Abstract
The 1000 Genomes Project was launched as one of the largest distributed data collection and analysis projects ever undertaken in biology. In addition to the primary scientific goals of creating both a deep catalogue of human genetic variation and extensive methods to accurately discover and characterize variation using new sequencing technologies, the project makes all of its data publicly available for community use. The project data coordination center has developed and deployed several tools to enable widespread data access.
Introduction
High throughput sequencing technologies including those created by Illumina (Illumina, Inc.), 454 (Roche Diagnostics Corp.) and SOLiD (Life Technologies), enable whole genome sequencing at an unprecedented scale and dramatically reduced costs over the gel capillary technology used in the human genome project. These technologies were at the heart of the decision in 2007 to launch the 1000 Genomes Project, an effort to comprehensively characterize human variation in multiple populations. In the pilot phase of the project the data helped create an extensive population-scale view of human genetic variation1.
The larger data volumes and shorter read lengths of next generation sequence technologies used by the project created substantial new requirements for the bioinformatics, analysis and data distribution methods. The project initially planned to collect 2x whole genome coverage for 1000 individuals, representing approximately 6 gigabasepairs of sequence per individual and 6 terabasepairs (Tbp) of sequence in total. Increasing sequencing capacity led to repeated revisions of these plans to the current project scale of collecting low coverage (~4x) whole genome and (~20x) whole exome sequence for 2500 individuals plus high coverage (~40x) for 500 individuals (an approximate 25 fold increase in sequence generation over original estimates). In fact, the pilot project itself collected 5Tbp of sequence data, resulting in 38,000 files and over 12 terabytes of data being available to the community. In March 2012 the still-growing project resources are more than 260 terabytes of data in more than 250,000 publicly accessible files.
As in previous efforts2–4, the 1000 Genomes Project recognized that data coordination would be critical to move forward productively and to ensure the data was available to the community in a reasonable time frame. Therefore, the Data Coordination Center (DCC) was set up jointly between the European Bioinformatics Institute (EBI) and the National Center for Biotechnology (NCBI) to manage project specific data flow, to ensure archival sequence data deposition and to manage community access through the FTP site and genome browser.
Here we describe the methods used by the 1000 Genomes Project to provide data resources to the community from raw sequence data to browseable project results. We provide examples drawn from the project’s data processing methods to demonstrate the key components of complex workflows.
Data Flow
Managing data flow in the 1000 Genomes Project such that the data is available within the project and to the wider community is the fundamental bioinformatics challenge for the DCC (Figure 1). With nine different sequencing centers and more than two dozen major analysis groups1, the most important initial challenges are (1) collating all the sequencing data centrally for necessary quality control (QC) and standardization; (2) exchanging the data between participating institutions; (3) ensuring rapid availability of both sequencing data and intermediate analysis results to the analysis groups; (4) maintaining easy access to sequence, alignment and variant files and their associated meta data; and (5) providing these resources to the community.
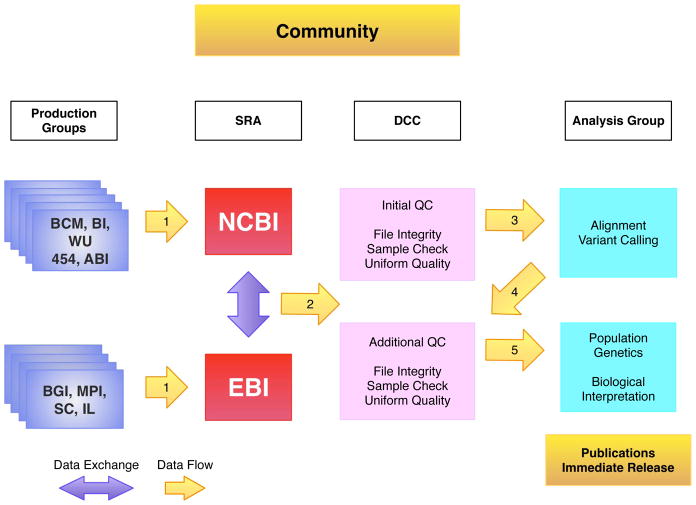
Figure 1.
