Abstract
We applied a dynamic Bayesian network method that identifies joint patterns from multiple functional genomics experiments to ChIP-seq histone modification and transcription factor data, and DNaseI-seq and FAIRE-seq open chromatin readouts from the human cell line K562. In an unsupervised fashion, we identified patterns associated with transcription start sites, gene ends, enhancers, CTCF elements, and repressed regions. Software and genome browser tracks are at http://noble.gs.washington.edu/proj/segway/.
Recently, the genomics community has seen an explosion in the availability of large-scale functional genomics data. Researchers have produced genome-wide data sets on the locations of transcription factor binding and histone modifications via chromatin immunoprecipitation (ChIP), open chromatin, and RNA transcription using sequence census assays. Consequently, our representation of the whole human genome has expanded from a sequence of nucleotides, occasionally annotated with discrete features, to a collection of numerical data tracks defined at almost every part of every chromosome. Simultaneously, we have moved beyond treating the cell state determined by functional genomics experiments as constitutive and universal. We will soon have access to data from dozens of cell types1. How can we make sense out of this multitude of data, unprecedented in experimental biology?
A segmentation procedure provides a conceptually simple approach to finding patterns in this kind of genomic data2,3,4,5,6,7. In this procedure, one partitions the genome into non-overlapping contiguous segments, assigning to each segment a label, such as “promoter” or “enhancer,” drawn from a finite set. One assigns the labels such that regions sharing the same segment label share certain properties in the observed data. Occam’s Razor implies the desirability of making the set of segment labels as small as possible, while still retaining their capability of accurately modeling the observations. The segmentation task involves finding segment boundaries while simultaneously assigning labels to the segments.
Approaches to segmentation using a hidden Markov model (HMM) suffer from some limitations. For example, HMMs handle missing data poorly, requiring interpolation and smoothing to process regions where data is not or cannot be reported. Similarly, hard or soft constraints on segment lengths prove complex and difficult to implement with a simple HMM.
Dynamic Bayesian networks (DBNs) provide a powerful framework for modeling the complex hidden relationships that explain observed data defined along an axis of arbitrary length. Automatic speech recognition researchers have long used DBNs8, and now scientists have started using them to solve biological problems9. A DBN is a graphical structure that depicts the conditional independence properties of multiple random variables. We can represent a standard HMM by a DBN with a hidden random variable for the HMM’s hidden state, and an observed random variable for the observations. With a DBN, however, we can easily incorporate arbitrarily complex relationships among variables at the current or nearby positions in the genome. The DBN allows us to include multiple hidden variables and model their interrelationships without flattening them into a single state variable and thereby ignoring the properties implied by these interrelationships. For example, the DBN can incorporate a structured state space, in which superlabels and sublabels augment a simple label set, where the superlabel might represent something like a region of active euchromatin, and the sublabel the 3′ end of an active gene within active euchromatin. In general, incorporating various types of prior knowledge, and interpreting a trained model, proves much easier with a DBN than with an HMM. For example, we have extended the model to include a principled view of heterogeneous missing data (Supplementary Discussion, Supplementary Fig. 1, and Supplementary Table 1).
In this communication, we describe a new segmentation method, called Segway, which uses DBN techniques to perform simultaneous segmentation and clustering of genomic data. We describe the application of Segway to human ChIP-seq, DNase-seq, and FAIRE-seq data from the ENCODE Project1.
We performed unsupervised training on 1% of the human genome using the Segway model and 31 ENCODE signal tracks. We arbitrarily fixed the number of labels at 25 so that the set of labels would be sufficiently small to remain interpretable by biologists. The use of 25 labels provides human convenience in the interpretation of results. Greater numbers of labels may have more statistical support, but yield difficult-to-interpret results. Our method aims to reduce complex data for easier interpretation, and using an arbitrarily small number of labels served this purpose. The discovered parameters characterize a diverse set of biological patterns (Fig. 1). We then Viterbi decoded the genome, identifying the most probable path of segment labels given the trained parameters and observed signal.
Fig. 1.
