Abstract
Cockayne syndrome is a segmental progeria most often caused by mutations in the CSB gene encoding a SWI/SNF-like ATPase required for transcription-coupled DNA repair (TCR). Over 43 Mya before marmosets diverged from humans, a piggyBac3 (PGBD3) transposable element integrated into intron 5 of the CSB gene. As a result, primate CSB genes now generate both CSB protein and a conserved CSB-PGBD3 fusion protein in which the first 5 exons of CSB are alternatively spliced to the PGBD3 transposase. Using a host cell reactivation assay, we show that the fusion protein inhibits TCR of oxidative damage but facilitates TCR of UV damage. We also show by microarray analysis that expression of the fusion protein alone in CSB-null UV-sensitive syndrome (UVSS) cells induces an interferon-like response that resembles both the innate antiviral response and the prolonged interferon response normally maintained by unphosphorylated STAT1 (U-STAT1); moreover, as might be expected based on conservation of the fusion protein, this potentially cytotoxic interferon-like response is largely reversed by coexpression of functional CSB protein. Interestingly, expression of CSB and the CSB-PGBD3 fusion protein together, but neither alone, upregulates the insulin growth factor binding protein IGFBP5 and downregulates IGFBP7, suggesting that the fusion protein may also confer a metabolic advantage, perhaps in the presence of DNA damage. Finally, we show that the fusion protein binds in vitro to members of a dispersed family of 900 internally deleted piggyBac elements known as MER85s, providing a potential mechanism by which the fusion protein could exert widespread effects on gene expression. Our data suggest that the CSB-PGBD3 fusion protein is important in both health and disease, and could play a role in Cockayne syndrome.
Keywords: Cockayne, CSB, TCR, interferon, immunity, piggyBac
1. Introduction
Cockayne syndrome (CS) is a devastating and ultimately fatal progeroid syndrome affecting hundreds of children and occasional adults worldwide. Although often apparently normal at birth, children affected by this multisystem disorder soon exhibit postnatal growth failure, wasting (cachexia), progressive neurological and retinal degeneration, mental retardation, skeletal abnormalities, gait defects, and sun sensitivity, but never an increase in skin cancer or other tumors.
Most cases of CS are caused by mutations in the Cockayne syndrome Group B gene (CSB, also known as ERCC6) encoding a SWI/SNF-like DNA-dependent ATPase that can wind DNA [1] and remodel chromatin both in vitro [2] and in vivo [3]. The remaining cases of CS are caused by mutations in the CSA gene [4] which is required for ubiquitin-dependent degradation of CSB [5–7], or by rare alleles of the xeroderma pigmentosum (XP) genes XPB, XPD, and XPG [8]. These three XP genes are required along with XPA, XPC, XPE, and XPF for nucleotide excision repair (NER), and loss of XP gene function results in susceptibility to skin cancer.
Significantly, all 5 genes that can cause CS (CSA, CSB, XPB, XPD, and XPG) are required for transcription-coupled repair (TCR or TC-NER) where XPB and XPD are subunits of TFIIH and XPG is required to stabilize TFIIH [9]. In TCR, actively transcribing RNA polymerase II (pol II) stalls at DNA damage, triggering assembly of an NER complex that repairs the transcribed strand of the DNA and allows transcription to proceed [10]. TCR is distinct from global genome repair (GGR) which detects and repairs DNA damage on both strands of the DNA independently of transcription throughout the cell cycle. Although the NER complexes formed in TCR and GGR contain the same core factors (XPA, XPB, XPD, XPF and XPG), the GGR complex requires two additional proteins (XPC and XPE) whose functions in recognizing and partially unwinding the DNA damage are performed by pol II in TCR. An emerging view is that CSB serves as an adaptor to assemble a stable NER complex wherever pol II has stalled at DNA damage, and CSA then removes CSB and pol II leaving an NER complex in place [6, 11]. Thus CSA, CSB, XPB, XPD, and XPG mutations that cause CS may do so not just directly by failing to carry out TCR, but also indirectly by trapping scarce CSB in stable nonfunctional TCR complexes; the resulting depletion of free CSB could then affect many genes whose transcription or chromatin structure is dependent on CSB in normal growth [3] and in hypoxia [12].
Although CS is usually recessive, complete loss of CSB function does not invariably cause CS. A 33 year old male, UVSS1KO, who expressed no CSB-related proteins as a result of a nonsense mutation at CSB codon 77, exhibited UV sensitive syndrome (UVSS) but no other CS symptoms [13]. A 47 year old woman, KPSX6, with a frameshift mutation at the same codon, was initially diagnosed with UVSS and did not exhibit late-onset progeria until age 45 [14]. Thus the complete absence of CSB protein can in fact be less harmful than expression of larger CSB nonsense fragments or full length missense mutants. Most recently, Laugel et al. [15] described two CS patients, CS539VI and CS548VI, in which identical homozygous mutations spanning the 5' UTR eliminate all CSB transcription, yet cause classical early-onset CS. Interestingly, all four of these unusual UVSS or CS individuals with complete loss of CSB expression appear to be consanguineous: The parents of UVSS1KO are first cousins; the parents of KPSX6 are said to be consanguineous; and patients CS539VI and CS548VI, although apparently unrelated, are both from the highly inbred population of Reunion Island consistent with a founder effect. Consanguinity in all four of these cases may not be coincidental, and suggests that genetic background might delay or accelerate the appearance of CS symptoms. Indeed, background effects could explain the unusually heterogeneous onset, severity, and multiplicity of CS symptoms [16] as well as the telling observation that the same CSB R735opal mutation can cause either CS or a form of XP known as DeSanctis-Cacchione syndrome [17].
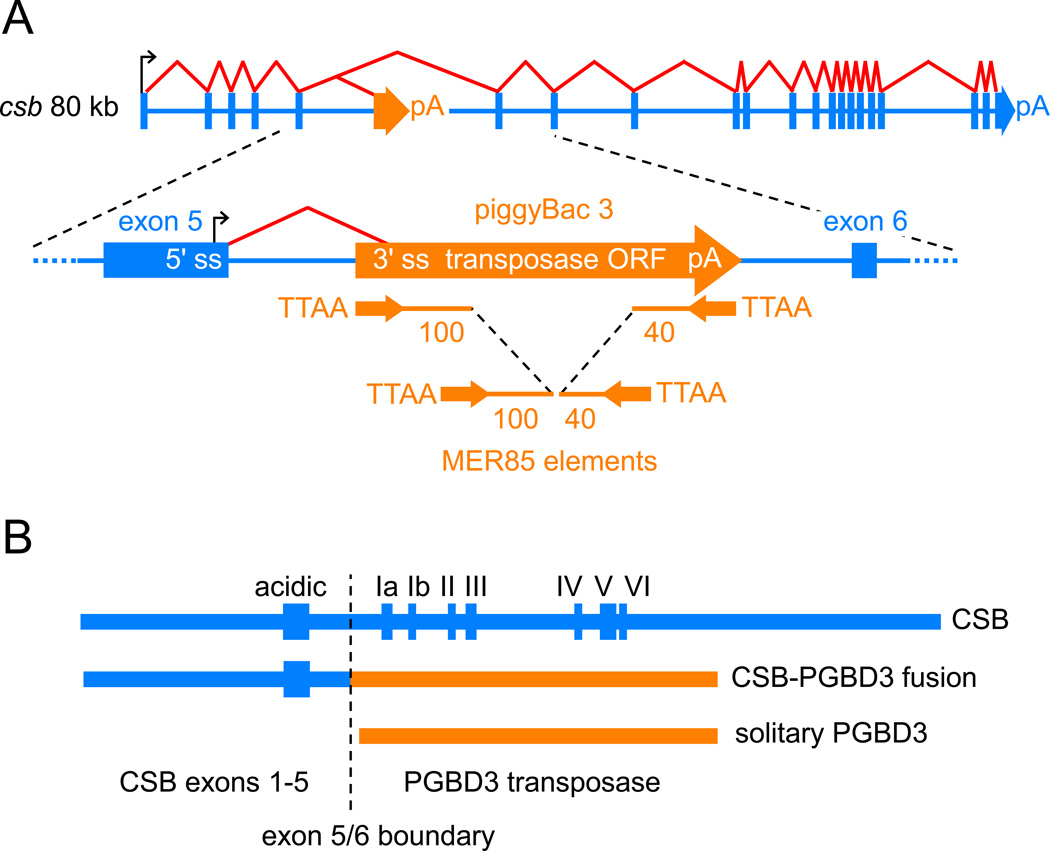
Several years ago, we found that the piggyBac transposable element PGBD3 had integrated into intron 5 of the primate CSB gene before marmosets diverged from humans >43 Mya [18]. As a result, the primate CSB gene now generates three proteins: intact CSB, a more abundant CSB-PGBD3 fusion protein in which the first 5 of the 22 CSB exons are alternatively spliced to the PGBD3 transposase, and most abundant of all, solitary PGBD3 transposase transcribed from an internal promoter in CSB exon 5 (Fig. 1). Conservation of the CSB-PGBD3 fusion protein for >43 My strongly suggests that the fusion protein is advantageous in the presence of functional CSB; and the shared N-terminal CSB domain suggests that CSB and the CSB-PGBD3 fusion protein may be functionally related.
Fig. 1.
piggyBacs are mobile DNA elements that survive as alternative 3' exons. (A) PGBD3 inserted into intron 5 of the primate CSB gene at least 43 Mya in the common ancestor of simian primates, with the result that the CSB gene now generates three proteins as shown in (B): full length CSB by default splicing of all 22 CSB exons, CSB-PGBD3 fusion protein by alternative splicing between CSB exon 5 and the PGBD3 alternative 3' terminal exon, and solitary PGBD3 driven by a cryptic promoter in CSB exon 5 [18]. The PGBD3 insertion generated a TTAA target site duplication. Immediately inside the subterminal inverted repeats of the mobile element, the transposase open reading frame (ORF) is flanked upstream by a 3' splice site (3' ss) and downstream by a polyadenylation site (pA). MER85 elements are nonautonomous internally-deleted PGBD3-derived elements that were last mobilized by a PGBD3-like transposase about 35 Mya [19]. CSB and PGBD3 sequences are indicated in blue and orange, respectively. The schematic not drawn to scale; the CSB gene spans 80 kb, the PGBD3 element 2.5 kb, and intact MER85s only 140 bp. (B) A comparison of the three proteins encoded by the CSB locus. The fusion protein joins the acidic 465 N-terminal residues of CSB exons 1–5, but none of the ATPase motifs (Roman numerals), to the 595 residue PGBD3 transposase.
CSB mutations that cause CS are uniformly distributed over the entire CSB coding region [8] but only those nonsense and frameshift mutants located downstream of intron 5 continue to make the CSB-PGBD3 fusion protein ([18] and Fig. 1). The implication is that CS usually reflects loss of functional CSB but does not require, and may even be unaffected by, continued expression of the CSB-PGBD3 fusion protein. A priori, however, the CSB-PGBD3 fusion protein could be advantageous, neutral, or disadvantageous in the absence of functional CSB.
