Abstract
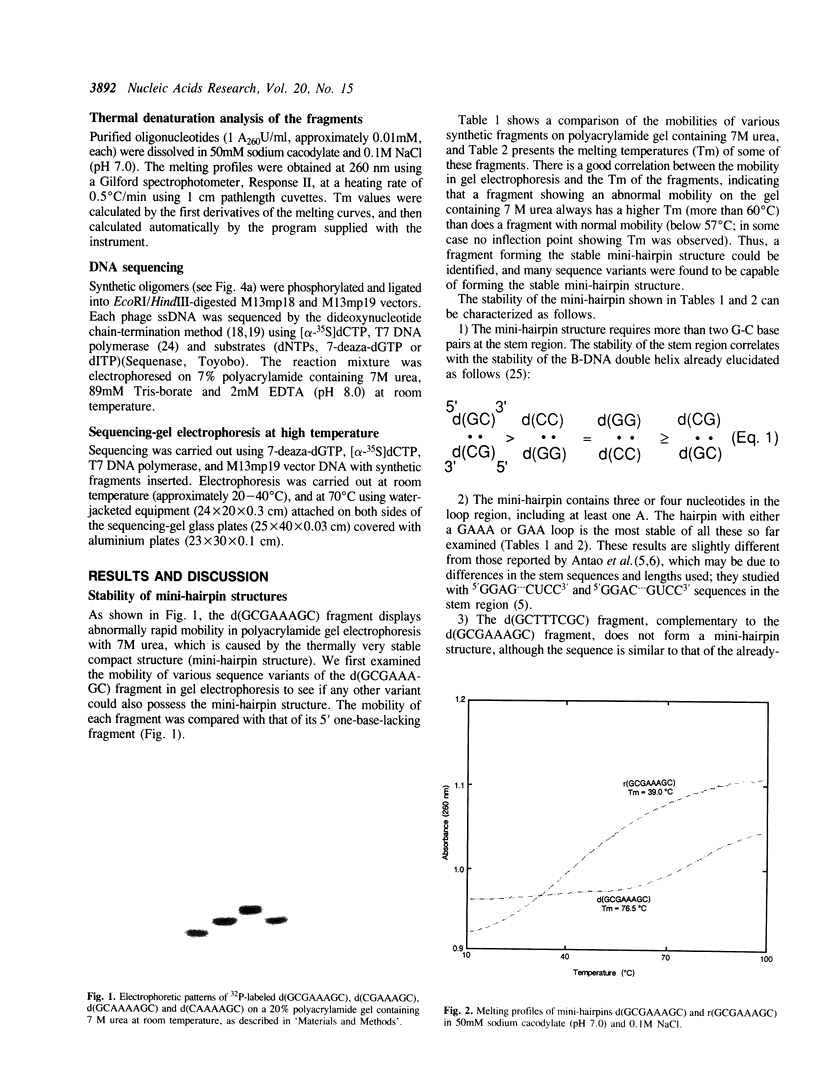
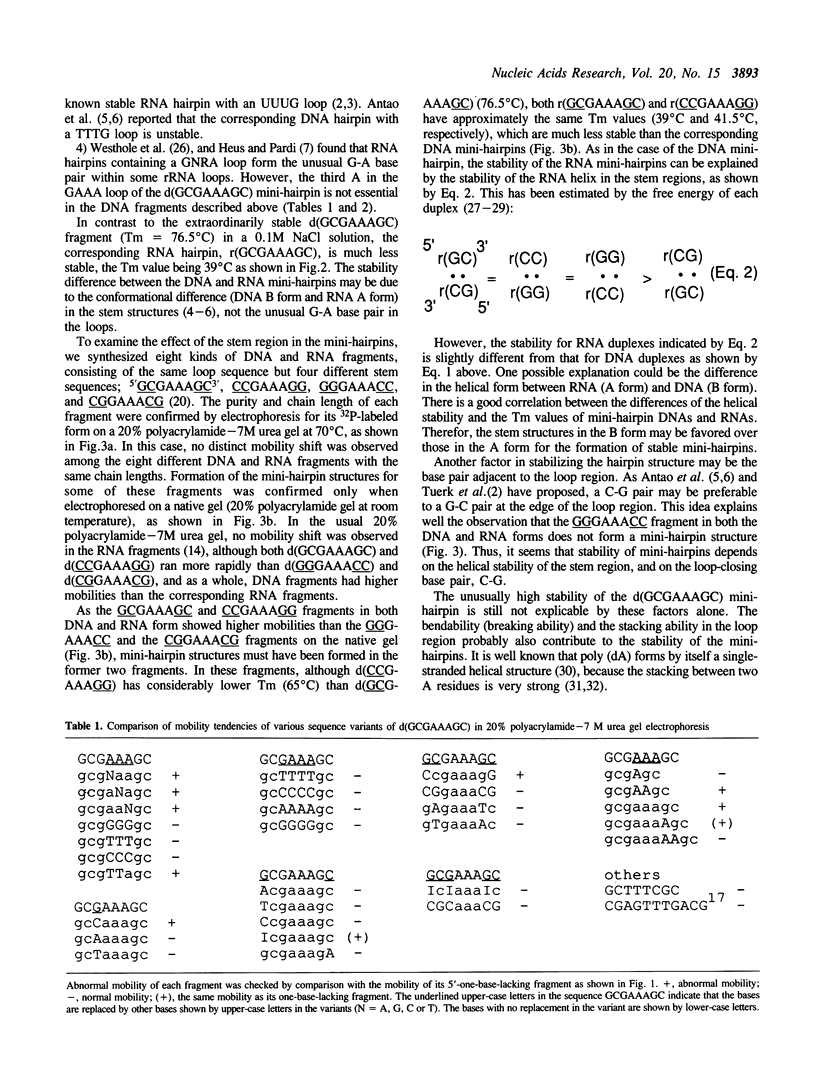
A small DNA fragment having a characteristic sequence d(GCGAAAGC) has been shown to form an extraordinarily stable mini-hairpin structure and to have an unusually rapid mobility in polyacrylamide gel electrophoresis, even when containing 7M urea. Here, we have studied the stability of the various sequence variants of d(GCGAAAGC) and the corresponding RNA fragments. Many such sequence variants form stable mini-hairpins in a similar manner to the d(GCGAAAGC) sequence. The RNA fragment, r(GCGAAAGC) also forms a mini-hairpin structure with less stability. The DNA mini-hairpins with GAAA or GAA loop are much more stable than DNA and RNA mini-hairpins with other loop sequence so far as has been examined. The stability difference between DNA and RNA mini-hairpins may be deduced to the stem structures formed by DNA (B form) and RNA (A form). The stable hairpins consisting of the GCGAAAGC sequence cause strong band compression on the sequencing gel. This phenomenon should be carefully considered in DNA sequencing.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhin M. R., Alblas J., van Duin J. Secondary structure at the 3' terminal region of RNA coliphages: comparison with tRNA. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):110–118. doi: 10.1016/0167-4781(90)90150-z. [DOI] [PubMed] [Google Scholar]
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao V. P., Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992 Feb 25;20(4):819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtereva T. A., Pavlov Y. I., Kramorov V. I., Migunova B., Kiselev O. I. DNA sequencing with thermostable Tet DNA polymerase from Thermus thermophilus. Nucleic Acids Res. 1989 Dec 25;17(24):10507–10507. doi: 10.1093/nar/17.24.10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Bowling J. M., Bruner K. L., Cmarik J. L., Tibbetts C. Neighboring nucleotide interactions during DNA sequencing gel electrophoresis. Nucleic Acids Res. 1991 Jun 11;19(11):3089–3097. doi: 10.1093/nar/19.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Approaches to rapid DNA sequence analysis. Anal Biochem. 1983 Dec;135(2):247–263. doi: 10.1016/0003-2697(83)90680-2. [DOI] [PubMed] [Google Scholar]
- Frank R., Köster H. DNA chain length markers and the influence of base composition on electrophoretic mobility of oligodeoxyribonucleotides in polyacrylamide-gels. Nucleic Acids Res. 1979;6(6):2069–2087. doi: 10.1093/nar/6.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Uhlenbeck O. C., Pardi A. Sequence-dependent structural variations of hammerhead RNA enzymes. Nucleic Acids Res. 1990 Mar 11;18(5):1103–1108. doi: 10.1093/nar/18.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao I., Ishida M., Watanabe K., Miura K. Unique hairpin structures occurring at the replication origin of phage G4 DNA. Biochim Biophys Acta. 1990 Oct 23;1087(2):199–204. doi: 10.1016/0167-4781(90)90205-g. [DOI] [PubMed] [Google Scholar]
