SUMMARY
Human LMNA gene mutations result in laminopathies that include Emery-Dreifuss Muscular Dystrophy (AD-EDMD) and Hutchinson-Gilford Progeria, the premature aging syndrome (HGPS). The Lmna null (Lmna−/−) and progeroid LmnaΔ9 mutant mice are models for AD-EDMD and HGPS respectively. Both animals develop severe tissue pathologies with abbreviated life spans. Like HGPS cells, Lmna−/− and LmnaΔ9 fibroblasts have typically misshapen nuclei. Unexpectedly, Lmna−/− or LmnaΔ9 mice that are also deficient for the inner nuclear membrane protein Sun1 show markedly reduced tissue pathologies and enhanced longevity. Concordantly, reduction of SUN1 over-accumulation in LMNA mutant fibroblasts and in HGPS cells corrected nuclear defects and cellular senescence. Collectively, these findings implicate Sun1 protein accumulation as a common pathogenic event in Lmna−/−, LmnaΔ9, and HGPS disorders.
Introduction
The nuclear lamina, that underlies the inner nuclear membrane (INM), is a meshwork of type V intermediate filament proteins consisting primarily of the A and B type lamins (Guttinger et al., 2009). Mammalian somatic cells express four major types of lamins, including A and C encoded by the Lmna gene (Burke and Stewart, 2006; Stuurman et al., 1998), and B1 and B2, each encoded by their own genes (Lmnb1 and 2) (Shimi et al., 2008; Stuurman et al., 1998). In addition to providing mechanical strength to the nucleus, recent discoveries in nuclear-lamina associated human diseases have established intimate connections between the nuclear envelope/lamina, and processes such as gene expression, DNA repair, cell cycle progression and chromatin organization (Liu et al., 2005; Nagano and Arahata, 2000; Chi et al., 2009a; Capell and Collins, 2006; Worman and Courvalin, 2004).
Some 28 diseases/anomalies (the nuclear envelopathies) are linked to mutations in proteins of the nuclear envelope and lamina with about half the diseases arising from mutations in the Lamin genes, predominately LMNA. These disease phenotypes range from cardiac and skeletal myopathies, lipodystrophies, peripheral neuropathies, to premature aging with early death (Burke and Stewart, 2002; Burke and Stewart, 2006; Burke et al., 2001; Chi et al., 2009a; Worman and Courvalin, 2004). Two notable laminopathies are the autosomal dominant form of Emery-Dreifuss Muscular Dystrophy (AD-EDMD) that results in muscle wasting and cardiomyopathy and Hutchinson-Gilford progeria syndrome (HGPS), a rare genetic premature aging disease, where affected individuals expire with a mean life span of 13 years (Kudlow et al., 2007). AD-EDMD is caused by missense mutations and/or deletions throughout the LMNA gene that generally disrupt the integrity of the lamina, resulting in mechanical weakening of the nucleus, making it more vulnerable to mechanical stress. With HGPS, most cases arise from a single heterozygous mutation at codon 1824 of LMNA. This mutation produces an in-frame deletion of 50 amino acids, generating a truncated form of LAΔ50 lamin A, termed progerin, which remains farnesylated (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003; Goldman et al., 2004). HGPS individuals are overtly normal at birth with the disease manifesting around 18 months (Merideth et al., 2008). The current view is that the permanently farnesylated progerin is affixed to the nuclear membrane, resulting in a toxic gain of function that elicits HGPS. How farnesylated progerin triggers HGPS is not understood.
Lmna−/− mice (Sullivan et al., 1999) were developed and found to model AD-EDMD. Subsequently, another mouse model was created with homozygous LmnaL530P/L530P mutations in the Lmna gene (later termed LmnaΔ9 mice; Hernandez et al., 2010; Mounkes et al., 2003) that expresses a deleted form of Lmna (deleted for exon 9 with the in-frame removal of 40 amino acids of lamin A/C). There are distinct differences between Lmna−/−, LmnaΔ9, and HGPS. The Lmna−/− mouse does not express a full length lamin A protein whereas the LmnaΔ9 mouse recapitulates many HGPS-associated pathologies including early death, skeletal anomalies and vascular smooth muscle defects (Mounkes et al., 2003) and homozygously expresses a farnesylated lamin A-ΔExon9 mutant protein that, though similar, is not-identical to the heterozygous expression of the LAΔ50 mutant protein in HGPS. Nonetheless, Lmna−/− and LmnaΔ9 mice and HGPS individuals share three significant features. All have Lmna mutations, significant dystrophic cellular, tissue, and organ changes, and markedly abbreviated life spans.
Currently, although aberrant LAΔ50 progerin expression is implicated as causing HGPS, the full understanding of this and other causal event(s) for lamin A-associated pathology is elusive. The lamins are proposed to interact with many INM proteins including Emerin, lamina-associated polypeptides (LAPs), MAN1, and the SUN domain proteins, SUN1 and SUN2 (Burke and Stewart, 2002; Mattioli et al., 2011; Crisp et al., 2006; Ostlund et al., 2009); a detailed biochemical understanding of these interaction is complicated by the relative insolubility of these proteins. The SUNs are components of the LINC (links the nucleoskeleton and cytoskeleton) complex that connect the nuclear lamina and envelope with the cytoskeleton (Crisp et al., 2006). The LINC complex is important in nuclear positioning and cellular migration in lower and higher eukaryotes (Malone et al., 1999). How the inner nuclear membrane SUN proteins function with lamins remains unclear, but currently there is no evidence that they are involved in laminopathies (Haque et al., 2010). Here we present evidence that Lmna−/−, LmnaΔ9, and HGPS dysfunctions converge at a common pathogenic over accumulation of the inner nuclear envelope Sun1 protein. Accordingly, loss of the Sun1 gene in Lmna−/− and LmnaΔ9 mice extensively rescues cellular, tissue, organ, and life span abnormalities. Similarly, the knock down of over accumulated SUN1 protein in primary HGPS cells corrected their nuclear defects and cellular senescence. Our results reveal Sun1 over accumulation as a potentially pivotal pathologic effector of some laminopathies.