In order to understand the roles of the CSB-PGBD3 fusion protein in health and disease, we set out to investigate the functions of the protein experimentally. We show here that (1) the fusion protein inhibits TCR of oxidative damage but facilitates TCR of UV damage — demonstrating that it can modulate DNA damage responses; (2) expression of the fusion protein in CSB-null UV-sensitive syndrome (UVSS) cells induces an interferon-like response resembling both the innate antiviral response as well as the prolonged interferon response normally maintained by unphosphorylated STAT1 (U-STAT1) — implying that the fusion protein may elevate basal levels of antiviral and antipathogen defenses; (3) coexpression of the fusion protein with CSB upregulates the insulin growth factor binding protein IGFBP5 and downregulates IGFBP7 — suggesting that the fusion protein may also confer a metabolic advantage; and finally (4) the fusion protein binds in vitro to a dispersed family of 900 internally deleted piggyBac elements known as MER85s — suggesting that the CSB-PGD3 fusion protein, when bound to MER85 or related elements, may regulate expression of nearby genes. Taken together, our data support the hypothesis that the CSB-PGBD3 fusion protein is important in health, and may also play a role in CS disease.
2. Materials and methods
2.1. DNA constructs
The parent for all expression constructs was the bicistronic pIREShyg3 vector (Clontech). A N-terminal 3 FLAG tag was inserted to generate pFLAG-IREShyg, followed by an HA tag to generate pFLAG-HA-IREShyg. Open reading frames for intact CSB (4.5 kb) and the CSB-PGBD3 fusion protein (3.2 kb) were inserted downstream of the tags to generate pFLAG-HA-CSB-IREShyg and pFLAG-HA-CSB-PGBD3-IREShyg. To generate pFLAG-CSB-PGBD3-IRESneo, the hygR gene of pFLAG-HA-IREShyg was replaced by the neoR gene from pIRESneo3, and the open reading frame of the CSB-PGBD3 fusion protein was inserted downstream of the tags. Details are available upon request.
2.2. Stably transfected pools
pFLAG-HA-CSB-IREShyg3, pFLAG-HA-CSB-PGBD3-IREShyg3, and the empty vector pFLAG-HA-IREShyg3 were linearized with XhoI just downstream from the poly(A) site, and transfected using TransIT-LT1 reagent (Mirus) into UVSS1KO cells grown in DMEM. Selection with 100, 150 and 200 µg/ml hygromycin (Invitrogen) was begun after 24 h, and both drug and media were refreshed every 48–72 h. Confluent wells containing 50–100 colonies were trypsinized and passaged thereafter as pools. To generate doubly-transfected cells for the HCR experiments, the singly-transfected hygromycin-resistant pools were transfected with either pFLAG-CSB-PGBD3-neo linearized by PvuI within the ampR gene, or with pFLAG-neo linearized with SpeI just upstream from the CMV promoter. G418 selection was increased from 200 to 600 µg/ml while continuing 200 µg/ml hygromycin selection. We were unable to obtain hyg + CSB-neo or fusion-hyg + CSB-neo cells that expressed readily detectable CSB protein. For clarity, the cell lines used in this and the previous study [3] are listed in Table 1.
Table 1. Cell lines used in this and the previous study.
The primary GM00739 fibroblast line derived from patient CS1AN was obtained from the Coriell Institute and transformed with retroviral hTERT [3]. UVSS1KO fibroblasts derived from patient UVSS1KO and transformed with a replication-defective SV40 [13] were the kind gift of Kiyoji Tanaka (Osaka University). EGFP, enhanced green fluorescent protein; hyg, hygromycin resistance; neo, neomycin resistance; fusion, CSB-PGBD3 fusion protein. Puromycin-resistant cells express untagged proteins from the bicistronic vector pIRESpuro (Clontech); hygromycin-resistant cells express FLAG-HA tagged proteins or the FLAG-HA tag alone; neomycin-resistant cells express FLAG-tagged proteins or the FLAG tag alone. Hyphens indicate transformation protocol for cell lines, antibiotic selection for transfection of genes and tags. The microarray experiments (Table S1) were performed using singly-transfected CSB-hyg and fusion-hyg pools, and a doubly-transfected CSB-hyg + fusion-neo line, all normalized for consistency to hyg alone. See Materials and methods for details.
| Cell line | Source | Use in this study |
|---|---|---|
| CS1AN-hTERT | Newman et al. [3] | |
| CS1AN-hTERT + eGFP-puro | Newman et al. [3] | Table 4 |
| CS1AN-hTERT + CSB-puro | Newman et al. [3] | Table 4 |
| UVSS1KO-SV40 + hyg | this study | Fig. 2A, Tables 4 and S1 |
| UVSS1KO-SV40 + fusion-hyg | this study | Fig. 2A, Tables 4 and S1 |
| UVSS1KO-SV40 + CSB-hyg | this study | Fig. 2A, Tables 4 and S1 |
| UVSS1KO-SV40 + hyg + neo | this study | Fig. 2B,C |
| UVSS1KO-SV40 + hyg + fusion-neo | this study | Fig. 2B,C |
| UVSS1KO-SV40 + fusion-hyg + neo | this study | Fig. 2B,C |
| UVSS1KO-SV40 + CSB-hyg + neo | this study | Fig. 2B,C |
| UVSS1KO-SV40 + CSB-hyg + fusion-neo | this study | Fig. 2B,C, Tables 4 and S1 |
2.3. Western blots
To assay CSB and CSB-PGBD3 expression (Fig. S1), subconfluent cells were harvested, resuspended in 2 × SDS sample loading buffer, and immediately heated to 100°C for 10 min. To assay STAT1 expression, cytoplasmic and nuclear fractions were prepared as described [20, 21]. To examine the UV response, adherent cells were washed in PBS, subjected to 40 J/m2 UV irradiation, and allowed to grow for 30 min in fresh medium before harvest [22]. SDS-PAGE and Western blots were as described previously [18]. CSB and the CSB-PGBD3 fusion protein were detected with an antigen-purified rabbit polyclonal raised against CSB residues 1–240 [23]. The β-actin loading and dilution control was detected with a mouse monoclonal antibody (Sigma-Aldrich A2228). STAT1, phospho-STAT1(Tyr701) and phospho-STAT1(Ser727) antibodies were used to identify STAT1 phosphorylation states (Cell Signaling Technology #9172, #9171 and #9177). HRP-conjugated secondary antibodies were goat anti-rabbit and anti-mouse (ThermoScientific #31460 and #31430).
2.4. Recovery of RNA synthesis (RRS) assays
Cells were grown in 24 well microtiter plates under 200 µg/ml hygromycin selection before irradiation and during recovery. The cells were washed in PBS, subjected to 10 J/m2 UV irradiation under a germicidal lamp, and immediately immersed in 1 ml unlabeled medium. For recovery times of 2, 6, 12 and 24 h as well as for unirradiated controls, unlabeled medium was replaced with DMEM containing 10 µCi/ml of [5,6-3H]-uridine (GE Healthcare) followed by pulse-labeling for 1 h at 37°C. Samples were processed as described [13]. Scintillation data were normalized to cell number, plotted, and standard errors of the mean calculated using Excel and GraphPad Prism. All assays were performed in triplicate. UV irradiation was calibrated using an Ultraviolet Meter (UVP).
2.5. Host cell reactivation (HCR) assays
UV- and OsO4-damaged pEGFP-IRESpuro plasmid templates were prepared as described [24]. Transfections using Fugene 6 (Roche) were performed in quadruplicate for each level of DNA damage. EGFP fluorescence was measured using a plate reader and the values corrected for cell number based on protein content as determined by the BCA Protein Assay Kit (Pierce). The corrected fluorescence measurements were averaged and normalized to the corresponding untreated controls. The data from two independent experiments were averaged, plotted, and standard errors of the mean calculated using Excel and GraphPad Prism.
2.6. Expression array protocol and data analysis
Sample preparation for expression array analysis, data generation by the Center for Expression Arrays (University of Washington), and RT-PCR validation have been described previously [3]. Three independent preparations of total RNA from the CSB, CSB-PGBD3, CSB + CSB-PGBD3, and tag-only cells (Table 1) were quality-controlled, labeled, and used to probe Affymetrix GeneChip® Human Genome U133 Plus 2.0 Arrays. The 12 datasets were normalized using the Probe Logarithmic Intensity Error (PLIER) program of the Affymetrix Expression Console v1.1.1. Using the Significance Analysis for Microarrays (SAM) program of the MeV v4.4 software suite (www.TM4.org), fold changes for expression of individual probes were calculated by comparing the 3 datasets for one pool to the 3 datasets for another, and the resulting 9 pairwise fold changes were averaged to give the fold change for that probe.
2.7. Mobility shift assays
Open reading frames encoding the PGBD3 transposase and CSB-PGBD3 fusion protein [3] were cloned into the pFastBAC HT baculovirus vector (Invitrogen Bac-to-Bac® Baculovirus Expression System). Virus production and protein expression in SF9 cells were performed as recommended by the supplier. Soluble hexahistidine tagged protein was partially purified over a TALON® resin (Clontech), eluted with imidazole HCl, desalted, and concentrated by Centricon filtration. Six different MER85s that closely matched the 140 bp Repbase consensus (www.girinst.org/repbase/) were amplified by genomic PCR using an upstream primer with a BamHI site and a downstream primer with EcoRI; the upstream flank varied from 82–189 bp, the downstream flank from 187–326 bp (Fig. S2). The PCR fragments were cloned between the BamHI and EcoRI sites of pBluescript, excised by restriction digestion, and [32P]-labeled by filling in the ends. Mobility shift assays were performed as described [25].
