Background and Scope
The brain contains vast numbers of interconnected neurons that constitute anatomical and functional networks. Structural descriptions of neuronal network elements and connections make up the “connectome” of the brain (Hagmann, 2005; Sporns et al., 2005; Sporns, 2011), and are important for understanding normal brain function and disease-related dysfunction. A long-standing ambition of the neuroscience community has been to achieve complete connectome maps for the human brain as well as the brains of non-human primates, rodents, and other species (Bohland et al., 2009; Hagmann et al., 2010; Van Essen and Ugurbil, 2012). A wide repertoire of experimental tools is currently available to map neural connectivity at multiple levels, from the tracing of mesoscopic axonal connections and the delineation of white matter tracts (Saleem et al., 2002; Van der Linden et al., 2002; Sporns et al., 2005; Schmahmann et al., 2007; Hagmann et al., 2010), the mapping of neurons organized into functional circuits (Geerling and Loewy, 2006; Ohara et al., 2009; Thompson and Swanson, 2010; Ugolini, 2011), to the identification of cellular-level connections, and the molecular properties of individual synapses (Harris et al., 2003; Arellano et al., 2007; Staiger et al., 2009; Micheva et al., 2010; Wouterlood et al., 2011). But despite the numerous connectivity studies conducted through many decades we are still far from achieving comprehensive descriptions of the connectome across all these levels. There is increasing awareness that new neuroinformatics tools and strategies are needed to achieve the goal of compiling the brain’s connectome, and that any such effort will require systematic, large-scale approaches (Bohland et al., 2009; Akil et al., 2011; Zakiewicz et al., 2011; Van Essen and Ugurbil, 2012).
Systematic literature mining to compile and share complete overview of known connections in the macaque brain was pioneered by Rolf Kötter and co-workers (Stephan et al., 2001, 2010). While yielding promising results (Kötter, 2004; Bota et al., 2005; van Strien et al., 2009), more coordinated efforts are needed to collect, organize, and disseminate connectome data sets. To this end, there is an urgent need to develop and identify neuroinformatics approaches that allow different levels of connectivity data to be described, integrated, compared, and shared within the broader neuroscience community.
This Research Topic of Frontiers in Neuroinformatics, dedicated to the memory of Rolf Kötter (1961–2010) and his pioneering work in the field of brain connectomics, comprises contributions that elucidate different levels of connectivity analysis (from MRI-based methods, through axonal tracing techniques, to mapping of functional connectivity in relation to detailed 3-D reconstructions of individual neurons), and point to several recent methodological advances and neuroinformatics-related challenges at the level of data mining, management, and integration. In this Editorial, we review the advances represented in these reports, and discuss some of the grand challenges in this emerging field.
Advances in Multi-Level Connectivity Mapping
Sophisticated neuroimaging techniques have opened up new possibilities to infer structural and functional connectivity at a macroscopic scale. Through measurement of oriented water diffusion restricted by cellular elements in the brain, non-invasive methods based on diffusion magnetic resonance imaging (dMRI, Figures 1A,B) play a key role in current neuroanatomical efforts to explore the human connectome (Hagmann et al., 2010; Van Essen and Ugurbil, 2012). The different dMRI tractography methods proposed so far still require time-consuming manual intervention and supervision that may compromise reliability. To overcome this problem, Yendiki et al. (2011)1 present a method for automated probabilistic reconstruction of white matter pathways that incorporates a priori anatomical knowledge, and demonstrate automatic tractography analyses in schizophrenia patients and healthy subjects (Figure 1B). The ability to perform dMRI tractography without manual intervention will greatly facilitate studies with very large populations, which will be essential for establishing a connectome for the human brain (Marcus et al., 2011) as well as for improving early diagnostic imaging in brain disease.
