Abstract
Human bone marrow stem cell (BMSC) may be directed to differentiate into multiple cell types, including adipocyte, chondrocyte, osteocyte and photoreceptor, among others. At present, little is known about the features of the BMSC and the protein control mechanism underlying their differentiation into photoreceptor-like cells. In the present study, BMSCs are induced to differentiate into photoreceptor-like cells in an in vitro model simulating the in vivo microenvironment. Up to 32 proteins are identified and differentially expressed through two-dimensional difference gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to establish a differential protein database for photoreceptor-like cells from BMSC-induced differentiation. Western blot analysis further confirms the expression of some of the identified proteins. The present study proposes the total protein expression and possible molecular mechanism during the differentiation of BMSCs into photoreceptor cells.
Keywords: bone marrow stem cell, induced to differentiate, photoreceptor-like cells
INTRODUCTION
Human bone marrow stem cells (BMSCs) are multipotent stem cells. They can be directionally differentiated into fat, cartilage, bone, and muscle cells, and so on[1]-[4]. Recently, many scholars have induced BMSC to express a specific marker in photoreceptor cells[5]-[6]. BMSC differentiation into photoreceptor cells is complex and requires a series of protein changes and information control. Currently, information regarding the features of BMSCs and the underlying protein control mechanism of their differentiation into photoreceptor cells is scarce. With emerging technology and the development of the proteome, proteomics technologies have been widely applied to study of the induction of BMSC differentiation. In the present study, BMSCs are induced and differentiated into photoreceptor-like cells of the retina by simulating the microenvironment of the retina in vivo. Two-dimensional difference gel electrophoresis (2D-DIGE) technology is adopted to compare the proteins expressed during the in vitro BMSC differentiation into photoreceptor-like cells to establish a differential protein expression profile. A total of 37 protein spots with significant differential expression are found. Up to 32 differential proteins are completely identified through matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis.
MATERIALS AND METHODS
Human BMSCs strains cultured and differentiated to photoreceptor-like cell
Human BMSC strains were incubated in mesenchymal stem cell medium (MSCM) in an incubator with 100% humidity cultured. The liquid was changed every three days. When the cultures reached 80%-85% confluence, the solution was digested with 0.25% trypsin and subcultured in the proportion of 1:2.
P4 human retinal pigment epithelial (RPE) cells were cultured in 10% fetal bovine serum + RPMI 1640 culture medium. When the RPE cells reached approximately 95% confluence, the cultivation liquid was collected and filtered using a 0.22 µm mesh and mixed with MSCM containing 20ng/mL each of basic fibroblast growth factor, epidermal growth factor, and brain-derived neurotrophic factor at a ratio of 1:4 to prepare the conditioned medium[6].
Human BMSC strains from P2 to P4 were obtained for experimentation. The cultured BMSCs in the conditioned medium were used as inducted group, whereas the MSCM culture was used as the negative control. An inverted phase contrast microscope was used to observe cellular morphology and differentiation in both groups.
Immunocytochemistry
The cell adhesion sheets were collected at 3, 5, and 7 days after induction, fixed, and then lysed with 1% Triton-100. Primary anti-rhodopsin antibodies were added at a ratio of 1:200. The sheets were maintained overnight at 4°C. The sheets were washed with phosphate-buffered saline (PBS). Biotinylated, horseradish peroxidase (HRP)-conjugated anti-immunoglobulin secondary antibodies were added to the culture for 30 minutes at 37°C, and then they were washed with PBS. The samples were stained with 3,3′-diaminobenzidine and hematoxylin at room temperature for color development. For each index, 5 microscope fields were randomly chosen to calculate the percentage of positively staining.
Real-time PCR
Primer express 2.0 software was adopted to design the primer and probe. An ABI 3900 table-type high-flux DNA synthesizer was used to synthesize the primer. The cells with maximum induced positive staining (7 days) were adopted as the experimental group. BMSCs cultured with MSCM were considered as the control group. Then, 1×105 cells were collected for each group. Trizol was used to extract total RNA from the cells. A Revert AidTM First Strand cDNA Synthesis Kit was adopted to reverse-transcribe the total RNA and synthesize first strand cDNA using an SYBR® Premix Ex Taq-™ kit. A Thermal Cycler Dice® Real-Time System was used for real-time PCR. Predenaturation was performed at 95°C for 30 seconds and at 95°C for 5 seconds. Annealing was conducted for 40 cycles at 60°C for 30 seconds.