RESULTS
Loss of Sun1 ameliorates Lmna−/− and LmnaΔ9 pathologies
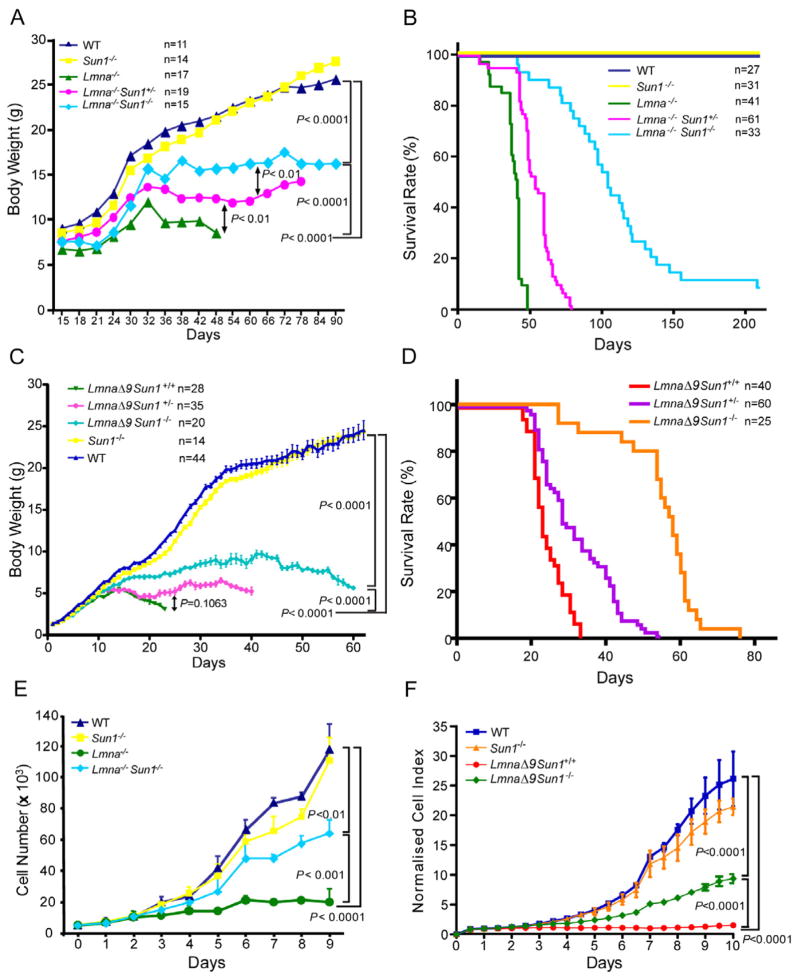
To gain insight into the cooperativity, if any, between INM proteins and the underlying lamina in disease development, we bred Sun1+/− (Chi et al., 2009b) and Lmna+/− (Sullivan et al., 1999) mice to produce Lmna−/−Sun1−/− offspring. A priori, it was anticipated that inactivating both the Lmna and Sun1 genes in Lmna−/−Sun1−/− mice would lead to a more severe pathological phenotype than that seen for Lmna−/− animals. Surprisingly, we observed the opposite. In the Lmna−/− context, the removal of Sun1, rather than exacerbating pathology, unexpectedly ameliorated deficits in body weight (Figure 1A; P < 0.0001) and longevity (Figure 1B; P < 0.01). This rescue of Lmna−/− mice by loss of Sun1 was verified in a second laminopathy model, the LmnaΔ9 mutant mouse (Mounkes et al., 2003; Hernandez et al., 2010). The body weight and longevity deficits in LmnaΔ9 mice were also rescued in the LmnaΔ9Sun1 −/− counterparts (Figure 1C, D). Remarkably, while all LmnaΔ9 mice expired by 30 days after birth, their LmnaΔ9Sun1 −/− littermates thrived past this date, most achieving lifespans more than twice this duration (Figure 1D). At the cellular level, the severely reduced proliferation of Lmna−/− and LmnaΔ9 fibroblasts was also substantially corrected in Lmna−/−Sun1−/− and LmnaΔ9Sun1 −/− cells (Figure 1E, F).
Figure 1. Defects in body weight and longevity in Lmna−/− and LmnaL530P/L530P (LmnaΔ9 mice) mice are ameliorated in homozygous Sun1 knockout Lmna−/−Sun1−/− and LmnaΔ9Sun1 −/− animals.
(A) Body weights are averages from mice with the indicated genotypes. The number (n) of animals used is indicated.
(B) Kaplan-Meier graph showing increased life span of Lmna−/−Sun1−/− compared to Lmna−/− mice. Median survival of wild type or Sun1−/− is >210 days in a 7 month follow up; Lmna−/− mice have median survival of 41 days; Lmna−/−Sun1+/− mice have a median of 54 days; Lmna−/−Sun1−/− mice have a median of 104 days (P <0.01 comparing Lmna−/− and Lmna−/−Sun1−/−).
(C) Body weights of LmnaΔ9 mice that are wild type, heterozygous, or homozygous for Sun1 deficiency. Wild type and Sun1−/− cohorts are graphed for comparison. Values are averages+/−SEM from animals in each cohort. Number (n) of animals is indicated. (P <0.0001 comparing LmnaΔ9Sun1 +/+ and LmnaΔ9Sun1 −/−).
(D) Kaplan-Meier graph showing increased life span of LmnaΔ9Sun1 −/− compared to LmnaΔ9Sun1 +/+ mice. LmnaΔ9Sun1 +/− mice are also graphed. (P <0.0001 comparing LmnaΔ9Sun1 +/+ and LmnaΔ9Sun1 −/−).
(E) Cell proliferation of the indicated MEFs. Curves are averages+/−SD, representative of >3 independent isolates from embryos of the indicated genotypes.
(F) Proliferation curves of MAFs (mouse adult fibroblasts) from WT, Sun1−/−, LmnaΔ9Sun1 +/+ and LmnaΔ9Sun1 −/− mice. MAFs were seeded at a density of 1000 cells per well. Growth was measured, and normalized cell indexes (averages+/−SD) are presented.
Tissue pathologies are improved in Sun1−/−Lmna−/− mice
Lmna−/− and Lmna−/−Sun1−/− animals grow to a greater size and live longer than their corresponding LmnaΔ9 and LmnaΔ9Sun1 −/− counterparts (Figure 1A–D). Cultured Lmna−/− and Lmna−/−Sun1−/− cells proliferated well while LmnaΔ9 and LmnaΔ9Sun1 −/− cells are challenging, requiring extracellular matrices or hypoxic conditions for propagation (Hernandez et al., 2010). For detailed characterizations, we chose to study the Lmna−/− and Lmna−/−Sun1−/− animals and their cells.
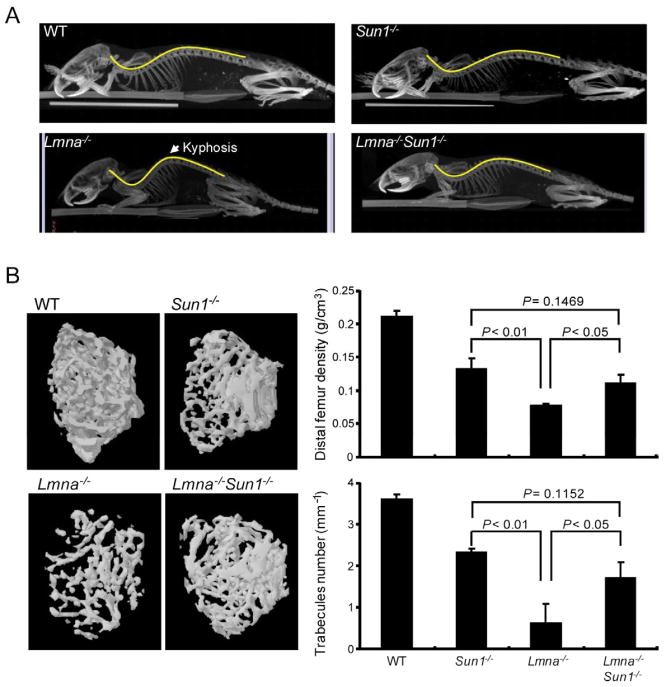
We compared tissue changes in Lmna−/− to Sun1−/−Lmna−/− mice. The spine of Lmna−/− mice by microcomputerized tomography was grossly lordokyphotic; this defect was absent in WT and Sun1−/− mice and was corrected in Lmna−/−Sun1−/− animals (Figure 2A). The femoral bone of 40-day-old Lmna−/− mice showed trabecular and bone densities that were notably sparser and thinner than Sun1−/− or WT mice; in Lmna−/−Sun1−/− animals the deficits were markedly improved (Figure 2B). In other tissues such as cardiac and skeletal muscle, pathologies, previously described in the Lmna−/− mice, were corrected and improved in the Lmna−/−Sun1−/− mice (Figure S1).
Figure 2. Correction of the Lmna−/− skeletal defects in the Lmna−/−Sun1−/− double knock out mouse.
(A) Micro-CT scans of the indicated mice. Lmna−/− mice display a lordokyphosis (curvature of the spine) phenotype corrected in Lmna−/−Sun1−/− mice.
