Abstract
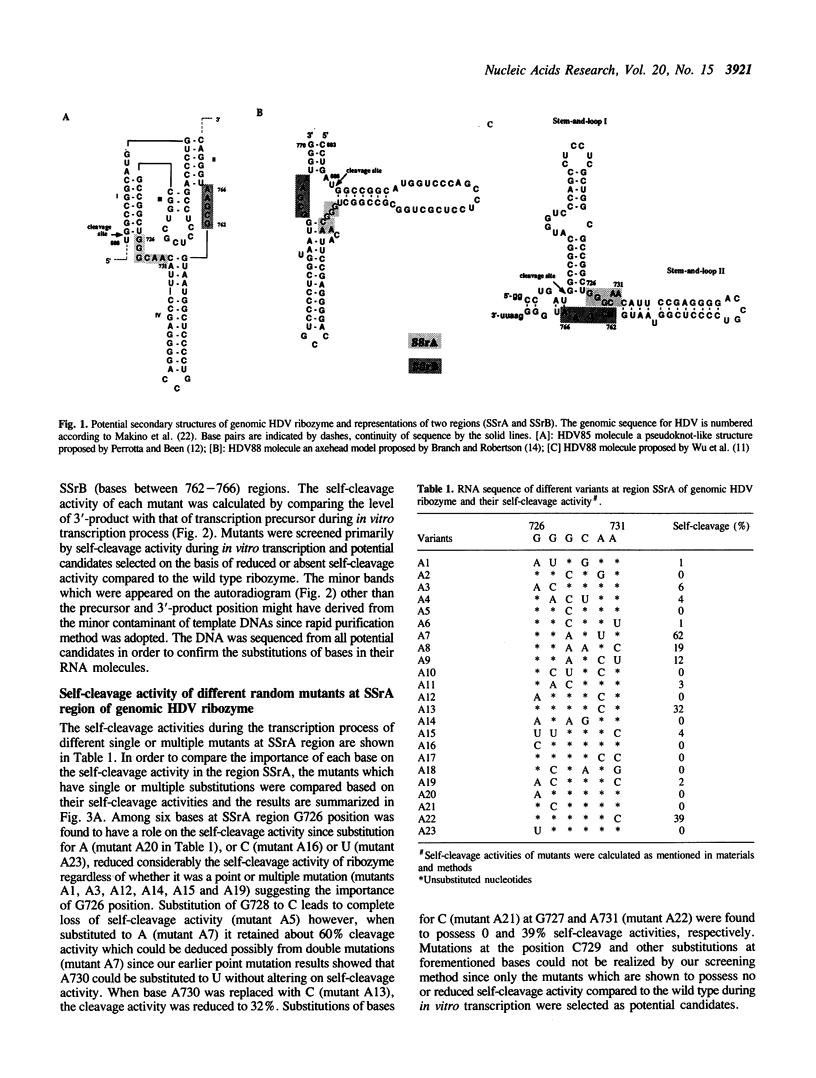
In elucidating function of two important single-stranded regions [SSrA (726-731 nt) and SSrB (762-766 nt)] derived mainly from three secondary structure models in genomic hepatitis delta virus (HDV) ribozyme possessing self-cleavage activity, we have constructed several random mutants at those two regions on the HDV88 molecule (683-770 nt) by oligonucleotide-directed mutagenesis. When self-cleavage activities were compared among mutants, at the region SSrA, G726 was found to play an important role during cleavage reaction since substitutions of the base to A (mutant A20) or C (mutant A16) or U (mutant A23), reduced the ribozyme activity to very low levels suggesting the importance of G726 position. C763 at SSrB region was found to play a more significant role during catalysis than G726 (at region SSrA) since any substitutions at C763 completely inactivated the ribozyme. Other bases located in these two regions could be substituted to other bases at the expense of some self-cleavage activity. The results presented here together with our previous deletion analysis indicate that these two regions may play an important role during cleavage process.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branch A. D., Robertson H. D. Efficient trans cleavage and a common structural motif for the ribozymes of the human hepatitis delta agent. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10163–10167. doi: 10.1073/pnas.88.22.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldstein P. A., Buzayan J. M., van Tol H., deBear J., Gough G. R., Gilham P. T., Bruening G. Specific association between an endoribonucleolytic sequence from a satellite RNA and a substrate analogue containing a 2'-5' phosphodiester. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2623–2627. doi: 10.1073/pnas.87.7.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Govindarajan S., Chin K. P., Redeker A. G., Peters R. L. Fulminant B viral hepatitis: role of delta agent. Gastroenterology. 1984 Jun;86(6):1417–1420. [PubMed] [Google Scholar]
- Hampel A., Tritz R., Hicks M., Cruz P. 'Hairpin' catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990 Jan 25;18(2):299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. Y., Sharmeen L., Dinter-Gottlieb G., Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol. 1988 Dec;62(12):4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Chang M. F., Shieh C. K., Kamahora T., Vannier D. M., Govindarajan S., Lai M. M. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature. 1987 Sep 24;329(6137):343–346. doi: 10.1038/329343a0. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991 Apr 4;350(6317):434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. Cleavage of oligoribonucleotides by a ribozyme derived from the hepatitis delta virus RNA sequence. Biochemistry. 1992 Jan 14;31(1):16–21. doi: 10.1021/bi00116a004. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 1990 Dec 11;18(23):6821–6827. doi: 10.1093/nar/18.23.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein S. P., Been M. D. Evidence that genomic and antigenomic RNA self-cleaving elements from hepatitis delta virus have similar secondary structures. Nucleic Acids Res. 1991 Oct 11;19(19):5409–5416. doi: 10.1093/nar/19.19.