2D-DIGE and MALDI-TOF-MS analysis
Protein Extraction
Approximately 1×107 cells were collected from the experimental and control groups. The protein extraction was conducted as follows: The cultivation liquid was removed and the cells were washed thrice with PBS. The cells were disintegrated with a cell lysis solution of ACK lysis buffer and ultrasonicated in an ice bath. The cells were fractionated through centrifugation at 12 000rpm for 20 minutes at 4°C. The supernate was collected and precipitated overnight at -20°C after the addition of acetone. Then, the solution was centrifuged at 12000 rpm for 2 minutes at 4°C and was then allowed to precipitate. The protein content was determined using a 2D-Quant kit based on the manufacturer's instructions.
2D-DIGE two-dimensional electrophoresis
To determine the intensity of DIGE fluorescence at 4°C, 50µg of proteins were obtained from the experimental and control groups, respectively, and placed into two expanded Eppendorf tubes and 25µg each of the samples were mixed in one tube. Then, Cy3 and Cy5 dyes were added to each tube, respectively, and Cy2 dye was added into the mixed sample. Lysine (1µL) was added when ice reaction without any radiation was made, and to terminate ice reaction. After mixing the loading solutions evenly, they were centrifuged for 8 minutes at 12 000×g, and the upper layer was obtained for isoelectric focusing. From 300 V for 45 minutes, 700 V for 45 minutes, 1500 V for 1.5 hours, and then increased to 9 000 V, to reach a total of 9 000 Vh. Then, the immobilized pH gradient strip equilibration was performed. The conditions for the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) were as follows: S1) 2-2.5 W/gel for 45 minutes, S2) 17 W/gel for around 4.5 hours, and S3) bromophenol blue was at the bottom at 10°C without any radiation. For staining, the proteins were separated based on the variations in isoelectric point and molecular weight (MW). The 2-DE gel was stained with Deep Purple™ Total Protein Stain. Then, the images were sanned with a Typhoon 9 400 at maximum gray value of 60 000-90 000.
Treatment of Protein Spots
Imagemaster 7.0 was used to analyze the spots that had more than 1.5-fold differences in intensity. The selected protein spots from the gel were placed in Eppendorf tubes for dehydration after Coomassie staining and destaining. Then, the spots were swelled in 20ng/µL of ice-cold trypsin for 30 minutes, and 10µL of covering liquid was added at 37°C for enzyme cleavage for 12-16 hours. The spots were ultrasonicated for 15 minutes. Then, the covering liquid was aspirated, and an extraction solution was added. The specimens were shaken for 10 minutes to dehydrate and aspirate the extraction solution. The solution was then vacuum-dried and kept at -20°C. Then, 1.5µL of heavy solution was added into the dried sample to dissolve completely the peptide segments. Samples were repeatedly aspirated into the small hole of a steel target several times. The specimens were fed into a mass spectrometer for detection after 0.5µL of ground substance was added into samples and were completely dried.
Protein Determination
An ABI 4800 MALDI-TOF Mass Spectrometer from US was used to conduct the mass spectrographic analyses. The conditions were as follows: UV light at 355nm, a repetition rate of 200Hz, an accelerating voltage of 20 KV, and an optimum resolution of 1500Da. The signals were recorded within a mass scanning range of 700 to 3200Da. The autonomous peak of trypsinogen was used as the internal standard to normalize the mass spectrometer. All mass spectrograms were obtained in default mode.
The Mascot distiller software was used to filter the baseline peak and to identify the signal peak. The Mascot software from Matrix Science was used to search the IPI database and to identify related proteins and their functions. The search conditions were as follows: the peptide mass was limited to within 800-4000Da and the maximum allowable MW error of peptide fragment was controlled in ±50ppm. The enzymatic fragment was selected incompletely as one, and the error ranges of the apparent PI and Mr were unlimited. The species was human, and the MH and monoisotopic were selected as ions. The fixed modification was set to carbamidomethyl whereas the variable modification was set to oxidation.
Western blot analysis
Protein concentration was adjusted based on the total protein concentrations of the experimental and control groups. Equivalent protein samples were obtained and separated by SDS-PAGE. The specimens were transferred onto polyvinylidene difluoride membranes and blocked with 5% skim milk powder. Rabbit anti-human zyxin antibodies and rabbit anti-human β-tropomyosin antibodies were added. Then, HRP-labeled sheep anti-mouse secondary antibodies (Zhongshan Goldenbridge Biotechnology Co., Ltd.) were added. Mouse anti-human β-actin monoclonal antibodies were used as the internal control, following the 2 procedures used above. Quantity One was used to conduct a semiquantitative analysis of the reaction bank and record the measured gray values.