(B) Three-dimensional micro-CT images of the femoral trabeculae from 40-day-old mice (left panels). Thinner trabecular formation was observed in the Lmna−/− mouse compared to the other genotypes. Right panels quantify bone density+/−SD (upper) and the number of trabeculaes/mm+/−SD (lower). P values are shown. (see also Figure S1)
Sun1 accumulates at the nuclear envelope (NE) and the Golgi of Lmna−/− MEFs
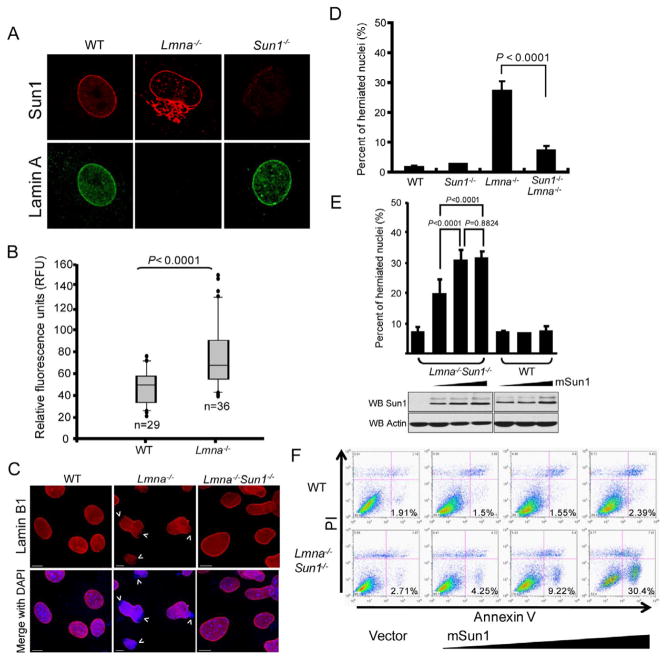
To seek a molecular explanation for loss-of-lamin A changes and their correction by Sun1 depletion, we investigated Sun1 expression in lamin A WT and Lmna−/− MEFs. Sun1 and lamin A co-localize at the NE in WT MEFs (Figure 3A, left panels). By contrast in Lmna−/− MEFs, Sun1 is found in the NE and at increased levels in the Golgi (Figure 3A, middle panels; and Figure S2A), based on co-staining with Golgi marker GM130 (Figure S2A, right) but not with ER marker calnexin (Figure S2A, left). NE localization and Golgi over accumulation of Sun1 were also seen in LmnaΔ9 mouse fibroblasts (Figure S2A, right). That Sun1 localizes with Golgi constituents in Lmna−/− cells was supported by biochemical fractionation of mouse tissues that detected Sun1 and GM130 in the same sucrose density fractions (Figure S2B). When Lmna−/− cells were examined for the relative distribution of Sun1 in the NE versus the Golgi, the amount in the latter increased proportionally with its level in the former (Figure S2C), suggesting that increased levels of Sun1 protein, in a Lmna−/− context, first occupies and saturates NE sites before “spilling” into the Golgi compartment. The average Sun1 expression level in individual Lmna−/− MEFs was significantly higher than that in WT MEFs (Figure 3B, Lmna−/− n=36, WT n=29, P < 0.0001) with the highest expressing former cells having approximately 8 fold greater levels of Sun1 than the lowest expressing latter counterparts; by contrast, in Lmna−/− cells other NE proteins such as Sun2 and Nup153 were unchanged in distribution or amounts while Emerin and Nesprin1 were not significantly increased but showed modest increases in ER relocalization (Figure S2D, E). The increase in Sun1 protein (Figure S2E) was not due to elevated Sun1 mRNA levels (compare WT and Lmna−/−; Figure S2F). This result together with heightened Sun1 accumulation (Figure S3A) when WT and Lmna−/− MEFs were treated with proteasome inhibitor lactacystin and the prolonged half life of Sun1 protein in Lmna−/− vs. WT MEFs (Figure S3B) suggests that Sun1 over accumulation in Lmna−/− cells is due to reduced protein turnover.
Figure 3. Extranuclear Sun1 is accumulated in the Golgi of Lmna−/− MEFs.
(A) Cells were immunostained with anti-lamin A (green) and anti-Sun1 (red) antibodies. Extranuclear Golgi localization of Sun1 is seen in Lmna−/− MEFs (also see Figure S2).
(B) Quantification of Sun1 in MEFs. Mean±SD reflects collective results from two separate experiments with n = 29 (WT) and n = 36 (Lmna−/−) MEFs. Difference between WT and Lmna−/− is statistically significant (P <0.0001). See also Figure S3.
(C) WT, Lmna−/− and Lmna−/−Sun1−/− MEFs were stained with anti-Lamin B1 (red) and DAPI (blue). Lamin B1 nuclear envelope staining is intact in WT and Lmna−/−Sun1−/− MEFs, with the staining being irregular with herniations in Lmna−/− nuclei. Arrows point to disruptions in nuclear envelope. Bars: 10 μm.
(D) Quantification of prevalence of cells with nuclear envelope disruptions. Values are averages+/−SD from three independently isolated MEFs of the indicated genotype (each counted for 300 nuclei). Prevalence of nuclear disruptions between Lmna−/− and Lmna−/−Sun1−/− MEFs is significantly different (P < 0.0001).
(E) (Upper) Sun1 over expression in the absence of lamin A exacerbates nuclear herniations. WT and Lmna−/−Sun1−/− MEFs were transfected with increasing mouse Sun1 (mSun1) expression vector. Nuclei were stained 48 hours later. Values are averages+/−SD from three experiments (each sample was counted for 300 nuclei per experiment). (Lower) Transfected cells were Western blotted for Sun1 expression and actin (as loading control).
(F) WT (top) or Lmna−/−Sun1−/− (bottom) MEFs were transfected with vector-alone (left) or increasing amounts of mSun1 (right three panels) and analyzed 48 hours later by FACS for propidium iodide (PI; Y-axis) and annexin V (X-axis). Percentage of apoptotic cells (in the lower right quadrant) is indicated.
Sun1 over accumulation increases nuclear defects
WT MEFs have circular or slightly ovoid nuclei while Lmna−/− nuclei are irregularly shaped with frequent herniations and blebs (Figure 3C) (Sullivan et al., 1999). Intriguingly, Lmna−/− nuclear abnormalities are significantly (P < 0.0001) reduced in Lmna−/−Sun1−/− cells (Figure 3C, D) suggesting that the nuclear irregularities are not explained simply by loss-of-lamin A which is equally absent in Lmna−/− and Lmna−/−Sun1−/− cells. On the other hand, because both Lmna−/− and LmnaΔ9 cells show Sun1 accumulation in the Golgi (Figure 3A; Figure S2A), this event could possibly account for the observed pathologies. This view, if correct, provides a parsimonious explanation for why Lmna−/− and LmnaΔ9 diseases in mice are alleviated when Sun1 levels are reduced (Figure 1).
The above reasoning predicts that deliberate Sun1 over expression in a Lmna−/− context should exacerbate nuclear aberrancies. To test this, we transfected increasing amounts of a mouse Sun1 (mSun1) expression vector into either Lmna−/−Sun1−/− or WT MEFs. The over expression of Sun1 progressively increased the prevalence of nuclear herniations in Lmna−/−Sun1−/− MEFs, without significantly affecting WT MEFs (Figure 3E). The transfections also elicited dose-dependent increases in the apoptosis of Lmna−/−Sun1−/− cells (Figure 3F).
