Abstract
Brca1 is required for DNA repair by homologous recombination (HR) and normal embryonic development. Here we report that deletion of the DNA damage response factor 53BP1 overcomes embryonic lethality in Brca1-nullizygous mice, and rescues HR deficiency, as measured by hypersensitivity to PARP (polyADP-ribose polymerase) inhibition. However, Brca1,53BP1 double-deficient cells are hypersensitive to DNA interstrand cross-links (ICLs), indicating that BRCA1 has an additional role in DNA cross-link repair that is distinct from HR. Disruption of the non-homologous end-joining (NHEJ) factor, Ku, promotes DNA repair in Brca1-deficient cells; however deletion of either Ku or 53BP1 exacerbates genomic instability in cells lacking FANCD2, a mediator of the Fanconi Anemia pathway for ICL repair. BRCA1 therefore has two separate roles in ICL repair, whereas FANCD2 provides a key activity that can not be bypassed by ablation of 53BP1 or Ku.
Introduction
In mammalian cells, homologous recombination (HR) and non-homologous end joining (NHEJ) are the two major pathways involved in the repair of DNA double-strand breaks (DSBs) (Kass and Jasin, 2010). HR is initiated by DNA end resection, which involves the production of recombinogenic 3’ single-stranded DNA by the action of several proteins including BRCA1, Mre11, CtIP, Exo1 and Blm (Gravel et al., 2008; Sartori et al., 2007; Stracker and Petrini, 2011; Yun and Hiom, 2009). Following end resection, single-stranded DNA is stabilized by binding of replication protein A (RPA). Rad51 subsequently replaces RPA on single-stranded DNA, enabling strand invasion at an intact homologous DNA region, which is used as a template for repair (Kass and Jasin, 2010). In contrast to HR, NHEJ directly re-ligates broken DNA. This process is initiated by the Ku70/Ku80 (Ku) heterodimer which binds directly to the break and recruits the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), stabilizing and aligning the ends (Getts and Stamato, 1994; Rathmell and Chu, 1994; Taccioli et al., 1994). End rejoining is then completed by activities of the XRCC4/DNA ligase IV (Lig4) complex (Critchlow and Jackson, 1998).
The importance of double-strand break (DSB) repair in mammalian cells is demonstrated by the tumor predisposition in humans and mice associated with mutation of the HR gene, Brca1. Rescue of homologous recombination in Brca1-deficient mice, which can be achieved by deletion of the DNA damage response factor, 53BP1, causes a significant reduction in genomic instability and tumor incidence (Bouwman et al., 2010; Bunting et al., 2010; Cao et al., 2009). Although 53BP1 is not a core NHEJ component, it is required for V(D)J recombination, class switch recombination and fusion of uncapped telomeres, all of which are dependent on NHEJ (Difilippantonio et al., 2008; Dimitrova et al., 2008; Manis et al., 2004; Ward et al., 2003). Rescue of homologous recombination in Brca1-deficient cells by deletion of 53BP1 correlates with a significant increase in exonuclease-mediated resection of DNA double-strand breaks (Bothmer et al., 2011; Bunting et al., 2010), highlighting the importance of regulation of DNA end resection in determining DSB repair pathway choice.
Besides its essential role in repairing DSBs that occur spontaneously during DNA replication, HR is also important for repair of DSBs that arise during processing of DNA interstrand cross-links (ICLs) (Kee and D'Andrea, 2010). ICLs, which are produced by the reaction of certain metabolites and drugs with DNA, activate a repair pathway comprising at least 15 gene products. Mutation of any of these genes causes the human disease, Fanconi Anemia (FA), which is associated with pancytopenia, tumor predisposition and hypersensitivity to DNA crosslinking agents (Kee and D'Andrea, 2010; Wang et al., 2007). Several recent reports have indicated that genomic instability in FA cells is dependent on the activity of NHEJ factors (Adamo et al., 2010; Pace et al., 2010). For example, it was reported that loss of Ku in FANCC-mutant chicken or human cells relieved their sensitivity to agents that cause ICLs. Furthermore, the activity of NHEJ has been shown to negatively affect DNA repair in cells lacking the HR factor, BRCA2 (Patel et al., 2011).
To gain further insight into how DNA repair involving BRCA1 or factors of the FA pathway is affected by the activity of NHEJ proteins, we have tested the effects of deleting Ku or 53BP1 in Brca1- and FANCD2-deficient mice. Surprisingly, we find that 53BP1 deletion does not affect the sensitivity of Brca1-deficient cells to DNA cross-linking agents, despite the previous finding that HR is restored in Brca1,53BP1 double-deficient cells (Bouwman et al., 2010; Bunting et al., 2010). By contrast, Ku depletion reduces-but does not abrogate-the sensitivity of Brca1-deficient cells to both PARP (polyADP-ribose polymerase) inhibitor and cisplatin, suggesting important differences in the roles of Ku70/80 and 53BP1 in response to ICLs. Contrary to the case with Brca1-deficient cells, we found that deletion of either Ku80 or 53BP1 causes an increase in the sensitivity of FANCD2−/− cells to DNA cross-linking drugs. Thus, loss of NHEJ proteins can either cause additive repair defects, or suppression of repair defects in response to ICLs.
Results
Genomic instability and cell death in Brca1-deficient cells after PARP inhibitor treatment is dependent on Ku and 53BP1
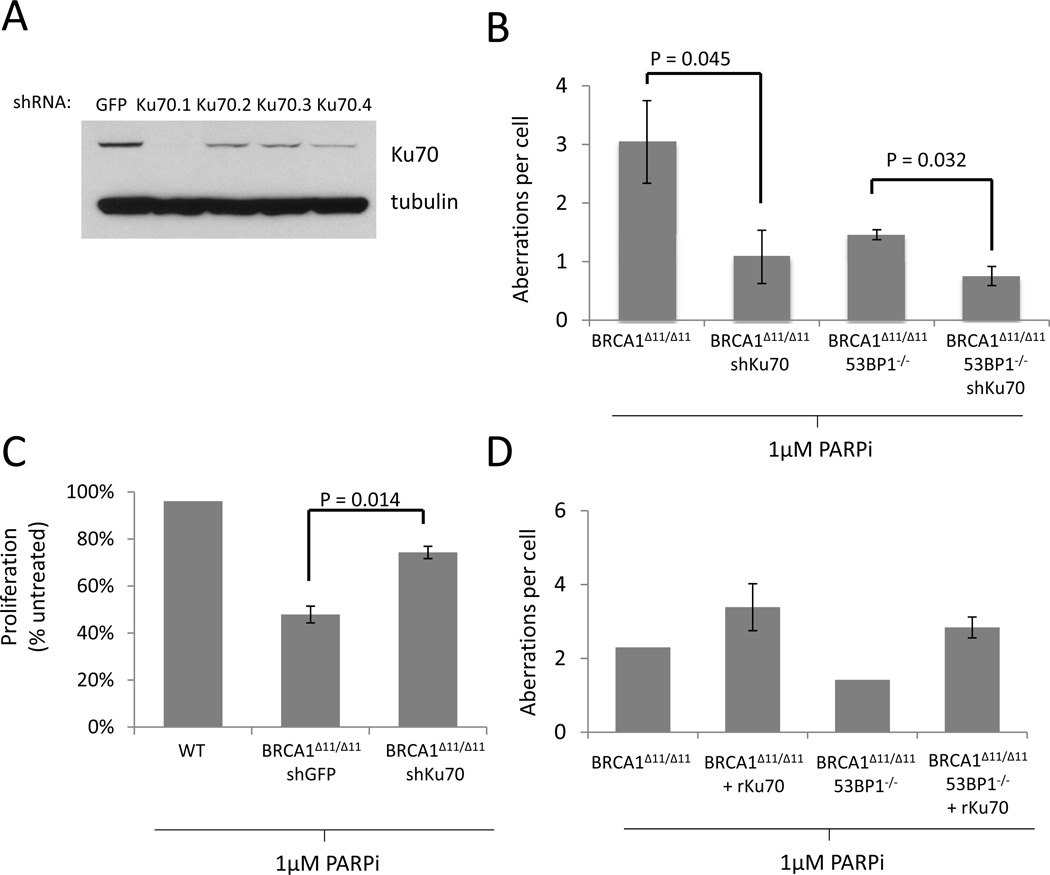
To investigate how Ku contributes to genomic instability in Brca1-deficient cells, we knocked down Ku70 in Brca1Δ11/Δ11 mouse embryonic fibroblasts (MEFs). The Brca1Δ11 allele encodes a mutant isoform of Brca1 lacking exon 11, which encodes ~50% of the WT protein (Xu et al., 1999b). As we found previously in primary Brca1Δ11/Δ11 lymphocytes, Brca1Δ11/Δ11 MEFs are highly sensitive to PARP inhibitor, an agent that is toxic to cells deficient in HR (Bunting et al., 2010) (Figure S1A). We identified a shRNA (Ku70.1) that significantly ablated Ku70 expression in Brca1Δ11/Δ11 MEFs, as determined by Western blotting (Figure 1A). We found that in Brca1Δ11/Δ11 cells, knockdown of Ku70 caused a significant decrease in the level of genomic instability (chromosome and chromatid breaks, radial chromosomes and translocations) induced by PARP inhibitor treatment (Figure 1B) and improved proliferation relative to cells expressing a control shRNA (Figure 1C). To further test the importance of Ku70 in modulating the degree of genomic instability in Brca1-deficient cells, we prepared cells carrying a stably integrated retroviral construct expressing rat Ku70 (rKu70). Overexpression of Ku70 in these cells led to an increase in genomic instability in Brca1Δ11/Δ11 and Brca1Δ11/Δ11,53BP1−/− MEFs (Figure 1D, Figure S1B). Altogether, these results suggest that Ku contributes to genomic instability in Brca1Δ11/Δ11 cells.