RESULTS
Morphologic Changes in BMSCs during Differentiation into Photoreceptor-like Cells
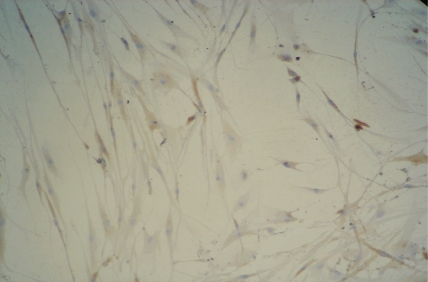
BMSCs from P2 to P8 had uniformly fusiform configurations and grew rapidly. After subculturing for 2 days, the number of cells increased logarithmically. After 4-5 days, the cells grew as clone-like cluster or fish stock with up to 80%-90% confluence. In the control group, after the BMSCs were induced for 2 days, the cellular morphology gradually changed. Their cells shrank, and neurites formed. The cells evidently partially shrank after 5 days, and the cellular appendages presented triangular or simple bipolar and multipolar cells. After 7 days, some cells were fusiform, and the bipolar and multipolar cells became evidently longer(Figure 1,2). Synapses, similar to those in cone and rod cells, were found in the protruding end. For 1 to 3 days, the cell propagation rate was slower than that of the control group, which evidently began slowly after 4-5 days. After 7 days, some cells started undergoing apoptosis. There was no evident change in the BMSC configuration in the control group.
Figure 1. P3 BMSCs showed cluster and fish block-like morphology, and growth of clony forming unit-fibroblastic 7d after passage.
Figure 2. BMSCs displayed bipolar or multipolar in the morphology 7d after induction (×200).
Immunohistochemistry of induced cells and real-time PCR identification
Immunohistochemistry was used to visualize rhodopsin expression in the experimental group.
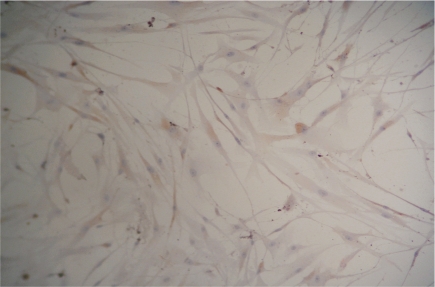
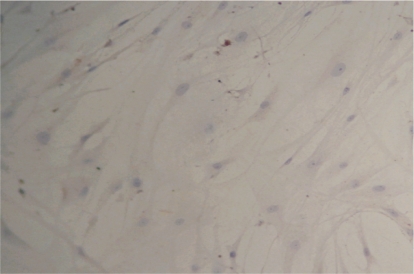
Induction rates of the positive cells at 3, 5, and 7 days were 4.83%±0.29%, 25.36%±0.32%, and 28.87%±0.43%, respectively(Figure 3,4). In the control group, rhodopsin expression was negative(Figure 5). The rhodopsin and recoverin mRNA were clearly expressed in the experimental group, whereas no expression was observed in the control group(Figure 6,7). This shows that photoreceptor-like cells from BMSC differentiation express the distinctive marker of normal photoreceptor cells, and the process can be used as model for culturing human photoreceptor cells in vitro (Table 1).
Figure 3. ICH staining detected the expression of rhodopsin 5d after induction.
Figure 4. ICH staining detected the expression of rhodopsin 7d after induction.
Figure 5. ICH staining for the expression of rhodopsin in control BMSCs.
Figure 6. Amplification curves for expression of rhodopsin in the inducted and non-inducted BMSCs.
Figure 7. Amplification curves for expression of recoverin in the inducted and non-inducted BMSCs.
Table 1. Primers for real-time RT-PCR.
