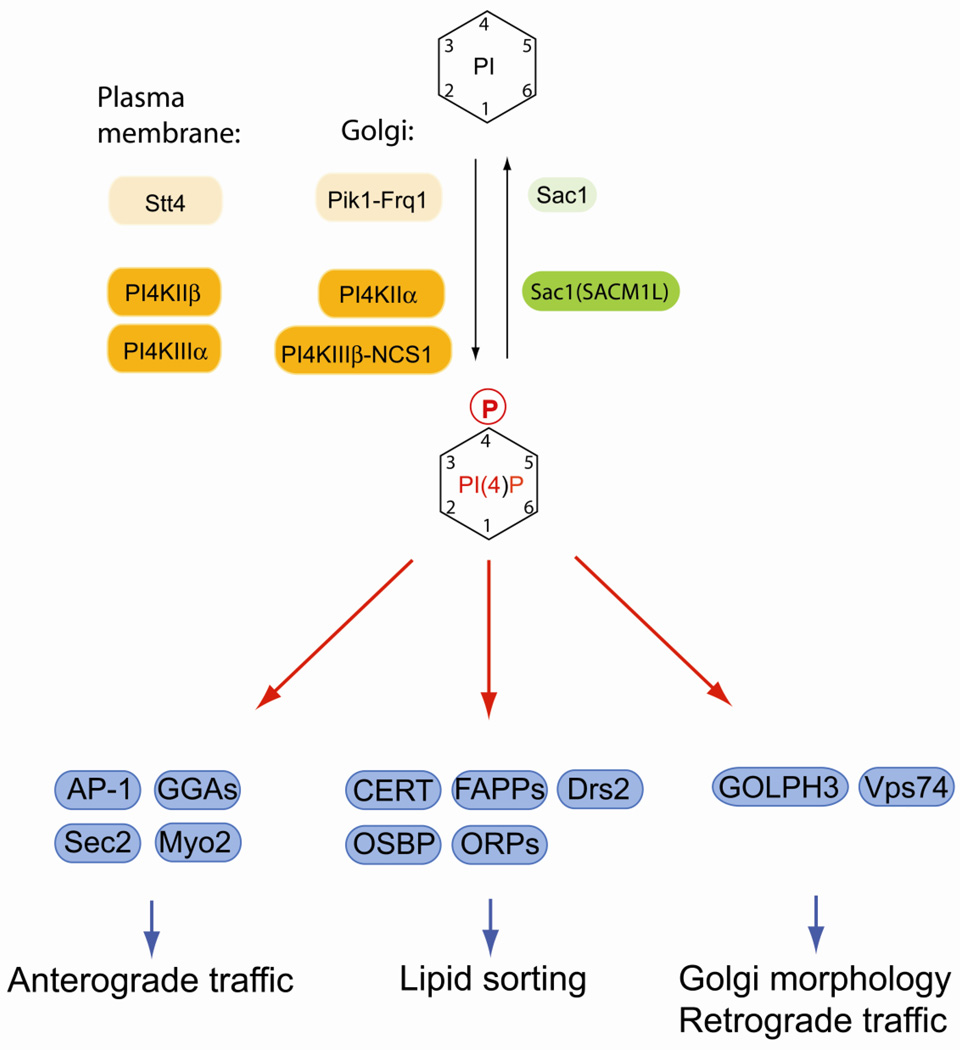
Fig. 3.
Regulation of PI(4)P and its essential function at the Golgi. In yeast, PI(4)P is synthesized by two essential PI 4-kinases, the Golgi-localized Pik1-Frq1 complex and plasma membrane Stt4. In mammalian cells, PI4KIIα and PI4KIIIβ generate Golgi PI(4)P, whereas PI4KIIβ and PI4KIIIα are involved in plasma membrane PI(4)P synthesis. Turnover and spatial control of PI(4)P is achieved by the evolutionary conserved lipid phosphatase Sac1 that also responds to nutrients and growth signals. Many effectors at the Golgi bind PI(4)P and regulate anterograde trafficking from the Golgi, resident enzyme recycling, lipid dynamics and sphingolipid biosynthesis. Mammalian lipid kinases or kinase complexes are marked by orange boxes, lipid phosphatases are in green boxes. Alternative names for certain enzymes are in parentheses. Yeast homologs are indicated by lighter shades of the same colors. Effector proteins are in blue boxes.
PI(4)P is synthesized by type II and type III PI 4-kinases that are unrelated in primary sequences and have distinct structural and enzymatic properties [82]. Mammalian cells have four PI4Ks, namely PI4KIIα, PI4KIIβ, PI4KIIIα, and PI4KIIIβ (Fig. 3) [82]. PI4KIIβ localizesto endosomes and translocates to the plasma membrane after growth factor stimulation [83]. PI4KIIIα is localized at the ER, but synthesizes a plasma membrane pool of PI(4)P possibly at plasma membrane/ER contact sites [84]. Both PI4KIIα and PI4KIIIβ localize to Golgi membranes and generate Golgi PI(4)P. PI4KIIα is not restricted to the Golgi and is also present at the ER, the plasma membrane and late endosomal compartments [83, 85, 86]. PI4KIIα is modified by palmitoylation at a cysteine-based motif within the catalytic domain, which is important for membrane association and targeting to the Golgi [87, 88]. The activity of PI4KIIα is regulated by cholesterol levels and biochemical fractionation experiments suggest that this enzyme may preferentially associate with lipid-raft domains [88, 89]. Specific enrichment of PI4KIIα at cholesterol-rich Golgi regions may explain the spatial segregation of PI4KIIα and PI4KIIIβ within the Golgi that was observed in polarized cells [90].
Targeting of PI4KIIIβto the Golgi requires complex formation with neuronal calcium sensor-1 (NCS-1), a myristoylated co-factor that is also involved in modulating Ca2+-dependent processes in neuronal cells [91–93]. NCS-1 binds to a variety of proteins involved in signal transduction, including ion channels, G-protein-coupled receptors and inositol trisphosphate receptors, but the exact role of this protein in neurotransmission and synaptic plasticity remains to be determined [94]. In yeast, the PI4KIIIβhomolog Pik1 forms a similar complex with the myristoylated co-factor Frq1 [95, 96]. Recruitment of PI4KIIIβ to the Golgi is regulated by the small GTPase Arf1 that interacts with both PI4KIIIβ and NCS-1 [92, 97]. The enzymatic activity of PI4KIIIβ at the Golgi is controlled by protein kinase D (PKD). Phosphorylation of PI4KIIIβ by PKD1 or PKD2 triggers recruitment of 14-3-3 proteins, which prevents dephosphorylation and causes sustained activity of PI4KIIIβ [98, 99]. Yeast Pik1 is phosphorylated by unknown kinases and phosphorylated Pik1 binds to 14-3-3 proteins like its mammalian counterpart. [100]. 14-3-3 binding to Pik1 regulates the distribution of Pik1 between cytosol, nucleus and Golgi membranes [100]. In Drosophila melanogaster, the PI4KIIIβ homolog four wheel drive (Fwd) binds and recruits the recycling endosomal Rab11 to Golgi membranes during cytokinesis, which is required for proper cell division [101]. Overall these results indicate that Golgi PI 4-kinases generate functionally distinct pools of PI(4)P, perhaps in response to specific physiological stimuli, and future work will be required to dissect these different functions.