Figure 1. Ku expression correlates with genomic instability and reduced proliferation in Brca1-deficient cells treated with PARP inhibitor.
(A) Ku70 expression in cells expressing Ku70 shRNAs. First lane (GFP) shows expression of Ku70 in cells expressing a control shRNA specific for GFP. The GFP and KU70.1 shRNAs were selected for integration into Brca1Δ11/Δ11 MEFs. (B) Average genomic instability observed in metaphase spreads from MEF cells treated overnight with 1µM PARP inhibitor (C) Proliferation of MEFs growing in the presence of 1µM PARP inhibitor, relative to untreated cells. (D) Genomic instability in metaphases from MEFs overexpressing rat Ku70. Error bars show standard deviation in each case. See also Figure S1.
As reported previously, deletion of 53BP1 reduced the level of genomic instability in Brca1Δ11/Δ11 cells (Figure 1B) and significantly increased the proliferation of these cells in the presence of PARP inhibitor (Figure S1A). Knockdown of Ku70 further reduced the number of chromosome aberrations observed in Brca1Δ11/Δ11,53BP1−/− cells (Figure 1B), indicating that both Ku70 and 53BP1 contribute to genomic instability in Brca1-deficient cells.
Brca1Δ11/Δ11 and Brca1-null mice die in utero (Ludwig et al., 1997; Xu et al., 1999b). However, embryonic lethality in Brca1Δ11/Δ11 mice can be overcome by additional deletion of 53BP1 (Cao et al., 2009). To test whether deletion of Ku could enable an equivalent rescue of embryonic lethality in Brca1Δ11/Δ11 mice, we bred Brca1Δ11/+ mice to Ku80+/− animals. Whereas we were able to obtain Brca1+/+Ku80−/− and Brca1Δ11/+Ku80−/− pups, we found that Brca1Δ11/Δ11Ku80−/− double-deficient mice did not survive to birth (Table 1). We were also unable to obtain Brca1Δ11/Δ11Ku80−/− embryos at E13.5 (Table 1). Thus, in contrast to deletion of 53BP1, targeting of Ku is not sufficient to overcome the embryonic lethality phenotype seen in Brca1Δ11/Δ11 mice.
Table 1. Impact of deletion of Ku or 53BP1 on the survival of Brca1Δ11/Δ11 and Brca1-null embryos.
Frequency of embryos at day E13.5 and live-born pups of the indicated genotypes is shown. See also Table S1.
| Brca1Δ11/+ Ku80+/− × Brca1Δ11/+ Ku80+/− intercross: | |||||
|---|---|---|---|---|---|
| Brca1Δ11/Δ11Ku80+/+ or Brca1Δ11/Δ11Ku80+/− |
Brca1+/+Ku80−/− or Brca1Δ11/+Ku80−/− |
Brca1Δ11/Δ11Ku80−/− | Other genotypes |
||
| E13.5 embryos | Expected: | 9 | 9 | 3 | 27 |
| (48 screened) | Observed: | 4 | 12 | 0 | 32 |
| Live pups | Expected: | 30 | 30 | 10 | 90 |
| (160 screened) | Observed: | 0 | 13 | 0 | 147 |
| Brca1+/− 53BP1+/− × Brca1+/− 53BP1+/− intercross: | |||||
|---|---|---|---|---|---|
| Brca1−/− 53BP1+/+ or Brca1−/− 53BP1+/− |
Brca1+/+ 53BP1−/− or Brca1+/− 53BP1−/− |
Brca1−/− 53BP1−/− | Other genotypes |
||
| Live pups | Expected: | 21.75 | 21.75 | 7.25 | 65.25 |
| (116 screened) | Observed: | 0 | 22 | 4 | 90 |
| Brca1+/− 53BP1−/− × Brca1+/− 53BP1−/− intercross: | |||||
|---|---|---|---|---|---|
| Brca1−/− 53BP1+/+ or Brca1−/− 53BP1+/− |
Brca1+/+ 53BP1−/− or Brca1+/− 53BP1−/− |
Brca1−/− 53BP1−/− | Other genotypes |
||
| Live pups | Expected: | 0 | 66 | 22 | 0 |
| (88 screened) | Observed: | 0 | 72 | 16 | 0 |
53BP1 deletion does not affect the sensitivity of Brca1-deficient cells to cisplatin
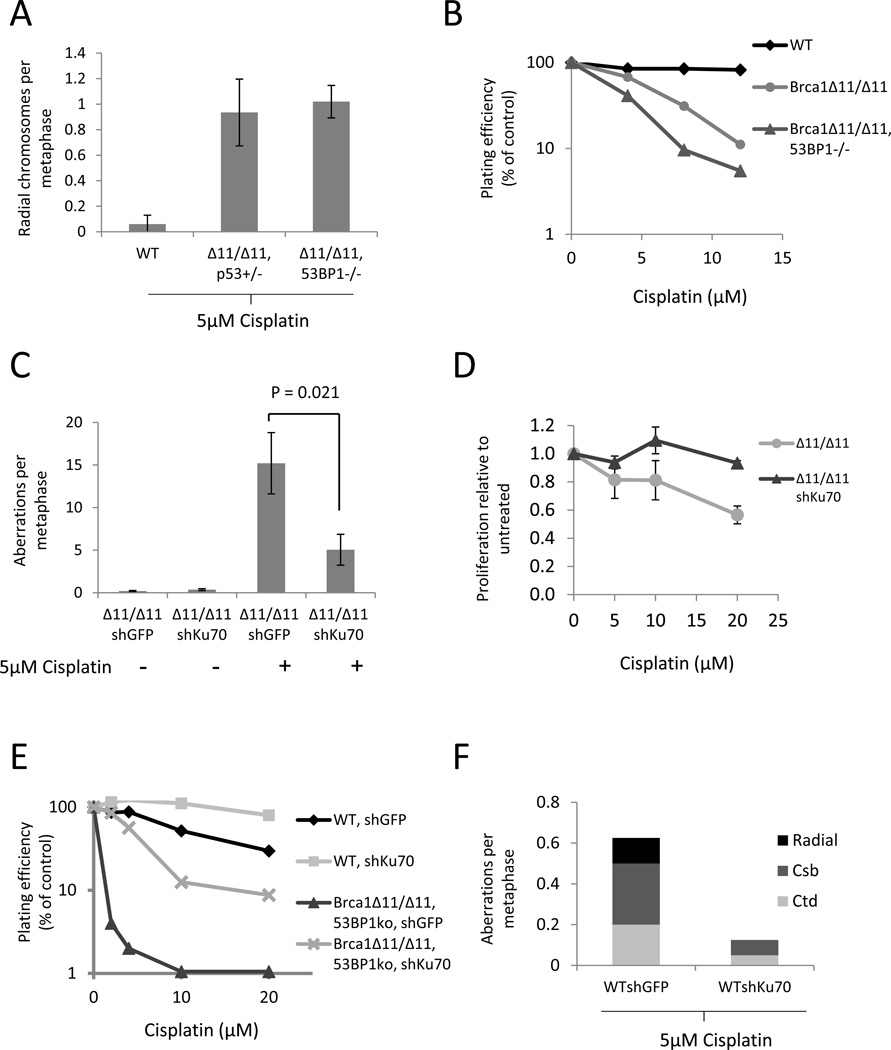
Platinum-based drugs such as cisplatin and carboplatin are clinically important agents for the treatment of breast cancer (Cobleigh, 2011). We tested Brca1Δ11/Δ11 and Brca1Δ11/Δ1153BP1−/− cells to determine their sensitivity to cisplatin. Whereas Brca1Δ11/Δ1153BP1−/− cells were resistant to PARP inhibitor (Figures 1B, Figure S1A), we found that Brca1Δ11/Δ1153BP1−/− cells were just as sensitive as Brca1Δ11/Δ11 to the effects of cisplatin. This equivalent sensitivity was seen by measurements of both genomic instability in lymphocyte metaphases, and colony formation assays using MEFs (Figures 2A and 2B). The high sensitivity of Brca1Δ11/Δ1153BP1−/− cells to cisplatin was unexpected, because HR is restored to near WT levels in these cells (Bunting et al., 2010).
Figure 2. Reduced genomic instability and increased survival of Brca1-deficient cells with Ku70 knockdown after cisplatin treatment.