| Gene name | Primer sequence (forward and reverse) | Product size |
| Rhodopsin | 5′-CGAGCGGTACGTGGTGGTGT-3′ | 445bp |
| 5′-GGAGCCCTGGTGGGTGAAGA-3′ | ||
| Recoverin | 5′-TGTGTTCCGCAGCTTCGATT-3′ | 369bp |
| 5′-TGAGGCTCAAACTGGATCAG-3′ | ||
| β-actin | 5′-AAAGACCTGTACGCCAACACAG-3′ | 556bp |
| 5′-TTTTAGGATGGCAAGGGACTTC-3′ |
Identification of protein by 2D-DIGE dimensional electrophoresis
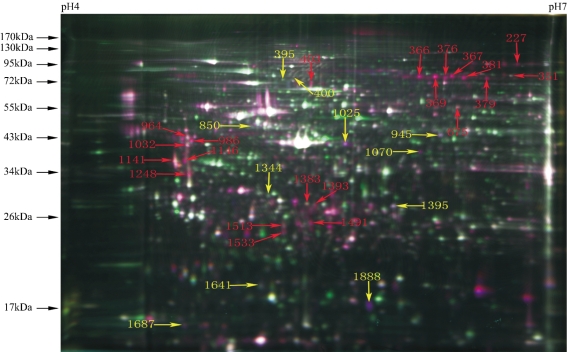
To ensure the repeatability of the 2D-DIGE, the experiments were performed in triplicate. The results indicate that the experiment is highly repeatable. A total of 1914 isoelectric points were obtained in 4-7 proteins after the 2D-DIGE gel was scanned at three distinct wavelengths. Based on the statistical analysis, there were 37 protein spots with more than 1.5-fold differences, and 32 differential proteins were identified through mass spectrometry. Among them, 11 proteins were upregulated and 21 proteins were downregulated during BMSC differentiation (Figure 8). Table 2 lists the related information about the 32 protein spots and the MW gi of the protein spots from left to right.
Figure 8. MS identification of 32 differentially expressed protein spots (Numbers represent the Spot ID of identified proteins, Red for down-regulated and yellow for up-regulated).
Table 2. The information on the 32 proteins identified by MALDI-TOF-MS.
| Spot ID | Protein name | MASCOT Algorithm |
Peptides | Sequence Coverage |
MW | PI | Accession NO | ||
| MASCOT score | P (%) | (%) | |||||||
| 227 | zyxin | 138 | 3.7e-09 | 5 | 1.10E+01 | 62436 | 6.22 | 4508047 | |
| 351 | stress-induced-phosphoprotein 1 | 182 | 1.5e-13 | 6 | 9% | 63227 | 6.4 | 5803181 | |
| 366 | NAG22 protein | 168 | 3.7e-12 | 5 | 17% | 56486 | 5.73 | 13186201 | |
| 367 | NAG22 protein | 339 | 2.9e-29 | 5 | 14% | 56486 | 5.73 | 13186201 | |
| 369 | NAG22 protein | 415 | 7.4e-37 | 6 | 15% | 56486 | 5.73 | 13186201 | |
| 376 | NAG22 protein | 196 | 5.8e-15 | 4 | 14% | 56486 | 5.73 | 13186201 | |
| 379 | prelamin-A/C isoform 1 precursor | 257 | 4.6e-21 | 18 | 32% | 74380 | 6.57 | 27436946 | |
| 381 | zyxin | 203 | 1.2e-15 | 4 | 9% | 62436 | 6.22 | 4508047 | |
| 395 | unnamed protein product | 133 | 4.8e-23 | 3 | 10% | 53642 | 5.08 | 10438296 | |
| 400 | PREDICTED: hypothetical protein isoform 4 | 284 | 9.3e-24 | 9 | 22% | 61784 | 4.93 | 114581740 | |
| 403 | unnamed protein product | 314 | 9.3e-27 | 20 | 32% | 65037 | 5.47 | 221039676 | |
| 675 | T-complex protein 1 subunit beta isoform 2 | 316 | 5.8e-27 | 5 | 15% | 53027 | 6 | 311771535 | |
| 850 | thioredoxin domain-containing protein 5 isoform 3 | 403 | 1.2e-35 | 7 | 22% | 36725 | 5.32 | 224493972 | |
| 945 | GDP dissociation inhibitor 2, isoform CRA_a | 616 | 5.8e-57 | 21 | 49% | 48680 | 7.51 | 119606836 | |
| 964 | reticulocalbin-1 precursor | 167 | 4.6e-12 | 6 | 22% | 38866 | 4.86 | 4506455 | |
| 986 | Chain A, Bm-40, FsEC DOMAIN PAIR | 313 | 1.2e-26 | 9 | 35% | 27852 | 5.53 | 2624793 | |
| 1032 | reticulocalbin-1 precursor | 268 | 3.7e-22 | 8 | 31% | 38866 | 4.86 | 4506455 | |
| 1070 | PREDICTED: gelsolin-like capping protein isoform 9 | 205 | 7.4e-16 | 7 | 21% | 38779 | 5.88 | 55597035 | |
| 1141 | beta tropomyosin | 391 | 1.8e-34 | 17 | 38% | 29980 | 4.7 | 6573280 | |
| 1146 | tropomyosin alpha-4 chain isoform 1 | 405 | 7.4e-36 | 13 | 31% | 32874 | 4.69 | 223555975 | |
| 1025 | cytochrome b-c1 complex subunit1, mitochondrial precursor | 341 | 1.8e-29 | 7 | 31% | 53297 | 5.94 | 46593007 | |
| 1248 | Chain C, Human Pcna | 159 | 2.9e-11 | 9 | 42% | 29072 | 4.53 | 2914385 | |
| 1344 | EF-hand domain-containing protein D2 | 551 | 1.8e-50 | 13 | 38% | 26795 | 5.15 | 20149675 | |
| 1383 | cathepsin D preproprotein | 267 | 4.6e-22 | 6 | 15% | 45037 | 6.1 | 4503143 | |
| 1393 | cathepsin D preproprotein | 214 | 9.3e-17 | 7 | 15% | 45037 | 6.1 | 4503143 | |