Golgi PI(4)P cooperates with activated Arf1 to regulate protein and lipid sorting at this organelle. The clathrin adaptors adaptor protein complex 1 (AP-1) and γ-ear-containing, ADP-ribosylation factor-binding (Gga) proteins bind directly to PI(4)P at the Golgi (Fig. 3) [102–104]. Efficient Golgi association of these clathrin adaptors requires additional binding to Arf1-GTP [102–104]. In yeast, Golgi PI(4)P can regulate the sequential activation of GTPases from the Rab family. The Rab GEF Sec2 binds simultaneously to Golgi PI(4)P and the Rab GTPase Ypt32 [105]. Sec2 can also activate the Rab GTPase Sec4, which in turn recruits the exocyst effector Sec15, but this can only occur at post-Golgi membranes with low PI(4)P, which allows Sec15 to replace Ypt31 on Sec2 [105]. Another effector in this mechanism is Myo2, a myosin responsible for transporting secretory vesicles to sites of cell growth [106]. Myo2 binds to PI(4)P at late Golgi membranes and also interacts with Ypt31 and Sec4 and thus receives inputs from both phosphoinositides and Rab proteins for membrane association and facilitating vesicular transport [107].
In mammalian cells, Golgi phosphoprotein 3 (GOLPH3) is a PI(4)P effector required for recruiting the unconventional myosin MYO18A. Depletion of GOLPH3 results in trafficking defects and aberrant Golgi morphology [108]. GOLPH3 is an oncoprotein involved in regulating mTOR ([109], which suggests an interesting functional link between the control of Golgi function and regulation of cell growth. The yeast homologue of GOLPH3 is Vps74, a protein required for Golgi enzyme trafficking and localization. [110]. Vps74 binds to PI(4)P and also interacts with Golgi glycosylation enzymes and regulates their recruitment into retrograde COP-I transport vesicles [110–112].
Another functionally distinct group of PI(4)P effectors at the Golgi is comprised of proteins involved in non-vesicular transport of lipids (Fig. 3). Ceramide transfer protein (CERT), four-phosphate adaptor proteins (FAPPs) and oxysterol-binding proteins (OSBP) have PH domains that bind to both PI(4)P and Arf1-GTP. CERT shuttles ceramide that is synthesized at the ER to the Golgi, where it is used to synthesize sphingolipids [113]. FAPP2 binds to glucosylceramide and is required for glycosphingolipid biosynthesis [114, 115]. Interestingly, FAPP proteins play also a role in anterograde trafficking from the Golgi [116–118]. Although it is not entirely clear how FAPP2 functions in these different processes, recent studies show that FAPP2 can dimerize, which triggers membrane tubulation in vitro [119, 120]. FAPP2 may therefore coordinate local sphingolipid distribution at the Golgi and promote assembly of trafficking structures. [118–120].
OSBP and OSBP related proteins (ORP) play a role in both lipid homeostasis and membrane traffic. It is possible that some of these proteins catalyze sterol transfer between different compartments at specific membrane contact sites, which may be important for organellar integrity and trafficking [121–123]. In yeast, the homologous Osh proteins are involved in regulating the localization of Rho GTPases Arf1, Cdc42, Rho1 and the Rab GTPase Sec4 [124, 125]. It is also possible that OSBPs and ORPs function as sterols and posphoinositide sensor at organellar contact sites. Recent studies in yeast provided new exciting insights into the mechanism by which yeast Osh proteins may control lipid homeostasis. A recent report showed that Osh3 interacts simultaneously with the transmembrane adaptor Scs2 at the ER and binds to plasma membrane PI(4)P at sites that are in close proximity of the cortical ER [126]. This pool of PI(4)P is synthesized by the PI 4-kinase Stt4 that forms a stable complex with two cofactors, Ypp1 and Efr3, at spatially restricted membrane regions [127]. In turn Osh3 binds and activates Sac1, a lipid phosphatase that is mainly retained at the ER in proliferating cells [126]. Biochemical assays determined that activation by Osh3 allows Sac1 to act in trans at closely apposed lipid membranes and such a mechanism could account for Sac1-dependent regulation of Stt4-dependent PI(4)P pools at the plasma membrane [126]. In an alternative mechanism, Osh4 (also known as Kes1) may play a central role in PI(4)P homeostasis [128]. Osh4 also interacts with Sac1 and is implicated in regulating trafficking from the Golgi [124, 129]. A recent study showed that Osh4 can bind to PI(4)P in exchange for sterols in vitro and may thus function in delivering the phosphoinositide substrate to the Sac1 phosphatase [130]. Although, the source of the PI(4)P pool that is recognized by Osh4 is unknown, there is evidence that Osh4 associates with exocytic vesicles and is required for their docking at the plasma membrane during polarized growth [131]. Whether Osh4 can deliver PI(4)P from these docking sites at the plasma membrane to the ER for degradation by Sac1 remains to be determined.
Local regulation of sphingolipids and PI(4)P may be important for other aspects of Golgi membrane lipid regulation as well. Drs2, a flippase required for regulating phosphatidylserine distribution between membrane bilayer leaflets is activated by PI(4)P at the Golgi [132]. PI(4)P binds to a C-terminal regulatory domain that resembles a split PH domain [132]. In addition, ArfGEF Gea2 binds to the same C-terminal domain and also stimulates flippase activity [132]. In yeast, lipid flippases are regulated by the protein kinase Fpk1 [133]. Interestingly, the upstream kinase Ypk1 that inhibits Fpk1 is negatively regulated by complex sphingolipids [133]. These findings underline the central role of PI(4)P and sphingolipid dynamics in Golgi membrane remodeling.
The Golgi is characterized by continuous influx and exit of membranes and proteins, which requires a specific control system to prevent the random equilibration of PI(4)P pools. The major factor in controlling the steady state distribution of PI(4)P is the evolutionary conserved lipid phosphatase Sac1 that was originally discovered by genetic analyses in yeast [134, 135]. Sac1 is a type II transmembrane protein with a large cytosolic domain containing the catalytic lipid phosphatase motif [44]. In both yeast and mammals, Sac1 shuttles between ER and Golgi membranes [136]. In proliferating mammalian cells, Sac1 is mainly localized at the ER and at cisternal Golgi region, which creates a steep gradient of PI(4)P across the Golgi resulting in a particularly high concentration at the TGN (trans-Golgi network) [137]. The relevance of this distribution is not entirely clear, but deficiency in Sac1 function causes early embryonic lethality in mice and leads to aberrant Golgi structures, mitotic spindle defects and Golgi enzyme mislocalization in cultured cells [137, 138]. It is therefore possible that the PI(4)P gradient at the Golgi provides directionality for Golgi enyzme localization and anterograde trafficking. Sac1 is also required for downregulation of secretion after starvation and during quiescence. This mechanism was first discovered in yeast, where Sac1 accumulates at the Golgi and downregulates PI(4)P when cells run out of carbon sources and proliferation is terminated [139, 140]. These starvation conditions also cause dissociation of PI 4-kinase Pik1 from the Golgi [100, 139]. When starved cells are stimulated with glucose, Sac1 rapidly translocates back to the ER and Pik1 reassociates with Golgi membranes [100, 139]. A similar mechanism is also present in mammalian cells. Mammalian Sac1 orthologues have the ability to oligomerize during serum starvation, which triggers recruitment into COP-II vesicles and accumulation of Sac1 at the TGN [141, 142]. Downregulation of Golgi PI(4)P in quiescent cells produces a reduction in secretory capacity [141]. Stimulation with growth factors induces rapid shuttling of Sac1 back to the ER [141]. The nutrient and growth-factor regulated traffic of Sac1 is therefore an important mechanism to synchronize secretion and Golgi function with growth and proliferation.