(A) Frequency of radial chromosome formation in lymphocytes of the indicated genotypes after overnight treatment with 5µM cisplatin. (B) Colony formation in MEFs treated for 2 hrs with cisplatin. (C) Genomic instability in MEFs expressing stably integrated shRNA against either GFP or Ku70, treated overnight with 5µM cisplatin. (D) Growth of BRCA1Δ11/Δ11 cells with stably integrated Ku70 shRNA in the presence of cisplatin. Cisplatin was applied for 24 hrs and growth assayed with CellTiter-Glo after a total 5 days. (E) Colony formation of MEF lines treated for 2hrs with cisplatin. (F) Genomic instability in control (WTshGFP) or Ku70 knockdown MEFs after overnight treatment with 5µM cisplatin. Error bars show standard deviation in each case. See also Figure S2.
Genomic instability in cells treated with cisplatin arises from the ability of cisplatin to form mutagenic intra- and interstrand DNA cross-links (Kee and D'Andrea, 2010; Wang, 2007). To determine whether intra- or interstrand cross-links were responsible for cisplatin toxicity in Brca1Δ11/Δ11 and Brca1Δ11/Δ1153BP1−/− cells, we tested two additional agents that produce a high proportion of DNA interstrand cross-links: nitrogen mustard and mitomycin C (Figures S2A and S2B). Brca1Δ11/Δ11 cells were hypersensitive to both of these drugs, and 53BP1 deletion did not affect sensitivity in either case (Figures S2A and S2B). Thus, even though HR proceeds efficiently, Brca1Δ11/Δ1153BP1−/− cells are hypersensitive to a variety of drugs that induce DNA interstrand cross-links.
The sensitivity of Brca1Δ11/Δ1153BP1−/− cells to ICLs indicates that BRCA1 has a function in ICL repair that is separate from its known role in HR. To examine this further, we measured the assembly of nuclear Rad51 foci in cells treated with MMC. Rad51 nucleoprotein assembly is considered to be an essential step in DNA repair by HR (Kass and Jasin, 2010). We observed that, as is the case with ionizing radiation, Brca1Δ11/Δ11 cells showed defective Rad51 foci formation after MMC treatment but Brca1Δ11/Δ1153BP1−/− cells showed Rad51 foci formation at close to wild-type levels (Figure S2C). As Brca1Δ11/Δ1153BP1−/− cells are nonetheless highly sensitive to DNA cross-linking agents (Figures 2A and 2B), these data indicate that BRCA1 has a role in DNA cross-link repair that is independent of its previously known role in mediating Rad51 loading at DNA break sites.
Rescue of embryonic lethality in Brca1-null mice by 53BP1 deletion
To ensure that our results were not specific to cells with the hypomorphic Brca1Δ11 allele, we tested the effect of cross-linking drugs in Brca1-null cells. Brca1 nullizygosity has a much more severe phenotype than the Brca1Δ11/Δ11 mutation, with embryonic lethality at E5.5–E8.5 in Brca1−/− mice compared to E12.5–E18.5 in Brca1Δ11/Δ11 homozygotes (Ludwig et al., 1997; Xu et al., 1999b). Furthermore, whereas Brca1Δ11/Δ11p53+/− animals are viable, embryonic lethality in Brca1-null animals cannot be overcome by deletion of p53 (Xu et al., 2001). Consistent with this, we found that it was not possible to generate Brca1−/−53BP1+/+ or Brca1−/−53BP1+/− pups (Table 1). Strikingly, however, double-null Brca1−/−53BP1−/− pups were obtained at a frequency only slightly lower than the expected Mendelian ratio (Table 1). 53BP1 deletion, in contrast to p53 deletion, is therefore able to rescue embryonic lethality in mice that are null for Brca1.
Brca1−/−53BP1−/− mice appeared normal in all respects except that males were sterile and had small testes (Figure S3A). As was the case with Brca1Δ11/Δ1153BP1−/− cells, Brca1−/−53BP1−/− cells showed resistance to PARP inhibitor (Figure S3B) but were hypersensitive to cisplatin and mitomycin C (Figures S3C–F). Hypersensitivity to DNA cross-linking drugs is therefore a common feature of Brca1-null and Brca1Δ11/Δ11 cells that cannot be rescued by 53BP1 deletion. Male specific sterility in Brca1−/−53BP1−/− mice further supports an HR-independent role for BRCA, in this case during spermatogenesis. Female mice, by contrast, showed normal ovaries and were fertile, suggesting a differential requirement for BRCA1 in male and female gametogenesis. Although Brca1-deficient mice were previously shown to have a defect in mammary development (1et al., 1999a), we observed no difference in mammary ductal morphogenesis in female WT, 53BP1−/− or Brca1−/−53BP1−/− mice at pregnancy day P8.5 (Figure S3G).