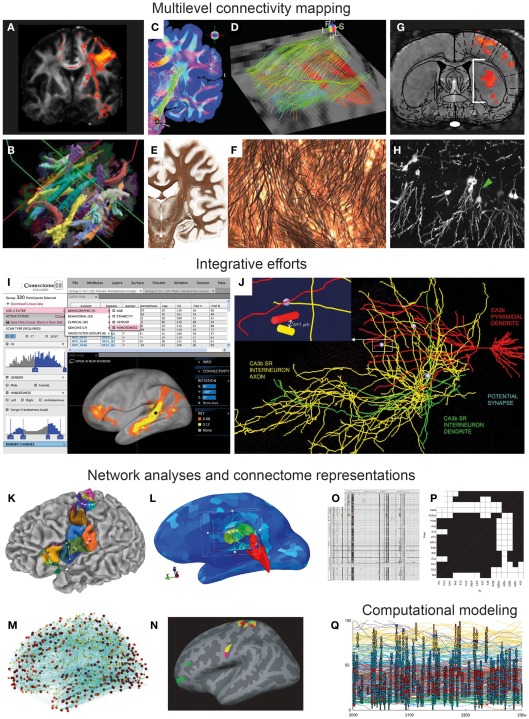
Figure 1.
From multi-modal connectivity data to integrated connectomes. Image panels illustrating recent progress, selected and modified from contributions to the Research Topic “Mapping the Connectome” as specified below. (A–F) Connectivity mapping methods in the human brain span from indirect in vivo assessment of fiber tracts from diffusion MRI data [(A,B) Gorbach et al., 2011; Yendiki et al., 2011] to ex vivo mapping of detailed fiber architectures [(C,E) Axer et al., 2011b; (E,F) Annese, 2012]. (G–H) Novel experimental methods in animal models include combined optogenetic and functional MRI mapping of specific connections (G) (Lee, 2011) and high-resolution histological imaging [(H) Chung et al., 2011]. (I–J) Examples of recent efforts to accumulate, integrate, and share connectivity data, represented by the graphical user interface of the Human Connectome Project [(I) Marcus et al., 2011], and a data mining effort combining 3-D reconstructions of hippocampal neurons to explore potential synaptic connections [(J) Ropireddy and Ascoli, 2011]. (K–N) Recent sophisticated approaches to network analysis, based on connectivity-based cortical parcelation [(K) Gorbach et al., 2011], different connectivity visualizations using open-source tools [(L,M) Gerhard et al., 2011], and identification of structural network motifs [(N) Echtermeyer et al., 2011]. (O–P) Updated connectome matrix representations from large-scale data mining efforts for the whole rat brain [(O) Bota et al., 2012] and for the rat hippocampal region [(P) Sugar et al., 2011]. (Q) Visualization of computational model of a tadpole spinal cord connectome (Borisyuk et al., 2011).
Estimates of “functional networks” described on the basis of statistical associations derived from time series data (neuronal recordings) represent another important category of approaches to define the human brain connectome. The relationship of anatomical to functional networks is explored by Daffertshofer and van Wijk (2011). Using computational modeling of large-scale neural networks these authors argue that patterns of synchronization should be analyzed in the context of changes in local amplitude to improve prediction of brain dynamics from structure. In a related paper, Segall et al. (2012) also employ statistical methods and independent component analysis to describe spatial correspondences between gray matter density measurements and resting state functional MRI signal fluctuations recorded from a very large group of healthy subjects. But while associations between several structural and functional features can be observed (Segall et al., 2012), the anatomical substrates underlying such indirect in vivo measurements remain obscure and require further investigation.