PI(4,5)P2 regulates membrane dynamics at the plasma membrane
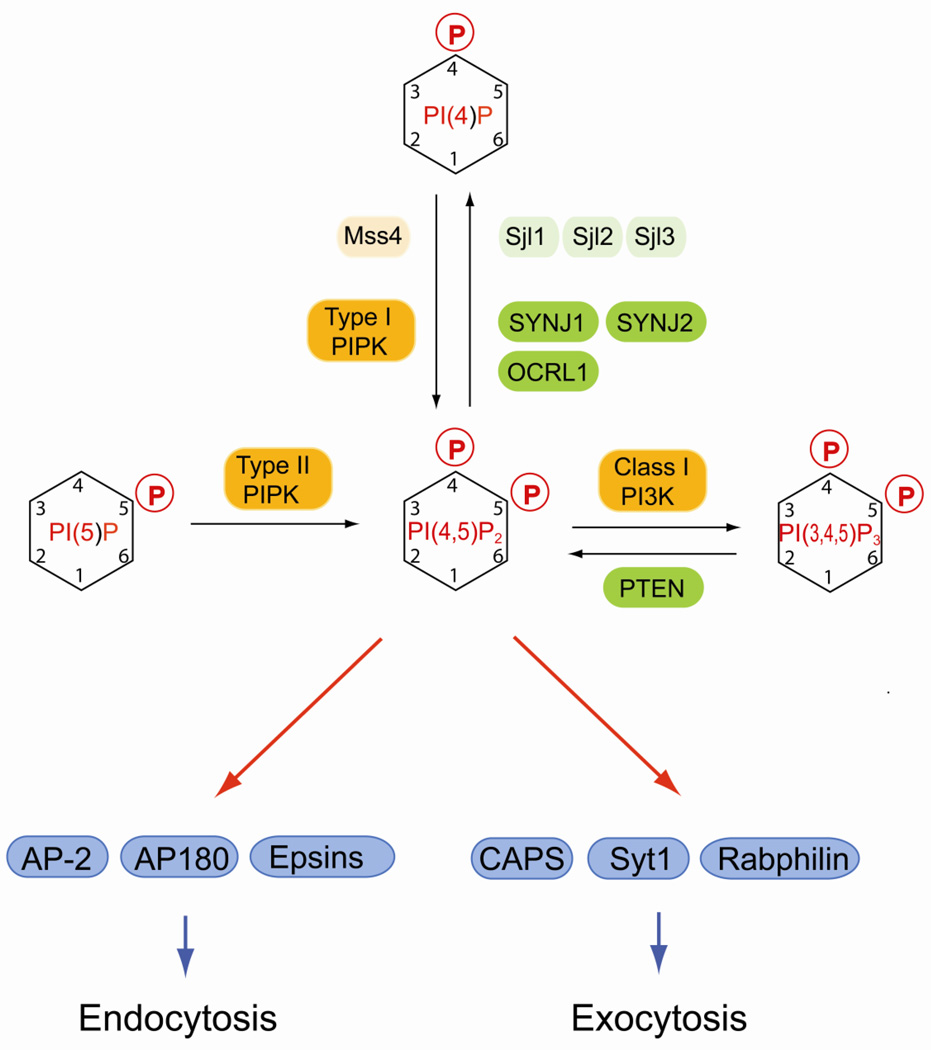
PI(4,5)P2 is enriched at the plasma membrane and regulates many distinct processes at this location [143]. Small fractions of PI(4,5)P2 were also detected at the Golgi and the nuclear envelope [144]. PIP5K activity appears be associated with Golgi membrane, however, the enzyme responsible for this activity was not identified [145].
Plasma membrane PI(4,5)P2 is produced by two distinct classes of lipid kinases (Fig. 4). Type I PIP kinase phosphorylates PI(4)P and this reaction is the major route for plasma membrane for PI(4,5)P2 biosynthesis [146]. However, PI(4,5)P2 can also be generated from PI(5)P by type II PIP kinase [146]. In yeast cells, all PI(4,5)P2 is generated by Mss4 utilizing two distinct PI(4)P pools that are generated by Pik1 and Stt4 at the Golgi and the plasma membrane respectively [147, 148]. Interestingly, pik1 and stt4 mutants have distinct phenotypes either in endocytosis or in actin cytoskeletal arrangement that are likely caused by PI(4,5)P2 deficiency [148], indicating that PI(4,5)P2 generated at the plasma membrane may form functionally distinct pools. This idea is supported by a study that showed anisotropic distribution of plasma membrane PI(4,5)P2 in yeast cells during mating that depends on Stt4 but is independent of Pik1 [149].
Fig. 4.
Regulation of PI(4,5)P2 and its function in vesicular traffic. In yeast, PI(4,5)P2 is generated by the PIP 5-kinase Mss4. In mammals, PI(4,5)P2 is synthesized by type I and type II PIP kinases either from PI(4)P or from PI(5)P, respectively. PI(4,5)P2 is enriched at the plasma membrane, where it regulates clathrin-mediated endocytosis and exocytosis of synaptic vesicles and secretory granules. Rapid turnover of PI(4,5)P2 by lipid phosphatases from the synaptojanin family is important for rapid recycling of endocytic vesicles. Early endosomal trafficking and recycling is also regulated by the lipid phospatase OCRL1. PI(4,5)P2 regulates many additional processes including the actin cytoskeleton, ion channels, and signal transduction at the plasma membrane, which is not depicted in the figure. Mammalian lipid kinases are marked by orange boxes, lipid phosphatases are in green boxes. Yeast homologs are indicated by lighter shades of the same colors. Effector proteins are in blue boxes.
PI(4,5)P2 is important for classical signal transduction either via phospholipase C-mediated cleavage into inositol-trisphosphate and diacylglycerol or via PI 3-kinase-dependent phosphorylation to PI(3,4,5)P3 [2, 150]. PI(4,5)P2 is also important for interactions between the plasma membrane and the cytoskeleton [150]. PI(4,5)P2 plays additional critical roles in regulating exocytosis and endocytosis. During exocytosis, plasma membrane PI(4,5)P2 functions in immobilizing secretory granules and in controlling docking of synaptic vesicles prior to fusion [151, 152]. Several PI(4,5)P2 binding proteins play a role in this process including CAPS (calcium-activated protein for secretion), Syt1 and rabphilin (Fig. 4) [152].
PI(4,5)P2 is also directly involved in regulating clathrin-mediated endocytosis. Many of the clathrin adaptors required for endocytosis such as AP-2, AP180 and epsin bind directly to PI(4,5)P2 (Fig.4) [153–155]. The initial PI(4,5)P2-dependent plasma membrane recruitment of these factors is stabilized by additional interactions with transmembrane cargo proteins containing tyrosine based sorting motifs [156]. Plasma membrane PI(4,5)P2 is also important for regulating actin dynamics during the endocytic internalization reaction [157]. After internalization, PI(4,5)P2 is rapidly turned over by lipid phosphatases mainly from the synaptojanin family, which is essential for the disassembly of clathrin coats at endocytic vesicles (Fig. 4) [158]. Deletion of synaptojanin-1 in mice causes accumulation of clathrin-coated vesicles and leads to neurological defects and perinatal lethality [159]. Zebrafish synaptojanin-1 mutants have a less severe phenotype and show defective synaptic transmission at hair-cell synapses [160]. In contrast, a mutation in synaptojanin-2 results in progressive hearing loss and hair cell degeneration in mice [161]. In yeast, deletion of synaptojanin-like phosphatases causes defects in actin organization, endocytosis, and clathrin-mediated sorting between the Golgi and endosome that correlate with an accumulation of PI(4,5)P2 at the cell periphery [162]. Though these studies show species-dependent differences in phenotypes, it is clear that turnover of PI(4,5)P2 by synaptojanin lipid phosphatases is instrumental for rapid recycling of clathrin coated endocytic vesicles.