| 1395 | glutathione S-transferase omega-1 isoform 1 | 181 | 1.8e-13 | 5 | 19% | 27833 | 6.23 | 4758484 | |
| 1491 | cathepsin B | 172 | 1.5e-12 | 3 | 30% | 17428 | 5.44 | 741376 | |
| 1513 | keratin 1 | 127 | 4.6e-08 | 6 | 16% | 66198 | 8.16 | 11935049 | |
| 1533 | Chain A, Ternary Complex y Of An Crk Sh2 Domain, Crk-Derived Phophopeptide, And Abl Sh3 Domain B Nmr Spectroscopy | 81 | 0.0017 | 4 | 46% | 12122 | 6.79 | 253722243 | |
| 1641 | mitochondrial heat shock 60kD protein 1 variant 1 | 201 | 1.8e-15 | 5 | 15% | 60813 | 5.83 | 189502784 | |
| 1687 | myosin, light polypeptide 6, alkali, smooth muscle and non-muscle, isoform CRA_d | 120 | 2.3e-07 | 4 | 25% | 13264 | 4.47 | 119617308 | |
| 1888 | alpha-2-macroglobulin | 462 | 6.9e-040 | 6 | 5% | 168953 | 5.71 | 157954061 | |
| 1888 (2) | alpha-2-macroglobulin | 166 | 5.5e-11 | 4 | 4% | 168953 | 5.71 | 157954061 | |
Identification of functional classification of protein
The 32 differentially expressed proteins were classified into 7 categories: cytoskeletal proteins, molecular chaperones, kinases for energy metabolism, signal transduction pathway-related proteins, cell proliferation proteins, differentiation and apoptosis-related proteins, calcium-binding and ion channel proteins, and other proteins.
Verification of Western blot analysis results
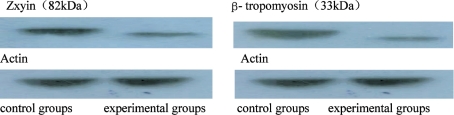
In the present study, Western blot analysis was performed to verify further the differential expression of some proteins. Positive bands at 82 and 33kDa in the experimental group were detected. These values were slower than that of the control group, indicating that zxyin and β-tropomyosin were downregulated after induction (Figure 9).
Figure 9. Expression of Zxyin and β-tropomyosin by Western blot.
DISCUSSION
Photoreceptor cells, namely cones and rods, consist of five parts, an outer segment, connecting cilium, inner segment, body, and synapses. Each outer segment covers more than 700 flat membranous discs. The outer segments of rod cells are cylindrical whereas they are cone-shaped in cone cells. The configurations of some cells in the experimental group were altered after induction for 2 days. After 5 days, some cells shrank with obvious proptosis. There were experimental in some cells after 7 days, and bipolar and multipolar cells became evidently longer. The synapse is found at the protruding end, similar to those in cone and rod cells. Immunohistochemistry and RT-PCR verified that the induced cells expressed markers specific to photoreceptor cells, e.g., rhodopsin and recoverin; it also showed the differentiation of the induced cells into photoreceptor cells.
BMSC differentiation into photoreceptor-like cells is a complex process that requires a series of protein changes and information adjustment. Proteomics technology can be used to probe the differential expression of proteins and the mechanism of BMSC differentiation into photoreceptor-like cells in the dynamic state and from an integral aspect. We established an in vitro induction system of human BMSC differentiation to photoreceptor-like cells. Up to 37 protein spots that are clearly differentially expressed were found using 2D-DIGE. Additionally, 32 differentially expressed proteins were identified by MALDI-TOF-MS analysis. Among these proteins, 11 proteins were upregulated, whereas 21 proteins were downregulated. A differentially expressed proteinogram was established after 7 days of BMSC differentiation into photoreceptor-like cells. In the proteinogram, the downregulation of p38, chain C, and human proliferating cell nuclear antigen (PCNA) was evident, and the upregulation of myosin, mitochondrial heat shock 60kDa protein 1 variant 1, thioredoxin domain-containing protein 5 isoform 3, Cytochrome B-C1 complex subunit 1, mitochondrial, and EF-hand domain-containing protein D2 was observed. These results indicate important adjustments during the induction.