In this respect, novel methodologies for characterizing microstructural aspects of the human and primate brains may provide new ways to bridge the gap between post mortem microscopic and in vivo macroscopic and functional measures reflecting neural connectivity. Polarized light imaging (PLI) of histological sections allows quantitative analysis of fiber orientations with very-high spatial resolution. Axer et al. (2011a) demonstrate automated 3-D reconstruction of fiber orientations across multiple histological sections in the human brain stem, yielding highly resolved datasets that are useful complements for both conventional histological stains and DTI data (Figures 1C,D). Using the same approach, Axer et al. (2011b) show how 3-D PLI derived fiber orientation vectors can subsequently be used as a basis for high-resolution tractography of fiber tracts, potentially suitable for bridging microscopic and macroscopic connectome representations. The importance of correlating various non-invasive MRI derived measurements to cellular-level morphological data is also emphasized by Annese (2012), presenting the perspective that whole-brain histological maps (Figures 1E,F) created using large-scale digital microscopy spanning several histological modalities will support the analysis and interpretation of MRI-based connectivity studies. The potential of high-throughput and very-high-resolution histological methods for creating multiscale representations of brain data is further demonstrated by Chung et al. (2011), who use knife-edge scanning microscopy to section and reconstruct microscopic brain data at sub-micrometer resolution (Figure 1H), within volumes that can span an entire mouse brain. The inherently multiscale nature of the acquired data sets demand sophisticated visualization and analysis tools for integrating cellular to systems scales, a challenge that is addressed with the introduction of a web-based neuroinformatics platform (Chung et al., 2011). It will be a further, substantial challenge to extract actual connectivity information from these histological representations.
Recent advances in optogenetic functional MRI (ofMRI) allow non-invasive, selective mapping of brain circuit elements that are triggered on the basis of genetic markers, anatomic location, or axonal projection target. In a perspective article, Lee (2011) outlines the potential of ofMRI to enhance system level mapping and our understanding of neural circuits. The combination of the unique ability of optogenetics to selectively control cellular activity and of the mapping of hemodynamic responses with fMRI, opens exiting new possibilities for in vivo functional circuit analysis in animals (Figure 1G), as well as diverse models of neurological disease. The approach potentially has considerable advantages over classical axonal tracing methods, by allowing the investigation of network dynamics and longitudinal investigations of development and aging, as well as monitoring of neural network changes occurring in disease models.
Integrative Efforts: Assembling Connectomes
The methodological diversity of current (functional and structural) approaches to mapping connectomes represents a major challenge to the field, and comparison of data across scales, modalities, and species remains a formidable problem (Sporns, 2011). It is therefore a long-standing ambition of the neuroinformatics community to provide new tools and approaches for integrating neuroscience data (Akil et al., 2011; Van Essen and Ugurbil, 2012). Several of the contributions included in the “Mapping the Connectome” Research Topic present novel ways to visualize and assemble connectome data.
In their technology report, Marcus et al. (2011) of the Human Connectome Project consortium centered at Washington University and the University of Minnesota2 present their plans to non-invasively assess the long range connections of the human brain using diffusion MRI data from large numbers of subjects, supplemented by structural MRI, genetic, and behavioral data. An important first step for this ambitious project is to develop an informatics platform for storing, visualizing, and analyzing massive amounts of data (Figure 1I). A key element will be to share the open-source platform and associated tools with the community, to allow further hypothesis-driven analysis and data mining in this multi-modal pool of data. A particular challenge for future data integration and knowledge synthesis will be to achieve interoperability with other neuroinformatics tools and data collections.
The challenge of integrating and visualizing human connectome data of different modalities has been addressed by Gerhard et al. (2011). Their flexible open-source Connectome Viewer Toolkit3 allows integrated storage and viewing of different types of imaging data and structured metadata. The toolkit is based on a container file format, and linked to a suite of existing tools libraries allowing data mining, viewing, and comparison. The authors present several example analyses using diffusion MRI data that are integrated, manipulated, and analyzed (Figures 1L,M). This freely available open-source toolkit stands out as a powerful workbench platform for future integration of human connectome data coming from multi-modal neuroimaging.
Another approach to combine and integrate various neuroanatomical data is to use a common reference space (Bjaalie, 2002; Hawrylycz et al., 2011). In their contribution to this Research Topic, Ropireddy and Ascoli (2011) demonstrate how potential connections (defined by close apposition of axonal and dendritic segments) can be inferred from a collection of detailed 3-D reconstructions of neuronal extensions that were accumulated in a common 3-D reconstruction of the rat hippocampus on the basis of positional values (Figure 1J). The major innovation lies in estimating this aspect of neuronal connectivity at high-resolution not only over a narrow field of view, but across a macroscopic brain region, the rat hippocampus. The paper reports on anatomical differences across different locations in the hippocampus and points to possible generalizations of the approach to other regions of the brain.