Data Flow in the 1000 Genomes Project. The sequencing centers submit their raw data to one of the two SRA databases (arrow 1), which exchange data. The DCC retrieves FASTQ files from the SRA (arrow 2) and performs QC steps on the data. The analysis group access data from the DCC (arrow 3), aligns the sequence data to the genome and uses the alignments to call variants. Both the alignment files and variant files are provided back to the DCC (arrow 4). All the data is publically released as soon as possible. Sequencing center names are provided in supplementary table 1.
In recent years, data transfer speeds using TCP/IP-based protocols such as FTP have not scaled with increased sequence production capacity. In response some groups have resorted to sending physical hard drives with sequence data5, although handling data this way is very labor intensive. At the same time data transfer requirements for sequence data remain well below those encountered in physics and astronomy, so building a dedicated network infrastructure was not justified. Instead, the project elected to rely on an Internet transfer solution from Aspera, Inc. (Emeryville, CA), a UDP-based method that achieves data transfer rates 20–30 times faster than FTP in typical usage. Using Aspera, the combined submission capacity of the EBI and NCBI currently approaches 30 Terabytes per day, with both sites poised to grow as global sequencing capacity increases.
The 1000 Genomes Project was responsible for the first multi-terabase submissions to the sequence read archives (SRA): the EBI SRA provided as a service of the European Nucleotide Archive (ENA) and NCBI SRA6. Over the course of the project, the major sequencing centers developed automated data submission methods to either the EBI or the NCBI, while both SRA databases developed generalized methods to search and access the archived data. The data formats accepted and distributed by both the archives and the project have also evolved from the expansive SRF (Sequence Read Format) files to the more compact BAM7 and FASTQ formats (see Table 1). This format shift was made possible by a better understanding of the needs of the project analysis group, leading to a decision to stop archiving raw intensity measurements from read data in order to focus exclusively on base calls and quality scores.
Table 1.
File Formats Used in the 1000 Genomes Project
| File Format | Description | Further information/Citation |
|---|---|---|
| SRF | Container format for data from sequencing machines based on ZTR | http://srf.sourceforge.net/ |
| FASTQ | Text based format for sequence and quality values | http://en.wikipedia.org/wiki/FASTQ_format |
| SAM/BAM | Sequence Alignment and Map format. A compact Alignment format for placement of short read data with respect to a reference genome. The consortium designed this file format. | http://samtools.sourceforge.net/Reference7 |
| VCF | Variant Call Format a column based text format for storing variant calls and individual genotypes for those calls. This file format was designed by the consortium. | http://vcftools.sourceforge.net/Reference9 |
As a “community resource project”8, the 1000 Genomes Project publicly releases prepublication data as described below as quickly as possible. The project has mirrored download sites at the EBI (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp) and NCBI (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp) that provide project and community access simultaneously and efficiently increase the overall download capacity. The master copy is directly updated by the DCC at the EBI, and the NCBI copy is usually mirrored within 24 hours via a nightly automatic Aspera process. Generally users in the Americas will access data most quickly from the NCBI mirror, while users in Europe and elsewhere in the world will have better service from the EBI master.
The raw sequence data, as FASTQ files, appear on the 1000 Genomes FTP site within 48–72 hours after the EBI SRA has processed it. This processing requires that data be available in the EBI SRA, meaning that data originally submitted to the NCBI SRA must first be mirrored at the EBI. Project data is managed though periodic data freezes associated with a dated sequence.index file (supplementary note 1). These files were produced approximately every two months during the pilot phase, while for the full project the release frequency varies depending on the output of the production centers and the requirements of the analysis group.
Alignments based on a specific sequence.index file are produced within the project and distributed via the FTP site in BAM format, while the analysis results are distributed in VCF format9. Index files created by the Tabix software10 are also provided for both BAM and VCF files.
All data on the FTP site has been through an extensive QC process. For sequence data this includes syntax and quality checking of the raw sequence data and sample identity confirmation. For alignment data QC includes file integrity and metadata consistency checking (supplementary note 3).
Data Access
The entire 1000 Genomes Project data set is available and the most logical approach to obtain it is to mirror the contents of the FTP site, which is as of March 2012 more than 260 terabytes. Our experience is that most users are more interested in analysis results and targeted raw data or alignment slices from specific regions of the genome rather than the entire data set. Indeed, the analysis files are distributed via the FTP site in directories named for the sequence.index freeze date they are based on. (supplementary note 4). However, with hundreds of thousands of available files, locating and accessing specific project data by browsing the FTP directory structure can be extremely difficult.