Heat map of discovered Gaussian parameters in an unsupervised 25-label segmentation of 31 ENCODE signal tracks. Each row contains parameters for one signal track, and each column contains parameters for one segment label. Within each row, we performed an affine transformation such that the largest mean was 1 and the smallest 0. The color in each cell indicates the transformed mean parameter μ according to the color bar on the left. The width of the inner boxes is proportional to the square root of the variance parameter σ2, after multiplying by the linear factor used in the transformation of μ. Dendrograms show a hierarchical clustering by both rows and columns. We compared the model parameters and segmentation against existing literature to manually assigned functional categories to the segment labels (D: dead, F: FAIRE, R: repression, H3K9me1: histone 3 lysine 9 monomethylation, L: low, GE: gene end, TF: transcription factors, C: CTCF, TSS: transcription start site, GS: gene start, E: enhancer, GM: gene middle).
We refer to the resulting assignment of labels to non-overlapping genomic regions as the segmentation (Supplementary Results, Supplementary Fig. 2, and Supplementary Table 2). The labels allow one to easily identify other regions that exhibit similar signal patterns in the observation tracks. By examining the learned parameters directly (Fig. 1 and Supplementary Fig. 3) and comparing the segmentation to public annotations (Fig. 2 and Supplementary Figs. 3–5), we assigned functional categories to groups of segment labels based on notable features. Many of these labels recapitulated known patterns in the chromatin literature, and some represented novel patterns.
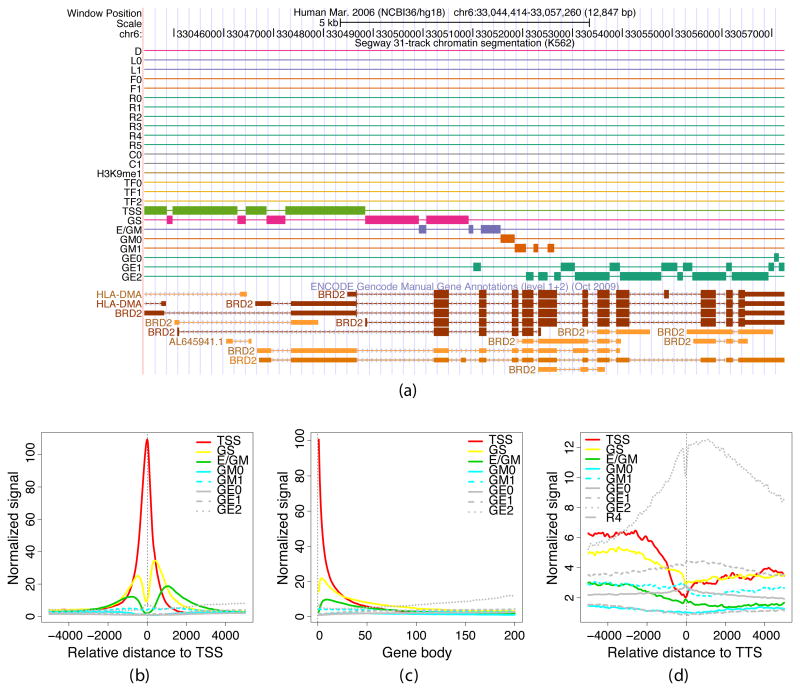
Fig. 2.
Gene structure in Segway labels. (a) Segmentation of the BRD2 locus, as shown in the University of California, Santa Cruz (UCSC) Genome Browser. The first three rows show the position along chromosome 6 and a scale bar. The next 25 rows correspond to the labels of the segmentation, with a thick bar present at any position where the segmentation has the corresponding label5. Labels are colored by functional category. The bottom rows show GENCODE manual gene annotations. (b)–(d) Aggregation of gene-related Segway labels with respect to the starts, middles and ends of protein-coding genes from GENCODE v7. Each panel plots the mean density of a set of gene-related Segway labels as a function of genomic location relative to (a) the TSS, (b) the gene body, extending from the TSS to the TTS, and (c) the TTS. The frequency of each label (vertical axis) is normalized by the relative proportion of the genome that is covered by that label. Genomic distances in the center panel have been scaled to a proportion of gene length.