Axonal connections have been investigated experimentally in animal models for many decades, yielding valuable data reported in a huge number of publications. While the textual format of such reports make it difficult to extract formal knowledge and compare connectivity data, this pool of laboriously collected experimental results remains important to utilize. Rolf Kötter’s pioneering work, particularly the creation of the CoCoMac database4, is a major milestone on the way to creating comprehensive and accessible online repositories for connectome data (Stephan et al., 2001, 2010). The availability of this repository of primate cortical connections has spurred a large number of projects in the analysis of complex brain connectivity (e.g., Modha and Singh, 2010) as well as the computational modeling of associated brain dynamics (e.g., Cabral et al., 2011; Deco and Jirsa, 2012).
Several groups have invested in further ambitious data mining projects to aggregate connectome data from legacy data (Sugar et al., 2011; Tallis et al., 2011; Bota et al., 2012). Over several years, Bota and co-workers have developed a publicly available neuroinformatics system, called the brain architecture knowledge management system (BAMS), which currently contains >50,000 connectivity reports from the rat brain (Figure 1O). The BAMS system5 provides a valuable framework in which curated connectivity data can be stored and retrieved. In their review article, Bota et al. (2012) provide an update on recently added data and functionality, and discuss general methodology and strategy for producing global connection matrices.
Building upon their earlier work in the hippocampus (van Strien et al., 2009), Sugar et al. (2011) present an interactive connectome of hippocampal and parahippocampal connections. The authors have extracted ∼2600 descriptions of hippocampal connections from 226 published reports, and assembled them into a versatile, searchable application providing a comprehensive description of all known network elements in this region (Figure 1P). Overcoming a range of challenges related to level of detail, incongruent and incomplete reports, and diverse use of nomenclatures, Sugar et al. provide the most comprehensive connectome description so far for a specific rat brain region. The efforts of these hippocampal experts present an excellent case study for how connectivity data should be assembled for other brain regions.
The process of extracting integrated knowledge representations from connectivity data requires a framework for standardized data descriptions, such as a common atlas space and consistent terminologies for neuroanatomical entities. Tallis et al. (2011) have combined two pre-existing rat brain atlas systems (Dashti et al., 2001; Burns et al., 2006) with a data management system (Russ et al., 2011) into a software system for synthesizing knowledge based on neural connectivity data. This system provides access to experimental tract-tracing data mapped onto atlas plates (Swanson, 2004) coupled to a semantic framework, and permits analyses and interpretation of connectivity patterns based on spatial and semantic views.
Taken together, these different attempts at assembling connectomes from existing and new data illustrate novel possibilities of gaining new knowledge through data systems that allow systematic integration and comparison of data based on standard nomenclatures, semantics, and spatial frameworks. To fulfill the long-term ambition of having reasonably complete connectomes we must also address the challenge of data exchange and interoperability across database systems of this kind.
Applied Connectomics: Network Analysis and Modeling
While new and sophisticated approaches to connectome mapping and data integration will be essential for the further advancement of this field, it is important to also investigate how accumulated connectome data can be utilized to further our understanding of relationships between brain structure and function. The ways in which connectome data are used will to a large extent also dictate priorities for data collection and assembly. In this Research Topic, five contributions illustrate how connectomics can be applied for network analysis and modeling.
A fundamental principle of brain organization is that functional systems are formed by specific anatomical connections. Knowledge about the specific hodological organization of different brain regions may thus predict the various functional properties of such regions. Gorbach et al. (2011) explore relationships between the functional and connectional “fingerprints” of cerebrocortical areas in the human brain, by using hierarchical information-based clustering of MRI-based connectivity measures. They propose an automated hierarchical parcelation approach to identify cortical subunits that are consistent with cytoarchitectonic maps and previous connectivity-based parcelation schemes (Figure 1K).