Cisplatin cytotoxicity in BRCA1-deficient and WT cells is dependent on Ku
As Ku depletion improved the survival of Brca1Δ11/Δ11 cells treated with PARPi (Figure 1C), we tested whether Ku similarly modulated their sensitivity to ICLs. Unlike 53BP1 deletion, we found that Ku70 depletion reduced genomic instability in Brca1Δ11/Δ11 MEFs treated with cisplatin, and enhanced their proliferative ability in short-term growth assays as well as in long-term clonogenic colony formation assays (Figures 2C and 2D, Figure S2D). Ku70 depletion was also able to improve the growth of Brca1Δ11/Δ11 53BP1−/− MEFs treated with cisplatin as measured by colony formation (Figure 2E). Interestingly, we noticed that Ku70 depletion also enabled WT cells to proliferate better after treatment with cisplatin (Figure 2E). This is consistent with a previous report, which showed that deletion of Ku80 afforded increased survival in WT cells treated with cisplatin (Jensen and Glazer, 2004). Furthermore, improved growth in Ku70-deficient cells after cisplatin treatment correlated with reduced genomic instability (Figure 2F). These findings demonstrate that the presence of Ku sensitizes WT, Brca1Δ11/Δ11, and Brca1Δ11/Δ1153BP1−/− cells to the cytotoxic effects of cisplatin.
Ku antagonizes HR without significantly affecting DSB resection
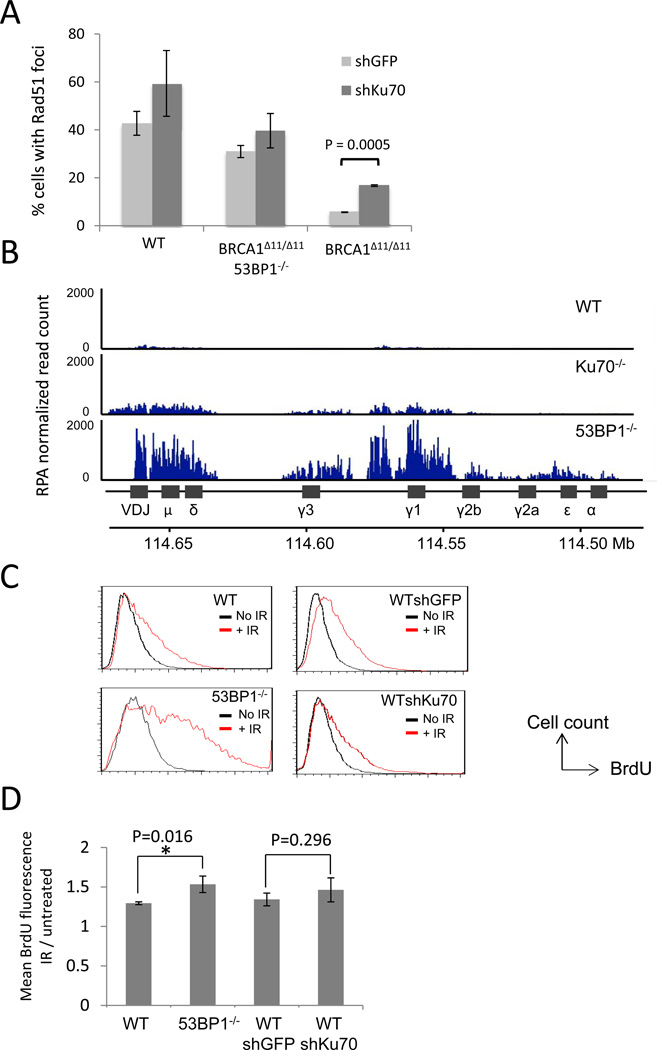
Rad51 loading at sites of DNA double-strand breaks is a critical step in repair of DNA damage by HR, and deletion of 53BP1 in Brca1-deficient cells was previously shown to increase the proportion of cells with Rad51 foci following DNA damage (Bouwman et al., 2010; Bunting et al., 2010). To address the mechanism by which Ku depletion promotes genome integrity in Brca1Δ11/Δ11 cells, we tested whether knockdown of Ku70 could cause an equivalent increase in irradiation-induced Rad51 foci. We found that although Rad51 foci formation was reduced in Brca1Δ11/Δ11 cells relative to WT, knocking down Ku70 caused a statistically significant (P=0.0005) increase in Rad51 foci (Figure 3A), consistent with an increase in DNA repair by HR in the absence of Ku70 (Pierce et al., 2001). An increase in the proportion of irradiated cells exhibiting Rad51 foci after Ku knockdown was also seen in WT and Brca1Δ11/Δ1153BP1−/− cells, although in these cases the increase did not reach the level of statistical significance.
Figure 3. Effect of 53BP1 and Ku on Rad51 foci and DSB resection.
(A) Quantification of Rad51 immunofluorescence in mouse embryonic fibroblasts of the indicated genotypes. Cell expressing either control shRNA against GFP or shRNA against Ku70 were irradiated (5Gy, 4hrs recovery) and stained with anti-Rad51 antibody. The average percentages of cells (+/− standard deviation) with more than 5 nuclear foci from three experiments are shown. (B) Anti-RPA ChIP-Seq in B cells. B cells were stimulated to undergo class switch recombination in vitro. Chromatin from B cells was harvested 48 hrs post-stimulation and used for RPA ChIP. RPA read count (normalized by the total library size per million) is shown at the IgH locus. (C) Non-denaturing anti-BrdU immunofluorescence in MEFs treated with ionizing radiation (30Gy, 2 hrs recovery), measured by flow cytometry. Resection is measured by detection of exposed BrdU (x-axis). (D) Quantification of mean BrdU fluorescence intensity of the irradiated population shown in (C), normalized to the untreated population. Average +/− standard deviation from 5 experiments. See also Figure S4.
Exonuclease resection of DNA double-strand breaks is considered to be a critical step in HR, because it generates single-stranded DNA that allows loading of RPA and Rad51 around the break site. To test the extent to which 53BP1 and Ku regulate resection of DNA double-strand breaks, we performed anti-RPA chromatin immunoprecipitation in B cells from WT, 53BP1−/− and Ku70−/− mice. Stimulation of B cells with LPS and IL4 generates multiple DSBs centered around the immunoglobulin heavy chain (IgH) Sμ, Sγ1, Sγ3 and Sε switch regions (Nussenzweig and Nussenzweig, 2010). RPA loads at the site of DNA double-strand breaks following exonuclease resection, hence a greater enrichment of DNA sequences in the anti-RPA ChIP fraction indicates a greater amount of DSB resection (Yamane et al., 2011). 53BP1−/− B cells showed a significantly increased extent of pull-down of IgH sequences in the anti-RPA ChIP fraction (Figure 3B), consistent with enhanced resection in these cells. This result is in accordance with previous reports, which suggested that loss of 53BP1 increases the extent of resection of DNA DSBs (Bothmer et al., 2010; Bunting et al., 2010; Difilippantonio et al., 2008). In comparison to 53BP1−/− cells, however, Ku70−/− cells showed only a minor increase in resection relative to WT, as measured by RPA ChIP at IgH (Figure 3B).
To further test the role of NHEJ factors in the regulation of DSB resection, we pulsed WT, 53BP1−/− and Ku70-depleted MEFs with BrdU to enable the measurement of single-stranded DNA at ionizing radiation-induced DSBs. We adapted an existing non-denaturing BrdU immunofluorescence protocol for measurement of DSB resection (Sartori et al., 2007) to allow quantification of the amount of resection in a population of cells by flow cytometry (Figure 3C). Notably, we found that 53BP1−/− cells showed a statistically significant increase in resection after irradiation compared to WT cells (Figures 3C and 3D). In contrast, Ku70-knockdown cells did not significantly increase resection compared to cells expressing a control shRNA. By staining for DNA content in the sample population, we observed that resection was significantly more extensive in cells in the S/G2 phases of the cell cycle (Figure S4A), consistent with previous findings (Huertas, 2010). Taken together, these results indicate that whereas 53BP1 has a major role in regulating resection of DNA DSBs, Ku plays a more limited role in this process. The increase in Rad51-dependent HR seen in Ku-deficient cells after DNA damage (Figure 3A) therefore arises from a mechanism other than increased resection (see discussion).