To assist in searching the FTP site we provide a file called current.tree at the root of the FTP site. This file was designed to enable mirroring the FTP site and contains a complete list of all files and directories including time of last update and file integrity information. We developed a web interface (http://www.1000genomes.org/ftpsearch) to provide direct access to the current.tree file using any user-specified sample identifier(s) or other information found in our data file names, which follow a strict convention to aid searching. The search returns full file paths to either the EBI or the NCBI FTP site and supports filters to exclude file types likely to produce a large number of results such as FASTQ or BAM files (supplementary note 5)
For users wanting discovered variants or alignments from specific genomic regions without downloading the complete files, subsections of BAM and VCF files can be obtained either directly with Tabix or via a web-based data-slicing tool (supplementary note 2). VCF files can be further divided by sample name or population using the data-slicer.
1000 Genomes data can be viewed in the context of extensive genome annotation such as protein coding genes and whole genome regulatory information though the dedicated 1000 Genomes browser based on the Ensembl infrastructure11 and available at http://browser.1000genomes.org. The browser displays project variants before they are processed by dbSNP or appear in genome resources such as Ensembl or the UCSC genome browser. The 1000 Genomes browser also provides Ensembl variation tools including the Variant Effect Predictor12 as well as SIFT13 and Polyphen14 predictions for all non-synonymous variants (supplementary note 6). The browser supports viewing of both 1000 Genomes Project and other web-accessible indexed BAM and VCF files in genomic context (Figure 2). A stable archival version of the 1000 Genomes browser based on Ensembl code release 60 and containing the pilot project data is available at http://pilotbrowser.1000genomes.org.
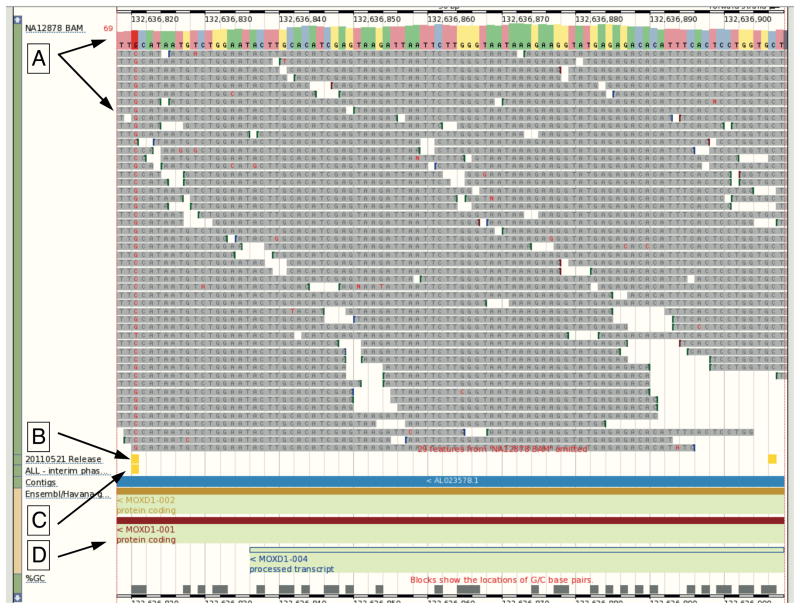
Figure 2.
Remote File Viewing. The 1000 Genomes Browser enables the attachment of remote files to allow accessible BAM and VCF files to be displayed within Location view. The tracks in the image from our October 2011 browser based on Ensembl version 63 are (A) NA12878 BAM file from EBI FTP site with consensus sequence noted by the upper arrow and sequence reads by the lower arrow. (B) Variants from 20110521 release VCF file show as a track with two variants in yellow (C) Variants from the 20101123 release database shown as a track with one variant in yellow (D) Gene annotation from Ensembl showing the genomic context. The ability for users to view data from files allows rapid access to new data before the database can be updated.
The underlying MySQL databases that support the project browser are also publicly available and these can be directly queried or accessed programmatically using the appropriate version of the Ensembl API (supplementary note 7).
Users may also explore and download project data using the NCBI data browser at http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/. The browser displays both sequence reads and individual genotypes for any region of the genome. Sequence for selected individuals covering the displayed region can be downloaded in BAM, SAM, FASTQ or FASTA format. Genotypes can likewise be downloaded in VCF format (supplementary note 8).