Golgi targeting of Sun1 elicits nuclear herniations
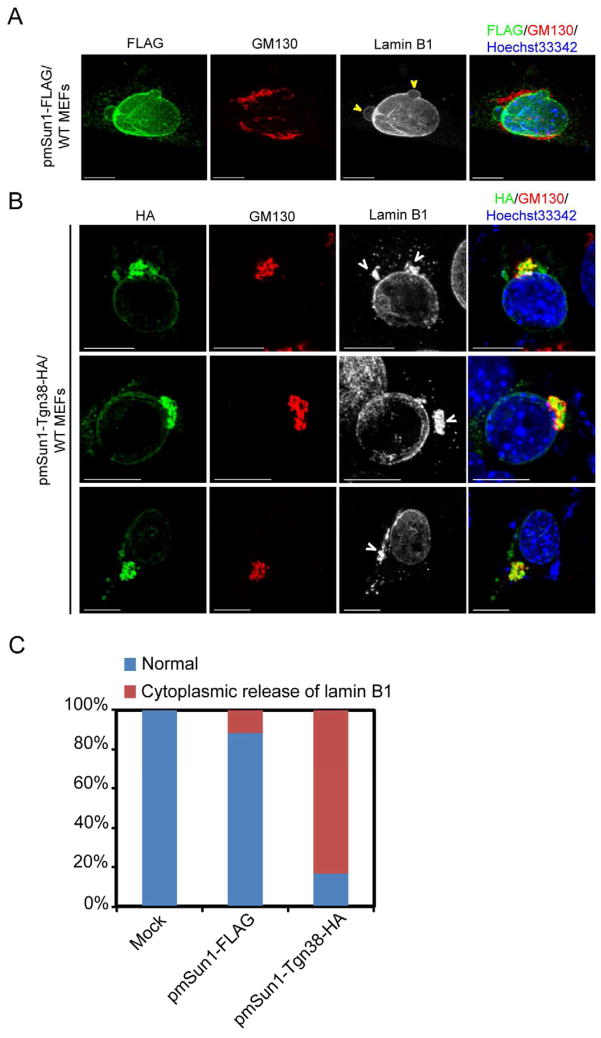
A remarkable feature of Sun1 expression in Lmna−/− MEFs is its mis-accumulation in the extranuclear Golgi apparatus (Figure 3A; Figure S2A). Protein mis-accumulation in human organelle storage disorders has been described for lysosomal storage diseases such as Fabry, Tay-Sachs, Gaucher, Niemann-Pick, Pompe, and Krabbe (Metz et al., 2011), and endoplasmic reticulum storage diseases such as cystic fibrosis, a1-antitrypsin deficiency, hereditary hypoparathyroidism, and procollagen type I, II, IV deficiency (Rutishauser and Spiess, 2002); however, to date, there are no good examples of Golgi storage diseases. To test if the deliberate Golgi-mis-accumulation of Sun1 is significantly pathogenic, we constructed an HA-tagged Tgn38-fused Golgi-targeting mSun1 expression vector [Tgn38 is an integral Golgi protein; (Szentpetery et al., 2010)]. Sun1 protein, when over expressed in WT MEFs, localized to the nuclear envelope and elicited barely discernable mild nuclear blebbings (Figure 4A), while transfected Tgn38-Golgi-targeted mSun1 dramatically increased Golgi-accumulation and nuclear herniations with obvious cytoplasmic accumulation of lamin B1 (Figure 4B) in 83% of Tgn38-Golgi-mSun1 expressing cells (Figure 4C). Recently, it was reported that the Sun1-related Sun2 protein has a Golgi-retrieval signal ensuring its transport from the Golgi back to the ER (Turgay et al., 2010). Although not yet determined, Sun1 may differ in the Golgi-retrival signal which could explain why a SUN1-mutant (human SUN1 a.a. 103–785) [Figure S4, (Haque et al., 2010; Chi et al., 2007)] and a wild type Sun1 protein that is expressed in the absence of cell endogenous lamin A (i.e. Lmna−/− cells; Figure 3A, Figure S2A) are both found in the Golgi. We also checked if the Golgi-localizing SUN1 (103–785) mutant elicits nuclear aberrations. Indeed, over-expression of the SUN1 (103–785) mutant increased nuclear envelope rupture and cytoplasmic redistribution of lamin B1 (Figure S4).
Figure 4. Over expression of Golgi-targeted Sun1 increased nuclear aberrations and cell death.
(A) WT MEFs were transfected with FLAG-tagged mouse Sun1 vector and stained with mouse anti-FLAG (green), rabbit anti-GM130 (red), and goat anti-lamin B1 (grey scale). A representative image of modest nuclear blebs and ruffles seen in some transfected cells is shown. Bars, 10 μm.
(B) A Golgi-targeted mouse Sun1 (fused with Tgn38, HA-tagged) expression plasmid was transfected into WT MEFs. Thirty hours later, cells were stained with mouse anti-HA (green), rabbit anti-GM130 (red), and goat anti-lamin B1 (grey scale). Aberrancies were visualized by cytoplasmic lamin B1 staining (see arrow heads) of pmSun1-Tgn38-HA transfected cells. Bars, 10 μm.
(C) Quantification of the cytoplasmic release of lamin B1 in MEFs transfected (for 30 hours) with either mSun1 (mSun1-FLAG) or the Golgi-targeted mSun1 (pmSun1-Tgn38-HA). One hundred cells were counted in each case. See also Figure S4.
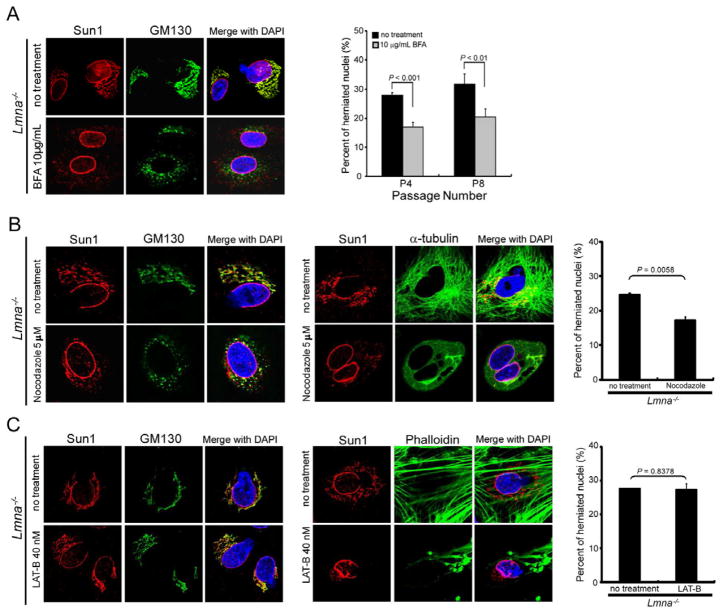
The above results suggest that reducing Sun1 accumulation in the Golgi might moderate Lmna−/− nuclear irregularities. Brefeldin A (BFA) is an antibiotic that reversibly interferes with the anterograde transport of macromolecules from the endoplasmic reticulum (ER) to the Golgi (Marie et al., 2008). We asked if BFA treatment of Lmna−/− cells would reduce Sun1 in the Golgi. Confocal imaging of Lmna−/− MEFs treated with BFA at 10 μg/mL for 24 hours showed a reduction in most, albeit not all, Golgi-trafficked Sun1 and GM130 proteins (Figure 5A, left panels) with statistically significant (P < 0.001; P < 0.01) reduction in nuclear aberrations in cells passaged four (P4) to eight (P8) times in culture (Figure 5A, right graph). We also treated Lmna−/− MEFs with nocodazole to block microtubule organization (Figure 5B) or latrunculin B to interrupt actin assembly (Figure 5C). Nocodazole disrupts the Golgi apparatus (Thyberg and Moskalewski, 1999), and its treatment of Lmna−/− MEFs led to a punctated redistribution of otherwise Golgi-associated Sun1 and GM130 (Figure 5B). This treatment also led to a moderate, but statistically significant, reduction of nuclear aberrations (Figure 5B, right graph). By contrast, latrunculin B did not affect Sun1 distribution in the Golgi nor ameliorated nuclear defects (Figure 5C). Collectively, the findings demonstrate that endogenous (Figure 5) or exogenous (Figure 4, S4) Sun1 mis-accumulation in the Golgi elicits substantial cellular pathologies and that reducing Sun1 Golgi accumulation restores cellular normalcy.