BRCA1 mediates FANCD2 foci formation after treatment with cross-linking agents
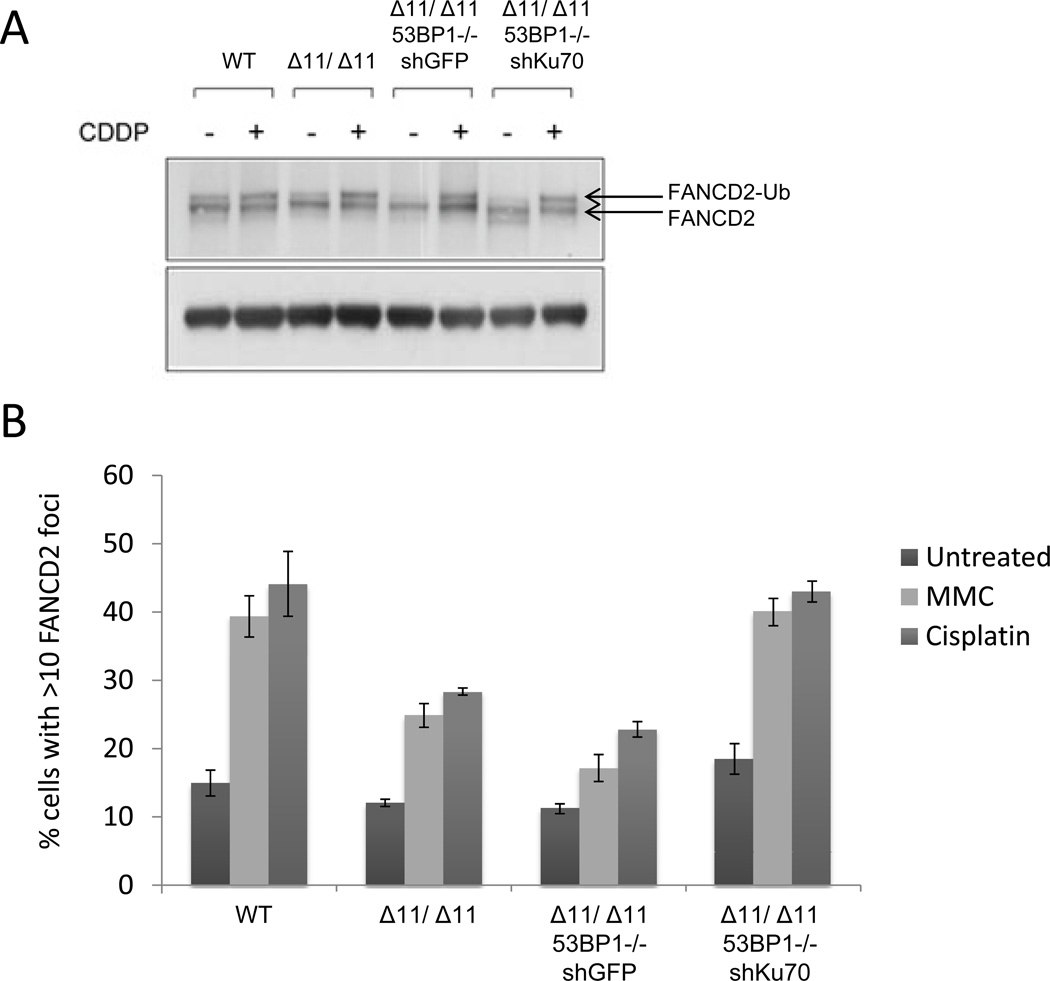
The sensitivity of Brca1Δ11/Δ1153BP1−/− cells to DNA cross-linking agents (Figures 2A and 2B, Figures S2A and S2B) strongly suggests that BRCA1 provides an activity that is required for DNA cross-link repair that is separate from its function in HR. FANCD2 ubiquitylation and recruitment to sites of DNA cross-links are considered to be essential steps in cross-link repair (Huang and D'Andrea, 2010; Knipscheer et al., 2009; Long et al., 2011). We therefore tested whether these steps are normal in Brca1Δ11/Δ11 and Brca1Δ11/Δ1153BP1−/− cells. BRCA1 was previously reported to be dispensable for FANCD2 ubiquitylation, but required for FANCD2 foci formation after DNA damage (Garcia-Higuera et al., 2001; Vandenberg et al., 2003). We found that FANCD2 ubiquitylation, as measured by Western blotting, was not significantly altered in either Brca1Δ11/Δ11 or Brca1Δ11/Δ1153BP1−/− cells (Figure 4A). By contrast, the number of cells showing FANCD2 foci after treatment with cisplatin or mitomycin C was reduced in both Brca1Δ11/Δ11 and Brca1Δ11/Δ1153BP1−/− cells (Figure 4B). Reduced FANCD2 foci in these cells was not caused by reduced growth rate, as Brca1Δ11/Δ11, Brca1Δ11/Δ1153BP1−/− and WT controls showed a similar cell cycle distribution (Figure S4B).
Figure 4. FANCD2 ubiquitylation and damage foci in Brca1-deficient cells.
(A) Western Blot showing FANCD2 ubiquitylation in WT, Brca1Δ11/Δ11 and Brca1Δ11/Δ1153BP1−/− MEFs with and without cisplatin (CDDP) treatment. Brca1Δ11/Δ1153BP1−/− MEFs expressed either control shRNA or shRNA against Ku70. (B) FANCD2 foci analysis in MEFs treated with either cisplatin or mitomycin C. Cells with more than 10 FANCD2 foci were scored as positive. Mean +/− SEM shown. See also Figure S3.
We found that depletion of Ku mitigates the toxic effects of DNA cross-linking agents (Figures 2C–F). We therefore extended our approach by testing whether FANCD2 foci are affected by the presence of Ku70. We found that depletion of Ku70 restored the formation of FANCD2 foci after cisplatin or MMC to a level equivalent to that seen in WT cells (Figure 4B). We conclude that Ku70/80 affects recruitment or retention of FANCD2 at sites of DNA cross-link repair.