3. Results
3.1. DNA repair assays
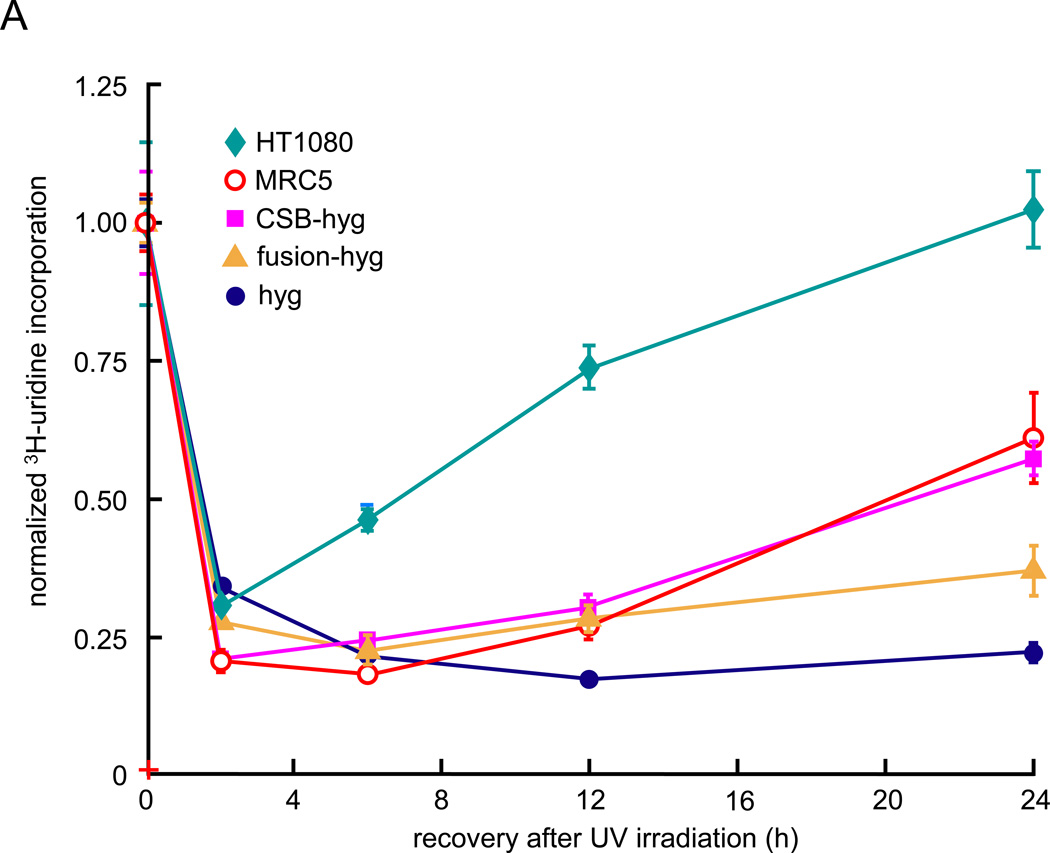
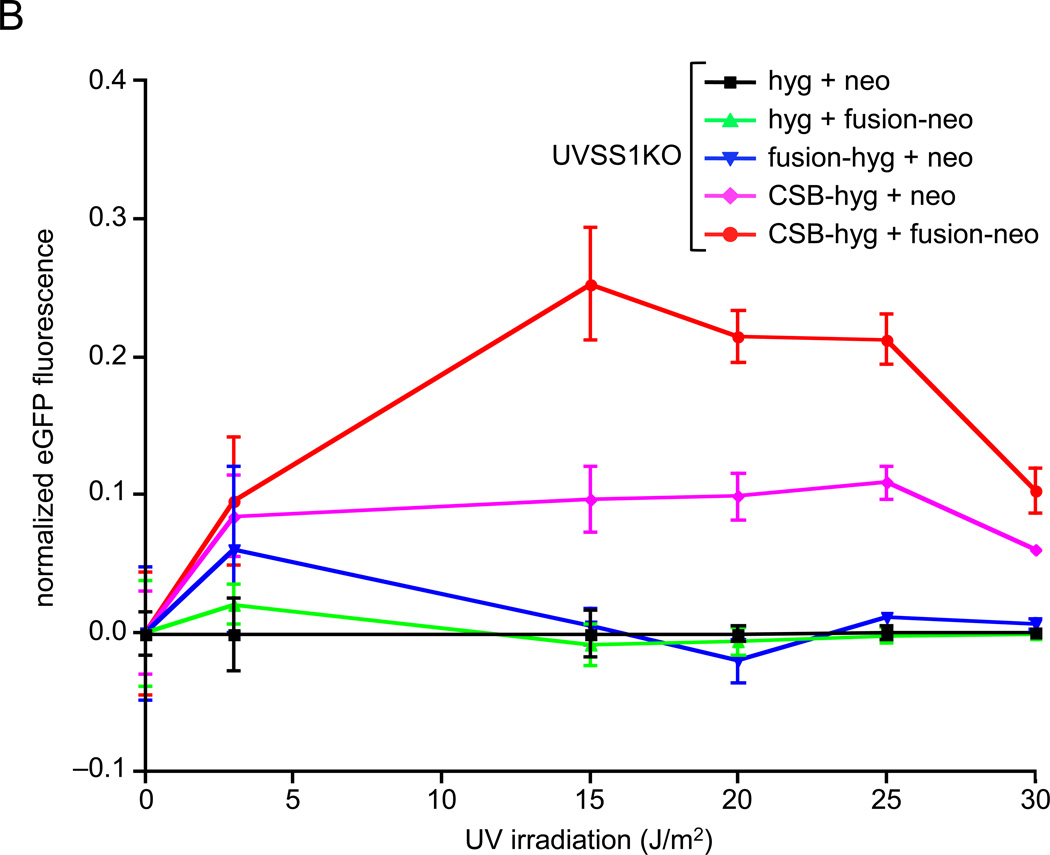
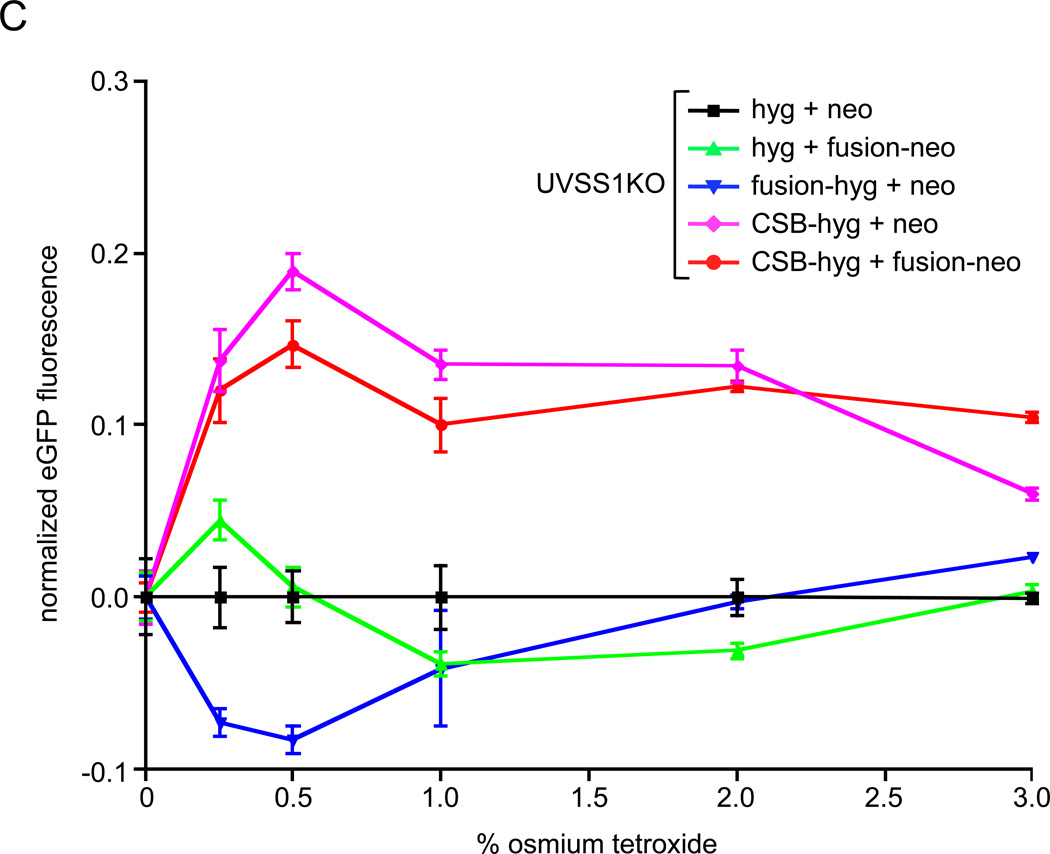
We assayed the effects of the CSB-PGBD3 fusion protein on DNA repair in CSB-null UVSS1KO cells using both a recovery of RNA synthesis (RRS) assay after UV irradiation of whole cells (Fig. 2A), and host cell reactivation (HCR) assays after UV irradiation (Fig. 2B) or osmium tetroxide oxidation (Fig. 2C) of an EGFP reporter plasmid prior to transfection. Importantly, osmium tetroxide (OsO4) oxidation generates thymine glycol damage, which is known to require CSB for efficient repair [24].
Fig. 2.
Recovery of RNA synthesis (RRS) following UV damage, and host cell reactivation (HCR) assays for repair of oxidation- and UV-induced DNA damage. Assays were performed using CSB-null UVSS1KO-derived pools stably expressing CSB, CSB-PGBD3 fusion protein (two independent pools selected with either hygromycin or neomycin), both proteins, or tags only (Table 1). (A) RRS assays monitoring 3H-uridine incorporation after UV irradiation of growing cells with 10 J/m2/min. The SV40-immortalized normal lung cell line MRC5-SV and the the HT1080 fibrosarcoma cell linewere included as a controls [26]. (B) HCR assays for UV damage. The EGFP expression construct was irradiated with 1 J/m2/min UV from a germicidal lamp for 5–30 min before transfection. EGFP fluorescence was measured 48 h after transfection and normalized to cell number at each time point to correct for cell growth. (C) HCR assays for oxidative damage. The reporter EGFP expression construct was pretreated with the indicated concentrations of osmium tetroxide before transfection. Assays were performed in triplicate (panel A) or octuplicate (panels B and C); error bars smaller than the datapoint icons are not shown.
In the UVSS1KO-derived cells used for the RRS and HCR assays (Table 1), CSB levels were 2- to 4-fold higher and CSB-PGBD3 fusion protein levels 4- to 8-fold higher than in human euploid HT1080 fibrosarcoma cells as determined by Western blots of a dilution series probed for the FLAG-HA tag (Fig. S1). We have consistently found that the natural ratio of CSB to CSB-PGBD3 fusion protein is about 1 to 4 in many human cell lines, whether the line expresses high levels of CSB (HT1080) or 8- to 10-fold lower levels (WI38). Thus the ratio of CSB to CSB-PGBD3 fusion protein is within the normal range for the cells used in the RRS and HCR assays. We deliberately did not resort to differential siRNA knockdown of CSB and CSB-PGBD3 in normal cells because knockdown is never complete [27], very low levels of CSB expression do not impair normal RRS in WI38 cells ([3]; J. C. Newman and A. D. Bailey, unpublished observations), and the CSB-PGBD3 fusion protein cannot be efficiently knocked down by siRNA without simultaneously knocking down CSB and/or solitary PGBD3 transposase (Fig. 1B).
Surprisingly, expression of the CSB-PGBD3 fusion protein alone was almost 40% as effective as expression of intact CSB protein in restoring RRS (Fig. 2A) although the fusion protein lacks all of CSB’s conserved ATPase motifs (Fig. 1B and [18]). Although it is difficult to imagine a direct role for the CSB-PGBD3 fusion protein in repair of UV damage, the effect could be indirect; for example, incorporation of the CSB-PGBD3 fusion protein in place of normal CSB might facilitate disassembly of stalled TCR, DNA repair, and/or chromatin remodeling complexes, thus allowing backup repair pathways access to the DNA. In any event, the ability of the CSB-PGBD3 fusion protein to accelerate RRS after UV irradiation of CSB-null cells was so intriguing that we revisited this result using the very different HCR assay for DNA repair.
To more clearly display the effect of the fusion protein on low (and presumably more physiologically relevant) levels of UV and oxidative DNA damage, we plotted the HCR data in a new way. The log of transcriptional activity is usually plotted against a linear measure of DNA damage, although a semi-log plot exaggerates differences between cell lines at high levels of damage while minimizing differences at low levels. Instead, we normalized the HCR data to control cells expressing only the drug resistance markers, allowing the relative transcriptional activity (and thus TC-NER) to be compared over the entire range of DNA damage (Fig. 2B,C).
More surprisingly, the CSB-PGBD3 fusion protein strongly synergized with CSB to stimulate UV repair by 200 to 250% in the HCR assay, whereas expression of the fusion protein alone had little effect and expression of the CSB protein alone rescued UV repair as expected (Fig. 2B).
Finally, expression of CSB protein in CSB-null UVSS1KO cells rescues repair of oxidative DNA damage in the HCR assay,although expression of the CSB-PGBD3 fusion protein alone only mildly inhibits both residual oxidative repair in the absence of CSB and normal oxidative repair in the presence of CSB (Fig. 2C). Curiously, another UV sensitive line Kps3SV13.3 that is also deficient in UV repair [28] has been shown to repair oxidative thymine glycol damage as proficiently as wild type [24].
We conclude that the CSB-PGBD3 fusion protein has significant biological activity based on its ability to stimulate UV repair in the RRS assay in the absence of CSB (FIg. 2A) and to synergize strongly with CSB in the HCR assay for repair of UV damage (Fig. 2B). These DNA repair activities presumably reflect the ability of the fusion protein to influence the mechanism or pathways of DNA repair, and could help to explain why the protein has been conserved in the hominid lineage for over 43 My [18]. Alternatively, as described below, conservation could reflect the ability of the fusion protein to induce major changes in gene expression that resemble an innate antiviral immune response.