Notably, Segway “rediscovered” protein-coding genes, as the unsupervised segmentation included chromatin states associated with the starts, middles, and ends of genes (Fig. 2) found without direct recourse to information traditionally used to find genes (primary sequence, similarity to mRNAs or genome sequences of other species). Several labels tended to reside in particular locations within protein-coding genes, and the discovered transition parameters of the model increased the probability of moving from the labels that tended to reside in 5′ ends of genes to the labels found more often in 3′ ends of genes. We also found expected patterns of transcription factor binding near the transcription start site (TSS), histone H2A.Z associated with the TSS10, and histone mark H3K79me2 associated with the gene start10 (Supplementary Fig. 6). The patterns of chromatin structure in protein-coding genes fit well with existing knowledge (Supplementary Results, and Supplementary Figs. 7 and 3).
Many of the chromatin states which, at first glance, seemed to associate with protein-coding genes, turned out to be associated only with genes active in the tissue type of the assays incorporated into the segmentation (Supplementary Results). Segway also discovered patterns associated with specific functional elements, such as enhancers, insulators, regions of repressed gene expression, and “dead” regions (Supplementary Results and Supplementary Figs. 5 and 8).
We used Segway to generate hypotheses about the role of individual sequences in promoting transcription, which we then tested experimentally. We began by identifying expressed and unexpressed genes using RNA data. To determine whether specific sequences identified by segmentation labels sufficed for these genes’ expression or non-expression, we tested small ~1-kbp constructs that overlapped the gene’s TSS and either a segmentation TSS label (24 positive predictions) or an R label (67 negative predictions) using transient transfection luciferase assays. Every positive prediction resulted in substantial expression of a reporter, a majority of the negative predictions resulted in no substantial expression, and a larger majority of negative predictions not overlapping CpG islands resulted in no substantial expression (Supplementary Fig. 9a). Positive predictions showed higher median expression activity than negative predictions.
Segway differs substantially from several previously described methods for jointly analyzing chromatin data (Supplementary Discussion). For example, Segway solves a fundamentally different problem than ChromaSig11, in the sense that ChromaSig does not attempt to fully partition the genome or integrate arbitrary combinations of functional genomics data. Segway is most similar to a hidden Markov model-based method called ChromHMM5. However, Segway and ChromHMM differ in several respects, chiefly the relative resolution of the two methods: Segway operates on the full data set, whereas ChromHMM works at 200 bp resolution and condenses each data track to a single binary datum within each 200-bp window. Consequently, Segway provides a finer-grained segmentation. Whereas the segments in a published ChromHMM segmentation5 had a minimum length of 200 bp, a mean of 4,862 bp and a median of 800 bp, Segway segments had a mean length of 168 bp and a median of 124 bp.
We directly compared the behavior of Segway and ChromHMM in three separate loci (Fig. 3 and Supplementary Fig. 10). Segway successfully identified the TSS of GATA1, followed by a series of gene-related labels that generally progress from “gene start” through “gene middle” labels (Fig. 3a). ChromHMM, in contrast, identified a large region that it identifies as “active promoter,” spanning thousands of base pairs. It labeled the rest of the gene “strong enhancer.” This example suggests that the 200 bp window size makes precise identification of fine-scale elements like promoters and internal gene structure difficult. We saw a similar effect at the HBG1 locus (Fig. 3b). Here, a known enhancer12 appeared in the midst of a large (~10 kbp) ChromHMM segment labeled “active promoter,” whereas Segway placed a small enhancer label in the middle of the known enhancer region. We saw this behavior genome-wide (Supplementary Results).
Fig. 3.
Comparison of Segway and ChromHMM segmentations in several locations. (a) The GATA1 locus. (b) The HBG1 locus and an upstream enhancer12.
We have shown that Segway successfully reduces heterogeneous datasets into understandable patterns with clear biological implications. In the future, the flexibility of the DBN suggests a solution to the problem of learning large numbers of segment labels while retaining comprehensibility. Extension to a hierarchical segmentation would allow the learning of many sublabel patterns, while keeping a higher-level yet smaller-order structure that a researcher could analyze and understand more readily. While effective hyperparameter setting for hierarchical segmentation requires further research, we have already implemented the capability for hierarchical training and identification in Segway.