Figure 5. Brefeldin A and nocodazole, but not latrunculin, treatment reduced nuclear irregularities in Lmna−/− MEFs.
(A) (Left) Staining of Sun1 (red) and GM130 (green) in Lmna−/− MEFs treated for 24 hours with brefeldin A (BFA, 10 μg/mL); note the reduction of Sun1 and GM130 from the Golgi. (Right) Quantification of BFA treatment on the nuclear morphology of Lmna−/− MEFs. Untreated and treated cells were stained with mouse Sun1-specific antibody or DAPI in cells passaged 4 (P4), and 8 (P8) times, respectively. The nuclear morphology was evaluated by observers blinded for genotype and by computerized image analyses. Nuclear irregularities are also seen in HGPS cells (see list in Table S1).
(B) (Left) Sub-cellular localization of Sun1 in Lmna−/− MEFs untreated or treated with 5 μM nocodazole for 4 hours. The Golgi complex was stained with mouse antibody against GM130 (green) and rabbit antibody against mouse Sun1 (red). (Middle) Cells untreated and treated with nocodazole and stained for α-tubulin are shown. (Right) Quantification of nocodazole treatment on the nuclear morphology of Lmna−/− MEFs. Difference between untreated and treated cells is P = 0.0058.
(C) (Left) Lmna−/− MEFs were untreated or treated with 40 nM of latrunculin (LAT-B) for 12 hours. Cells were fixed and stained for Sun1 and GM130. (Middle) Cells untreated and treated with latrunculin and visualized with fluorescent phalloidin for actin are shown. (Right) Quantification of LAT-B treatment on the nuclear morphology of Lmna−/− MEFs. Difference between untreated and treated cells was statistically insignificant (P = 0.8376). All values are mean+/−SD.
SUN1 over accumulation in HGPS cells correlates with dysfunction
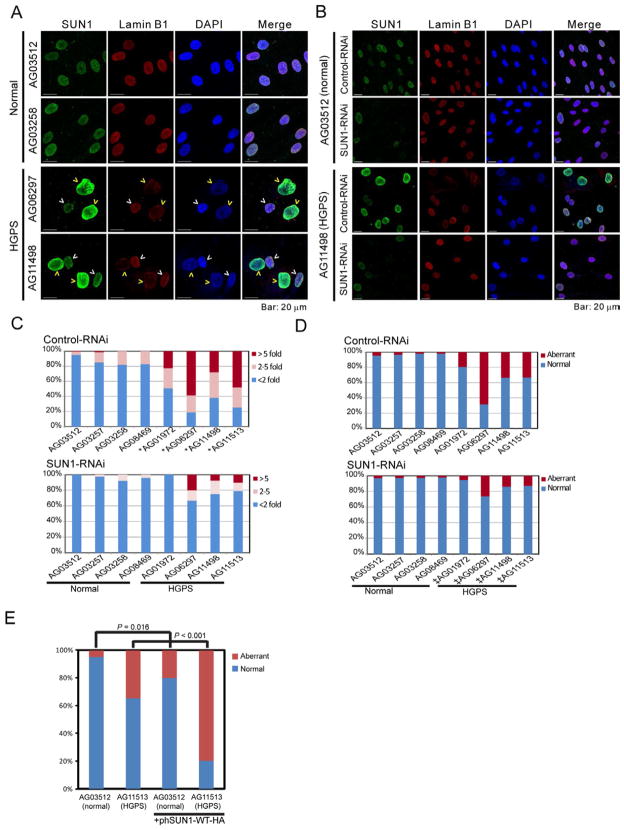
We next investigated SUN1 expression in HGPS cells querying if (and how) this protein might contribute to pathology. We immunostained SUN1 expression in human skin fibroblasts from seven independent HGPS [LMNA 1824C>T (G608G)] (Figure S5A and Table S1) and four normal individuals and verified LAΔ50 progerin expression (Goldman et al., 2004) in HGPS, but not, normal cells (Figure S5B). By immunofluorescence, brighter SUN1 staining was observed in HGPS cells compared to control cells (representative examples are in Figures 6A and S5A, Normal vs. HGPS) which is consistent with increased SUN1 expression by Western blotting (Figure S5B) and with an earlier report of SUN1 accumulation in HGPS cells (Haque et al., 2010). Of note, our stainings showed that not every HGPS cell had elevated SUN1, but cells that stained brightest for SUN1 were also ones that had larger nuclei and more severe nuclear morphological distortions (compare dim-SUN1 HGPS cells, white arrow heads to bright-SUN1 HGPS cells, yellow arrow heads; Figure 6A). SUN1 mRNA levels did not differ significantly in HGPS versus normal cells (Figure S5C) supporting the interpretation that reduced protein turnover (Figure S3B), not increased transcription, underlies SUN1 accumulation.
Figure 6. Nuclear irregularities in HGPS fibroblasts correlate with SUN1 expression.
(A) SUN1 and lamin B1 in normal (AG03512 and AG03258) and HGPS (AG06297 and AG11498) skin fibroblasts are stained with anti-human SUN1 (green) and anti-lamin B1 (red). DAPI is in blue. Yellow arrow heads point to cells expressing high-SUN1, white arrow heads to cells with low-SUN1.
(B) Nuclear morphologies and SUN1-staining of control (AG03512) and HGPS (AG11498) skin fibroblasts transfected with control- or SUN1-siRNA for 72 hours.
(C) Quantification of SUN1 immunofluorescent intensities in cells treated with Control or SUN1 siRNA. One hundred twenty to two hundred cells from each of the indicated samples were visualized and quantified for staining intensities. The intensities were normalized to the SUN1 intensity in AG03512 cells. Cells with SUN1 intensities less than 2 fold different from average are represented by blue bar; cells that are > 2 fold, but <5 fold are represented by pink bar; cells that stained >5 fold above average are represented by brown bar. *, P < 0.001 when compared to AG03512 cells (t-test).
(D) Quantification of the prevalence of cells from (B) with nuclear irregularities. ‡, P < 0.0001, when comparing the same cells treated with control RNAi or SUN1-RNAi (Fisher’s exact test). See also Figure S5.
(E) Aberrant nuclear morphology in normal and HGPS fibroblasts transfected with a HA-tagged human SUN1 expression plasmid. Two hundred mock transfected cells per sample and fifty transfected cells per sample were scored. P values, Fisher’s exact test.
To address if elevated SUN1 levels in HGPS result in pathologies, we asked if knocking down SUN1 alleviates nuclear defects. SUN1-specific or control siRNAs were transfected into HGPS or normal skin fibroblasts, and nuclear appearance was monitored (Figure S5D). The nuclear morphologies were unchanged in cells treated with control siRNA (Figure 6B, S5E); but SUN1-specific siRNA reduced the prevalence of bright-SUN1 HGPS cells (compare AG11498 upper to lower row, Figure 6B, C), and lowered the number of cells with aberrant nuclei (Figure 6B, 6D, S5E, Table S1). The contribution of SUN1 to nuclear morphology was assessed conversely by deliberately over expressing exogenous SUN1. Ectopic SUN1 over expression in HGPS and normal fibroblasts significantly increased aberrant nuclei (Figure 6E).