- Hirao I., Naraoka T., Kanamori S., Nakamura M., Miura K. Synthetic oligodeoxyribonucleotides showing abnormal mobilities on polyacrylamide gel electrophoresis. Biochem Int. 1988 Jan;16(1):157–162. [PubMed] [Google Scholar]
- Hirao I., Nishimura Y., Naraoka T., Watanabe K., Arata Y., Miura K. Extraordinary stable structure of short single-stranded DNA fragments containing a specific base sequence: d(GCGAAAGC). Nucleic Acids Res. 1989 Mar 25;17(6):2223–2231. doi: 10.1093/nar/17.6.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R. RNA sequencing with radioactive chain-terminating ribonucleotides. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5334–5338. doi: 10.1073/pnas.75.11.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Gilham P. T. Anomalous hairpin formation in an oligodeoxyribonucleotide. Nucleic Acids Res. 1985 Nov 25;13(22):8259–8274. doi: 10.1093/nar/13.22.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie K. K., Usman N., Nicoghosian K., Cedergren R. J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn C. S., Bostelaar L. J., Van Boom J. H., Altona C. Conformational characteristics of the trinucleoside diphosphate dApdApdA and its constituents from nuclear magnetic resonance and circular dichroism studies. Extrapolation to the stacked conformers. Eur J Biochem. 1980 Nov;112(1):95–110. doi: 10.1111/j.1432-1033.1980.tb04991.x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn C. S., Haasnoot C. A., Altona C. Circular dichroism studies of 6-N-methylated adenylyladenosine and adenylyluridine and their parent compounds. Thermodynamics of stacking. Eur J Biochem. 1980 May;106(1):85–95. doi: 10.1111/j.1432-1033.1980.tb05999.x. [DOI] [PubMed] [Google Scholar]
- Powell J. T., Richards E. G., Gratzer W. B. The nature of stacking equilibria in polynucleotides. Biopolymers. 1972 Jan;11(1):235–250. doi: 10.1002/bip.1972.360110118. [DOI] [PubMed] [Google Scholar]
- Sakata T., Hiroaki H., Oda Y., Tanaka T., Ikehara M., Uesugi S. Studies on the structure and stabilizing factor of the CUUCGG hairpin RNA using chemically synthesized oligonucleotides. Nucleic Acids Res. 1990 Jul 11;18(13):3831–3839. doi: 10.1093/nar/18.13.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior M. M., Jones R. A., Breslauer K. J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripkin E. A., Adhin M. R., de Smit M. H., van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990 Jan 20;211(2):447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G., van Boom J. DNA hairpin loops in solution. Correlation between primary structure, thermostability and reactivity with single-strand-specific nuclease from mung bean. Nucleic Acids Res. 1991 Apr 11;19(7):1505–1511. doi: 10.1093/nar/19.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]