Deletion of 53BP1 or Ku exacerbates genomic instability in FANCD2-deficient cells
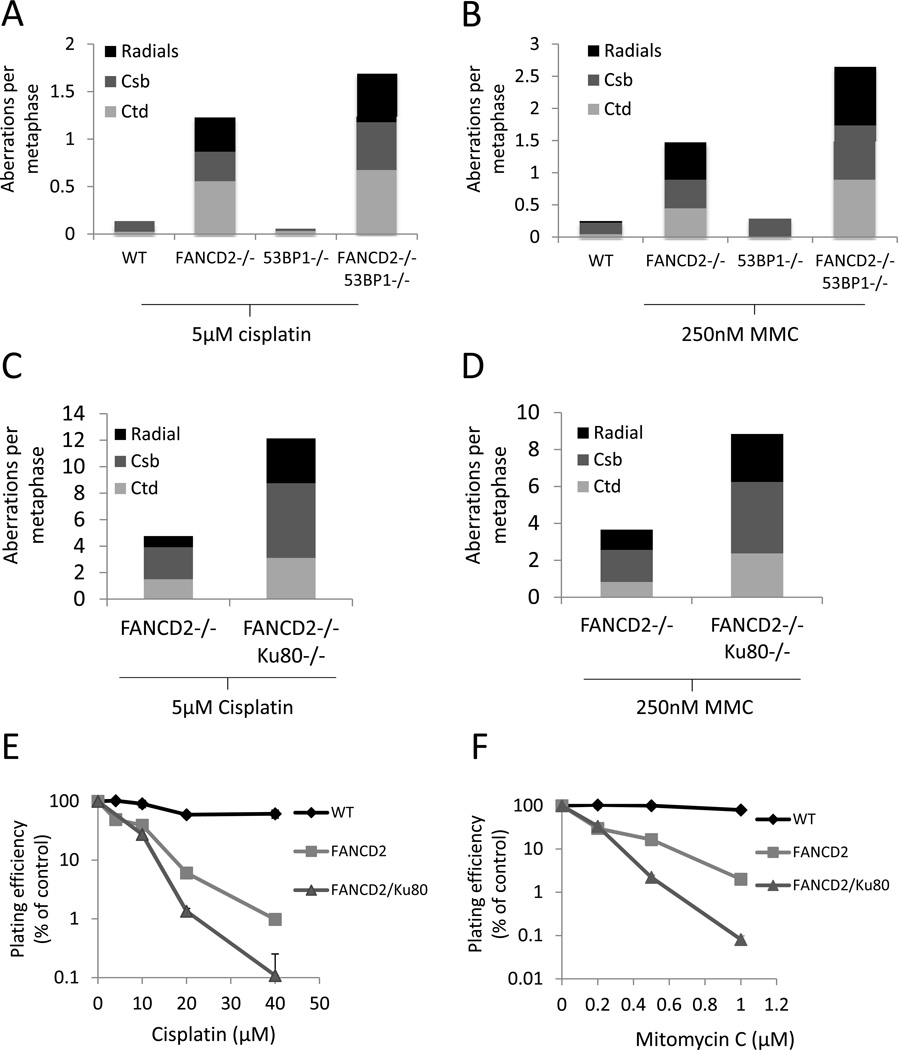
Hypersensitivity to DNA cross-linking agents is a key diagnostic test for genetic deficiency in components of the Fanconi Anemia (FA) pathway (Wang, 2007). Cells from FA patients, or knockout mice with deficiencies in the FA pathway, are unable to repair DNA interstrand cross-links, and tend to accumulate genomic instability with an increased risk of tumorigenesis (Kee and D'Andrea, 2010; Wang, 2007). Recent studies have reported that hypersensitivity of human and chicken DT40 cells to interstrand cross-linking agents can be rescued by knockdown, deletion or inhibition of NHEJ factors such as Ku, Lig4 or DNA-PKcs (Adamo et al., 2010; Pace et al., 2010). To test whether it was possible to prevent genomic instability in FA cells by genetic ablation of DNA damage response factors, we bred mice that were double-null for the key FA pathway component, FANCD2 (Kee and D'Andrea, 2010; Wang, 2007), and either 53BP1 or Ku80. Consistent with the reported sensitivity of FANCD2−/− cells to DNA cross-linking agents (Houghtaling et al., 2003), cells from FANCD2−/− mice were hypersensitive to cisplatin and mitomycin C (Figures 5A and 5B). We found that 53BP1 deletion in FANCD2−/− cells led to increased sensitivity to cisplatin (Figure 5A). Similar results were observed with mitomycin C treatment, which induced higher levels of genomic instability in FANCD2−/−53BP1−/− cells relative to FANCD2−/− (Figure 5B). To examine the impact of 53BP1 loss in vivo we examined hematopoietic stem cells (HSCs). FANCD2−/− mice have hematopoietic defects including a 50% reduction in the frequency of both HSCs and common lymphoid progenitors (CLPs) (Zhang et al., 2010). Quantification of HSC and CLP frequencies in double-mutant mice revealed that these defects were equivalent in the combined absence of FANCD2 and 53BP1 (Figures S5A and S5B). Thus, loss of 53BP1 renders FANCD2-deficient cells more sensitive to DNA cross-linking agents, and does not alleviate the severity of hematopoietic defects in mouse models of Fanconi Anemia.
Figure 5. Genomic instability in metaphases from cells treated overnight with drugs to induce DNA interstrand cross-links (ICLs).
(A) Genomic instability in B cells from WT, FANCD2−/− and FANCD2−/−53BP1−/− mice treated overnight with 5µM cisplatin. (B) Genomic instability in B cells from WT, FANCD2−/− and FANCD2−/−53BP1−/− mice treated overnight with 250 nM mitomyin C (MMC). (C) Total genomic aberrations in metaphases from FANCD2−/− and FANCD2−/− Ku80−/− mouse embryonic fibroblast cells after overnight treatment with 5µM cisplatin. (D) Genomic instability in metaphases from FANCD2−/− and FANCD2−/−Ku80−/− after overnight treatment with 250nM MMC. (E) Colony forming assay in WT, FANCD2−/− and FANCD2−/−Ku80−/− MEFs treated with cisplatin. (F) Colony forming assay WT, FANCD2−/− and FANCD2,Ku80 double-knockout MEFs treated with mitomycin C. (Mean +/− standard deviation shown.) See also Figure S5.
Although Ku80−/− and FANCD2−/− mice are viable, we were not able to obtain live mice that were double-null for FANCD2 and Ku80 (n= 98 pups screened), suggesting that Ku deficiency also exacerbates developmental defects in the absence of FANCD2. This observation is consistent with a previous report, which showed that cells deficient in both FANCD2 and the NHEJ factor DNA-PKcs had a diminished capacity to repair DNA damage compared to either single mutant (Houghtaling et al., 2005). Although we could not obtain double-null pups, we were able to isolate FANCD2−/−Ku80−/− MEFs, and we tested these for their ability to repair interstrand cross-links induced by cisplatin and mitomycin C. We found that, as compared to FANCD2−/− cells, FANCD2−/−Ku80−/− double-knockout MEFs showed increased chromosomal damage in response to either cisplatin (Figure 5C) or mitomycin C (Figure 5D). Colony formation assays with FANCD2−/−Ku80−/− MEFs revealed that these cells grew significantly worse than FANCD2−/− single knockout cells after treatment with cisplatin or mitomycin C (Figures 5E and 5F). These results indicate that FANCD2 provides an essential activity for repair of interstrand cross-links in murine cells that cannot be rescued by deletion of either 53BP1 or Ku. Indeed, the increased severity of the phenotypes observed after combining deficiency in FANCD2 with loss of either 53BP1 or Ku suggests that NHEJ partially compensates for FANCD2 deficiency in cisplatin-induced ICL repair.