Echtermeyer et al. (2011) examine changes in network structure that appear when the same brain is mapped at different spatial resolutions. Their emphasis is on network motifs and their composition and spatial distribution (Figure 1N). The study highlights the important roles of spatial scale and resolution for drawing conclusions based on network analysis.
Computational modeling represents an increasingly important approach aiming at combining available data on anatomical connectivity with a virtual exploration of functional properties that emerge from the interaction of structural coupling and neural dynamics. Using a model of a simple spinal cord system, the developing spinal cord of the Xenopus, Borisyuk et al. (2011) introduce a new approach toward characterizing connectomes by constructing the network on the basis of known developmental processes of neuronal and axonal growth. The resulting network (Figure 1Q) is then studied with a number of visualization and topological analysis tools, revealing relationships between sets of simple developmental rules and topological regularities. To meet the considerable computational challenges of simulating complex neural network models, Kunkel et al. (2011) have developed strategies for adapting network representations to reduce the memory consumption for simulation software. Such efforts are highly relevant for future up-scaling of computational modeling efforts, which will be necessary to incorporate more comprehensive structural and functional connectome data.
Finally, French et al. (2011) utilize data sets on rodent brain connectivity and gene expression patterns to explore possible inter-relations, and identify several aspects of connectivity and gene expression that are indeed correlated. This work highlights an integrative aspect of connectomics that links connectivity data with other non-connectomic data sources, e.g., from genomics and proteomics. The confluence of connectomics and genomics will likely be a major growth area in the not-too-distant future.
Progress Made and Main Challenges
The papers presented in this Research Topic demonstrate approaches to mapping, integrating, and utilizing connectivity data through structured neuroinformatics, in the spirit of the “Kötter School of Neuroinformatics.” The multiple modalities and levels of investigation represented in this collection of Research Topic papers illustrate the need for concerted and sustained efforts by several research communities to arrive at reasonably comprehensive connectome maps linking brain regions and neurons. The complexity and multi-faceted nature of brain connectivity clearly calls for cooperation, collaboration, and mutual understanding of methods, problems, and results.
A fundamental challenge for the establishment of a common description of brain connectivity will be to cross-validate the different methodologies. Some important efforts have already been made at different scales (Schmahmann et al., 2007; Bock et al., 2011; Briggman et al., 2011), but further work is needed to link methods such as structural and functional MRI (Segall et al., 2012), 3-D-PLI and tractography (Axer et al., 2011b), and optogenetics and fMRI (Lee, 2011). Since studies of the human connectome continue to rely on indirect connectivity measures, such as DTI and related imaging methods, we will need parallel experimental efforts in animal models and robust statistical methods to demonstrate the validity of connectivity data obtained by non-invasive means. This need for validation also underlines the continuing value of conventional connectivity data that are derived by tract-tracing in animal models and are compiled in databases such as CoCoMac or BAMS. A number of strategies may be imagined to further expand and enhance such compilations. For example, computational text mining approaches may develop to a level where they can be employed for the automated gathering of connectivity information from the published research literature6. Further, curated databases may collaborate more closely with field experts, in order to preserve the multi-faceted neuroanatomical knowledge acquired in many experimental labs over several decades (where knowledge is currently threatened by the upcoming retirement of many of the “traditional” anatomists), or new experimental efforts may be started for the systematic, “industrial” brain connectivity gathering in rodent models (Bohland et al., 2009; Zakiewicz et al., 2011).
The papers collected in this Research Topic offer a snapshot of diverse approaches pursued at the inception of a wide array of connectome studies across scales and species. The end goal of a comprehensive understanding of the network structure of complex nervous systems will require the integration of data and methods for mapping connectivity from neurons to systems. We have an exciting time ahead of us.
Footnotes
1Research Topic references are indicated by underlined text.