SUN1 expression correlates with HGPS heterochromatin profile and cellular senescence
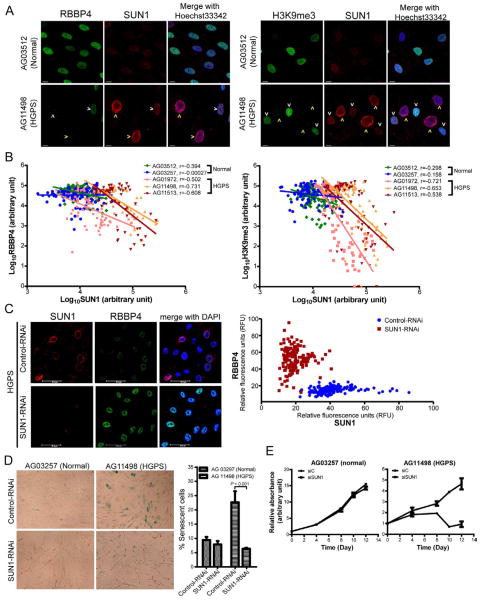
Chromatin disorganization and massive heterochromatin loss are correlated with nuclear shape alterations in HGPS cells (Shumaker et al., 2006; Goldman et al., 2004). Assays for HGPS heterochromatin loss have included markers such as the lamin A-associated NURD (nucleosome remodeling and deacetylase) component RBBP4 (Pegoraro et al., 2009) and the pan heterochromatin marker histone H3K9me3 (Shumaker et al., 2006; Scaffidi and Misteli, 2005). To corroborate the nuclear morphology findings (Figure 6), we investigated how SUN1 expression correlates with previously described HGPS heterochromatin changes. When HGPS or normal skin fibroblasts were stained for RBBP4 (Figure 7A, left) or H3K9me3 (Figure 7A, right), an inverse correlation was observed between the expression of SUN1 and RBBP4 (Figure 7B, left) or H3K9me3 (Figure 7B, right). In agreement with the results in Figure 6A, only a subset of HGPS cells were bright-SUN1 (yellow arrows = bright-SUN1, white arrows = dim-SUN1, Figure 7A); and interestingly the bright-SUN1 cells were also those with the larger more distorted nuclei as well as staining sparsely for RBBP4 (Figure 7A, B, left) or H3K9me3 (Figure 7A, B, right). Separately, we found that RBBP4 expression was substantially reduced in ~70% of Lmna−/− MEFs (Figure S6A) and in Lmna−/− mouse liver tissue (Figure S6B), further supporting an inverse relationship between Sun1 and NURD activity.
Figure 7. Knock down of SUN1 alleviated HGPS-associated loss of NURD complex and cellular senescence.
(A) Normal (AG03512) and HGPS (AG11498) skin fibroblasts were stained for heterochromatin markers (RBBP4 or H3K9me3; green) and SUN1 (red). Yellow arrow heads point to high-SUN1 cells; white arrow heads denote low-SUN1 cells. See also Figure S6.
(B) Expression levels of RBBP4 or H3K9me3 and SUN1 in two normal and three HGPS skin fibroblasts were quantified by MetaMorph software. Each dot represents fluorescence intensity (in Log10 scale) in a single cell of RBBP4 (left) or H3K9me3 (right) vs. SUN1. Linear curve fitting and correlation coefficient (r) are indicated. In HGPS cells, RBBP4 and H3K9me3 expression correlates negatively with SUN1 expression.
C) HGPS fibroblasts (AG03513) treated with control or SUN1 siRNA for 72 hours were stained with antibodies for SUN1 (red) and RBBP4 (green). Increased RBBP4 expression was observed in SUN1 siRNA-treated cells compared to control siRNA-treated cells. Graphic quantification of the staining intensities of RBBP4 vs. SUN1 in individual HGPS fibroblasts treated with control (blue) or SUN1 (brown) siRNA is shown (right); each dot represents a single cell (154 control and 157 SUN1 RNAi treated cells were quantified).
(D) Visualization (Left) and quantification (Right) of acidic senescence associated β-galactosidase (SA-β-Gal) in normal (AG03257) and HGPS (AG11498 at passage 8) fibroblasts transfected with Control- or SUN1-RNAi for 96 hours. Standard deviations are from three independent assays counting 1200 to 2000 cells in each experiment. Cell scoring was performed in a blinded fashion by an independent investigator. P value (Chi-square) is indicated.
(E) Cell proliferation in normal (AG03257) and HGPS (AG11498) cells transfected with control or SUN1 RNAi. Cells at ~50% confluency were transfected. When cells reached confluency, equal numbers were seeded into dishes and quantified for proliferation using Cell Counting Kit-8 24 hours after cell seeding (day 0) and after another 4, 8, 10, 12 days. Relative absorbance at 460nm was obtained by [(Absorbance460nm-background Absorbance460nm) at day N]/[(Absorbance460nm-background Absorbance460nm) at day 0]. Standard deviations are from triplicate experiments.
We next asked if knock down of SUN1 would reverse HGPS-associated heterochromatin changes. We compared control-RNAi and SUN1-RNAi transfected HGPS cells and found that the latter did recover RBBP4 expression relative to the former (Figure 7C). Because heterochromatin dysregulation is correlated with cellular senescence (Di et al., 2011) and because HGPS cells senesce prematurely (DeBusk, 1972; Eriksson et al., 2003), we queried how SUN1 affects HGPS senescence by knocking down SUN1 for 96 hours and examining acidic senescence associated β-galactosidase (SA-β-Gal) in control and HGPS cells (Figure 7D). In normal cells, the extent of senescence was similar (~9%) between control-siRNA or SUN1-siRNA samples (Figure 7D); however, in HGPS cells, the observed high level of ambient senescence (~22%) as measured by β-galactosidase was dramatically decreased (to ~6%) after SUN1 knock down. Moreover, HGPS fibroblasts when treated with SUN1-RNAi gained a proliferative advantage over control-RNAi treated cells (Figure 7E). These data collectively support the interpretation that increasing SUN1 accumulation is associated with HGPS pathology and removing over-expressed SUN1 restores normal cellular physiology.
DISCUSSION
Here, we show that aberrant Sun1 expression is a critical pathogenic event common to Lmna−/−, LmnaΔ9, and HGPS disorders. As noted here and elsewhere, Lmna−/− mice (Sullivan et al., 1999), LmnaΔ9 mice (Hernandez et al., 2010; Mounkes et al., 2003), and HGPS individuals (Merideth et al., 2008) share a constellation of disorders that include nuclear aberrations, dystrophic organ and tissue abnormalities, and abbreviated lifespan. A current view is that progerin is causal of the LAΔ50 HGPS disease (Burtner and Kennedy, 2010; Liu et al., 2005; Scaffidi and Misteli, 2005; Goldman et al., 2004). How progerin mechanistically signals cellular and tissue damage remains elusive. That said, the existence of the dystrophic and cardiomyopathic pathologies in Lmna−/− mice and multiple examples of Lmna mutations (Novelli et al., 2002; Plasilova et al., 2004; Sullivan et al., 1999) that do not synthesize progerin, but do produce degenerative-dystrophic diseases such as Emery-Dreifuss muscular dystrophy (Bonne et al., 1999), Charcot-Marie-Tooth (De Sandre-Giovannoli et al., 2002; Chaouch et al., 2003), Mandibuloacral dysplasia (Novelli et al., 2002), Dunnigan-type familial partial lipdystrophy (Cao and Hegele, 2000), atypical Werner’s syndrome (Chen et al., 2003), and limb girdle muscular dystrophy (Muchir et al., 2000; Kitaguchi et al., 2001), require an understanding of progerin- independent and dependent factors/cofactors underlying the pathologies.
The Sun1 protein connects the nucleoplasm with the cytoskeleton (Crisp et al., 2006) and has roles in nuclear anchorage, nuclear migration, and cell polarity. Deficits in Sun1 correlate with developmental retardation in neurogenesis, gametogenesis, myogenesis, and retinogenesis (Ding et al., 2007; Lei et al., 2009; Zhang et al., 2009; Yu et al., 2011; Chi et al., 2009b). To date, how an inner nuclear envelope protein like Sun1 fits into the pathogenesis of laminopathies is unknown (Stewart et al., 2007).
