Abstract
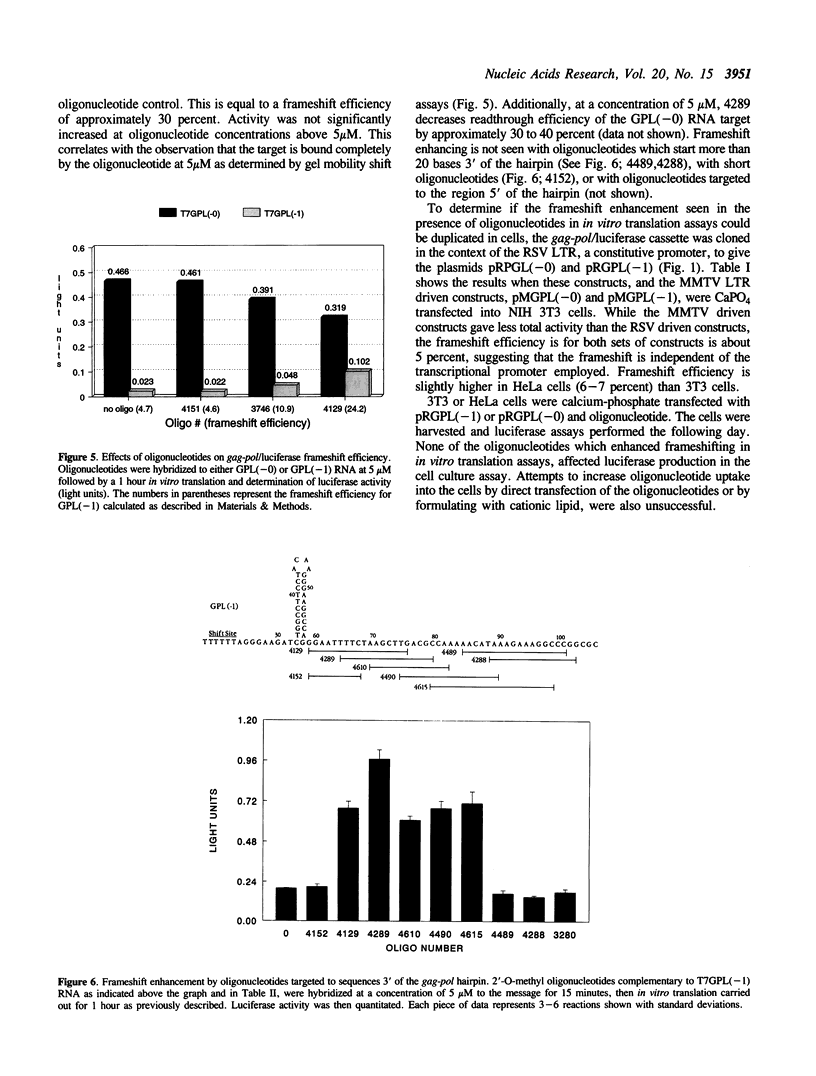
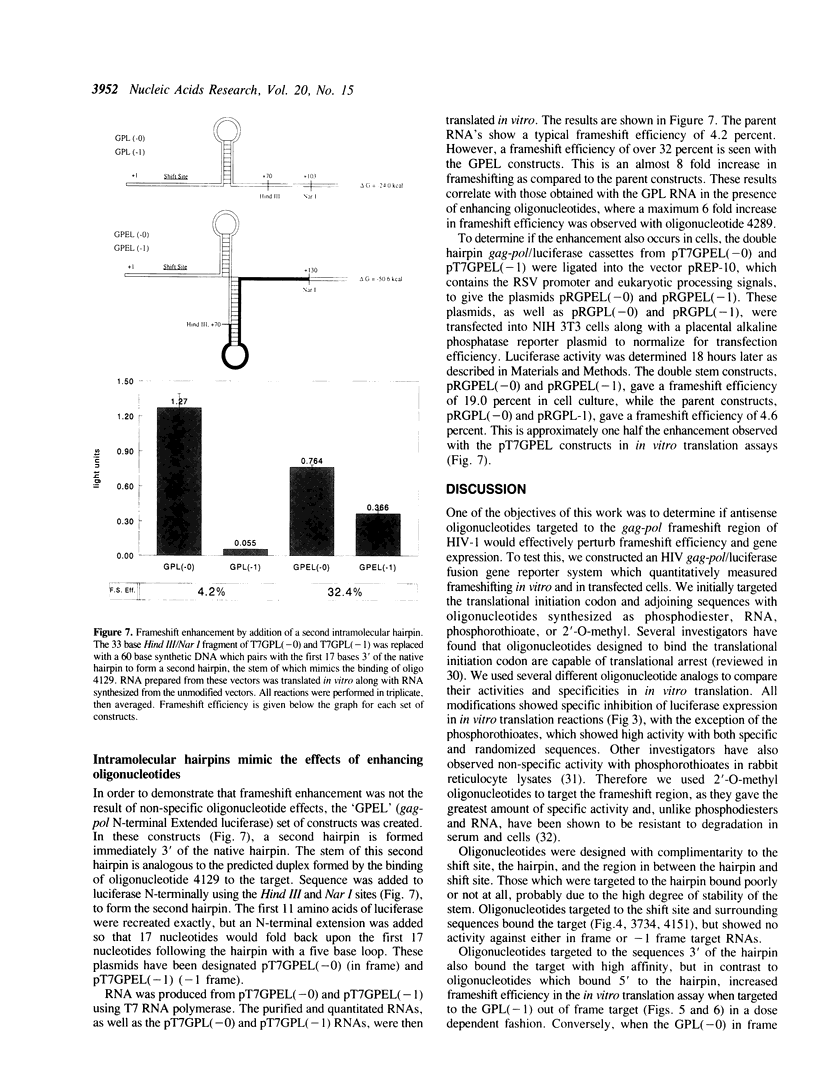
The pol gene of all retroviruses is expressed as a gag-pol fusion protein which is proteolytically processed to produce all viral enzymes. In the human immunodeficiency virus (HIV), the gag and pol genes overlap by 241 nucleotides with pol in the -1 phase with respect to gag. The gag-pol fusion is produced via a -1 ribosomal frameshifting event that brings the overlapping, out-of-phase gag and pol genes into translational phase. Frameshifting occurs at a so called 'shift site' 8-10 nucleotides upstream of a hairpin loop which may play a role in the regulation of frameshifting. We have fused this region of HIV-1 to the 5' end of the firefly luciferase reporter gene in order to quantitatively measure ribosomal frameshifting both in cells and by in vitro translation. A series of 2'-O-methyl oligonucleotides was designed to specifically bind the sequences which flank the gag-pol hairpin. Ribosomal frameshifting is enhanced up to 6 fold by those oligonucleotides which bind the area just 3 to the stem. Oligonucleotides which bind 5' to the stem have no effect on frameshift efficiency. In addition, we have constructed a series of fusion genes which mimic the effect of the bound oligonucleotides with intramolecular hairpins. The results suggest that increasing RNA secondary structure downstream of the shift site increases the frequency of ribosomal frameshifting, and that this effect can be mimicked by antisense oligonucleotides.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agrawal S., Goodchild J., Civeira M. P., Thornton A. H., Sarin P. S., Zamecnik P. C. Oligodeoxynucleoside phosphoramidates and phosphorothioates as inhibitors of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7079–7083. doi: 10.1073/pnas.85.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazenave C., Loreau N., Toulmé J. J., Hélène C. Anti-messenger oligodeoxynucleotides: specific inhibition of rabbit beta-globin synthesis in wheat germ extracts and Xenopus oocytes. Biochimie. 1986 Sep;68(9):1063–1069. doi: 10.1016/s0300-9084(86)80180-8. [DOI] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H. E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K. M., Goff S. P. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988 Jun;62(6):2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D. L., Levin J. G., Rein A., Oroszlan S. Translational suppression in retroviral gene expression. Adv Virus Res. 1992;41:193–239. doi: 10.1016/S0065-3527(08)60037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Feng Y. X., Lee B. J., Rein A., Levin J. G., Oroszlan S. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1989 Dec;173(2):736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henthorn P., Zervos P., Raducha M., Harris H., Kadesch T. Expression of a human placental alkaline phosphatase gene in transfected cells: use as a reporter for studies of gene expression. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6342–6346. doi: 10.1073/pnas.85.17.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988 Jan 21;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le S. Y., Chen J. H., Maizel J. V. Thermodynamic stability and statistical significance of potential stem-loop structures situated at the frameshift sites of retroviruses. Nucleic Acids Res. 1989 Aug 11;17(15):6143–6152. doi: 10.1093/nar/17.15.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre M., Bayard B., Lebleu B. Specific antiviral activity of a poly(L-lysine)-conjugated oligodeoxyribonucleotide sequence complementary to vesicular stomatitis virus N protein mRNA initiation site. Proc Natl Acad Sci U S A. 1987 Feb;84(3):648–652. doi: 10.1073/pnas.84.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. Translational frameshift suppression in mouse mammary tumor virus and other retroviruses. Enzyme. 1990;44(1-4):320–331. doi: 10.1159/000468768. [DOI] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Cabradilla C. D., Benton C. V., Lasky L. A., Capon D. J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985 Feb 7;313(6002):450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- Murphy E. C., Jr, Kopchick J. J., Watson K. F., Arlinghaus R. B. Cell-free synthesis of a precursor polyprotein containing both gag and pol gene products by Rauscher murine leukemia virus 35S RNA. Cell. 1978 Feb;13(2):359–369. doi: 10.1016/0092-8674(78)90204-0. [DOI] [PubMed] [Google Scholar]
- Philipson L., Andersson P., Olshevsky U., Weinberg R., Baltimore D., Gesteland R. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978 Jan;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Reil H., Hauser H. Test system for determination of HIV-1 frameshifting efficiency in animal cells. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):288–292. doi: 10.1016/0167-4781(90)90183-3. [DOI] [PubMed] [Google Scholar]
- Rice N. R., Stephens R. M., Burny A., Gilden R. V. The gag and pol genes of bovine leukemia virus: nucleotide sequence and analysis. Virology. 1985 Apr 30;142(2):357–377. doi: 10.1016/0042-6822(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Sproat B. S., Lamond A. I., Beijer B., Neuner P., Ryder U. Highly efficient chemical synthesis of 2'-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989 May 11;17(9):3373–3386. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers T., Baker B. F., Cook P. D., Zounes M., Buckheit R. W., Jr, Germany J., Ecker D. J. Inhibition of HIV-LTR gene expression by oligonucleotides targeted to the TAR element. Nucleic Acids Res. 1991 Jun 25;19(12):3359–3368. doi: 10.1093/nar/19.12.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B. Ribosomal frameshifting, jumping and readthrough. Curr Opin Cell Biol. 1991 Dec;3(6):1051–1055. doi: 10.1016/0955-0674(91)90128-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]