The p38/mitogen-activated protein kinase (MAPK) pathway is an important and highly conserved signal system for the external signaling of eukaryotic cells. This pathway belongs to the silk/serine-threonine protein kinase family, which is present in most cells. Members of the MAPK family consist of extracellular signal-regulated protein kinase (ERK), p38MAPK, C-jun amino terminal/stress activated protein kinase (JNK/SAPK), and enzymes in the mitogen-activated protein kinases1 (BMKl/ERK5) pathway[7]. The p38 signaling pathway, activated by an external stimulus (such as ultraviolet radiation, radioactive ray, heat shock and proinflammatory factor and physiological stress), is called the MAPK emergence signal channel. This pathway is one of the recent research hotspots in signal transduction. Through the cascade reaction of upstream multi-stage kinases, an extracellular signal is transferred intracellularly, resulting in the activation of p38. This signal pathway controls the activity of many downstream transcription factors, such as transcription factors involved in activation, NF-κB, and growth arrest, as well as DNA damage gene. Recent studies have shown that the p38 phosphorylated pathway is an important common signal pathway for cell differentiation. Bai[8] found that the p38 pathway plays a role in promoting MSC differentiation into epidermoid cells. The blockage of upstream signal Rho may increase the activation of the p38 pathway, and promotes MSC differentiation into epidermoid cells to a certain degree. The present study shows the evident downregulation of p38 during BMSC differentiation. The result shows that p38 signaling pathway plays an important role in regulating the differentiation of BMSC into photoreceptor cells.
Chain C and human PCNA
PCNA is a protein that participates in DNA replication during cell proliferation. This protein is localized in the cell nucleus and is a cyclic trimer that consists of three similar subunits. PCNA is not expressed in cells during G0-G1. PCNA upregulation occurs during late G1 and peaks during the S period, whereas rapid downregulation is observed during G2-M. At the start of eukaryotic DNA replication, PCNA enables DNA polymerase δ and uses the DNA slide in its loop to instruct the continuous synthesis of the guiding chain. PCNA, which is only present in normal proliferative and tumor cells, plays an important role in initiating cell proliferation and is a better index for reflecting the vegetative state of cells. In addition, apoptosis-related protein p21 competes for the binding site of DNA polymerase δ of PCNA to induce the cells to exit G1 and enter the S period, resulting in the direct blockage of DNA synthesis and inducing cell apoptosis. Recently, there has been increasing interest in PCNA studies. In the present experiment, evident PCNA downregulation was observed during the differentiation of BMSCs into photoreceptor-like cells. BMSCs have strong proliferative capacities. BMSCs increase logarithmically 2 d after the passage culture. After 4-5d, the BMSCs proliferated by up to 85%-90%, whereas the multiplication capacity of the induced cells was evidently decreased. These results conform to the trend of PCN expression.
Mitochondrial heat shock 60 kDa protein 1 variant 1
Heat shock proteins (HSPs), a group of highly conserved proteins, exist widely in prokaryotic and eukaryotic cells and they are also called stress proteins or molecular chaperones. Approximately 70%-80% HSP60 of eukaryotic cell are mainly located in the mitochondria, and 20%-30% is in the endochylema[9]. As two of the most important molecular chaperone proteins in the mitochondria, 2 HSP60 heptamer and 1 HSP10 heptamer form a high-molecular-weight polymer called large respirator, which is an important structure for mitochondrial protein refolding and restoration. Liang et al. found that HSP60 in the endochylema participated in the synthesis and protection of cytoskeletal proteins, such as microtubulin and actin. HSP60 has a dual regulatory function, anti-apoptosis and pro-apoptosis. HSP60 physiologically plays a role in anti-apoptosis by maintaining the normal conformation and function of anti-apoptotic factors, thereby inhibiting the activation of pro-apoptotic factors and reducing the formation of reactive oxygen species in the mitochondria. In the pathologic state, HSP60 expression and its subcellular localization, as well as its pro-apoptotic effects are changed. Many studies have indicated that HSP60 in is a stress protein released by BMSCs that are close to undergoing necrosis and apoptosis[10],[11]. HSP60 expression therefore indicates the presences of necrotic and apoptotic cells among the BMSCs. HSP60 protein 1 variant 1 was evidently upregulated in the mitochondria during the differentiation of BMSCs into photoreceptor-like cells, illustrating that there were more necrotic and apoptotic cells among the induced cells than the non-induced BMSCs. In the present experiment, we observed a reduction in the multiplication rate of the induced cells compared with that of the non-induced BMSCs. This may be caused by the increase in the number of necrotic and apoptotic cells among the BMSCs, which will lower the multiplication rate of cells. Thioredoxin domain-containing protein 5 isoform 3 (Trx).