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein S. P., Been M. D. Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry. 1990 Sep 4;29(35):8011–8016. doi: 10.1021/bi00487a002. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Sharmeen L., Kuo M. Y., Dinter-Gottlieb G., Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol. 1988 Aug;62(8):2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. B., Dinter-Gottlieb G. Antigenomic Hepatitis delta virus ribozymes self-cleave in 18 M formamide. Nucleic Acids Res. 1991 Mar 25;19(6):1285–1289. doi: 10.1093/nar/19.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y. A., Kumar P. K., Nishikawa F., Kayano E., Nakai S., Odai O., Uesugi S., Taira K., Nishikawa S. Deletion of internal sequence on the HDV-ribozyme: elucidation of functionally important single-stranded loop regions. Nucleic Acids Res. 1992 Feb 25;20(4):747–753. doi: 10.1093/nar/20.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. Self-cleavage of RNA in the replication of small pathogens of plants and animals. Trends Biochem Sci. 1989 Nov;14(11):445–450. doi: 10.1016/0968-0004(89)90103-5. [DOI] [PubMed] [Google Scholar]
- Taira K., Uchimaru T., Tanabe K., Uebayasi M., Nishikawa S. Rate limiting P-O(5') bond cleavage of RNA fragment: ab initio molecular orbital calculations on the base-catalyzed hydrolysis of phosphate. Nucleic Acids Res. 1991 May 25;19(10):2747–2753. doi: 10.1093/nar/19.10.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira K., Uebayasi M., Maeda H., Furukawa K. Energetics of RNA cleavage: implications for the mechanism of action of ribozymes. Protein Eng. 1990 Aug;3(8):691–701. doi: 10.1093/protein/3.8.691. [DOI] [PubMed] [Google Scholar]
- Thill G., Blumenfeld M., Lescure F., Vasseur M. Self-cleavage of a 71 nucleotide-long ribozyme derived from hepatitis delta virus genomic RNA. Nucleic Acids Res. 1991 Dec 11;19(23):6519–6525. doi: 10.1093/nar/19.23.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebayasi M., Uchimaru T., Tanabe K., Nishikawa S., Taira K. Preferential chelation of cationic ligands to axial-equatorial oxygens over equatorial-equatorial dianionic oxygens: implication to the mechanism of action of ribozymes. Nucleic Acids Symp Ser. 1991;(25):107–108. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Lai M. M. RNA conformational requirements of self-cleavage of hepatitis delta virus RNA. Mol Cell Biol. 1990 Oct;10(10):5575–5579. doi: 10.1128/mcb.10.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Lin Y. J., Lin F. P., Makino S., Chang M. F., Lai M. M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Wang Y. J., Hung C. F., Lee H. J., Lai M. M. Sequence and structure of the catalytic RNA of hepatitis delta virus genomic RNA. J Mol Biol. 1992 Jan 5;223(1):233–245. doi: 10.1016/0022-2836(92)90728-3. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Perreault J. P., Labuda D., Usman N., Cedergren R. Mixed DNA/RNA polymers are cleaved by the hammerhead ribozyme. Biochemistry. 1990 Dec 25;29(51):11156–11160. doi: 10.1021/bi00503a002. [DOI] [PubMed] [Google Scholar]