Discussion
Efficiency of ICL repair is modified by BRCA1 and Ku70/80
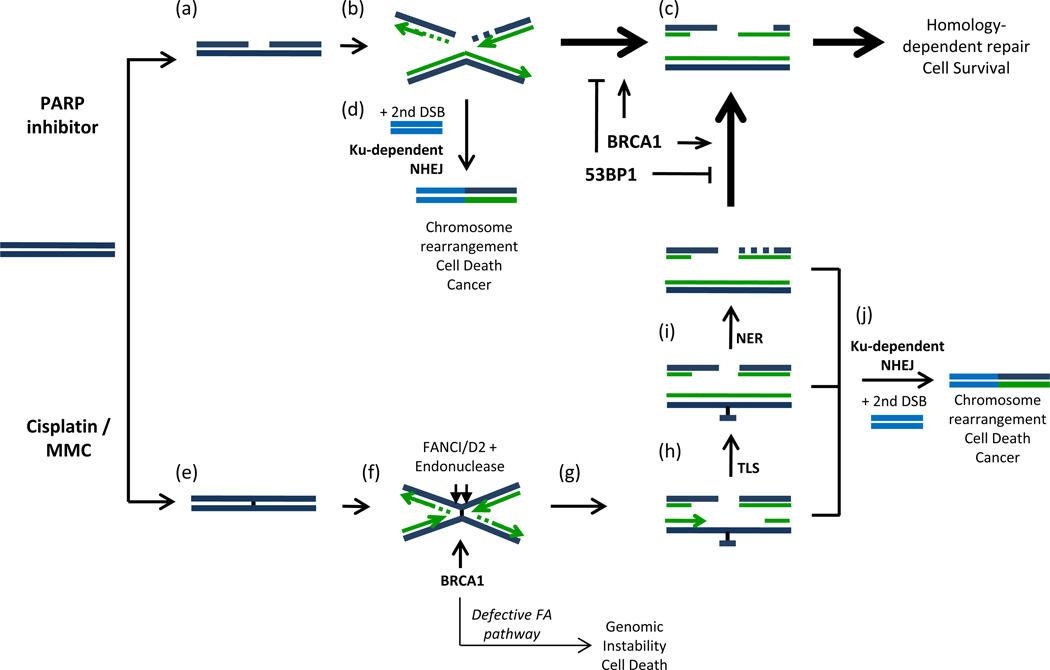
Our study reveals that the requirements for BRCA1 in Rad51 nucleoprotein assembly (Bhattacharyya et al., 2000) and FANCD2 retention at DNA damage sites (Vandenberg et al., 2003) represent two distinct activities of the BRCA1 protein. We propose that during ICL repair, BRCA1 functions early at the cross-link excision step, and later, during HR. 53BP1 only affects the function of BRCA1 during the later HR stage. Deletion of 53BP1 therefore has no effect on the hypersensitivity of Brca1-deficient cells to agents that cause ICLs. In contrast, Ku affects both steps where BRCA1 is active, hence deletion of Ku reduces the hypersensitivity of Brca1-deficient cells to both PARP inhibitors and DNA cross-linking agents. We summarize these results in a model showing the impact of BRCA1 and NHEJ factors in repairing different types of DNA damage (Figure 6).
Figure 6. Model for repair of DNA double-strand breaks (DSBs) induced by PARP inhibitor or DNA cross-linking agents.
(a) Treatment with PARP inhibitor stabilizes spontaneous DNA single-strand breaks, which are converted to DSBs during DNA replication (Bryant et al., 2005; Farmer et al., 2005). (b) DNA DSBs can be repaired either by NHEJ or HR. (c) 5′ – 3′ exonuclease resection commits repair to the error-free HR pathway. 53BP1 antagonizes double-strand break resection. (d) Ku70/80 can potentially join the DSB to a second DNA end present in the cells to cause chromosome rearrangements. (e) Treatment with cisplatin or MMC generates interstrand DNA cross-links. (f) Interstrand cross-links cause replication fork stalling and collapse. Accumulation of FANCI/D2, dependent on BRCA1, recruits endonucleases that cut DNA on either side of the interstrand cross-link to generate a double-strand break (g). (h) Trans-lesion synthesis (TLS) and (i) nucleotide excision repair (NER) re-generate duplex DNA on one sister chromatid, enabling homology-dependent repair of the DNA DSB. (j) Aberrant joining mediated by Ku70/80-competes with normal repair and can potentially generate chromosome rearrangements leading to cancer or cell death.
Increased DSB resection associated with 53BP1 deletion does not improve ICL repair
Treatment with PARP inhibitor stabilizes single-strand breaks (Figure 6a), which are converted to double-strand breaks during DNA replication following collapse of the replication fork at a single-strand break (Bryant et al., 2005; Farmer et al., 2005). According to our model, the double-strand break formed by this process can be directed to either HR or NHEJ. The key rate-limiting step is resection of the DSB, which commits repair to HR (Figure 6b). 53BP1 deletion significantly increases resection; hence in the absence of 53BP1, error-free HR becomes the principal repair pathway (Figure 6c). Cisplatin or MMC treatment produces ICLs. These ICLs cause replication fork collapse, but in this case, the double-strand break is not immediately available for HR, because the homologous template must first be repaired by translesion synthesis (TLS) and nucleotide excision repair (NER) (Figures 6g–i). Increased resection of DSBs mediated by ablation of 53BP1 may not be beneficial to ICL repair, and if so, only at a late stage when TLS and NER are complete. On the other hand, DSBs produced as a consequence of FANCD2-dependent endonuclease action at DNA cross-links can be inappropriately joined to other DSBs present in the cell by the action of Ku-dependent NHEJ prior to commitment to HR (Figure 6). Deletion or knockdown of Ku therefore promotes error-free repair at the sites of ICLs by inhibiting potentially mutagenic repair by NHEJ.
Ku deletion does not rescue the embryonic lethality observed in Brca1-deficient mice. This also represents a significant difference compared to deletion of 53BP1, which rescues the embryonic lethality of homozygous Brca1Δ11/Δ11 or Brca1-null mice (Table S1) (Bunting et al., 2010). Failure of Ku deletion to rescue the embryonic lethality of Brca1-deficient mice correlates with the minor impact on DSB resection that is achieved by targeting Ku relative to 53BP1 (Figure 3B–D). This difference in DSB resection likely reflects the distinct nature of the interaction between these repair factors and DNA. Whereas Ku binds directly to exposed DNA ends, 53BP1 binds to a histone mark that is present in the entire chromatin area around the double strand break (Botuyan et al., 2006; Huyen et al., 2004). Any impact that Ku has on resection is therefore likely to be in the immediate vicinity of the DNA end, whereas 53BP1 is capable of impacting resection throughout a much larger chromatin domain. We hypothesize that the ability to bypass the requirement for BRCA1 in mammalian development requires a large extent of recombinogenic single-stranded DNA during replication, which is afforded by 53BP1 deletion, but not by Ku deficiency. In summary, although both Ku and 53BP1 antagonize HR, the impact of these factors in repairing various types of lesions is distinct because Ku promotes mutagenic repair by NHEJ whereas 53BP1 inhibits DNA end processing.
Differing requirements for BRCA1 and FANCD2 in upstream ICL repair
Our finding that Brca1−/−53BP1−/− cells are HR competent but still hypersensitive to ICLs suggests that in addition to its established role in HR, BRCA1 has an upstream role in processing ICLs prior to DSB repair by HR (Figure 6f). BRCA1 has previously been reported to regulate the accumulation of FANCD2 into repair foci (Garcia-Higuera et al., 2001; Vandenberg et al., 2003). We found that FANCD2 foci are still impaired in Brca1Δ11/Δ1153BP1−/− cells (Figure 4B), which may explain why loss of 53BP1 does not reduce the sensitivity of Brca1-deficient cells to DNA cross-linking agents. The requirement of BRCA1 for optimal retention of FANCD2 at the sites of ICL repair may be dependent on the reported ability of BRCA1 to promote chromatin unfolding (Ye et al., 2001), which may facilitate foci formation by monoubiquitylated FANCD2. Alternatively, BRCA1 may regulate signaling pathways downstream of DNA damage (such as ATR activation) (Yarden et al., 2002) or transcriptional events required for ICL repair (Aiyar et al., 2005; Zhu et al., 2011).
Depletion of Ku70/80 was able to reverse the defect in FANCD2 accumulation in Brca1-deficient cells (Figure 4B), suggesting that Ku70/80 and BRCA1 have antagonistic effects in regulating FANCD2 accumulation. Although the exact mechanism by which loss of Ku increases FANCD2 accumulation is unclear, one possibility is that Ku binding to DNA ends produced either by endonuclease excision of DNA cross-links or by replication fork regression displaces FA gene products that are required for faithful repair at the cross-link site. Alternatively, Ku binding to DNA ends may prevent the action of a putative nuclease activity recently reported to be associated with FANCD2 (Pace et al., 2010).