Trx is a highly conservative and ubiquitously expressed small protein with a molecular weight 13kDa, and an active disulphide center was used to catalyze the oxidation-reduction reaction. Trx maintains the oxidation reduction equilibrium of cells by facilitating the scavenging of free radicals, enabling the cell to withstand H2O2 and its toxic effects to reduce apoptosis. This protein forms a complex with ASKl, thereby inhibiting ASKI and preventing apoptosis[12]. Additionally, Trx plays an important role in signal transduction by regulating MAPK, as well as the activation of transcription factor and other functions. In the present experiment, BMSCs were induced to differentiate into photoreceptor-like cells. The oxidation-reduction system of cells was activated because of the changes in metabolism. Trx was evidently upregulated, which indicates that it plays a role in maintaining the oxidation-reduction equilibrium of cells and the prevention of apoptosis.
Cytochrome BC1 complex subunit 1, mitochondrial
The mitochondrial respiratory chain consists of four complexes, including cytochrome C, and coenzyme Q. The cytochrome B-C1 complex refers to complex III, also known as the CoQH2-cytochrome C reductase complex. This complex transmits 4 H+ to the intermembrane space for oxidation-reduction when a pair of electrons is transmitted. This complex moves continuously in the mitochondrial inner membrane, so it has no stable structure. This protein was evidently upregulated, which indicates that it participates in the oxidation-reduction reaction.
EF-hand domain-containing protein D2
The EFh-domain containing protein D2 was first found in lymphocytes[13] and later in different tissues. It is highly expressed in marrow[14]. The B-cell receptor is adjusted to induce immature B-cell and apoptosis of its original B-cell, and BCL2L1 is adjusted to control natural apoptosis. Zhang et al[15] found that the EF-hand domain-containing protein D2 is upregulated during MSC differentiation into osteoblast and proved that it plays an important role during BMSC differentiation. In the present experiment, EFh-domain containing protein D2 clearly increased after induction, indicating that it plays an important role during BMSC differentiation.
Myosin, light polypeptide 6, alkali, smooth muscle and non-muscle, isoform CRA_d
Myosin, which exists widely in muscle and non-muscle cells, combines and interacts with actin to hydrolyze adenosine-5′-triphosphate, guanosine-5′-triphosphate, cytidine triphosphate, and ect enzyme. This protein supplies power for muscle contraction, cytoplasmic streaming, organelle movement, material transport, and mitosis. Myosin is also called the molecular motor of cytoskeleton. The present study found that myoglobulin significantly increased during the differentiation of BMSCs into photoreceptor-like cells, indicating that the cytoskeleton polymerizes and depolymerizes continuously with variations in the cellular morphology. Myosin increases significantly to supply additional power to adjust the polymerization of actin. In the present experiment, cytoskeletal proteins such as zyxin, NAG22, prelamin-A/C isoform 1 precursor, β-tropomyosin, tropomyosin alpha-4 chain isoform 1, and CK-1 significantly decreased. This may have resulted from the termination of the karyokinesis of mature cells and cell cycle retardation, thereby increasing myosin. The increase in myosin expression likely regulates actin polymerization. Based on the research analysis, the downregulation or upregulation of proteins enables the continuous polymerization and depolymerization of the cytoskeleton during BMSC differentiation. This behavior leads to the change in the cellular morphology of the BMSCs from the initial fusiform into the spindle-shaped photoreceptor-like cells. Studies have shown that mechanical tension is used to control the differentiation of stromal stem cells through the actin microfilament complex[16]. The proteins are closely related to the highly diverse biological action of cells, stabilization, and signal conduction. However, further research is needed to determine their mechanisms and their interactions.
In summary, the differential protein profile of photoreceptor-like cells from BMSC-induced differentiation was established by adopting a differential proteomics method. A total of 32 differentially expressed proteins were identified, among which 11 were upregulated and 21 proteins were downregulated. The present study provides important information for determining the exact mechanism of differentiation of BMSCs into photoreceptor-like cells. Further investigation should be performed to ascertain the functions of these proteins.