3.2. Microarray analysis of CSB-null UVSS1KO-derived cell pools expressing CSB, CSB-PGBD3 fusion protein, both proteins, or neither
We next examined the more general role of the fusion protein in gene expression and cell physiology using microarray analysis. RNA from UVSS1KO-derived pools expressing CSB, the CSB-PGBD3 fusion protein, both proteins, or neither (Table 1) was characterized using Affymetrix U133A Plus 2.0 GeneChips, and the raw data processed with the PLIER and SAM programs. As shown in Table S1A(a) and (b) — where (a) and (b) designate sheets in a workbook — genes exhibiting a robust expression change of 2-fold or more included 305 genes (388 probes) regulated by CSB, 581 genes (767 probes) regulated by CSB-PGDB3, and 1354 genes (1674 probes) regulated by CSB + CSB-PGBD3 together. The microarray data were validated by RT-PCR of selected genes (Table S2) which, as is often the case, largely confirmed but occasionally diverged from the microarray values [3].
To generate an initial overview of the processes and pathways implicated by these gene expression changes, we used the L2L software suite and microarray database ([29]; www.depts.washington.edu/l2l) to examine the complete lists of robustly regulated genes (Table S1A). Genes upregulated by the CSB-PGDB3 fusion protein were found to match strongly with genes upregulated by interferons (IFNs) and viral infection, more weakly with aspects of the immune response, and to a lesser extent with inflammation, apoptosis, and neural growth. In contrast, genes affected by expression of CSB alone or CSB + CSB-PGDB3 fusion protein exhibited fewer matches of comparable significance. Genes downregulated by CSB matched those regulated by IL-2 in both directions, and may reflect signaling pathways controlling proliferation (data not shown).
To carefully examine the interferon, viral, immune and other minor signatures identified by L2L analysis of all robustly regulated genes in the CSB-PGBD3 fusion protein dataset, we assembled a list of potentially relevant genes by manually interrogating the NCBI Gene Database using key phrases suggested by the L2L analysis: interferon-regulated, -induced, and -repressed; interferon α, β, γ; STAT1 and STAT; immune and inflammatory responses; apoptosis; and neural growth and development. We then assigned each of these potentially relevant genes to 1 of the 10 functional categories indicated in columns F through O of Tables S1B(a) and S1B(b) based on the gene description in the NCBI Gene Database. Lastly, we culled the complete list of robustly regulated genes (Table S1A) leaving only those genes that fall into at least 1 of the 10 functional categories (Table S1B).
3.3. The CSB-PGBD3 fusion protein induces a strong interferon-like response in CSB-null cells that is repressed by coexpression of CSB
Using the binomial distribution [29] and a human gene count of 17,506 [30], we calculated the significance of overlaps between genes regulated at least 2- or 4-fold by the fusion protein (Table S1A) and genes belonging to each of the 10 functional categories identified by L2L and listed in columns F through O of Tables S1B(a) and S1B(b). As shown in Table 2, expression of the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells induces a strong but atypical interferon response resembling a composite of the canonical responses to IFN-α, IFN-γ and, to lesser extent, IFN-β. For example, of 457 genes known to be regulated, induced, or repressed by an interferon, 63 overlapped with genes induced at least 2-fold by the CSB-PGBD3 fusion protein, and 37 overlapped with genes induced at least 4-fold [31].
Table 2.
Expression of the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells induces atypical interferon-like response. The binomial probability of overlap is shown between genes robustly regulated by the fusion protein (Table S1A) and genes belonging to each of the 10 functional categories indicated in columns F through O of Tables S1B(a) and S1B(b). The most significant p-values (p<6E-09) are highlighted in orange; somewhat less significant p-values (3E-04>p>7E-08) are highlighted in yellow.
| GO terms and processes | Genes in GO list |
CSB | CSB + fusion | fusion | |||
|---|---|---|---|---|---|---|---|
| fold induction genes affected |
≥ 2 fold 305 |
≥ 4 fold 93 |
≥ 2 fold 1354 |
≥ 4 fold 219 |
≥ 2 fold 581 |
≥ 4 fold 143 |
|
| IFN regulated/induced/repressed | 457 | 10 | 2 | 45 | 7 | 63 | 37 |
| 2.78E-01 | 6.98E-01 | 6.34E-02 | 3.48E-01 | 5.49E-21 | 8.36E-25 | ||
| IFN-a regulated/induced/repressed | 74 | 1 | 1 | 10 | 1 | 22 | 16 |
| 7.25E-01 | 3.26E-01 | 6.58E-02 | 6.05E-01 | 2.39E-14 | 3.83E-18 | ||
| IFN-β regulated/induced/repressed | 40 | 1 | 0 | 3 | 1 | 10 | 6 |
| 5.02E-01 | 1.00E+00 | 3.74E-01 | 3.94E-01 | 1.33E-06 | 1.16E-06 | ||
| IFN-γ regulated/induced/repressed | 242 | 6 | 2 | 22 | 6 | 29 | 18 |
| 2.49E-01 | 3.68E-01 | 2.52E-01 | 8.54E-02 | 5.96E-09 | 3.06E-12 | ||
| IFN-ε regulated/induced/repressed | 53 | 1 | 1 | 5 | 2 | 7 | 2 |
| 6.03E-01 | 2.46E-01 | 3.91E-01 | 1.43E-01 | 2.22E-03 | 7.04E-02 | ||
| IFN-κ regulated/induced/repressed | 97 | 2 | 0 | 7 | 1 | 10 | 8 |
| 5.04E-01 | 1.00E+00 | 6.23E-01 | 7.04E-01 | 1.77E-03 | 1.63E-06 | ||
| IFN-ω regulated/induced/repressed | 20 | 0 | 0 | 1 | 0 | 3 | 1 |
| 1.00E+00 | 1.00E+00 | 7.87E-01 | 1.00E+00 | 2.98E-02 | 1.51E-01 | ||
| STAT1 regulated/induced/repressed | 192 | 7 | 1 | 27 | 5 | 18 | 12 |
| 5.30E-02 | 6.40E-01 | 2.75E-03 | 9.50E-02 | 1.06E-04 | 8.39E-08 | ||
| STAT regulated/induced/represseda | 257 | 7 | 1 | 26 | 4 | 22 | 9 |
| 1.65E-01 | 7.46E-01 | 1.05E-01 | 4.01E-01 | 7.15E-05 | 3.10E-04 | ||
| U-STAT1 prolonged expressionb | 108 | 5 | 2 | 16 | 3 | 49 | 29 |
| 4.21E-02 | 1.13E-01 | 1.17E-02 | 1.54E-01 | 1.20E-38 | 7.16E-35 | ||
| STAT1 prolonged expressionb | 35 | 2 | 0 | 4 | 0 | 23 | 19 |
| 1.25E-01 | 1.00E+00 | 2.87E-01 | 1.00E+00 | 2.68E-22 | 8.66E-29 | ||
| Immune response | 1330 | 35 | 5 | 99 | 11 | 79 | 37 |
| 1.23E-02 | 8.34E-01 | 6.69E-01 | 9.43E-01 | 9.00E-07 | 3.23E-10 | ||
| Immune responsec | 1043 | 27 | 4 | 73 | 7 | 33 | 6 |
| 3.00E-02 | 8.04E-01 | 8.25E-01 | 9.75E-01 | 6.34E-01 | 8.53E-01 | ||
| Inflammatory response | 1190 | 82 | 16 | 21 | 8 | 75 | 29 |
| 8.74E-01 | 4.20E-01 | 5.06E-01 | 3.01E-01 | 1.84E-07 | 3.76E-07 | ||
| Regulation of apoptosis | 2129 | 48 | 18 | 165 | 32 | 96 | 29 |
| 4.66E-02 | 3.98E-02 | 5.01E-01 | 1.70E-01 | 2.02E-03 | 6.48E-03 | ||
| Induction of apoptosis | 1054 | 22 | 8 | 80 | 15 | 69 | 23 |
| 2.25E-01 | 2.02E-01 | 5.85E-01 | 3.43E-01 | 1.40E-07 | 3.16E-05 | ||
| Neural development | 432 | 13 | 5 | 38 | 8 | 24 | 7 |
| 4.22E-02 | 8.26E-02 | 2.33E-01 | 1.78E-01 | 1.11E-02 | 6.67E-02 | ||
| Neural growth | 270 | 13 | 5 | 29 | 8 | 26 | 7 |
| 1.07E-03 | 1.54E-02 | 5.20E-02 | 2.15E-02 | 2.13E-06 | 7.29E-03 | ||
The list of interferon-related genes that is upregulated 2-fold or more by the CSB-PGBD3 fusion protein (Table S1B(c)) includes many prominent interferon response genes: the JAK kinase-activated signal transducers and activators of transcription STAT1 (7-fold) and STAT2 (2-fold); the 2'-5'-oligoadenylate synthetases OAS1 (118,000-fold), OAS2 (129-fold), OAS3 (11-fold), and OASL (29-fold) that activate the antviral RNase L [32]; the interferon-stimulated genes ISG15 (11-fold) and ISG20 (12-fold); the interferon-inducible genes IFI6 (22-fold), IFI27 (239-fold), IFI44 (29-fold), IFI44L (338-fold), and IFH1 (36-fold); the IFI genes with tetratricopeptide repeats IFIT1 (8-fold), IFIT2 (6-fold), IFIT3 (4-fold), IFIT5 (2-fold), and IFITM1 (4-fold); and the IRF9 subunit (6-fold) of the ISGF3 transcription factor. In addition, the IFN-α, β, and ω receptor IFNAR2 and the receptor-activated kinase JAK1 are induced 2.35 and 2.30-fold by coexpression of CSB and the CSB-PGBD3 fusion protein though not by either CSB nor fusion protein alone. Moreover, as might be expected from conservation of the CSB-PGBD3 fusion protein since marmosets [18], the presence of functional CSB almost completely suppresses the interferon-like response induced by fusion protein alone, reducing the p-values for overlap between fusion-induced genes and the interferon-response genes by a dramatic 21–23 orders of magnitude (Tables 2 and S1C).
3.4. U-STAT1 and ISGF3 appear to mediate the interferon-like response induced by the CSB-PGBD3 fusion protein in CSB-null cells
The CSB-PGBD3 fusion protein does not induce any of the interferons (Table S1) and thus cannot generate an interferon-like response through the canonical JAK-STAT pathway in which interferons bind to transmembrane receptors, activating intracellular receptor-associated JAK and TYK kinases that phosphorylate STATs on tyrosine. Instead, as shown in Table S1B(c), expression of the fusion protein elevates the mRNA levels for STAT1 (7-fold), STAT2 (2-fold), and IRF9 (6-fold) which together constitute the heterotrimeric transcription factor ISGF3 (interferon-stimulated gene factor 3). ISGF3 binds to ISREs (interferon-stimulated response elements) and normally drives the IFN-α and IFN-β responses, while STAT1 homodimers bind to GAS elements (IFN-γ activated sequences) and drive the IFN-γ response.
Although tyrosine phosphorylation was long thought to be essential for STAT activity, more recent work has shown that this is only true early in the IFN-β and IFN-γ responses [31, 33, 34]. Ptyr-STAT1 initially drives expression of a large number of interferon response genes, but most of these return to basal levels within 6 to 8 h presumably because continued expression would be damaging. Interestingly, Ptyr-STAT1 also induces STAT1 transcription, causing accumulation of transcriptionally active but unphosphorylated STAT1 (U-STAT1) and sustaining expression of a subset of the initial interferon-induced genes for an additional 48 to 72 h [31, 34].