References
- Akil H., Martone M. E., Van Essen D. C. (2011). Challenges and opportunities in mining neuroscience data. Science 331, 708–712 10.1126/science.1199305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annese J. (2012). The importance of combining MRI and large-scale digital histology in neuroimaging studies of brain connectivity and disease. Front. Neuroinform. 6:13. 10.3389/fninf.2012.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano J. I., Benavides-Piccione R., Defelipe J., Yuste R. (2007). Ultrastructure of dendritic spines: correlation between synaptic and spine morphologies. Front. Neurosci. 1:1. 10.3389/neuro.01.1.1.010.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axer H., Beck S., Axer M., Schuchardt F., Heepe J., Flucken A., Axer M., Prescher A., Witte O. W. (2011a). Microstructural analysis of human white matter architecture using polarized light imaging: views from neuroanatomy. Front. Neuroinform. 5:28. 10.3389/fninf.2011.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axer M., Grässel D., Kleiner M., Dammers J., Dickscheid T., Reckfort J., Hütz T., Eiben B., Pietrzyk U., Zilles K., Amunts K. (2011b). High-resolution fiber tract reconstruction in the human brain by means of three-dimensional polarized light imaging. Front. Neuroinform. 5:34. 10.3389/fninf.2011.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjaalie J. G. (2002). Localization in the brain: new solutions emerging. Nat. Neurosci. Rev. 3, 322–325 10.1038/nrn790 [DOI] [PubMed] [Google Scholar]
- Bock D. D., Lee W. C., Kerlin A. M., Andermann M. L., Hood G., Wetzel A. W., Yurgenson S., Soucy E. R., Kim H. S., Reid R. C. (2011). Network anatomy and in vivo physiology of visual cortical neurons. Nature 471, 177–182 10.1038/nature09802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland J. W., Wu C., Barbas H., Bokil H., Bota M., Breiter H. C., Cline H. T., Doyle J. C., Freed P. J., Greenspan R. J., Haber S. N., Hawrylycz M., Herrera D. G., Hilgetag C. C., Huang Z. J., Jones A., Jones E. G., Karten H. J., Kleinfeld D., Kötter R., Lester H. A., Lin J. M., Mensh B. D., Mikula S., Panksepp J., Price J. L., Safdieh J., Saper C. B., Schiff N. D., Schmahmann J. D., Stillman B. W., Svoboda K., Swanson L. W., Toga A. W., Van Essen D. C., Watson J. D., Mitra P. P. (2009). A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput. Biol. 5, e1000334. 10.1371/journal.pcbi.1000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisyuk R., Al Azad A. K., Conte D., Roberts A., Soffe S. R. (2011). Modeling the connectome of a simple spinal cord. Front. Neuroinform. 5:20. 10.3389/fninf.2011.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M., Dong H. W., Swanson L. W. (2005). Brain architecture management system. Neuroinformatics 3, 15–48 10.1385/NI:3:1:015 [DOI] [PubMed] [Google Scholar]
- Bota M., Dong H. W., Swanson L. W. (2012). Combining collation and annotation efforts toward completion of the rat and mouse connectomes in BAMS. Front. Neuroinform. 6:2. 10.3389/fninf.2012.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman K. L., Helmstaedter M., Denk W. (2011). Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188 10.1038/nature09818 [DOI] [PubMed] [Google Scholar]
- Burns G. A., Cheng W. C., Thompson R. H., Swanson L. W. (2006). The NeuARt II system: a viewing tool for neuroanatomical data based on published neuroanatomical atlases. BMC Bioinformatics 7, 531. 10.1186/1471-2105-7-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral J., Hugues E., Sporns O., Deco G. (2011). Role of local network oscillations in resting-state functional connectivity. Neuroimage 57, 130–139 10.1016/j.neuroimage.2011.04.010 [DOI] [PubMed] [Google Scholar]