PI(4,5)P2 is also dephosphorylated by the 5-phosphatase OCRL1 that localizes to early endosomes and Golgi compartment. Mutations in OCRL1 cause Lowe syndrome, which is characterized by a triad of clinical symptoms consisting of congenital cataracts, mental retardation and renal proximal tubular dysfunction [163]. OCRL1 interacts with clathrin, the adaptor protein APPL and several proteins from the Rab GTPase family and is involved in the endocytic pathway and in trafficking between endosomes and the trans-Golgi network [164–167]. A recent report shows that OCRL deficiency causes accumulation of PI(4,5)P2 in early endosomes and induces an increase in endosomal Factin, which may be the cause for the observed defects in early endosomal dynamics and in receptor recycling at the plasma membrane [168].
6. PI(5)P plays distinct roles in nuclear signaling and in membrane dynamics
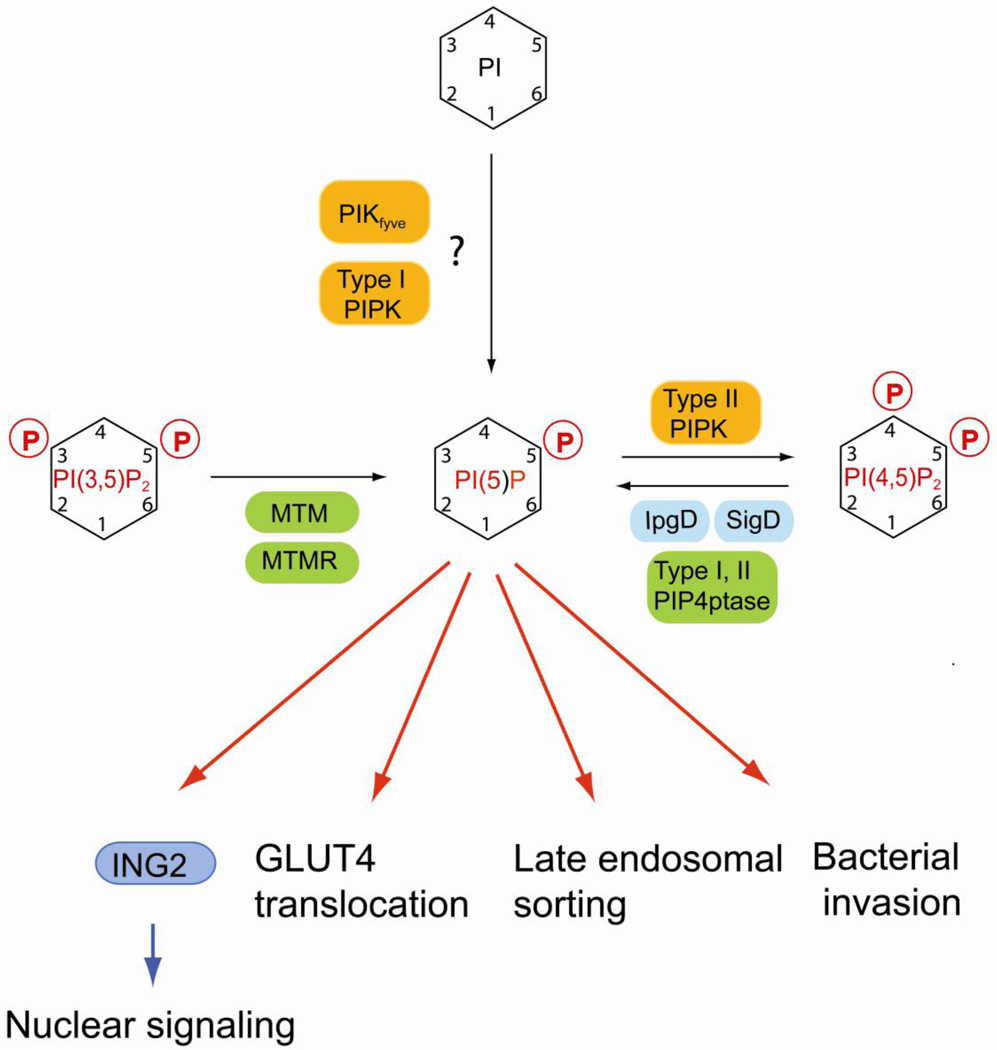
PI(5)P represents probably the least characterized phosphoinositide (Fig. 5). Although improved methods for the quantitative analysis of PI(5)P from cell extracts and subcellular fractions using HPLC are now available [169], it is unclear where this phosphoinositide localizes in live cells. PI(5)P appears to play a regulatory role in the nucleus and was shown to bind to the PHD finger motif in the nuclear protein inhibitor of growth protein-2 (ING2) [170]. This protein is involved in controlling histone acetylation and p53 during cellular stress [171].
Fig. 5.
Regulation of PI(5)P and its role in nuclear signaling and membrane dynamics. PI(5)P can be synthesized by MTM and MTMR phosphatases via dephosphorylation of PI(3,5)P2 Salmonella and Shigella bacterial pathogens express well characterized lipid phosphatases (IpgD and SigD) that convert PI(4,5)P2 to PI(5)P upon injection into the host cell which is important for the invasion mechanism. Recently, mammalian type I and II PIP 4-phosphatases have been identified that generate PI(5)P in vitro using PI(4,5)P2 as substrate. Biochemical assay suggest that PIKfyve and type I PIP kinase can directly phosphorylate PI to create PI(5)P, but it is unclear whether this reaction is relevant for the physiological regulation of PI(5)P. PI(5)P binds to the nuclear adaptor ING2 that regulates chromatin rearrangement in response to stress. But there is mounting evidence that PI(5)P may play an important role in regulating membrane dynamics in both endocytic and exocytic sorting. Mammalian lipid kinases are marked by orange boxes, lipid phosphatases are in green boxes. Bacterial lipid phosphatases are in cyan boxes. Effector proteins are in blue boxes.
The mechanisms for PI(5)P biosynthesis are not well characterized. Theoretically, PI(5)P can be generated either by dephosphorylation of PI(3,5)P2 and PI(4,5)P2 or by phosphorylation of PI (Fig.5) [172]. The most established pathway is the generation of PI(5)P from PI(3,5)P2 by the 3-phosphatase myotubularin [173]. Two lipid phosphatases, termed type I and type II PIP 4-phosphatase, have been described and these enzymes convert PI(4,5)P2 to PI(5)P in biochemical assays [174]. Both enzymes localize to endosomal and lysosomal membranes in cultured epithelial cells and it appears likely that these phosphatases contribute to the physiological regulation of PI(5)P. Finally, in vitro assays suggest that both type I PIP 5-kinase and PIKfyve can use PI as a substrate for PI(5)P synthesis [58, 175, 176], but it remains controversial whether these reactions occur in live cells [172].