Methods
Signal generation
We selected 31 ChIP-seq, DNase-seq, and FAIRE-seq assays performed in the chronic myeloid leukemia cell line K562 by the ENCODE Project1. We downloaded tagAlign files from the ENCODE Data Coordination Center (DCC)13. We used Wiggler (http://code.google.com/p/align2rawsignal/) to convert tag alignment positions into an extended reads per base (ERPB) signal for each position of the genome, pooling multiple replicates when available. We then loaded the signal data into a Genomedata14 archive. We include the URL for each of the tagAlign and signal files used in Supplementary Table 4.
Signal transformation
To reduce the distorting effects of high data values in sequence census assay data, Segway first transforms all data values with the inverse hyperbolic sine function asinh x = ln(x +√(x2 + 1)). This function has the compressing effect of a function like ln x for large values of x, but has much less of a compressing effect for small values. Additionally, it is defined on the entire real number line, preserving whether the input data values are positive or negative. One therefore avoids the discontinuities inherent in the use of a simple logarithmic transformation and the necessity to add some offset first to deal with zero values15.
The core model
Segway performs training and inference on a DBN model designed for genomic segmentation (Supplementary Fig. 11). The core of the default Segway DBN has some similarity to an HMM, with multiple observation tracks and a number of discrete hidden variables. The default Segway DBN has a segment label backbone defined for each base pair of a genomic region. This hidden variable emits an observation variable for each observation track present at that position. An observation track is a sequence of numerical observations, such as the number of sequence tags overlapping successive genomic positions, or the intensity of a microarray probe associated with a position. In the default model, the ith observation track is represented by the sequence of random variables X1:T(i) = (X1(i), …, XT(i)).
Some positions t may not correspond to a defined value of Xt(i). Examples include unmappable sequence in sequence census assays, or sequence not represented by a probe in a microarray assay. To explicitly model these missing data, we use an indicator variable to mark whether Xt(i) is defined (X˚t(i) = 1) or undefined (X˚t(i) = 0). The observation variables at every position have dependency only on the hidden segment label variable at that position Qt (Supplementary Fig. 11). When the indicator variable X˚t(i) = 0, then Xt(i) does not have a dependence on Qt. In that case, the conditional parent arc from Qt to Xt(i) is, for all intents and purposes, nonexistent in the DBN16.
Segway models the asinh-transformed data with an m × n matrix of scalar Gaussians, with one Gaussian for each combination of n observation tracks and m segment labels. Each Gaussian has a mean parameter μ, and variance parameter σ2, which control the signal distribution that the Gaussian emits. Segway ties variance parameters such that they are all equal for a given observation track.
Weighting
Segway weights the contribution of each observation variable such that tracks with different numbers of data points still have roughly the same contribution to the overall likelihood of the model. It does this by exponentiating during inference calculations every occurrence of an original probability P(Xt(i)|Qt) for some track i by the number of data points for that track N(i) = Σt X˚t(i) divided by the maximum number of data points for any particular track N* = maxj N(j). In other words, it replaces the original probability with a new probability
Since probabilities are always in the range [0, 1], an increase in weight will result in a decreased probability of the model overall, but when the weight is held constant, changes in the probabilities exponentiated to higher weights will affect the overall probability more than those exponentiated to lower weights.
The duration model
The Segway model includes additional random variables that allow tuning beyond the simple HMM-like core model. A discrete countdown variable Ct allows the specification of minimum or maximum segment length. This variable begins with an initial value dependent on Qt, and decreases by 1 at every position where the ruler marker variable Mt is 1 (“ruler marks”). Decreasing the countdown variable only upon the occurrence of an occasional ruler mark decreases the countdown variable state space while still allowing longer minimum or maximum segment lengths. This heuristic greatly decreases computational complexity. A binary segment transition variable Jt can either force the segment label to change at the current position (Jt =1), or prevent it from changing (Jt =0). Segway generates a conditional probability table P(Jt =1|Qt−1, Ct−1) that maps each (Qt−1, Ct−1) tuple to one of three rules that determine the value of Jt: (a) force: P(Jt = 1) = 1, (b) prevent: P(Jt = 0) = 1, or (c) allow: P(Jt = 1) = 1/(1 + L). The duration in which the segment label is unchanged following the point where the allow rule becomes active follows a geometric distribution with an expected length of the L value9,17.