Remarkably, expression of the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells induces 18 of the 20 genes (Table 3; also see Table 2) most strongly induced by overexpression of the unphosphorylatable Y701F-STAT1 mutant in normal BJ fibroblasts[34]. Although the precise mechanism by which U-STAT1 sustains expression of a subset of interferon-induced genes remains to be determined, an intriguing possibility is that U-STAT1, U-STAT2, and U-IRF9 may assemble into U-ISGF3 heterotrimers analogous to phosphorylated ISGF3 formed in the initial IFN-α or IFN-β responses (H. Cheon and G.R. Stark, personal communication).
Table 3.
Overlap between genes induced by the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells and genes induced by U-STAT1 in normal cells. Y701F-STAT1 (also known as U-STAT1) lacks Y701 and can only be phosphorylated on S727. Genes induced by overexpression of U-STAT1 in STAT1 normal cells are taken from Cheon et al. [34]; genes induced by CSB-PGBD3 fusion protein in CSB-null UVSS1KO background are from Table S1A.
| Gene | Gene Description | CSB + fusion | CSB | fusion |
|---|---|---|---|---|
| IFI27 | IFNa-inducible protein 27 | --- | --- | 238.83 |
| BST2 | bone marrow stromal cell antigen 2 | --- | --- | 26483.17 |
| OAS1 | 2',5'-oligoadenylate synthetase 1, 40/46kDa | --- | --- | 118155.11 |
| OAS2 | 2'-5'-oligoadenylate synthetase 2, 69/71kDa | --- | --- | 129.38 |
| OAS3 | 2'-5'-oligoadenylate synthetase 3, 100kDa | --- | --- | 11.03 |
| STAT1 | signal transducer and activator of transcription 1, 91kDa | 2.66 | --- | 6.92 |
| IFI44 | IFN-induced protein 44 | --- | --- | 29.45 |
| IFI44L | IFN-induced protein 44-like | --- | --- | 337.68 |
| IFIH1 | IFN induced with helicase C domain 1 | --- | --- | 35.60 |
| IFITM1 | IFN induced transmembrane protein 1 (9–27) | --- | --- | 4.34 |
| IFI35 | IFN-induced protein 35 | --- | 0.45 | 3.40 |
| IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | 2.36 | --- | 4.01 |
| MX1a | myxovirus (influenza virus) resistance 1 | --- | --- | 82.59 |
| IRF7 | IFN regulatory factor 7 | --- | 0.50 | --- |
| ISG15b | ISG15 ubiquitin-like modifier | 0.39 | --- | 11.32 |
| IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 | --- | --- | 8.35 |
| PLSCR1 | phospholipid scramblase 1 | 2.12 | --- | 6.23 |
| HERC6 | hect domain and RLD 6 | --- | --- | 4.30 |
| FLJ20035c | hypothetical protein FLJ20035 | --- | --- | 3.67 |
| EPSTI1 | epithelial stromal interaction 1 (breast) | --- | --- | --- |
also known as IFN-inducible protein p78 (mouse)
also known as G1P2
also known as DDX60
STAT phosphorylation on tyrosine (tyrosine 701 in STAT1 or the equivalent tyrosine in other STATs) was long thought to be required for subsequent phosphorylation on serine (serine 727 in STAT1 or the equivalent serine on other STATs) by one of several kinases that can fully activate nuclear STAT1 homo- and heterodimers in response to stressors of various kinds including UV, oxidative, or other kinds of DNA damage [32, 35]. However, instances are now beginning to emerge in which U-STATs play roles outside of the interferon response. For example, induction of apoptosis in cardiac myocytes by ischemia/reperfusion requires phosphorylation of STAT1 on serine 727 but not tyrosine 701 [36].
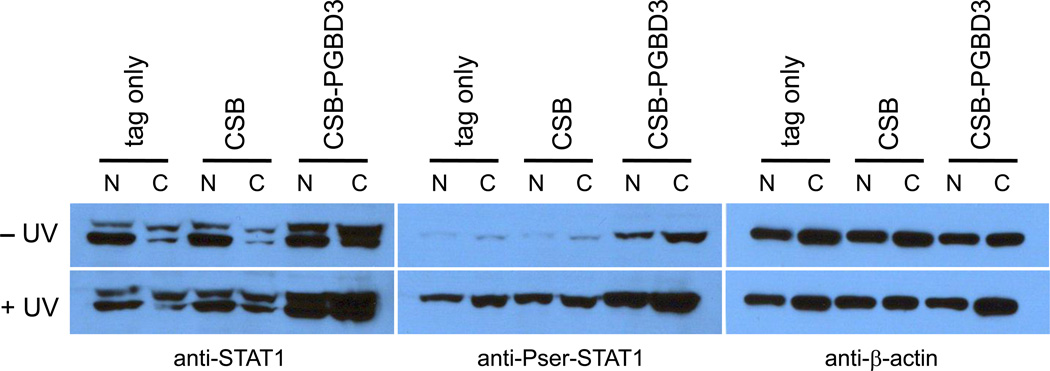
The strong correlation between genes induced by U-STAT1 in normal cells and by the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells (Table 3) led us to examine the phosphorylation state of STAT1 in CSB-null UVSS1KO cells stably transfected with CSB, the CSB-PGBD3 fusion protein, or neither (Table 1) with or without UV irradiation. As shown in Fig. 3, nuclear and cytoplasmic fractions were probed with antibodies against STAT1, Pser-STAT1, and β-actin as a loading control. No Ptyr-STAT1α or Ptyr-STAT1β was detected with anti-Ptyr-STAT1 antibody (Fig. S3, and data not shown), ruling out activation by the canonical JAK-STAT pathway, but anti-Pser-STAT1 antibody recognized Pser-STAT1β which was further induced by UV stress.
Fig. 3.
Expression of the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells induces U-STAT1, which can be phosphorylated on serine 727 and further induced by UV irradiation. Using CSB-null UVSS1KO cells stably transfected with CSB, CSB-PGBD3 fusion protein, or tags alone (Table 1), nuclear and cytoplasmic fractions were resolved by SDS-PAGE, blotted, and probed with anti-STAT1 (left panel), anti-Pser-STAT1 (middle panel), and anti-β-actin antibodies as a loading control (right panel). The STAT1 locus generates two proteins (left panel): full length STAT1α (upper band) and C-terminally deleted STAT1β (lower band) lacking the Ser727 phosphorylation site. STAT1β has been referred to as a splice variant or proteolysis product [37]; however, mRNAs annotated on the UCSC Genome Browser (build hg18) indicate that STAT1β reflects alternative polyadenylation within intron 23. This results in use of a TAA terminator immediately following exon 23, and a protein that retains Tyr701 but lacks the Ser727 phosphorylation site.
3.5. The CSB-PGBD3 fusion protein upregulates the RIG-I and MDA5 effectors of the innate, intracellular antiviral defense in CSB-null UVSS1KO cells
The ability of the CSB-PGBD3 fusion protein to induce an interferon-like response in CSB-null UVSS1KO cells without inducing interferon mRNAs suggested that the fusion protein might activate an innate cytoplasmic antiviral response such as those mediated by the RIG-I (aka DDX58) and/or MDA5 proteins [38, 39]. Indeed, as shown in Table S1B, expression of the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells strongly upregulates MDA5 (36-fold) and RIG-I (6- to 8-fold). RIG-I and MDA5 are normally activated by intracellular double-stranded or uncapped RNA indicative of viral infection, and are known to signal through the mitochondrial adaptor protein IPS-1 (IFN-β promoter stimulator 1, also called Signaling can stimulate the IFN-α and/or IFN-β promoters, generating secreted interferons that activate STAT1 through an autocrine circuit involving interferon cell-surface receptors and the canonical JAK-STAT pathway. However, RIG-I can also activate STAT1 through a newly discovered noncanonical pathway that is independent of cell-surface interferon receptors and possibly of the receptor-associated kinases JAK1, JAK2, and TYK2 as well [40].
Consistent with induction of an innate cytoplasmic antiviral response by the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells, one of the proteins most strongly induced by the fusion protein is BST2/tetherin. Originally known as bone marrow stromal antigen 2 (BST2), tetherin is a trans-membrane protein of the innate immune response that interferes with budding and release of enveloped viruses [41, 42]. BST2/tetherin is induced 26,500-fold as judged by expression array analysis (Table S1A) and 143-fold by the RT-PCR assay (Table S2); moreover, like many other genes induced by the CSB-PGBD3 fusion protein, BST2/tetherin is repressed >200-fold by coexpression of intact CSB (Tables S1A and S2).
Upregulation of RIG-I and MDA5 by the CSB-PGBD3 fusion protein could potentially be explained in many different ways, but one intriguing scenario would be that the CSB-PGBD3 fusion protein may deregulate CSB-dependent chromatin remodeling [3, 18], thus leading to aberrant transcription, generation of double-stranded or uncapped cytoplasmic RNA, activation of RIG-I and/or MDA5, and intracellular induction of the observed atypical interferon response. Admittedly, noncanonical activation of STAT1 by RIG-I overexpression in the U937 acute myeloid leukemia (AML) cell line results in STAT1 phosphorylation on both Tyr 701 and Ser 727 [40] whereas expression of the CSB-PGBD3 fusion protein in UVSS1KO fibroblasts induces STAT1 phosphorylated on Ser 727 alone (Figs. 3 and S3) as well as many of the same genes induced by the unphosphorylatable Y701F STAT1 mutant in normal BJ fibroblasts (Tables 2, 3 and S3). These differences in STAT1 phosphorylation state may however be cell type-specific like so many other aspects of the interferon response [43]. The significance of the innate, intracellular antiviral response induced by the CSB-PGBD3 fusion protein in the UVSS1KO background is further supported by a direct comparison of the CS1AN and UVSS1KO microarray datasets as described below (Section 3.6, and Tables 4 and S5).
Table 4.
CSB and CSB-PGBD3 fusion protein induce closely related antiviral responses in different genetic backgrounds. When analyzed using MSigDB instead of L2L, 15 interferon-related genes are upregulated by expression of CSB in the CSB compound heterozygote CS1AN line, 12 of which are also upregulated by expression of CSB-PGBD3 fusion protein in the CSB-null UVSS1KO line (Table S4B, highlighted in blue), and 11 of which belong to 3 related functional themes — viral RNA recognition, protein degradation, and membrane-mediated antiviral activities. (A) Regulation of the 15 genes (leftmost column) in the seven interferon-related MSigDB datasets. The correlation is mainly in the UP direction except for the dataset of [47] for STAT3 which often suppresses the interferon response. (B) Regulation of 12 of the 15 genes in the UVSS1KO datasets (Table S1). CSB-PGBD3 fusion protein is denoted as "fusion" in the tables below. UP and DOWN indicate a >2-fold difference in gene expression when CSB, fusion, or CSB + fusion are compared to the tags-only control). For convenience, vignettes of the 15 genes are provided in Table S6.
| A. MSigDB IFN-Related Datasets | |||||||
|---|---|---|---|---|---|---|---|
| DDX58 | MOSERLEa | DAUERb | BROWNEc | BROWNEd | DERe | DERf | DERg |
| DDX58 | UP | DN | |||||
| GBP1 | UP | UP | UP | UP | UP | ||
| HERC5 | UP | DN | |||||
| IFIH1 | UP | DN | |||||
| IFIT1 | UP | DN | UP | ||||
| IFIT2 | UP | UP | UP | ||||
| IFITM1 | UP | UP | UP | UP | |||
| ISG15 | DN | UP | UP | UP | UP | UP | |
| PHLDA1 | UP | UP | |||||
| PLSCR1 | UP | ||||||
| PMAIP1 | UP | UP | UP | UP | |||
| RBBP6 | UP | ||||||
| RSAD2 | UP | UP | UP | ||||
| SAMD9 | UP | DN | |||||
| TRIM14 | DN | UP | UP | UP | |||
| B. UVSS1KO datasets | |||
|---|---|---|---|
| Gene | fusiona | CSB | CSB + fusion |
| DDX58 | UPb | ||
| GBP1 | UP | UP | UP |
| HERC5 | |||
| IFIH1 | UP | ||
| IFIT1 | UP | ||
| IFIT2 | UP | ||
| IFITM1 | UP | ||
| ISG15 | UP | ||
| PHLDA1 | UP | ||
| PLSCR1 | UP | UP | |
| PMAIP1 | UP | UP | |
| RBBP6 | |||
| RSAD2 | UP | ||
| SAMD9 | UP | UP | |
| TRIM14 | |||
MOSERLE_IFNA_RESPONSE [46]
DAUER_STAT3_TARGETS_DN [47]. Note that STAT3 represses IFN-inducible genes involved in wound healing and cancer.