Interestingly, several bacterial enzymes convert plasma membrane PI(4,5)P2 to PI(5)P in vivo. Shigella flexneri is a bacterial pathogen that uses a type 3 secretion system to inject virulence factors into host cells to facilitate invasion [177]. Among these factors is invasion plasmid gene D (IpgD) a protein containing domains homologous to mammalian inositol 4-phosphatases. IpgD-mediated conversion of PI(4,5)P2 to PI(5)P induces membrane and actin cytoskeletal rearrangements that are required for the invasion process [178]. Recent evidence suggest that PI(5)P inhibits selectively the degradation of epidermal growth factor receptor (EGFR) by delaying its trafficking to late endosomal and lysosomal compartments, which in turn enhances signaling through the pathways downstream of EGFR in particular via Akt [179]. This mechanism for bacterial invasion is also found in other bacteria. Salmonella typhimurium contains the IpgD homolog SigD, which appears to function in the invasion process by breakdown of PI(4,5)P2 coupled to PI(5)P production [180].
Stimulation of cultured adipocytes causes a transient increase in PI(5)P levels, which promotes Akt phosphorylation and GLUT4 translocation to the plasma membrane [181, 182]. Together with the results obtained for bacterial pathogen-mediated control of PI(5)P, these findings suggest that this phosphoinositide plays an important but as yet incompletely defined role in membrane trafficking and dynamics.
7. Perspectives and conclusions
Phosphoinositides are intimately involved in controlling virtually all vesicular transport pathways. Recent evidence suggests that specific phosphoinositides act in concert with small GTPases mainly from the Arf1 and Rab family. A combinatorial code consisting of compartment-specific sets of phosphoinositides and activated Arfs and Rabs is responsible for high affinity recruitment of effector proteins to specific sites. In addition, phosphoinositides control membrane-cytoskeletal interactions, ion transporters and signal transduction at the plasma membrane, all of which is critical for proper control of membrane dynamics and organellar function. How the localization of certain phosphoinositides at specific compartments is regulated is not well understood. One limitation in the experimental approach to elucidate these question involves the probes that are commonly used to visualize phosphoinositides by fluorescence microscopy. It is likely that highly dynamic and functionally important phosphoinositide pools exist that cannot be detected using these tools. A given phosphoinositide can be rapidly converted to another phosphoinositide species, which either terminates or changes its physiological role. It is clear that lipid kinases and phosphatases play a paramount role in this regulation. However, the localization of these enzymes does not always explain the localization profile of the various phosphoinositides and future work will be required to resolve these questions. Finally, even small disturbances in the turnover or steady state levels of phosphoinositides lead to specific human disorders and it will be a future challenge to define how these lipid-based pathways intersect with other cellular regulation systems.
Highlights.
> Phosphoinositide lipids are critical regulators of membrane traffic. > Important roles in exocytosis, endocytosis, endosomal traffic and Golgi function. > Additional roles in autophagy and organellar dynamics. > Relationship to human disease is discussed.
Acknowledgements
We thank T. Nicolson for comments on the manuscript. P.M is funded by grants from the National Institutes of Health (GM071569 and GM084088).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol Life Sci. 2008;65:2833–2841. doi: 10.1007/s00018-008-8353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 4.Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 5.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 6.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 7.Stephens L, Hawkins PT, Downes CP. Metabolic and structural evidence for the existence of a third species of polyphosphoinositide in cells: D-phosphatidyl-myo-inositol 3-phosphate. Biochem J. 1989;259:267–276. doi: 10.1042/bj2590267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 9.Falasca M, Maffucci T. Rethinking phosphatidylinositol 3-monophosphate. Biochim Biophys Acta. 2009;1793:1795–1803. doi: 10.1016/j.bbamcr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 11.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. Embo J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269:31552–31562. [PubMed] [Google Scholar]
- 13.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. Embo J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H. FYVE finger proteins as effectors of phosphatidylinositol 3-phosphate. Chem Phys Lipids. 1999;98:87–94. doi: 10.1016/s0009-3084(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 16.Sato TK, Overduin M, Emr SD. Location, location, location: membrane targeting directed by PX domains. Science. 2001;294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- 17.Simonsen A, Stenmark H. PX domains: attracted by phosphoinositides. Nat Cell Biol. 2001;3:E179–E182. doi: 10.1038/35087112. [DOI] [PubMed] [Google Scholar]
- 18.Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 19.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 20.Callaghan J, Simonsen A, Gaullier JM, Toh BH, Stenmark H. The endosome fusion regulator early-endosomal autoantigen 1 (EEA1) is a dimer. Biochem J. 1999;338(Pt 2):539–543. [PMC free article] [PubMed] [Google Scholar]
- 21.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 22.Simonsen A, Gaullier JM, D'Arrigo A, Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J Biol Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- 23.Lawe DC, Patki V, Heller-Harrison R, Lambright D, Corvera S. The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J Biol Chem. 2000;275:3699–3705. doi: 10.1074/jbc.275.5.3699. [DOI] [PubMed] [Google Scholar]
- 24.Lawe DC, Chawla A, Merithew E, Dumas J, Carrington W, Fogarty K, Lifshitz L, Tuft R, Lambright D, Corvera S. Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J Biol Chem. 2002;277:8611–8617. doi: 10.1074/jbc.M109239200. [DOI] [PubMed] [Google Scholar]
- 25.Mishra A, Eathiraj S, Corvera S, Lambright DG. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1) Proc Natl Acad Sci U S A. 2010;107:10866–10871. doi: 10.1073/pnas.1000843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 27.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 2002;3:416–427. doi: 10.1034/j.1600-0854.2002.30605.x. [DOI] [PubMed] [Google Scholar]
- 28.Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D'Arrigo A, Stang E, Stenmark H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- 29.Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 30.Vieira OV, Harrison RE, Scott CC, Stenmark H, Alexander D, Liu J, Gruenberg J, Schreiber AD, Grinstein S. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Mol Cell Biol. 2004;24:4593–4604. doi: 10.1128/MCB.24.10.4593-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong Q, Lazar CS, Tronchere H, Sato T, Meerloo T, Yeo M, Songyang Z, Emr SD, Gill GN. Endosomal localization and function of sorting nexin 1. Proc Natl Acad Sci U S A. 2002;99:6767–6772. doi: 10.1073/pnas.092142699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Weering JR, Verkade P, Cullen PJ. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Semin Cell Dev Biol. 2010;21:371–380. doi: 10.1016/j.semcdb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9:133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 34.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 35.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 36.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 37.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 38.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Ma Z, Jiao X, Fariss R, Kantorow WL, Kantorow M, Pras E, Frydman M, Riazuddin S, Riazuddin SA, Hejtmancik JF. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am J Hum Genet. 2011;88:827–838. doi: 10.1016/j.ajhg.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon MS, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 2011;195:435–447. doi: 10.1083/jcb.201107033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 44.Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 45.Hughes WE, Woscholski R, Cooke FT, Patrick RS, Dove SK, McDonald NQ, Parker PJ. SAC1 Encodes a Regulated Lipid Phosphoinositide Phosphatase, Defects in Which Can Be Suppressed by the Homologous Inp52p and Inp53p Phosphatases. J Biol Chem. 2000;275:801–808. doi: 10.1074/jbc.275.2.801. [DOI] [PubMed] [Google Scholar]