In this study, we set Ct =10 and the force rule at the beginning of each segment. We also set the ruler marker every ten positions. This setting allows the enforcement of large minimum segment lengths with only a small increase in state space and computational complexity. When Ct decreases to 0, we set the allow rule with L =100,000 in order to model broader behavior when the observation variables do not control the segment label, as they usually do. Despite the enforcement of a minimum duration before allowing a change in segment label, it is still possible to have very short segments at the end of input sequences, such as at the 3′ ends of genome assembly scaffolds.
DBN inference
Segway uses the Graphical Models Toolkit (GMTK)18 to perform expectation–maximization (EM) training19, and Viterbi decoding20 on the Segway models. Segway uses model structure files in the GMTKL specification language, and users may modify them or replaced them with a DBN of arbitrary complexity.
For decoding, Segway divides large sequences into non-overlapping windows of no more than 2 Mbp in size each so that the necessary computation can fit into available memory.
Training and parameters
We performed ten separate instances of EM training, each with different initial parameters, and each instance had no more than 100 iterations. Segway picks an initial mean parameter μ for each track uniformly from the values in μ̃ ± 0.2σ̃ for the empirical mean μ̃ and standard deviation σ̃ of the training data. It sets variance parameters σ2 to the difference between the maximum and minimum data values within a particular observation track. We performed training on the ENCODE pilot regions (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/encodeRegions.txt.gz), which represent 1% of the human genome (30 Mbp).
Segway sets the probabilities P(Q1 = q) of starting a sequence with each segment label q ∈ [1, n] to 1/n, and does not adjust these probabilities during training. The conditional probability table P(Qt|Qt−1), which Segway only uses when the segment transition variable Jt = 1, has only zeroes in its diagonal. This prevents a “transition” that does not change the segment label. Segway initially sets the other values equally, but EM training will adjust them.
A training instance continues until the difference between the difference between the current likelihood Ln and the likelihood from the previous round Ln−1 diminishes such that
We designated the training instance with the highest final likelihood as the winning training instance, and used its parameters to create the primary segmentation discussed in this article.
Segment identification and visualization
We performed Viterbi decoding on 92% of the genome (2,828 Mbp), excepting excluded blacklist regions downloaded from the ENCODE Data Coordination Center1 (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/encodeDCC/wgEncodeMapability/wgEncodeDukeRegionsExcluded.bed6.gz). These regions contain repetitive elements that cause artifactually high signal in sequence census assays. We created the primary segmentation discussed in this article using the winning training instance parameters, but performed Viterbi decoding using parameters from the other nine training instances for comparison purposes. Segway produces segmentations in BED format, designed for easy uploading and display on the UCSC Genome Browser21.
Distributed computing
Segway uses the Grid Engine or Platform LSF distributed computing systems through the Distributed Resource Management Application API (http://www.drmaa.org/) interface. Using these systems, Segway trains or decodes on multiple sequences and multiple initial parameter values at once, leading to a considerable savings in wall clock time. Segway automatically tunes memory usage by first queuing jobs with a small amount of memory (such as 2 GiB), and then repeating with ever-larger amounts of memory if GMTK cannot complete inference with the smaller amount. This approach allows efficient resource management even when one cannot easily predict the memory usage in advance.
Genome assembly and annotations
We performed all training, segmentation, and analysis on the NCBI36 assembly of the human genome (hg18). We used GENCODE22 version 3c gene annotations (http://www.gencodegenes.org/releases/3c.html) for aggregation and overlap analyses, as well as track display. We used ENCODE Project CAGE23 data ftp://genome.crg.es/pub/Encode/data_analysis/TSS/Gencodev3cTSS.gff) for overlap analyses. We downloaded PhastCons scores for multiple alignments of 45 vertebrates to the human genome24 from the UCSC Genome Browser. We downloaded nucleosome positioning signal data collected in K562 by MNase digestion and high-depth sequencing from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeSydhNsome).