The major unexpected finding here is that while Lmna−/− mice and LmnaΔ9 mice thrive poorly and die prematurely, the removal of Sun1 creating Lmna−/−Sun1−/− and LmnaΔ9Sun1 −/− mice rescued pathologies and dramatically improved longevity (Figures 1, 2). To better understand these results, we observed that at the cellular level Lmna−/− and LmnaΔ9 fibroblasts have uniformly increased Sun1 expression with significant protein mis-accumulation in the Golgi (Figure 3A and Figure S2). Furthermore, approximately one in three LAΔ50 HGPS fibroblasts (Figure 6, 7; Figure S5; Table S1) was elevated for SUN1 expression with the bright (high)-SUN1, but not the dim (low)-SUN1, cells exhibiting abnormal nuclear size and shape, heterochromatin RBBP4 and H3K9me3 markers, and cellular senescence (Figure 6, 7). While one cannot do a SUN1 knock out experiment in LAΔ50 HGPS individuals, the knock down of SUN1 in LAΔ50 HGPS cells considerably improved nuclear size/shape defects, heterochromatin loss, and cellular senescence (Figure 6, 7). Thus, although the approaches (knock out and knock down) and disease models (Lmna−/−, LmnaΔ9, and LAΔ50 HGPS) are not identical, a parsimonious interpretation consistent with the collective results is that Sun1 over-accumulation represents a common effector of Lmna−/−, LmnaΔ9, and LAΔ50 HGPS pathologies.
How does Sun1 over-accumulate in Lmna−/−, LmnaΔ9, and LAΔ50 HGPS cells? Sun1 is normally located in the NE, positioned by mechanisms that are still obscure but may depend on interaction with lamin A filaments underlying the nuclear matrix (Haque et al., 2006; Mattioli et al., 2011; Ostlund et al., 2009). As noted above, a SUN1 protein deleted in its N-terminal (~100 amino acids) lamin A-interacting domain relocates from the NE to the Golgi [Figure S4; (Haque et al., 2010; Chi et al., 2007)]. Emerging evidence suggests that the SUN1-related SUN2 protein has a Golgi-retrieval sequence (Turgay et al., 2010) which is required for retrieval of SUN2 from the Golgi to the ER. Differences between the two proteins may explain why Sun1, but not Sun2, expressed in the absence of cell endogenous lamin A (i.e. Lmna−/− cells; Figure 3, Figure S2) accumulates in the Golgi. Our findings show that Sun1 accumulation arises from reduced protein turnover (Figure S3) and not increased transcription (Figure S2F, S5C), suggesting that approaches to enhance protein degradation might be therapeutically beneficial (Cao et al., 2011).
We did not discern obvious Golgi over accumulation of endogenous SUN1 in LAΔ50 HGPS cells. While the explanation for this remains unclear, it could be that Golgi-over accumulation of SUN1 in human cells is highly toxic and selects rapidly against the viability of cultured HGPS cells, or that the heterozygous expression of wild type lamin A in LAΔ50 HGPS cells is sufficient to locate endogenous SUN1 to the NE preventing overt Golgi mis-accumulation. Relevant to the former explanation, we observed that early passage Lmna−/− MEFs show more dramatic Sun1-Golgi mis-accumulation than late passaged counterparts, consistent with selection against cells with high Sun1-Golgi mis-accumulation.
What might be consequences of Sun1 mislocation in the Golgi? Our Golgi targeting experiments with mSun1-Tgn38 (Figure 4) and SUN1 (103–785) mutant protein (Figure S4) showed that Golgi-storage of Sun1 is cytotoxic. This toxicity may be akin to that elicited in abnormal human lysosomal- (Metz et al., 2011) or ER- (Rutishauser and Spiess, 2002) storage diseases. Aside from organelle storage disorders, other types of protein aggregation maladies like Alzheimer’s (Gouras et al., 2010) exist. In Alzheimer’s disease, evidence now suggests that it is the small soluble amyloid-β oligomers, not the large easily visualized amyloid-β fibrils/plaques, which produce neurotoxicity (Crews and Masliah, 2010). As mentioned above, we currently do not exclude that Golgi accumulation of SUN1 may indeed occur in LAΔ50 HGPS cells in vivo and that such cells may have rapidly succumbed and therefore are not represented in the mostly late passage repository-deposited HGPS fibroblasts (Table S1). However, like soluble amyloid-3 β oligomers which need not present as gross aggregates to be cytotoxic, it may be that the degree of SUN1 over expression in LAΔ50 HGPS cells (Figure 6E) is sufficient to functionally trigger pathology without having to reach levels required for overt Golgi-spillage. In LAΔ50 HGPS cells, increased SUN1 accumulation in the nuclear envelope may sufficiently create dysfunction by rendering a more rigid meshwork.
Progerin underlies LAΔ50 HGPS disease development (Eriksson et al., 2003; Scaffidi and Misteli, 2005; Goldman et al., 2004). How does SUN1 fit into the picture where progerin synthesis is the initial event inciting cellular dysfunction? In primary LAΔ50 HGPS cells or LmnaΔ9 mice where progerin (Figure S5B) or lamin A-ΔExon9 protein is expressed, Sun1 knock down is sufficient to remedy cellular aberrancies, and senescence and longevity defects (Figures 1, 6, 7). A cogent interpretation of these results is that SUN1 accumulation is positioned downstream of progerin or lamin A-ΔExon9 such that the depletion of SUN1 sufficiently interrupts pathologic signaling. In Lmna−/− mice where no progerin protein is synthesized, our data show that Sun1 accumulation remains an important trigger of loss-of-lamin A pathology. Future experiments are needed to clarify in non-codon 1824 (i.e. non LAΔ50) forms of HGPS (Plasilova et al., 2004) and in the many rare dystrophic human diseases (Burke et al., 2001; Chi et al., 2009a; Capell and Collins, 2006; Kudlow et al., 2007) where no gain-of-function progerin-like protein is synthesized, whether SUN1 (or other nuclear envelope protein) misaccumulation is similarly important to pathogenesis. Our current findings do suggest that at least in the Lmna−/−, LmnaΔ9, and LAΔ50 HGPS diseases, Sun1 over accumulation is critical to pathologies. If this notion can be broadly applied, it suggests that future clinical trials and therapies for laminopathies that treat disease upstream events (i.e. targeting progerin) without resolving the downstream pathogenic events (i.e. Sun1 misaccumulation) may be ineffective.
EXPERIMENTAL PROCEDURES
Animals
Knockout mice were created using standard procedures. Because Sun1−/− and Lmna−/− mice are reproductively defective (Ding et al., 2007; Alsheimer et al., 2004; Chi et al., 2009b), Sun1+/− mice were crossed with Lmna+/− mice to generate Lmna−/−Sun1−/− mice; or Sun1+/− mice were crossed with LmnaL530P/+ mice (Mounkes et al., 2003) to generate LmnaΔ9Sun1−/− mice. Mouse genotypes were verified by PCR. All animal experiments were conducted according to animal study protocols approved by the NIH or the Singapore Animal Use Committee.
Immunofluorescence and confocal microscopy
Cells, fixed in 4% paraformaldehyde in PBS for 30 minutes and permeabilized with 0.1% TritonX-100 for 5 minutes at room temperatures, were incubated with 1% BSA in PBS for 30 minutes to block nonspecific binding. Antibodies were diluted at 1:100 to 1:1000 and incubated for 1.5 hours at room temperature. After three washes with PBS, cells were probed with fluorescent (Alexa-488, Alexa-594 or Alexa-647)-conjugated secondary antibodies. Nuclei were counterstained with Hoechst33342 or DAPI (Invitrogen), and Sun1 intensity was visualized using a Leica TCS SP5 microscope and quantified by the ImageJ 1.42q (NIH) or MetaMorph (Molecular Devices) software. Other procedures are in the Supplementary material.