- 46.Nemoto Y, Kearns BG, Wenk MR, Chen H, Mori K, Alb JG, Jr, De Camilli P, Bankaitis VA. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J Biol Chem. 2000;275:34293–34305. doi: 10.1074/jbc.M003923200. [DOI] [PubMed] [Google Scholar]
- 47.Clague MJ, Lorenzo O. The myotubularin family of lipid phosphatases. Traffic. 2005;6:1063–1069. doi: 10.1111/j.1600-0854.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 48.Begley MJ, Dixon JE. The structure and regulation of myotubularin phosphatases. Curr Opin Struct Biol. 2005;15:614–620. doi: 10.1016/j.sbi.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Cao C, Laporte J, Backer JM, Wandinger-Ness A, Stein MP. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 2007;8:1052–1067. doi: 10.1111/j.1600-0854.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 50.Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger-Ness A. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Biol Cell. 2008;19:3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson FL, Niesman IR, Beiswenger KK, Dixon JE. Loss of the inactive myotubularin-related phosphatase Mtmr13 leads to a Charcot-Marie-Tooth 4B2-like peripheral neuropathy in mice. Proc Natl Acad Sci U S A. 2008;105:4916–4921. doi: 10.1073/pnas.0800742105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson FL, Dixon JE. The phosphoinositide-3-phosphatase MTMR2 associates with MTMR13, a membrane-associated pseudophosphatase also mutated in type 4B Charcot-Marie-Tooth disease. J Biol Chem. 2005;280:31699–31707. doi: 10.1074/jbc.M505159200. [DOI] [PubMed] [Google Scholar]
- 53.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 54.Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323(Pt 3):597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 56.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikonomov OC, Sbrissa D, Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J Biol Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 58.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J Biol Chem. 1999;274:21589–21597. doi: 10.1074/jbc.274.31.21589. [DOI] [PubMed] [Google Scholar]
- 59.Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, Parker PJ, Lemmon MA. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. Embo J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barth H, Meiling-Wesse K, Epple UD, Thumm M. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 2001;508:23–28. doi: 10.1016/s0014-5793(01)03016-2. [DOI] [PubMed] [Google Scholar]
- 61.Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 62.Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denis V, Cyert MS. Internal Ca(2+) release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J Cell Biol. 2002;156:29–34. doi: 10.1083/jcb.200111004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palmer CP, Zhou XL, Lin J, Loukin SH, Kung C, Saimi Y. A TRP homolog in Saccharomyces cerevisiae forms an intracellular Ca(2+)-permeable channel in the yeast vacuolar membrane. Proc Natl Acad Sci U S A. 2001;98:7801–7805. doi: 10.1073/pnas.141036198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q, Cheng X, Zhang Y, Weisman LS, Delling M, Xu H. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. Embo J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Camilli P, Chen H, Hyman J, Panepucci E, Bateman A, Brunger AT. The ENTH domain. FEBS Lett. 2002;513:11–18. doi: 10.1016/s0014-5793(01)03306-3. [DOI] [PubMed] [Google Scholar]
- 68.Chidambaram S, Mullers N, Wiederhold K, Haucke V, von Mollard GF. Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J Biol Chem. 2004;279:4175–4179. doi: 10.1074/jbc.M308667200. [DOI] [PubMed] [Google Scholar]
- 69.Eugster A, Pecheur EI, Michel F, Winsor B, Letourneur F, Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol Biol Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duncan MC, Costaguta G, Payne GS. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- 71.Duex JE, Tang F, Weisman LS. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol. 2006;172:693–704. doi: 10.1083/jcb.200512105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz D, Meisler MH, Weisman LS. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. Embo J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem. 2009;284:35794–35806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sbrissa D, Ikonomov OC, Fenner H, Shisheva A. ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J Mol Biol. 2008;384:766–779. doi: 10.1016/j.jmb.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell. 2008;19:4273–4286. doi: 10.1091/mbc.E08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, Meisler MH, Weisman LS. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci U S A. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katona I, Zhang X, Bai Y, Shy ME, Guo J, Yan Q, Hatfield J, Kupsky WJ, Li J. Distinct pathogenic processes between Fig4-deficient motor and sensory neurons. Eur J Neurosci. 2011;33:1401–1410. doi: 10.1111/j.1460-9568.2011.07651.x. [DOI] [PubMed] [Google Scholar]
- 79.Nicholson G, Lenk GM, Reddel SW, Grant AE, Towne CF, Ferguson CJ, Simpson E, Scheuerle A, Yasick M, Hoffman S, Blouin R, Brandt C, Coppola G, Biesecker LG, Batish SD, Meisler MH. Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P phosphatase FIG4. Brain. 2011;134:1959–1971. doi: 10.1093/brain/awr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 81.D'Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P - not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- 82.Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 83.Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem. 2002;277:20041–20050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- 84.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J, Wenk MR, Pellegrini L, Onofri F, Benfenati F, De Camilli P. Phosphatidylinositol 4-kinase type IIalpha is responsible for the phosphatidylinositol 4-kinase activity associated with synaptic vesicles. Proc Natl Acad Sci U S A. 2003;100:3995–4000. doi: 10.1073/pnas.0230488100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Minogue S, Waugh MG, De Matteis MA, Stephens DJ, Berditchevski F, Hsuan JJ. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci. 2006;119:571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- 87.Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Sudhof TC, Albanesi JP. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J Biol Chem. 2001;276:7705–7708. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- 88.Barylko B, Mao YS, Wlodarski P, Jung G, Binns DD, Sun HQ, Yin HL, Albanesi JP. Palmitoylation controls the catalytic activity and subcellular distribution of phosphatidylinositol 4-kinase II{alpha} J Biol Chem. 2009;284:9994–10003. doi: 10.1074/jbc.M900724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Waugh MG, Minogue S, Chotai D, Berditchevski F, Hsuan JJ. Lipid and peptide control of phosphatidylinositol 4-kinase IIalpha activity on Golgi-endosomal Rafts. J Biol Chem. 2006;281:3757–3763. doi: 10.1074/jbc.M506527200. [DOI] [PubMed] [Google Scholar]
- 90.Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- 91.Zhao X, Varnai P, Tuymetova G, Balla A, Toth ZE, Oker-Blom C, Roder J, Jeromin A, Balla T. Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J Biol Chem. 2001;276:40183–40189. doi: 10.1074/jbc.M104048200. [DOI] [PubMed] [Google Scholar]