Footnotes
Foundation items: National Natural Science Foundation of China(No.81070715); Innovative Platform Foundation of Fujian Province, China(No.2010Y2003)
REFERENCES
- 1.Xie MS, Xu GX, Huang Y. Rat Model for Anterior Segment Intraocular Surgery Induced Blood-Retinal Barrier Breakdown. Ophthalmologica. 2008;222:42–47. doi: 10.1159/000109278. [DOI] [PubMed] [Google Scholar]
- 2.Xie MS, Xu GX. Culture in vitro Modulation of human leukocyte antigen molecules and costimulatory molecules on human retinal pigment epithelium. Ophthalmologica. 2008;222:48–52. doi: 10.1159/000109279. [DOI] [PubMed] [Google Scholar]
- 3.Sasportas LS, Kasmieh R, Wakimoto H, Hingtgen S, van de Water JA, Mohapatra G, Figueiredo JL, Martuza RL, Weissleder R, Shah K. Assessment of therapeutic efficacy and fate of engineered human mesenchymal stem cells for cancer therapy. Proc Natl Acad Sci USA. 2009;106:4822–4827. doi: 10.1073/pnas.0806647106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amado LC, Saliaris AP, Schuleri KH, St M, Xie JS, Cattaneo S, Durand DJ, Fitton T, Kuang JQ, Stewart G, Lehrke S, Baumgartner WW, Martin BJ, Heldman AW, Hare JM. Cardiac repair with intramyocardial injection of allogeneic mesenchymal stem cells after myocardial infarction. Proc Natl Acad Sci USA. 2005;102:11474–11479. doi: 10.1073/pnas.0504388102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SR, Kim YR, Kang HS, Yim SH, Park CI, Min YH, Lee BH, Shin JC, Lim JB. Functional recovery after the transplantation of neurally differentiated mesenchymal stem cells derived from bone barrow in a rat model of spinal cord injury. Cell Transplant. 2009;18:1359–1368. doi: 10.3727/096368909X475329. [DOI] [PubMed] [Google Scholar]
- 6.Ladak A, Olson J, Tredget EE, Gordon T. Differentiation of mesenchymal stem cells to support peripheral nerve regeneration in a rat model. Exp Neurol. 2011;228:242–252. doi: 10.1016/j.expneurol.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Abdallah BM, Kassem M. Human mesenchymal stem cells: from basic biology to clinical applications. Gene Ther. 2008;15:109–116. doi: 10.1038/sj.gt.3303067. [DOI] [PubMed] [Google Scholar]
- 8.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 9.Wilson A, Shehadeh LA, Yu H, Webster KA. Age-related molecular genetic changes of murine bone marrow mesenchymal stem cells. BMC Genomics. 2010;11:229. doi: 10.1186/1471-2164-11-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop DJ, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 11.Halfon S, Abramov N, Grinblat B, Ginis I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011;20:53–66. doi: 10.1089/scd.2010.0040. [DOI] [PubMed] [Google Scholar]
- 12.Boucher SE. Simplified PCR assay for detecting early stages of multipotent mesenchymal stromal cell differentiation. Methods Mol Biol. 2011;698:387–403. doi: 10.1007/978-1-60761-999-4_28. [DOI] [PubMed] [Google Scholar]
- 13.Vuadens F, Gasparini D, Déon C, Sanchez JC, Hochstrasser DF, Schneider P, Tissot JD. Identification of specific proteins in different lymphocyte populations by proteomic tools. Proteomics. 2002;2(1):105–111. [PubMed] [Google Scholar]
- 14.Vuadens F, Rufer N, Kress A, Corthésy P, Schneider P, Tissot JD. Identification of swiprosin 1 in human lymphocytes. Proteomics. 2004;(8):2216–2220. doi: 10.1002/pmic.200300779. [DOI] [PubMed] [Google Scholar]
- 15.Zhang AX, Yu WH, Ma BF, Yu XB, Mao FF, Liu W, Zhang JQ, Zhang XM, Li SN, Li MT. Proteomic identification of differently expressed proteins responsible for osteoblast differentiation from human mesenchymal stem cells. Mol Cell Biochem. 2007;304(1-2):167–179. doi: 10.1007/s11010-007-9497-3. [DOI] [PubMed] [Google Scholar]
- 16.Rowena McB, Dana M P, Celeste M N, Kiran B, Christopher SC. Cell shape cytoskeletal tension and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]