BROWNE_HCMV_INFECTION_4HR_UP [48]
BROWNE_INTERFERON_RESPONSIVE_GENES [48]
DER_IFN_BETA_RESPONSE_UP [49]
DER_IFN_ALPHA_RESPONSE_UP [49]
DER_IFN_GAMMA_RESPONSE_UP [49]
CSB-PGBD3 fusion protein
UP indicates a difference of 2-fold or more when fusion, CSB, or CSB + fusion is compared to the UVSS1KO control.
3.6. CSB and the CSB-PGBD3 fusion protein both induce the innate intracellular immune response to viral infection
3.6.1
We had observed previously that addition of CSB to CS1AN cells (a CS patient-derived compound heterozygote expressing intact CSB-PGBD3 fusion protein and N-terminal CSB fragments) induced a chromatin remodeling signature [18]; however, the genetically equivalent addition of CSB to UVSS1KO cells expressing intact fusion protein induced an interferon-like instead of a chromatin remodeling signature. As described in Appendix A (Sections A1 and A2, Tables S4A and S4B), the apparent discrepancy actually reflects database bias, i.e. under- and over-representation of particular datasets depending on the interests of those who compiled the databases, when the databases were first compiled, and how actively the databases have been curated.
To avoid the complications of microarray database bias, we compared the raw CS1AN [3] and UVSS1KO datasets directly to each other. We used MSigDB to convert our probe lists to gene lists in a consistent fashion, and then — because MSigDB can only compare datasets from the MSigDB database — we wrote PERL scripts to compare our datasets directly and used Excel to calculate the binomial statistics for all overlaps between genes that are regulated 2-fold or more by CSB in the compound heterozygote CS1AN, and 2-fold or more by expression of CSB, the CSB-PGBD3 fusion protein, or both proteins in CSB-null UVSS1KO cells. The control datasets were CS1AN expressing EGFP [3] and UVSS1KO expressing tags only (Table 1). The comparisons are shown in Table S5.
Three conclusions from this comparison are straightforward: CSB regulates many genes independently of the CSB-PGBD3 fusion protein — as expected if regulation of these genes requires functional CSB protein; the CSB and the CSB-PGBD3 fusion protein can work synergistically — as might be expected if the N-terminal CSB domain of the fusion protein partially mimics or modulates normal CSB functions; and (3) CSB can reverse many effects of the CSB-PGBD3 fusion protein including the induction of interferon-related genes (Table S1C) — suggesting that functional CSB can displace the fusion protein from shared binding sites, and potentially explaining why the fusion protein does not behave as a dominant negative in normal individuals. Two other conclusions were unexpected, and potentially more exciting:
3.6.2
Expression of many genes requires coregulation by both CSB and the CSB-PGBD3 fusion protein — seemingly at odds with the dominance of CSB over the fusion protein as proposed above. A particularly intriguing instance of coregulation is the 7-fold induction of insulin growth factor binding protein 5 (IGFBP5) by CSB + fusion but not by CSB or fusion alone, and the concurrent 3-fold repression of IGFBP7 by CSB + fusion but not by CSB or fusion alone (Table S1). IGFBPs bind insulin and related proteins, modulating or inhibiting their action. These coregulation data therefore suggest that the CSB-PGBD3 fusion protein can modulate the IGF1/insulin pathway in the presence of functional CSB, and may have been conserved not only for a role in DNA repair or chromatin remodeling [3] but for the ability to confer a metabolic advantage. These data are also consistent with induction of IGFBP1 in both aged mice and an XPF-ERCC1 progeria, and with the hypothesis that organismal resources are reallocated by the IGF1/insulin pathway from growth to somatic preservation in response to unrepaired DNA damage [44, 45].
3.6.3
Most intriguingly, as shown in Table S5, expression of CSB in the CS1AN compound heterozygote (which should restore the normal genotype) resembled expression of the CSB-PGBD3 fusion protein in the CSB-null UVSS1KO line (which should resemble the majority of CSB mutants). While trying to understand why one cell line with a nominally normal CSB genotype (CS1AN expressing CSB) would partially resemble another with a nominally mutant CSB genotype (UVSS1KO expressing CSB-PGBD3), we noticed that 15 of the 20 overlapping upregulated genes most closely matched interferon-related lists in the MSigDB database (Table 4A,B). In contrast, no functional themes emerged from the 15 downregulated genes (data not shown).Moreover, of the 15 upregulated genes that accounted for the overlaps, 11 belong to just 3 related functional themes — viral RNA recognition, protein degradation, and membrane-mediated antiviral activities: 4 recognize various aspects of intracellular viral RNA (RIG-I aka DDX58, MDA5 aka IFIH1, IFIT1, IFIT2); 4 others are associated with protein degradation through ubiquitin-like or RING finger pathways (HERC2, ISG15, RBBP6, and TRIM14 — a possible member of the TRIM5α, TRIM6, TRIM22, and TRIM34 antiretroviral gene superfamily [50]; 3 others participate in membrane-related antiviral restriction (RSAD2 aka viperin or cig5 — which localizes to cytoplasmic lipid bodies and facilitates signaling through cell surface TLR7 and TLR9 nucleic acid receptors; PLSCR1 (phospholipid scramblase) — which induces a subset of interferon-stimulated genes (ISGs) including ISG15 and guanylate binding proteins known as GBPs; and GBP1 — a dynamin family protein involved in vesicle scission [51, 52]. For a more detailed description of these innate immunity genes and functions, see Table S6. Taken together, these data suggest that CSB and the CSB-PGBD3 fusion protein both contribute to the cellular antiviral state and interferon-like response. Moreover, upregulation of the innate antiviral proteins RIG-I, MDA5, and BST2/tetherin by the CSB-PGBD3 fusion proteinbut not by CSB alone (Section 3.5 and Table S1B) further suggests that CSB and the fusion protein have both overlapping and complementary functions in the innate antiviral immune response.
3.7. The CSB-PGBD3 fusion protein binds MER85 elements in vitro
Autonomous inverted terminal repeat transposons often give rise to internally deleted nonautonomous transposable elements, known as MITEs or miniature inverted terminal repeat elements, which are mobilized in trans by proteins encoded within the autonomous element [53]. Over 35 Mya, an autonomous 2.5 kb PGBD3 transposon (or a closely related piggyBac transposon) gave rise to MER85 elements [19] — nonautonomous 140 bp elements that retain the terminal inverted repeats of the autonomous PGBD3 elements but have lost the internal transposase ORF (Fig. 1A). These MER85s were mobilized in trans by the PGBD3 transposase and dispersed throughout the human genome in nearly 900 copies before mobility ceased ([19]; L.T. Gray, K.K. Fong, T. Pavelitz, and A.M. Weiner, manuscript in preparation).
We previously speculated that the CSB-PGBD3 fusion protein might bind MER85 elements through the C-terminal transposase domain, and that the acidic N-terminal CSB domain of the fusion protein might then influence expression of nearby genes either directly or through an effect on local chromatin structure [18]. To explore this hypothesis, we asked whether the CSB-PGBD3 fusion protein and/or solitary PGBD3 transposase are capable of binding MER85 elements in vitro.
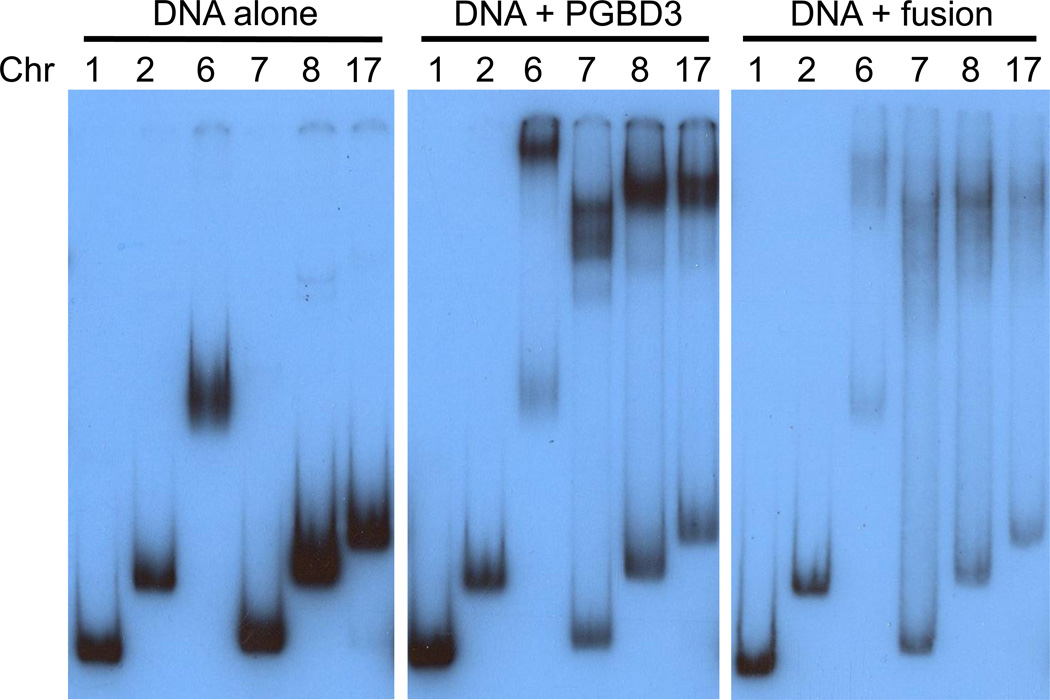
We used the Repbase MER85 consensus (www.girinst.org/repbase/) to find all homologous elements in the March 2006 assembly of the human genome sequence (build hg18). We then PCR amplified and cloned the 6 most highly conserved MER85s (Fig. S2, and Methods). We expressed hexahistidine-tagged CSB-PGBD3 fusion protein and PGBD3 transposase in the baculovirus system, and partially purified the proteins by cobalt chelate affinity chromatography. We assayed binding of the two proteins to the panel of MER85s by electrophoretic mobility shift assay (EMSA) as described [25]. The recombinant proteins were coincubated with end-labeled MER85 DNA fragments in the presence of nonspecific poly(dI-dC) competitor, and the resulting protein/DNA complexes resolved by native gel electrophoresis (Fig. 4).
Fig. 4.