- 92.Haynes LP, Thomas GM, Burgoyne RD. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J Biol Chem. 2005;280:6047–6054. doi: 10.1074/jbc.M413090200. [DOI] [PubMed] [Google Scholar]
- 93.de Barry J, Janoshazi A, Dupont JL, Procksch O, Chasserot-Golaz S, Jeromin A, Vitale N. Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-beta interaction in regulated exocytosis of PC12 cells. J Biol Chem. 2006;281:18098–18111. doi: 10.1074/jbc.M509842200. [DOI] [PubMed] [Google Scholar]
- 94.Weiss JL, Hui H, Burgoyne RD. Neuronal calcium sensor-1 regulation of calcium channels, secretion, and neuronal outgrowth. Cell Mol Neurobiol. 2010;30:1283–1292. doi: 10.1007/s10571-010-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hendricks KB, Wang BQ, Schnieders EA, Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- 96.Huttner IG, Strahl T, Osawa M, King DS, Ames JB, Thorner J. Molecular interactions of yeast frequenin (Frq1) with the phosphatidylinositol 4-kinase isoform, Pik1. J Biol Chem. 2003;278:4862–4874. doi: 10.1074/jbc.M207920200. [DOI] [PubMed] [Google Scholar]
- 97.Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 98.Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hausser A, Link G, Hoene M, Russo C, Selchow O, Pfizenmaier K. Phospho-specific binding of 14-3-3 proteins to phosphatidylinositol 4-kinase III beta protects from dephosphorylation and stabilizes lipid kinase activity. J Cell Sci. 2006;119:3613–3621. doi: 10.1242/jcs.03104. [DOI] [PubMed] [Google Scholar]
- 100.Demmel L, Beck M, Klose C, Schlaitz AL, Gloor Y, Hsu PP, Havlis J, Shevchenko A, Krause E, Kalaidzidis Y, Walch-Solimena C. Nucleocytoplasmic shuttling of the Golgi phosphatidylinositol 4-kinase pik1 is regulated by 14-3-3 proteins and coordinates Golgi function with cell growth. Mol Biol Cell. 2008;19:1046–1061. doi: 10.1091/mbc.E07-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Polevoy G, Wei HC, Wong R, Szentpetery Z, Kim YJ, Goldbach P, Steinbach SK, Balla T, Brill JA. Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J Cell Biol. 2009;187:847–858. doi: 10.1083/jcb.200908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci U S A. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Demmel L, Gravert M, Ercan E, Habermann B, Muller-Reichert T, Kukhtina V, Haucke V, Baust T, Sohrmann M, Kalaidzidis Y, Klose C, Beck M, Peter M, Walch-Solimena C. The Clathrin Adaptor Gga2p Is a Phosphatidylinositol 4-phosphate Effector at the Golgi Exit. Mol Biol Cell. 2008;19:1991–2002. doi: 10.1091/mbc.E06-10-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mizuno-Yamasaki E, Medkova M, Coleman J, Novick P. Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev Cell. 2010;18:828–840. doi: 10.1016/j.devcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Govindan B, Bowser R, Novick P. The role of myo2, a yeast class-v myosin, in vesicular transport. J. Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev Cell. 2011;20(1):47–59. doi: 10.1016/j.devcel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, Fuchs GJ, Meerloo T, Farquhar MG, Zhou H, Field SJ. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, Huang J, Saci A, Widlund HR, Fisher DE, Xiao Y, Rimm DL, Protopopov A, Wong KK, Chin L. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol. 2009;187:967–975. doi: 10.1083/jcb.200909063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tu L, Tai WC, Chen L, Banfield DK. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321:404–407. doi: 10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- 112.Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yasuda S, Kitagawa H, Ueno M, Ishitani H, Fukasawa M, Nishijima M, Kobayashi S, Hanada K. A novel inhibitor of ceramide trafficking from the endoplasmic reticulum to the site of sphingomyelin synthesis. J Biol Chem. 2001;276:43994–44002. doi: 10.1074/jbc.M104884200. [DOI] [PubMed] [Google Scholar]
- 114.D'Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 115.Halter D, Neumann S, van Dijk SM, Wolthoorn J, de Maziere AM, Vieira OV, Mattjus P, Klumperman J, van Meer G, Sprong H. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J Cell Biol. 2007;179:101–115. doi: 10.1083/jcb.200704091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Godi A, Campli AD, Konstantakopoulos A, Tullio GD, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 117.Vieira OV, Verkade P, Manninen A, Simons K. FAPP2 is involved in the transport of apical cargo in polarized MDCK cells. J Cell Biol. 2005;170:521–526. doi: 10.1083/jcb.200503078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yui N, Okutsu R, Sohara E, Rai T, Ohta A, Noda Y, Sasaki S, Uchida S. FAPP2 is required for aquaporin-2 apical sorting at trans-Golgi network in polarized MDCK cells. Am J Physiol Cell Physiol. 2009;297:C1389–C1396. doi: 10.1152/ajpcell.00098.2009. [DOI] [PubMed] [Google Scholar]
- 119.Cao X, Coskun U, Rossle M, Buschhorn SB, Grzybek M, Dafforn TR, Lenoir M, Overduin M, Simons K. Golgi protein FAPP2 tubulates membranes. Proc Natl Acad Sci U S A. 2009;106:21121–21125. doi: 10.1073/pnas.0911789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lenoir M, Coskun U, Grzybek M, Cao X, Buschhorn SB, James J, Simons K, Overduin M. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 2010;11:279–284. doi: 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ngo M, Ridgway ND. Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell. 2009;20:1388–1399. doi: 10.1091/mbc.E08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed-membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187:889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kozminski KG, Alfaro G, Dighe S, Beh CT. Homologues of oxysterol-binding proteins affect Cdc42p- and Rho1p-mediated cell polarization in Saccharomyces cerevisiae. Traffic. 2006;7:1224–1242. doi: 10.1111/j.1600-0854.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 126.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 127.Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Levine TP. Lipid traffic: Osh4p makes an unexpected exchange. J Cell Biol. 2011;195:927–929. doi: 10.1083/jcb.201111074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MKY, Bankaitis VA. Kes1p shares homology with human oxysterol binding-protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 130.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alfaro G, Johansen J, Dighe SA, Duamel G, Kozminski KG, Beh CT. The sterol-binding protein Kes1/Osh4p is a regulator of polarized exocytosis. Traffic. 2011;12:1521–1536. doi: 10.1111/j.1600-0854.2011.01265.x. [DOI] [PubMed] [Google Scholar]
- 132.Natarajan P, Liu K, Patil DV, Sciorra VA, Jackson CL, Graham TR. Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol. 2009;11:1421–1426. doi: 10.1038/ncb1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci U S A. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blagoveshchenskaya A, Mayinger P. SAC1 lipid phosphatase and growth control of the secretory pathway. Mol Biosyst. 2009;5:36–42. doi: 10.1039/b810979f. [DOI] [PubMed] [Google Scholar]
- 137.Cheong FY, Sharma V, Blagoveshchenskaya A, Oorschot VM, Brankatschk B, Klumperman J, Freeze HH, Mayinger P. Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity. Traffic. 2010;11:1180–1190. doi: 10.1111/j.1600-0854.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Y, Boukhelifa M, Tribble E, Morin-Kensicki E, Uetrecht A, Bear JE, Bankaitis VA. The Sac1 Phosphoinositide Phosphatase Regulates Golgi Membrane Morphology and Mitotic Spindle Organization in Mammals. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-12-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Faulhammer F, Kanjilal-Kolar S, Knodler A, Lo J, Lee Y, Konrad G, Mayinger P. Growth control of Golgi phosphoinositides by reciprocal localization of sac1 lipid phosphatase and pik1 4-kinase. Traffic. 2007;8:1554–1567. doi: 10.1111/j.1600-0854.2007.00632.x. [DOI] [PubMed] [Google Scholar]