Whereas FANCD2-deficient cells treated with DNA cross-linking agents accumulate additional genomic instability in the absence of Ku, Brca1-deficient cells show improved genomic stability and survival when Ku is depleted (Figures 2C–E, Figures 5C–D). These data suggest that the role of BRCA1 in upstream ICL processing is not essential; BRCA1 rather has an accessory role in mediating optimal FANCD2 accumulation, as in the absence of BRCA1, FANCD2 foci after DNA cross-linking are reduced but not absent (Figure 4B). In contrast, FANCD2 is an essential player in ICL repair, and a deficiency in FANCD2 can therefore not be compensated by deletion of 53BP1 or Ku.
Two recent reports indicated that deletion of Ku promotes survival and restores genome integrity in cells deficient in the FA pathway (Adamo et al., 2010; Pace et al., 2010). Nevertheless, discordant results were obtained when testing the effects of various NHEJ mutants. For example, in chicken cells, it was found that loss of DNA ligase IV (Lig4) in FANCC-mutant cells caused additive repair defects whereas loss of Ku suppressed the repair defects (Pace et al., 2010). The difference between our data and previously reported results may reflect the fact that DT40 cells used in earlier studies utilize HR for repair at a much higher frequency than other cell types, suggesting that different mechanisms could regulate DSB repair pathway choice in different model systems (Buerstedde and Takeda, 1991). Moreover, the function of FA proteins in mice and humans may be distinct, as several mouse models do not typically exhibit as severe congenital and hematopoietic abnormalities and cancer predisposition as in human patients. Nevertheless, based on our data on Ku deficiency and a corresponding study with DNA-PKcs−/− mice crossed with FANCD2−/− (Houghtaling et al., 2005) (Table S1), we do not believe it will be possible to relieve the FA phenotype by targeting NHEJ.
Secondary mutations in 53BP1 and Ku as potential contributors to chemoresistance
PARP inhibitors have shown considerable promise as targeted therapies for tumors with deficiencies in BRCA1 or BRCA2, and recent profiling of ovarian tumors has indicated that up to 50% of such cancer cases could be amenable to treatment with PARP inhibitors based on the presence of mutations that affect HR (TCGA-Network, 2011). Chemo resistance may arise in cancer cases treated with PARP inhibitor because of the presence of secondary mutations that reduce the sensitivity of HR-deficient tumor cells to PARP inhibition. We propose that mutations in 53BP1 and Ku70/80 are candidates for altered drug sensitivity in HR-deficient tumors, and that characterization of the status of these genes is likely to have prognostic value in planning treatment with PARP inhibitors or platinum-based chemotherapy.
Methods
Mice
Mice carrying the Brca1Δ11 allele were obtained from the NIH mouse repository. Brca1- and Ku80-null mice were obtained as described (Ludwig et al., 1997; Nussenzweig et al., 1996).
shRNA and growth assays
shRNA constructs were obtained from Open Biosystems. 24 hours after lentiviral infection, stable integrants were selected with puromycin. Western blotting was performed using anti-Ku70 mouse monoclonal antibody (mab-Ku70, 3114-500, Abcam used at 1:500). For overexpression, Rat Ku70 was subcloned (Yang et al., 1996) into the pMX retroviral vector for infection into passage-immortalized MEFs. For short-term growth assays, cells were grown continuously with PARP inhibitor or for 24 hours in the presence of cisplatin. Proliferation was assayed after 5 days with CellTiterGlo (Promega) according to the manufacturer’s instructions. For colony formation, cells were grown for 14 days, then fixed with methanol and stained with crystal violet. Metaphase preparation and telomere FISH was as described (Callen et al., 2007). PARP inhibitor (KU55993) was obtained from Astra Zeneca.
Native BrdU detection and RPA ChIP
Exponentially growing cells were pulsed with 1µM 5-bromo-2’-deoxyuridine (BrdU, Sigma) for 36 hrs, irradiated (30Gy, 2hrs recovery at 37°C); then fixed with methanol at −20°C for 20 minutes. Cells were blocked in 3% BSA in PBS for 30 mins, then stained with anti-BrdU monoclonal antibody for 1hr. Before analysis, propidium iodide was added to a final concentration of 20µg/ml. RPA ChIP was performed as previously described (Yamane et al., 2011).
Highlights.
DNA crosslink repair is regulated by two separate activities of BRCA1
Ku antagonizes HR by promoting NHEJ whereas 53BP1 inhibits DNA end resection
53BP1 deletion rescues embryonic development in BRCA1-nullizygous mice
Ku deletion exacerbates development defects and instability in FANCD2−/ − mice
Supplementary Material
Acknowledgements
We thank Drs Laura Niedernhofer, Jeremy Daniel, Elise Kohn for helpful discussions and suggestions, and Drs. Michael Eckhaus and Mark Bryant for assistance with histology. The work was supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research, a Department of Defense grant to A.N. (BC102335), and a K99/R00 grant (1K99CA160574-01) to S.B. Research was conducted in compliance with the Animal Welfare Act Regulations and other Federal statutes relating to animals and experiments involving animals and adheres to the principles set forth in the Guide for Care and Use of Laboratory Animals, National Research Council, 1996.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
GEO: ChIP-seq data for RPA, GSE35698.
References
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Aiyar S, Sun JL, Li R. BRCA1: a locus-specific "liaison" in gene expression and genetic integrity. J Cell Biochem. 2005;94:1103–1111. doi: 10.1002/jcb.20386. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobleigh MA. Other options in the treatment of advanced breast cancer. Semin Oncol. 2011;38(Suppl 2):S11–S16. doi: 10.1053/j.seminoncol.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends in biochemical sciences. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Getts RC, Stamato TD. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling S, Newell A, Akkari Y, Taniguchi T, Olson S, Grompe M. Fancd2 functions in a double strand break repair pathway that is distinct from non-homologous end joining. Hum Mol Genet. 2005;14:3027–3033. doi: 10.1093/hmg/ddi334. [DOI] [PubMed] [Google Scholar]
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, D'Andrea AD. A new nuclease member of the FAN club. Nat Struct Mol Biol. 2010;17:926–928. doi: 10.1038/nsmb0810-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Jensen R, Glazer PM. Cell-interdependent cisplatin killing by Ku/DNA-dependent protein kinase signaling transduced through gap junctions. Proc Natl Acad Sci U S A. 2004;101:6134–6139. doi: 10.1073/pnas.0400051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell WK, Chu G. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol Cell Biol. 1994;14:4741–4748. doi: 10.1128/mcb.14.7.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- TCGA-Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg CJ, Gergely F, Ong CY, Pace P, Mallery DL, Hiom K, Patel KJ. BRCA1-independent ubiquitination of FANCD2. Mol Cell. 2003;12:247–254. doi: 10.1016/s1097-2765(03)00281-8. [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999a;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999b;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Nussenzweig A, Yang WH, Kim D, Li GC. Cloning and characterization of rat Ku70: involvement of Ku autoantigen in the heat-shock response. Radiat Res. 1996;146:603–611. [PubMed] [Google Scholar]
- Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- Ye Q, Hu YF, Zhong H, Nye AC, Belmont AS, Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol. 2001;155:911–921. doi: 10.1083/jcb.200108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QS, Marquez-Loza L, Eaton L, Duncan AW, Goldman DC, Anur P, Watanabe-Smith K, Rathbun RK, Fleming WH, Bagby GC, et al. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood. 2010;116:5140–5148. doi: 10.1182/blood-2010-04-278226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.