The PGBD3 piggyBac transposase and the CSB-PGBD3 fusion protein bind to consensus MER85 elements in vitro. Electrophoretic mobility shift assays were performed [25] for binding of the recombinant PGBD3 and CSB-PGBD3 fusion proteins to 6 different genomic MER85 elements that closely match the 140 bp Repbase MER85 consensus. The multiple sequence alignment and genomic primers are shown in Fig. S2. Chromosome number is indicated above the lanes. MER85 elements from chromosomes 1 and 7 are MseI/MseI and AseI/HincII fragments, respectively, lacking flanking sequence; MER85s from chromosomes 2, 6, 8, and 17 are BamHI/EcoRI fragments that include the flanks. Although both proteins shift efficiently, the piggyBac transposase generates sharper bandshifts than the larger fusion protein on low percentage gels (6% 29:1 acrylamide:bisacrylamide for PGBD3 and 5% 80:1 acrylamide:bisacrylamide for CSB-PGBD3).
Although all of the MER85s conformed well to the Repbase consensus, only 4 of the 6 shifted strongly in vitro. Interestingly, the same 4 MER85s shifted with both solitary PGBD3 transposase (middle panel) and the CSB-PGBD3 fusion protein (right panel) indicating that the acidic N-terminal CSB domain did not interfere with DNA binding. Using antibodies against the N- and C-terminus of CSB, we found by ChIP-seq that all 6 MER85s bind the CSB-PGBD3 fusion protein in vivo (L.T. Gray, K.K. Fong, T. Pavelitz, and A.M. Weiner, manuscript in preparation). Thus the fusion protein may regulate gene expression both locally (by influencing gene expression near genomic binding sites) and more globally (by mimicking, modulating, or interfering with CSB functions).
4. Discussion
The CSB-PGBD3 fusion protein has been conserved for >43 My from marmoset to humans, and is as highly conserved as full length CSB protein [18]. Such striking conservation suggests that the fusion protein confers a selective advantage in the presence of functional CSB. What are these advantageous functions, what mechanisms are involved, and does the fusion protein contribute to CS disease in individuals who lack functional CSB but continue to express the fusion protein? As a first step toward answering these questions, we have examined the consequences of reintroducing the CSB-PGBD3 fusion protein, with or without functional CSB, into CSB-null UVSS1KO-derived cells which do not express either stable CSB fragments [13] or the fusion protein[3].
4.1 The CSB-PGBD3 fusion protein is biologically active
We have presented evidence that the fusion protein is biologically active in many respects: It modulates repair of UV and oxidative DNA damage as judged by RRS and HCR assays (Fig. 2); it regulates expression of many genes, and coregulates additional genes together with CSB (Table S1); when coexpressed with CSB, it induces insulin growth factor binding protein 5 (IGFBP5) and represses IGFBP7 (Table S1), consistent with an effect on the IGF1/insulin pathway [44, 45, 54, 55]; it induces a strong interferon-like response in the absence of interferons (Table 2) through a U-STAT1-mediated pathway (Figs. 3 and S3) that resembles the sustained response to interferon stimulation (Table 3); it induces an MDA5- and RIG-I-dependent innate antiviral response in the absence of RNA virus infection (Table 4); it binds to a large family of MER85 repetitive elements that are dispersed throughout the genome, potentially providing a mechanism for regulating expression of nearby genes (Fig. 4); as expected from conservation of the CSB-PGBD3 fusion protein, the interferon-like and innate antiviral responses are both dramatically repressed by coexpression of intact CSB (Tables 2 and S1C); and finally, expression of CSB in CS1AN cells that naturally express the fusion protein induces many of the same antiviral proteins [3] as expression of the fusion protein in CSB-null UVSS1KO cells (Tables 4A,B and S5), suggesting that CSB and CSB-PGBD3 fusion protein both contribute to the normal cellular antiviral state and interferon response.
4.2 Induction of interferon-like and antiviral responses without interferons or viral infection
How does the CSB-PGBD3 fusion protein induce interferon-like and antiviral responses without inducing interferons or activating the JAK/TYK pathway? Our current data favor two of many imaginable mechanisms: First, coinduction of all three components (STAT1, STAT2, and IRF9) of the heterotrimeric transcription factor ISGF3 (interferon-stimulated gene factor 3) suggest that the fusion protein activates a common node in the interferon response located downstream of the JAK/TYK kinases that increases Pser727-U-STAT1 and STAT2 but not Ptyr701-STAT1 (Tables S1 and S3, Figs. 3 and S3). Alternatively, as discussed above (section 3.5), loss of CSB chromatin-remodeling activity could lead to aberrant transcription, generation of dsRNA, and induction of the RIG-I and MDA5 innate immunity pathways that are normally induced by infection with RNA viruses or by JAK/TYK-independent pathways [40]. Whatever the mechanism(s), we speculate that the CSB-PGBD3 fusion protein may have been conserved for 43 My because it is able to prime or poise the interferon and/or innate immune or antiviral responses in which speed may be critical for success.
The phosphorylation of STAT1 on serine 727 without prior phosphorylation of tyrosine 701 (Fig. 3 and Fig. S3) is unusual but not unprecedented. STAT phosphorylation is known to be regulated by several serine kinases including ERK (extracellular signal-regulated protein kinase), p38, JNK (JUN N-terminal kinase), PKCδ (protein kinase Cδ), and possibly CAMK2 (calcium/calmodulin dependent kinase II); and JAK-STAT signalling can be regulated by a variety of cellular signaling pathways through SOCS proteins (suppressors of cytokine signalling), PIAS family proteins (protein inhibitors of activated STAT), and various PTPs (protein tyrosine phosphatases)[56, 57]. Perhaps most surprisingly, the innate immune response to cytoplasmic dsRNA is severely attenuated in human embryonic stem cells because certain key proteins are absent and others cannot be activated [58]. The developmental and tissue specificity of STAT activation, as well as the diversity of signaling inputs, are almost certain to increase the variety of regulatory nodes downstream of JAK/TYK kinases by which CSB-PGBD3 expression could activate interferon-like and antiviral responses in the absence of interferons and viral infection.
4.3 Possible relevance of the CSB-PGBD3 fusion protein to CS disease
Many nonsense and frameshift mutations within the N-terminal CSB domain of the CSB-PGBD3 fusion protein are known that prevent synthesis of the fusion protein, yet still cause CS; and there does not appear to be any correlation between continued expression of the CSB-PGBD3 fusion protein [8] and the surprisingly heterogeneous clinical presentation of CS patients [16]. Nevertheless, the ability of the fusion protein to modulate DNA repair and to induce an interferon-like innate antiviral response in UVSS1KO cells suggest that the fusion protein could contribute to CS especially in patients from consanguineous backgrounds where (as discussed in the Introduction) partial homozygosity uncovers some of the most divergent CS phenotypes [13–15].
Alternatively, Brooks et al. [59] have noted that several neurodegenerative diseases including Trichothiodystrophy (TTD), Aicardi-Goutières syndrome (AGS), and CS exhibit characteristic dysmyelination, calcification, and microcephaly. In TTD, causative mutations in the XPD component of TFIIH reduce TFIIH coactivator function on myelin-related genes. In AGS, mutations in the TREX1 or RNASEH2 nucleases cause accumulation of S-phase DNA fragments that induce a type I interferon response through the STING-dependent innate antiviral response[60]; and we show here that expression of the CSB-PGBD3 fusion protein in CSB-null UVSS1KO cells, or expression of intact CSB in patient-derived CS1AN cells that naturally express the fusion protein, both induce a similar cohort of innate antiviral genes (Tables 4 and S6). As all 5 genes that cause CS (CSA, CSB, XPB, XPD, and XPG) are subunits of TFIIH [9], and expression of the CSB-PGBD3 fusion protein or intact CSB can induce an interferon-like antiviral response in certain genetic backgrounds, it is possible that transcriptional dysregulation and/or an inappropriate interferon response may contribute to the remarkable heterogeneity in CS onset and symptoms.
4.4 The CSB-PGBD3 fusion protein may confer a metabolic advantage
Niedernhofer et al. [45] and van der Pluijm et al. [44] have suggested that CS may reflect reallocation of resources from growthto somatic preservation by the IGF1/insulin pathway in response to unrepaired DNA damage. Our data may support this hypothesis. We find that CSB and the CSB-PGBD3 fusion protein together, but neither protein alone, induce IGFBP5 and repress IGFBP7 (Table S1A) implying that the fusion protein can modulate the IGF1/insulin pathway in normal cells which have functional CSB. Similarly, IGFBP1 is induced in aged mice and an XPF-ERCC1 progeria [44, 45]; and the Drosophila IGFBP7 homolog binds insulin-like peptides in vivo, downregulates insulin/IGF signaling, and prolongs lifespan [61]. The discovery that CSB and CSA are required for mitochondrial as well as nuclear base excision repair (BER) potentially explains why CSB mutations increase cellular reactive oxygen species (ROS) and suggests that the effects of defective nuclear TCR, and activation of the IGF1/insulin pathway, may be compounded by mitochondrial dysfunction [62]. In any event, modulation of the IGF1/insulin pathway by the CSB-PGBD3 fusion protein in the presence of functional CSB may suggest that the conserved fusion protein confers a metabolic advantage rather than, or in addition to, effects on DNA repair and/or chromatin remodeling.
4.5 A cautionary note regarding the emergent functions of other human fusion proteins
Although the CSB-PGBD3 fusion protein has been conserved for >43 My, shares the same N-terminal 465 residues as CSB, and is coexpressed with CSB by alternative splicing [3, 18], the selective advantage of the fusion protein need not be related to CSB function in normal cells. Thus, although our data suggest that the fusion protein may compete with or modulate CSB functions in normal cells and/or affect CSB-related functions in individuals lacking functional CSB protein, the fusion protein could also have emergent functions that differ from the normal functions of the component proteins.
Consider the two other human fusion proteins that have been studied in some detail: (1) The NUP98-HOXA9 fusion protein, which is generated by a chromosomal rearrangement joining the N-terminal FG-repeat domain of nucleoporin NUP98 to the C-terminal DNA-binding domain of the HOXA9 homeodomain transcription factor, causes acute myeloid leukemia (AML) [63]. Although the fusion protein can function as a transcriptional activator targeted by the C-terminal DNA-binding HOXA9 domain [64], this is not the cause of AML. Rather, the N-terminal NUP98 FG-repeat domain of NUP98-HOXA9 forms intranuclear aggregates that sequester the exportin CRM1 [65] resulting in constitutive expression of transcription factors such as NFAT and NFκB that are normally downregulated by nuclear export. (2) The SETMAR fusion protein (aka Metnase) emerged 40–58 Mya and is generated by splicing of a functional histone methyltransferase (SET) domain to a mariner transposase (MAR) which retains specific binding to mariner terminal inverted repeats (TIRs) in vitro [66]. Strikingly, SETMAR/Metnase tethers a chromatin-altering histone methylase (SET) domain to dispersed mariner inverted repeats, just as the CSB-PGBD3 fusion protein tethers the potentially chromatin-altering acidic N-terminal domain of CSB to dispersed MER85 repeats. Yet the only known activity of SETMAR/Metnase is to facilitate nonhomologous end joining (NHEJ), a global repair function that requires both the histone methyltransferase of the SET domain [67] and an endonuclease activity of the mariner transposase but not the capacity for site-specific DNA binding [68]. Thus the ability of SETMAR/Metnase to facilitate NHEJ, and NUP98-HOXA9 to cause AML, both reflect emergent functions — and the same could be true for the CSB-PGBD3 fusion protein.
Highlights DNAREP_1667.
Cockayne syndrome is a devasting childhood progeria most often caused by defects in the CSB gene.
The CSB gene encodes a repair and chromatin remodeling SWI/SNF ATPase.
The CSB gene also generates an abundant CSB-PGBD3 fusion protein that joins the N-terminus of CSB to a piggyBac transposase.