We produced genome-wide visualizations and summary statistics using Segtools25. We based precision and recall statistics on the overlap of the bases within a segment against some annotation, where true positives (TP) are positions where the segment label overlaps the annotation, false positives (FP) are positions where the segment label does not overlap the annotation, and false negatives (FN) are positions where the annotation does not overlap the segment label. We expanded point annotations (such as TSSes) by 500 bp on each side before calculating these statistics. Precision (also known as positive predictive value) is defined as TP/(TP+FP). Recall (also known as sensitivity) is defined as TP/(TP+FN). We used precision and recall for evaluation instead of receiver operating characteristic curves, because precision and recall can provide a more informative measurement of performance26. We defined TSSs with reproducible CAGE support as those with at least 2 cytoplasmic CAGE tags.
Transient transfection luciferase assays
We predicted the promoter activity of sequences in the segmentation selected by the following procedure. We started with the segments that overlapped both GENCODE manually annotated protein-coding TSSs and a catalog of existing constructs from SwitchGear Genomics. We then excluded the segments that overlapped RepeatMasker 3.27 regions downloaded from the UCSC Genome Browser (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/). We defined positive predictions as those segments with a TSS label that overlapped a GENCODE TSS with at least 2 K562 cytosolic poly(A)+ CAGE tags and a GENCODE TSS for a gene with at least K562 RNA-seq reads per kilobase per million reads (RPKM) above the 90th percentile. We defined negative predictions as those segments with R labels that overlapped a GENCODE TSS with no CAGE tags in any K562 cellular compartment, and a GENCODE TSS for a gene with 0.0 RPKM K562 RNA-seq reads. We further divided negative predictions by whether they had GM12878 RNA-seq read RPKM over the 10th percentile or not. This process resulted in a candidate set of 26 positive predictions and 74 negative predictions.
SwitchGear Genomics performed transient transfection luciferase assays in triplicate on 24 positive predictions, 67 negative predictions, and 5 control constructs, narrowing from the candidate set based on constructs in stock. They mixed 100 ng luciferase reporter DNA, 0.4 μm Lipofectamine LTX (Invitrogen), and 0.1 μL PLUS Reagent (Invitrogen), and incubated it for 30 min. SwitchGear then thoroughly mixed this suspension with 25,000 freshly counted K562 cells resuspended in warm, complete media. They aliquotted 100 μL of the combined suspension into replicate white 96-well assay plates and incubated at 37 °C for 24 h. They then removed the plates from the incubator, added SteadyGlo reagent (Promega) to each well, and kept the plates in the dark for >30 min. They then read the plates on an LmaxII-384 luminometer (Molecular Devices). We considered the resulting data points substantial when their activity exceeds a threshold of the mean control activity plus 6 times the standard deviation control activity.
Gene Ontology enrichment analysis
We analyzed the sets of GENCODE TSSs that overlapped with each segment label for the enrichment of GO terms. We used the Ensembl27 Perl API to convert GENCODE transcript identifiers to UniProt28 identifiers. We then used FuncAssociate29 to find enrichment in 25 resulting gene sets, using 10,000 resamplings for each to determine hypothesis-wise P values. We then multiplied these P values by 25 as a Bonferroni correction for testing multiple hypotheses.
Transcription factor motif analysis
We obtained position specific scoring matrices (PSSMs) from the TRANSFAC 10.230 and JASPAR CORE 200931 databases. Using FIMO32, we scanned the human genome for significant matches to the PSSM (q < 0.1). From these candidate motifs, we selected known vertebrate cis-regulatory motifs with at least 1,000 significant matches in the genome. We used the Genome Structure Correction marginal region overlap test in block bootstrap33, as implemented in Statmap (http://www.statmap-bio.org), to test each segment label for enrichment in matches to each of the selected motifs. We then Bonferroni corrected the resulting P values.
Statistical analysis
We performed statistical analysis with R 2.11 (http://www.rproject.org/).
Supplementary Material
Supplementary Figure 1. A heterogeneous pattern of missing data due to ambiguously mappable sequence tags in 31 ENCODE data sets along a 10,000-bp region of human chromosome 22.
Supplementary Figure 2. Heat map of the overlap between the reference segmentation presented in this article and nine alternate segmentations generated by Segway with the same data.
Supplementary Figure 3. Graph of transitions between labels in the model learned by Segway.
Supplementary Figure 4. Aggregation plot of enrichment of the observed overlap between segmentation labels and position quantiles along components of 22, 428 GENCODE protein-coding gene annotations.
Supplementary Figure 5. Evolutionary conservation associated with Segway labels.