- 140.Faulhammer F, Konrad G, Brankatschk B, Tahirovic S, Knodler A, Mayinger P. Cell growth-dependent coordination of lipid signaling and glycosylation is mediated by interactions between Sac1p and Dpm1p. J Cell Biol. 2005;168:185–191. doi: 10.1083/jcb.200407118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knodler A, Nicolson T, Boehmelt G, Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem. 2003;278:52689–52699. doi: 10.1074/jbc.M307983200. [DOI] [PubMed] [Google Scholar]
- 143.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 144.Watt SA, Kular G, Fleming IN, Downes CP, Lucocq JM. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem J. 2002;363:657–666. doi: 10.1042/0264-6021:3630657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jones DH, Morris JB, Morgan CP, Kondo H, Irvine RF, Cockcroft S. Type I phosphatidylinositol 4-phosphate 5-kinase directly interacts with ADP-ribosylation factor 1 and is responsible for phosphatidylinositol 4,5-bisphosphate synthesis in the golgi compartment. J Biol Chem. 2000;275:13962–13966. doi: 10.1074/jbc.c901019199. [DOI] [PubMed] [Google Scholar]
- 146.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 147.Desrivieres S, Cooke FT, Parker PJ, Hall MN. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem. 1998;273:15787–15793. doi: 10.1074/jbc.273.25.15787. [DOI] [PubMed] [Google Scholar]
- 148.Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci U S A. 2010;107:11805–11810. doi: 10.1073/pnas.1005817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fukami K, Inanobe S, Kanemaru K, Nakamura Y. Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res. 2010;49:429–437. doi: 10.1016/j.plipres.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 151.Holz RW, Axelrod D. Localization of phosphatidylinositol 4,5-P(2) important in exocytosis and a quantitative analysis of chromaffin granule motion adjacent to the plasma membrane. Ann N Y Acad Sci. 2002;971:232–243. doi: 10.1111/j.1749-6632.2002.tb04467.x. [DOI] [PubMed] [Google Scholar]
- 152.Osborne SL, Wen PJ, Meunier FA. Phosphoinositide regulation of neuroexocytosis: adding to the complexity. J Neurochem. 2006;98:336–342. doi: 10.1111/j.1471-4159.2006.03892.x. [DOI] [PubMed] [Google Scholar]
- 153.Gaidarov I, Keen JH. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J Cell Biol. 1999;146:755–764. doi: 10.1083/jcb.146.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 155.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 156.Boll W, Rapoport I, Brunner C, Modis Y, Prehn S, Kirchhausen T. The mu2 subunit of the clathrin adaptor AP-2 binds to FDNPVY and YppO sorting signals at distinct sites. Traffic. 2002;3:590–600. doi: 10.1034/j.1600-0854.2002.30808.x. [DOI] [PubMed] [Google Scholar]
- 157.Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haffner C, Di Paolo G, Rosenthal JA, de Camilli P. Direct interaction of the 170 kDa isoform of synaptojanin 1 with clathrin and with the clathrin adaptor AP-2. Curr Biol. 2000;10:471–474. doi: 10.1016/s0960-9822(00)00446-2. [DOI] [PubMed] [Google Scholar]
- 159.Cremona O, Di Paolo G, Wenk MR, Luthi A, Kim WT, Takei K, Daniell L, Nemoto Y, Shears SB, Flavell RA, McCormick DA, De Camilli P. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- 160.Trapani JG, Obholzer N, Mo W, Brockerhoff SE, Nicolson T. Synaptojanin1 is required for temporal fidelity of synaptic transmission in hair cells. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000480. e1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Manji SS, Williams LH, Miller KA, Ooms LM, Bahlo M, Mitchell CA, Dahl HH. A mutation in synaptojanin 2 causes progressive hearing loss in the ENU-mutagenised mouse strain Mozart. PLoS One. 2011;6:e17607. doi: 10.1371/journal.pone.0017607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stefan CJ, Audhya A, Emr SD. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Loi M. Lowe syndrome. Orphanet J Rare Dis. 2006;1:16. doi: 10.1186/1750-1172-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A, Williams C, Konstantakopoulos A, Lucocq J, Johannes L, Rabouille C, Greene LE, Lowe M. Lowe Syndrome Protein OCRL1 Interacts with Clathrin and Regulates Protein Trafficking between Endosomes and the Trans-Golgi Network. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. Embo J. 2006;25:3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell. 2007;13:377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M. Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem. 2009 doi: 10.1074/jbc.M807442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vicinanza M, Di Campli A, Polishchuk E, Santoro M, Di Tullio G, Godi A, Levtchenko E, De Leo MG, Polishchuk R, Sandoval L, Marzolo M, De Matteis MA. OCRL controls trafficking through early endosomes via PtdIns4,5P(2)-dependent regulation of endosomal actin. Embo J. 2011 doi: 10.1038/emboj.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J. 2010;428:375–384. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 171.Bua DJ, Binda O. The return of the INGs, histone mark sensors and phospholipid signaling effectors. Curr Drug Targets. 2009;10:418–431. doi: 10.2174/138945009788185112. [DOI] [PubMed] [Google Scholar]
- 172.Lecompte O, Poch O, Laporte J. PtdIns5P regulation through evolution: roles in membrane trafficking? Trends Biochem Sci. 2008;33:453–460. doi: 10.1016/j.tibs.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 173.Tronchere H, Laporte J, Pendaries C, Chaussade C, Liaubet L, Pirola L, Mandel JL, Payrastre B. Production of phosphatidylinositol 5-phosphate by the phosphoinositide 3-phosphatase myotubularin in mammalian cells. J Biol Chem. 2004;279:7304–7312. doi: 10.1074/jbc.M311071200. [DOI] [PubMed] [Google Scholar]
- 174.Ungewickell A, Hugge C, Kisseleva M, Chang SC, Zou J, Feng Y, Galyov EE, Wilson M, Majerus PW. The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc Natl Acad Sci U S A. 2005;102:18854–18859. doi: 10.1073/pnas.0509740102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Roberts HF, Clarke JH, Letcher AJ, Irvine RF, Hinchliffe KA. Effects of lipid kinase expression and cellular stimuli on phosphatidylinositol 5-phosphate levels in mammalian cell lines. FEBS Lett. 2005;579:2868–2872. doi: 10.1016/j.febslet.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 176.Tolias KF, Rameh LE, Ishihara H, Shibasaki Y, Chen J, Prestwich GD, Cantley LC, Carpenter CL. Type I phosphatidylinositol-4-phosphate 5-kinases synthesize the novel lipids phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 5-phosphate. J Biol Chem. 1998;273:18040–18046. doi: 10.1074/jbc.273.29.18040. [DOI] [PubMed] [Google Scholar]
- 177.Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 178.Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J, Sheetz MP, Parsot C, Sansonetti PJ, Payrastre B. Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S.flexneri effector IpgD reorganizes host cell morphology. Embo J. 2002;21:5069–5078. doi: 10.1093/emboj/cdf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchere H, Payrastre B. Shigella flexneri infection generates the lipid PI5P to alter endocytosis and prevent termination of EGFR signaling. Sci Signal. 2011;4:ra61. doi: 10.1126/scisignal.2001619. [DOI] [PubMed] [Google Scholar]
- 180.Mason D, Mallo GV, Terebiznik MR, Payrastre B, Finlay BB, Brumell JH, Rameh L, Grinstein S. Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J Gen Physiol. 2007;129:267–283. doi: 10.1085/jgp.200609656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Grainger DL, Tavelis C, Ryan AJ, Hinchliffe KA. Involvement of phosphatidylinositol 5-phosphate in insulin-stimulated glucose uptake in the L6 myotube model of skeletal muscle. Pflugers Arch. 2011;462:723–732. doi: 10.1007/s00424-011-1008-4. [DOI] [PubMed] [Google Scholar]
- 182.Sbrissa D, Ikonomov OC, Strakova J, Shisheva A. Role for a novel signaling intermediate, phosphatidylinositol 5-phosphate, in insulin-regulated F-actin stress fiber breakdown and GLUT4 translocation. Endocrinology. 2004;145:4853–4865. doi: 10.1210/en.2004-0489. [DOI] [PubMed] [Google Scholar]