The project submits all called variants to the appropriate repositories using the handle “1000GENOMES”. Pilot project SNPs and small indels were submitted to dbSNP15, while structural variation data was submitted to DGVa16. Full project variants will be similarly submitted.
For users of Amazon Web Services (AWS), all currently available project BAM and VCF files are provided as a public data set via s3://1000genomes.s3.aws.com (supplementary note 9).
Finally, all links and announcements of the project data can be found on the project web site http://www.1000genomes.org and announcements are made available via rss (http://www.1000genomes.org/announcements/rss.xml), Twitter @1000genomes and via an email list 1000announce@1000genomes.org (supplementary note 10).
Discussion
Methods of data submission and access developed to support the 1000 Genomes Project offer benefits to all large scale sequencing projects and the wider community. The streamlined archival process takes advantage of the two synched copies of the SRA, which distribute the resource intensive task of submission processing. In addition, the close proximity of the DCC to the SRA ensures that all 1000 Genomes data is made available to the community as quickly as possible and allowed the archives to benefit from the lessons learned by the DCC.
Large-scale data generation and analysis projects can benefit from an organized and centralized data management activity2–4. The goals of such activities are to provide necessary support and infrastructure to the project while ensuring that data is made available as rapidly and widely as possible. In supporting the 1000 Genome Project analysis, an extensive data flow was established that includes multiple tests to ensure data integrity and quality (Figure 1). As part of this process, data is made available to members of the consortium and members of the public simultaneously at specific points in the data flow including at the collection of sequence data and the completion of alignments.
Beyond directly supporting the needs of the project, centralized data management ensures that resources targeted to users outside the consortium analysis group are created. These include the 1000 Genomes Browser at http://browser.1000genomes.org, submission of both preliminary and final variant data sets to dbSNP and to dbVar/DGVa, provisioning of alignment and variant files in the AWS cloud, and centralized variation annotation services.
The experiences of data management employed by the project reflect in part the difficulty of adopting existing bioinformatics systems to new technologies and in part the challenge of data volumes much larger than previously encountered. The rapid evolution of analysis and processing methods is indicative of the community effort to provide effective tools for understanding the data.
Supplementary Material
Acknowledgments
For early work and support to the DCC we thank Z. Iqbal, H. Khouri, F. Cunningham, Y. Chen, W. McLaren, V. Zalunin, R. Radhakrishnan, D. Smirnov, J. Paschall, Z. Belaia, R. Sanders, C. O’Sullivan, S. Keenan, G. Ritchie, G. Cochrane. For maintenance of the EBI computer infrastructure we acknowledge J. Barker, V. Silventoinen, G. Kellman and P. Jokinen. Funding support at the EBI is provided by the Wellcome Trust (grant number WT085532) and the European Molecular Biology Laboratory. This research was supported in part by the Intramural Research Program of the NIH National Library of Medicine.
References
- 1.1000 Genomes Project Consortium. Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Washington NL, Stinson EO, Perry MD, Ruzanov P, et al. The modENCODE Data Coordination Center: lessons in harvesting comprehensive experimental details. Database (Oxford) 2011;2011:bar023. doi: 10.1093/database/bar023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker M. Next-generation sequencing: adjusting to data overload. Nature Methods. 2010;7:495–499. [Google Scholar]
- 6.Shumway M, Cochrane G, Sugawara H. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:D870–D871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Handsaker B, Wysoker A, Fennell T, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toronto International Data Release Workshop Authors. Birney E, Hudson TJ, Green ED, et al. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. The Toronto Agreement describes a set of best practices for prepublication data sharing. These practices have been adopted by the 1000 Genomes Project and have helped drive the widespread use of the data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danecek P, Auton A, Abecasis G, Albers CA, et al. The Variant Call Format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flicek P, Amode MR, Barrell D, Beal K, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaren W, Pritchard B, Rios D, Chen Y, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. The Ensembl Variant Effect Predictor (VEP) provides a flexibel and regularly updated method to annotate all newly discovered variants and provides information about how such variants impact genes, regulatory regions and other key genomic features. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 14.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foelo ML, Sherry ST. In: Genetic Variation: A Laboratory Manual. Weiner MP, Gabriel SB, Stephens JC, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2007. pp. 41–61. [Google Scholar]
- 16.Church DM, Lappalainen I, Sneddon TP, Hinton J, et al. Public data archives for genomic structural variation. Nat Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.