Supplementary Figure 6. Aggregation of gene-related labels with respect to protein binding and histone marks.
Supplementary Figure 7. Enrichment of transcription factor binding site motifs in segment labels.
Supplementary Figure 8. Nucleosome positioning signal in the vicinity of CTCF sites identified by Segway.
Supplementary Figure 9. Box plot of luciferase activity in transient transfection assays for three replicates of 96 constructs.
Supplementary Figure 10. Comparison of Segway and ChromHMM segmentations
Supplementary Figure 11. Graphical model representation of the default Segway DBN.
Supplementary Table 1. Number of positions at which various numbers of signal tracks have missing data within the ungapped regions of the genome assembly.
Supplementary Table 2. Segment summary statistics by label.
Supplementary Table 3. Gene Ontology terms enriched in GENCODE TSSs overlapping with each segment label.
Supplementary Table 4. List of observation tracks and corresponding data identifiers.
Supplementary Results. Unsupervised segmentation of 31 ENCODE data sets. The rediscovery of protein-coding genes. Discriminating between active and inactive genes. Discovery of other genomic elements. Comparison with ChromHMM.
Supplementary Discussion. Segway reduces heterogeneous datasets into understandable patterns with clear biological implications. Segway differs conceptually from other related tools. DBN architecture allows a superior model of functional genomics data.
Supplementary References. References for Supplementary Information.
Acknowledgments
We thank P.J. Collins for assistance with the transient transfection assays, S. Djebali for processing CAGE data, C.E. Grant for motif analysis and A. Kundaje for helpful suggestions. We also thank the ENCODE Project Consortium, the ENCODE DCC, and the National Human Genome Research Institute (NHGRI) for providing early public access to the unpublished data used in this work. This work used data produced by the laboratories of B.E. Bernstein (Broad Institute of MIT and Harvard), M.P. Snyder (Stanford University), R.M. Myers (HudsonAlpha Institute for Biotechnology), P.J. Farnham (University of Southern California), V.R. Iyer (University of Texas at Austin), G.E. Crawford (Duke University), J.D. Lieb (University of North Carolina at Chapel Hill), J.A. Stamatoyannopoulos (University of Washington), T.S. Furey (University of North Carolina at Chapel Hill), P. Carninci (RIKEN), T.R. Gingeras (Cold Spring Harbor Laboratory), and A. Sidow. This publication was made possible by Grant Numbers 004695, 004561, and 006259 from NHGRI.
Footnotes
Author contributions
M.M.H., W.S.N., and J.A.B. conceived the project; M.M.H., W.S.N, and Z.W. designed computational and biological experiments. M.M.H., J.A.B., O.J.B., and J.W. developed software used in this work; M.M.H., O.J.B., and J.W. performed computational experiments and analyzed data; M.M.H., W.S.N., Z.W., J.A.B., O.J.B., and J.W. wrote the manuscript.
Bibliography
- 1.ENCODE Project Consortium. A user’s guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS. Unsupervised segmentation of continuous genomic data. Bioinformatics. 2007;23:1424–1426. doi: 10.1093/bioinformatics/btm096. [DOI] [PubMed] [Google Scholar]
- 3.Erdman C, Emerson JW. A fast Bayesian change point analysis for the segmentation of microarray data. Bioinformatics. 2008;24:2143–2148. doi: 10.1093/bioinformatics/btn404. [DOI] [PubMed] [Google Scholar]
- 4.Jaschek R, Tanay A. Spatial clustering of multivariate genomic and epigenomic information. In: Batzoglou S, editor. Research in Computational Molecular Biology, Lecture Notes in Computer Science. Vol. 5541. Springer; Berlin: 2009. pp. 170–183. [Google Scholar]
- 5.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilmes J, Bartels C. Graphical model architectures for speech recognition. IEEE Signal Process Mag. 2005;22:89–100. [Google Scholar]
- 9.Reynolds SM, Käll L, Riffle ME, Bilmes JA, Noble WS. Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLoS Comput Biol. 2008;4:e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr Opin Genet Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King DC, et al. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raney BJ, et al. ENCODE whole-genome data in the UCSC Genome Browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman MM, Buske OJ, Noble WS. The Genomedata format for storing large-scale functional genomics data. Bioinformatics. 2010;26:1458–1459. doi: 10.1093/bioinformatics/btq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson NL. Systems of frequency curves generated by methods of translation. Biometrika. 1949;36:149–176. [PubMed] [Google Scholar]