Supplementary Material
Highlights.
Progeric and dystrophic early deaths in mice are ameliorated by removal of Sun1.
Sun1 over-accumulation in the Golgi is pathogenic.
Cellular senescence of HGPS fibroblasts is corrected by depletion of SUN1.
Acknowledgments
Work was supported by NIAID intramural funds, the IATAP, NIAID contract to SoBran, Inc., the NHRI, Taiwan (NHRI 99A1-CSPP11-014), and NSC, Taiwan (NSC 98-2320-B-400-009-MY3), and the Singapore Biomedical Research Council and Agency for Science, Technology and Research (A*STAR). We thank S-Y Chuang, E. Miyagi, J. M. Ward, L.I. Cheng,, Z.J. Chen, J. Kabat, S. Becker, S. Bradford, Q. Su, and D. Donahue for assistance; and W. Leonard, J. Hanover, A. Dayton, Y.B.Shi, and D. Camerini-Otero for reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alsheimer M, Liebe B, Sewell L, Stewart CL, Scherthan H, Benavente R. Disruption of spermatogenesis in mice lacking A-type lamins. J Cell Sci. 2004;117:1173–1178. doi: 10.1242/jcs.00975. [DOI] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Burke B, Mounkes LC, Stewart CL. The nuclear envelope in muscular dystrophy and cardiovascular diseases. Traffic. 2001;2:675–683. doi: 10.1034/j.1600-0854.2001.21001.x. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet. 2006;7:369–405. doi: 10.1146/annurev.genom.7.080505.115732. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in hutchinson-gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Chaouch M, Allal Y, De Sandre-Giovannoli A, Vallat JM, mer-el-Khedoud A, Kassouri N, Chaouch A, Sindou P, Hammadouche T, Tazir M, Levy N, Grid D. The phenotypic manifestations of autosomal recessive axonal Charcot-Marie-Tooth due to a mutation in Lamin A/C gene. Neuromuscul Disord. 2003;13:60–67. doi: 10.1016/s0960-8966(02)00196-7. [DOI] [PubMed] [Google Scholar]
- Chen L, Lee L, Kudlow BA, Dos Santos HG, Sletvold O, Shafeghati Y, Botha EG, Garg A, Hanson NB, Martin GM, Mian IS, Kennedy BK, Oshima J. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- Chi YH, Chen ZJ, Jeang KT. The nuclear envelopathies and human diseases. J Biomed Sci. 2009a;16:96. doi: 10.1186/1423-0127-16-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Cheng LI, Myers T, Ward JM, Williams E, Su Q, Faucette L, Wang JY, Jeang KT. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 2009b;136:965–973. doi: 10.1242/dev.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Haller K, Peloponese JM, Jr, Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem. 2007;282:27447–27458. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- Crews L, Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer’s disease. Hum Mol Genet. 2010;19:R12–R20. doi: 10.1093/hmg/ddq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le MM, Levy N. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat JM, Tazir M, Kassouri N, Szepetowski P, Hammadouche T, Vandenberghe A, Stewart CL, Grid D, Levy N. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am J Hum Genet. 2002;70:726–736. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBusk FL. The Hutchinson-Gilford progeria syndrome. Report of 4 cases and review of the literature. J Pediatr. 1972;80:697–724. doi: 10.1016/s0022-3476(72)80229-4. [DOI] [PubMed] [Google Scholar]
- Di MR, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, dal ZR, Matti V, d’Ario G, Montani E, Mercurio C, Hahn WC, Gorgoulis V, Minucci S, dda di FF. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouras GK, Tampellini D, Takahashi RH, Capetillo-Zarate E. Intraneuronal beta-amyloid accumulation and synapse pathology in Alzheimer’s disease. Acta Neuropathol. 2010;119:523–541. doi: 10.1007/s00401-010-0679-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L, Roux KJ, Wong ES, Mounkes LC, Mutalif R, Navasankari R, Rai B, Cool S, Jeong JW, Wang H, Lee HS, Kozlov S, Grunert M, Keeble T, Jones CM, Meta MD, Young SG, Daar IO, Burke B, Perantoni AO, Stewart CL. Functional coupling between the extracellular matrix and nuclear lamina by Wnt signaling in progeria. Dev Cell. 2010;19:413–425. doi: 10.1016/j.devcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaguchi T, Matsubara S, Sato M, Miyamoto K, Hirai S, Schwartz K, Bonne G. A missense mutation in the exon 8 of lamin A/C gene in a Japanese case of autosomal dominant limb-girdle muscular dystrophy and cardiac conduction block. Neuromuscul Disord. 2001;11:542–546. doi: 10.1016/s0960-8966(01)00207-3. [DOI] [PubMed] [Google Scholar]
- Kudlow BA, Kennedy BK, Monnat RJ., Jr Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009;106:10207–10212. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, Pei D, Pendas AM, Cadinanos J, Lopez-Otin C, Tse HF, Hutchison C, Chen J, Cao Y, Cheah KS, Tryggvason K, Zhou Z. Genomic instability in laminopathy-based premature aging. Nat Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Marie M, Sannerud R, Avsnes DH, Saraste J. Take the ‘A’ train: on fast tracks to the cell surface. Cell Mol Life Sci. 2008;65:2859–2874. doi: 10.1007/s00018-008-8355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli E, Columbaro M, Capanni C, Maraldi NM, Cenni V, Scotlandi K, Marino MT, Merlini L, Squarzoni S, Lattanzi G. Prelamin A-mediated recruitment of SUN1 to the nuclear envelope directs nuclear positioning in human muscle. Cell Death Differ. 2011;18:1305–1315. doi: 10.1038/cdd.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO, III, Gahl WA, Introne WJ. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz TF, Mechtler TP, Orsini JJ, Martin M, Shushan B, Herman JL, Ratschmann R, Item CB, Streubel B, Herkner KR, Kasper DC. Simplified Newborn Screening Protocol for Lysosomal Storage Disorders. Clin Chem. 2011 doi: 10.1373/clinchem.2011.164640. [DOI] [PubMed] [Google Scholar]
- Mounkes LC, Kozlov S, Hernandez L, Sullivan T, Stewart CL. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature. 2003;423:298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- Muchir A, Bonne G, van der Kooi AJ, van MM, Baas F, Bolhuis PA, de VM, Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum Mol Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- Nagano A, Arahata K. Nuclear envelope proteins and associated diseases. Curr Opin Neurol. 2000;13:533–539. doi: 10.1097/00019052-200010000-00005. [DOI] [PubMed] [Google Scholar]
- Novelli G, Muchir A, Sangiuolo F, Helbling-Leclerc A, D’Apice MR, Massart C, Capon F, Sbraccia P, Federici M, Lauro R, Tudisco C, Pallotta R, Scarano G, Dallapiccola B, Merlini L, Bonne G. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am J Hum Genet. 2002;71:426–431. doi: 10.1086/341908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasilova M, Chattopadhyay C, Pal P, Schaub NA, Buechner SA, Mueller H, Miny P, Ghosh A, Heinimann K. Homozygous missense mutation in the lamin A/C gene causes autosomal recessive Hutchinson-Gilford progeria syndrome. J Med Genet. 2004;41:609–614. doi: 10.1136/jmg.2004.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser J, Spiess M. Endoplasmic reticulum storage diseases. Swiss Med Wkly. 2002;132:211–222. doi: 10.4414/smw.2002.09861. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, Goldman RD. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Varnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci U S A. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Courvalin JC. How do mutations in lamins A and C cause disease? J Clin Invest. 2004;113:349–351. doi: 10.1172/JCI20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.