The fusion protein affects DNA repair, and induces an interferon-like response in CSB-null cells.
We speculate regarding the function of the conserved fusion protein in health and disease.
Supplementary Material
Acknowledgements
This work was supported by NIH awards R01GM41624 (AMW), the Cell and Molecular Biology Training Program T32GM007270 (LTG), the Medical Scientist Training Program T32GM007266 (JCN), and a Rosetta Inpharmatics Fellowship from Merck Research Laboratories (JCN).
We thank Choli (Charlie) Lee and Jay Shendure (UW Department of Genome Sciences) for guidance, instruction, and deep sequencing on the Illumina/Solexa 1G Sequencer. We also thank Olivia Perwitasari and Michael Gale (UW Department of Immunology) for IFN-β treated human cell extracts; Roger Bumgarner of the UW Department of Microbiology and Center for Expression Arrays for collegial assistance; HyeonJoo Cheon and George Stark (Lerner Research Institute and Department of Genetics, Case Western Reserve University) for sharing the full list of genes that are regulated by overexpression of YF-STAT1 and WT-STAT1 in BJ cells [31] and form the basis for the analysis presented in Table S3; and Priscilla Cooper (UC Berkeley) for providing UVSS1KO cells originally obtained from Professor Kiyoji Tanaka (Osaka University).
Abbreviations
- CSB
Cockayne syndrome group B protein
- fusion protein
short for CSB-PGBD3 fusion protein and used interchangeably
- HCR
host cell reactivation
- IFN
interferon
- IGFBP5 and IGFBP7
insulin growth factor binding proteins 5 and 7
- My
million years
- Mya
million years ago
- NER
nucleotide excision repair
- PGBD3
piggyBac-derived element 3
- RRS
recovery of RNA synthesis
- STAT
signal transducer and activator of transcription
- TCR
transcription-coupled repair
- UVSS
UV-sensitive syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Appendix A. Consequences of microarray database bias.
Appendix B. Supplementary Figure and Table Legends.
Supplementary data associated with this article can be found, in the online version, at doi: XXXXXXXXX.
References
- 1.Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. J Biol Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 2.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci U S A. 2006;103:9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, Orioli D, Laugel V, Stary A, Hanawalt PC, Sarasin A, Stefanini M. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci U S A. 2009;106:6209–6214. doi: 10.1073/pnas.0902113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anindya R, Mari PO, Kristensen U, Kool H, Giglia-Mari G, Mullenders LH, Fousteri M, Vermeulen W, Egly JM, Svejstrup JQ. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol Cell. 2010;38:637–648. doi: 10.1016/j.molcel.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW, Svejstrup JQ. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A. 2009;106:20705–20710. doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Cormier-Daire V, Gener B, Tobias ES, Tolmie JL, Martin-Coignard D, Drouin-Garraud V, Heron D, Journel H, Raffo E, Vigneron J, Lyonnet S, Murday V, Gubser-Mercati D, Funalot B, Brueton L, Sanchez Del Pozo J, Munoz E, Gennery AR, Salih M, Noruzinia M, Prescott K, Ramos L, Stark Z, Fieggen K, Chabrol B, Sarda P, Edery P, Bloch-Zupan A, Fawcett H, Pham D, Egly JM, Lehmann AR, Sarasin A, Dollfus H. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 9.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 11.Gray LT, Weiner AM. Ubiquitin recognition by the Cockayne syndrome group B protein: binding will set you free. Mol Cell. 2010;38:621–622. doi: 10.1016/j.molcel.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Filippi S, Latini P, Frontini M, Palitti F, Egly JM, Proietti-De-Santis L. CSB protein is (a direct target of HIF-1 and) a critical mediator of the hypoxic response. EMBO J. 2008;27:2545–2556. doi: 10.1038/emboj.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horibata K, Iwamoto Y, Kuraoka I, Jaspers NG, Kurimasa A, Oshimura M, Ichihashi M, Tanaka K. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci U S A. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto S, Suga T, Kudo E, Ihn H, Uchino M, Tateishi S. Adult-onset neurological degeneration in a patient with Cockayne syndrome and a null mutation in the CSB gene. J Invest Dermatol. 2008;128:1597–1599. doi: 10.1038/sj.jid.5701210. [DOI] [PubMed] [Google Scholar]
- 15.Laugel V, Dalloz C, Stary A, Cormier-Daire V, Desguerre I, Renouil M, Fourmaintraux A, Velez-Cruz R, Egly JM, Sarasin A, Dollfus H. Deletion of 5' sequences of the CSB gene provides insight into the pathophysiology of Cockayne syndrome. Eur J Hum Genet. 2008;16:320–327. doi: 10.1038/sj.ejhg.5201991. [DOI] [PubMed] [Google Scholar]
- 16.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 17.Colella S, Nardo T, Botta E, Lehmann AR, Stefanini M. Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-cacchione variant of xeroderma pigmentosum. Hum Mol Genet. 2000;9:1171–1175. doi: 10.1093/hmg/9.8.1171. [DOI] [PubMed] [Google Scholar]
- 18.Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000031. e1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 21.Maziere C, Dantin F, Dubois F, Santus R, Maziere J. Biphasic effect of UVA radiation on STAT1 activity and tyrosine phosphorylation in cultured human keratinocytes. Free Radic Biol Med. 2000;28:1430–1437. doi: 10.1016/s0891-5849(00)00264-1. [DOI] [PubMed] [Google Scholar]
- 22.Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy DE, Muller M, Decker T. Specificity of signaling by STAT1 depends on SH2 and C-terminal domains that regulate Ser727 phosphorylation, differentially affecting specific target gene expression. EMBO J. 2001;20:91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu A, Fan HY, Liao D, Bailey AD, Weiner AM. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- 24.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Ares M, Jr, Chung JS, Giglio L, Weiner AM. Distinct factors with Sp1 and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1987;1:808–817. doi: 10.1101/gad.1.8.808. [DOI] [PubMed] [Google Scholar]
- 26.Vermeulen W, Rademakers S, Jaspers NG, Appeldoorn E, Raams A, Klein B, Kleijer WJ, Hansen LK, Hoeijmakers JH. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- 27.Horibata K, Saijo M, Bay MN, Lan L, Kuraoka I, Brooks PJ, Honma M, Nohmi T, Yasui A, Tanaka K. Mutant Cockayne syndrome group B protein inhibits repair of DNA topoisomerase I-DNA covalent complex. Genes Cells. 2011;16:101–114. doi: 10.1111/j.1365-2443.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 28.Itoh T, Yamaizumi M. UVs syndrome: establishment and characterization of fibroblastic cell lines transformed with simian virus 40 DNA. J Invest Dermatol. 2000;114:101–106. doi: 10.1046/j.1523-1747.2000.00843.x. [DOI] [PubMed] [Google Scholar]
- 29.Newman JC, Weiner AM. L2L: A simple tool for discovering the hidden signficance in microarray expression data. Genome Biol. 2005 doi: 10.1186/gb-2005-6-9-r81. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. Embo J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou YC, Liou KT, Chern CM, Wang YH, Liao JF, Chang S, Chou YH, Shen YC. Preventive effect of silymarin in cerebral ischemia-reperfusion-induced brain injury in rats possibly through impairing NF-kappaB and STAT-1 activation. Phytomedicine. 2010;17:963–973. doi: 10.1016/j.phymed.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Stephanou A, Scarabelli TM, Brar BK, Nakanishi Y, Matsumura M, Knight RA, Latchman DS. Induction of apoptosis and Fas receptor/Fas ligand expression by ischemia/reperfusion in cardiac myocytes requires serine 727 of the STAT-1 transcription factor but not tyrosine 701. J Biol Chem. 2001;276:28340–28347. doi: 10.1074/jbc.M101177200. [DOI] [PubMed] [Google Scholar]
- 37.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 38.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerlier D, Avice T. Use of an automatic cell harvester in a cellular radioimmunoassay. J Immunol Methods. 1984;75:159–166. doi: 10.1016/0022-1759(84)90235-7. [DOI] [PubMed] [Google Scholar]
- 40.Jiang LJ, Zhang NN, Ding F, Li XY, Chen L, Zhang HX, Zhang W, Chen SJ, Wang ZG, Li JM, Chen Z, Zhu J. RA-inducible gene-I induction augments STAT1 activation to inhibit leukemia cell proliferation. Proc Natl Acad Sci U S A. 2011;108:1897–1902. doi: 10.1073/pnas.1019059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauter D, Specht A, Kirchhoff F. Tetherin: holding on and letting go. Cell. 2010;141:392–398. doi: 10.1016/j.cell.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 42.Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 2010;18:388–396. doi: 10.1016/j.tim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Boxel-Dezaire AH, Stark GR. Cell type-specific signaling in response to interferon-gamma. Curr Top Microbiol Immunol. 2007;316:119–154. doi: 10.1007/978-3-540-71329-6_7. [DOI] [PubMed] [Google Scholar]
- 44.van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, Beems R, Niedernhofer LJ, Velasco S, Friedberg EC, Tanaka K, van Steeg H, Hoeijmakers JH, van der Horst GT. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 46.Moserle L, Indraccolo S, Ghisi M, Frasson C, Fortunato E, Canevari S, Miotti S, Tosello V, Zamarchi R, Corradin A, Minuzzo S, Rossi E, Basso G, Amadori A. The side population of ovarian cancer cells is a primary target of IFN-alpha antitumor effects. Cancer Res. 2008;68:5658–5668. doi: 10.1158/0008-5472.CAN-07-6341. [DOI] [PubMed] [Google Scholar]
- 47.Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, Enkemann S, Jove R, Haura EB. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 48.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong B, Zhou Q, Zhao J, Zhou A, Harty RN, Bose S, Banerjee A, Slee R, Guenther J, Williams BR, Wiedmer T, Sims PJ, Silverman RH. Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J Virol. 2004;78:8983–8993. doi: 10.1128/JVI.78.17.8983-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivieccio MA, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee SC, Brosnan CF. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. 2006;177:4735–4741. doi: 10.4049/jimmunol.177.7.4735. [DOI] [PubMed] [Google Scholar]
- 53.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garinis GA, Uittenboogaard LM, Stachelscheid H, Fousteri M, van Ijcken W, Breit TM, van Steeg H, Mullenders LH, van der Horst GT, Bruning JC, Niessen CM, Hoeijmakers JH, Schumacher B. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11:604–615. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 57.Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008;19:383–394. doi: 10.1016/j.cytogfr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Chen LL, Yang L, Carmichael GG. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle. 2010;9:3552–3564. doi: 10.4161/cc.9.17.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7:834–848. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alic N, Hoddinott MP, Vinti G, Partridge L. Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell. 2011;10:137–147. doi: 10.1111/j.1474-9726.2010.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sykora P, Wilson DM, 3rd, Bohr VA. Repair of persistent strand breaks in the mitochondrial genome. Mech Ageing Dev. 2011 doi: 10.1016/j.mad.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–6257. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghannam G, Takeda A, Camarata T, Moore MA, Viale A, Yaseen NR. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J Biol Chem. 2004;279:866–875. doi: 10.1074/jbc.M307280200. [DOI] [PubMed] [Google Scholar]
- 65.Takeda A, Sarma NJ, Abdul-Nabi AM, Yaseen NR. Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins. J Biol Chem. 2010;285:16248–16257. doi: 10.1074/jbc.M109.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beck BD, Lee SS, Williamson E, Hromas RA, Lee SH. Biochemical characterization of metnase's endonuclease activity and its role in NHEJ repair. Biochemistry. 2011;50:4360–4370. doi: 10.1021/bi200333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.