- 16.Bilmes J. Dynamic Bayesian multinets. In: Boutilier C, Goldszmidt M, editors. UAI ’00: Proceedings of the 16th Conference on Uncertainty in Artificial Intelligence. Morgan Kaufmann; San Francisco: 2000. pp. 38–45. [Google Scholar]
- 17.Grundy WN, Bailey TL, Elkan CP, Baker ME. Meta-MEME: motif-based hidden Markov models of protein families. Comput Appl Biosci. 1997;13:397–406. doi: 10.1093/bioinformatics/13.4.397. [DOI] [PubMed] [Google Scholar]
- 18.Bilmes J, Bartels C. On triangulating dynamic graphical models. In: Meek C, Kjærulff U, editors. UAI ’03, Proceedings of the 19th Conference in Uncertainty in Artificial Intelligence. Morgan Kaufmann Publishers; San Francisco: 2003. pp. 47–56. [Google Scholar]
- 19.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B. 1977;39:1–22. [Google Scholar]
- 20.Viterbi AJ. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans Inf Theory. 1967;13:260–269. [Google Scholar]
- 21.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7:S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi H, Kato S, Murata M, Carninci P. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol Biol. 2012;786:181–200. doi: 10.1007/978-1-61779-292-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buske OJ, Hoffman MM, Ponts N, Roch KGL, Noble WS. Exploratory analysis of genomic segmentations with Segtools. BMC Bioinformatics. 2011;12:415. doi: 10.1186/1471-2105-12-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis J, Goadrich M. The relationship between precision-recall and ROC curves. Proceedings of the 23rd International Conference on Machine Learning; New York: ACM; 2006. pp. 233–240. [Google Scholar]
- 27.Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingender E, et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandelin A, Alkema W, Engstrom P, Wasserman W, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bickel PJ, Boley N, Brown JB, Huang H, Zhang NR. Subsampling methods for genomic inference. Ann Appl Stat. 2010;4:1660–1697. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A heterogeneous pattern of missing data due to ambiguously mappable sequence tags in 31 ENCODE data sets along a 10,000-bp region of human chromosome 22.
Supplementary Figure 2. Heat map of the overlap between the reference segmentation presented in this article and nine alternate segmentations generated by Segway with the same data.
Supplementary Figure 3. Graph of transitions between labels in the model learned by Segway.
Supplementary Figure 4. Aggregation plot of enrichment of the observed overlap between segmentation labels and position quantiles along components of 22, 428 GENCODE protein-coding gene annotations.
Supplementary Figure 5. Evolutionary conservation associated with Segway labels.
Supplementary Figure 6. Aggregation of gene-related labels with respect to protein binding and histone marks.
Supplementary Figure 7. Enrichment of transcription factor binding site motifs in segment labels.
Supplementary Figure 8. Nucleosome positioning signal in the vicinity of CTCF sites identified by Segway.
Supplementary Figure 9. Box plot of luciferase activity in transient transfection assays for three replicates of 96 constructs.
Supplementary Figure 10. Comparison of Segway and ChromHMM segmentations
Supplementary Figure 11. Graphical model representation of the default Segway DBN.
Supplementary Table 1. Number of positions at which various numbers of signal tracks have missing data within the ungapped regions of the genome assembly.
Supplementary Table 2. Segment summary statistics by label.
Supplementary Table 3. Gene Ontology terms enriched in GENCODE TSSs overlapping with each segment label.
Supplementary Table 4. List of observation tracks and corresponding data identifiers.
Supplementary Results. Unsupervised segmentation of 31 ENCODE data sets. The rediscovery of protein-coding genes. Discriminating between active and inactive genes. Discovery of other genomic elements. Comparison with ChromHMM.
Supplementary Discussion. Segway reduces heterogeneous datasets into understandable patterns with clear biological implications. Segway differs conceptually from other related tools. DBN architecture allows a superior model of functional genomics data.
Supplementary References. References for Supplementary Information.