- Chung J. R., Sung C., Mayerich D., Kwon J., Miller D. E., Huffman T., Keyser J., Abbott L. C., Choe Y. (2011). Multiscale exploration of mouse brain microstructures using the knife-edge scanning microscope brain atlas. Front. Neuroinform. 5:29. 10.3389/fninf.2011.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffertshofer A., van Wijk B. C. (2011). On the influence of amplitude on the connectivity between phases. Front. Neuroinform. 5:6. 10.3389/fninf.2011.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti A., Burns G. A., Simmons D., Swanson L. W., Ghandeharizadeh S., Shahabi C., Stone J., Jia S. (2001). “The neuroanatomical rat brain viewer (NeuART),” in Computing the Brain: A Guide to Neuroinformatics, eds Grethe J. S., Arbib M. A. (San Diego: Academic Press; ), 189–202 [Google Scholar]
- Deco G., Jirsa V. K. (2012). Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. J. Neurosci. 32, 3366–3375 10.1523/JNEUROSCI.2523-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtermeyer C., Han C. E., Rotarska-Jagiela A., Mohr H., Uhlhaas P. J., Kaiser M. (2011). Integrating temporal and spatial scales: human structural network motifs across age and region of interest size. Front. Neuroinform. 5:10. 10.3389/fninf.2011.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L., Tan P. P., Pavlidis P. (2011). Large-scale analysis of gene expression and connectivity in the rodent brain: insights through data integration. Front. Neuroinform. 5:12. 10.3389/fninf.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerling J. C., Loewy A. D. (2006). Aldosterone-sensitive neurons in the nucleus of the solitary tract: bidirectional connections with the central nucleus of the amygdala. J. Comp Neurol. 497, 646–657 10.1002/cne.20993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard S., Daducci A., Lemkaddem A., Meuli R., Thiran J. P., Hagmann P. (2011). The connectome viewer toolkit: an open source framework to manage, analyze, and visualize connectomes. Front. Neuroinform. 5:3. 10.3389/fninf.2011.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbach N. S., Schutte C., Melzer C., Goldau M., Sujazow O., Jitsev J., Douglas T., Tittgemeyer M. (2011). Hierarchical information-based clustering for connectivity-based cortex parcellation. Front. Neuroinform. 5:18. 10.3389/fninf.2011.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P. (2005). From Diffusion MRI to Brain Connectomics. Ecole Polytechnique Fédérale de Lausanne (Lausanne: EPFL; ), 1 [Google Scholar]
- Hagmann P., Cammoun L., Gigandet X., Gerhard S., Grant P. E., Wedeen V., Meuli R., Thiran J. P., Honey C. J., Sporns O. (2010). MR connectomics: principles and challenges. J. Neurosci. Methods 194, 34–45 10.1016/j.jneumeth.2010.01.014 [DOI] [PubMed] [Google Scholar]
- Harris K. M., Fiala J. C., Ostroff L. (2003). Structural changes at dendritic spine synapses during long-term potentiation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 745–748 10.1098/rstb.2002.1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M., Baldock R. A., Burger A., Hashikawa T., Johnson G. A., Martone M., Ng L., Lau C., Larsen S. D., Nissanov J., Puelles L., Ruffins S., Verbeek F., Zaslavsky I., Boline J. (2011). Digital atlasing and standardization in the mouse brain. PLoS Comput. Biol. 7, e1001065. 10.1371/journal.pcbi.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötter R. (2004). Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics 2, 127–144 10.1385/NI:2:2:127 [DOI] [PubMed] [Google Scholar]
- Kunkel S., Potjans T. C., Eppler J. M., Plesser H. E., Morrison A., Diesmann M. (2011). Meeting the memory challenges of brain-scale network simulation. Front. Neuroinform. 5:35. 10.3389/fninf.2011.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H. (2011). Tracing activity across the whole brain neural network with optogenetic functional magnetic resonance imaging. Front. Neuroinform. 5, 21. 10.3389/fninf.2011.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus D. S., Harwell J., Olsen T., Hodge M., Glasser M. F., Prior F., Jenkinson M., Laumann T., Curtiss S. W., Van Essen D. C. (2011). Informatics and data mining tools and strategies for the human connectome project. Front. Neuroinform. 5:4. 10.3389/fninf.2011.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva K. D., Busse B., Weiler N. C., O’Rourke N., Smith S. J. (2010). Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron 68, 639–653 10.1016/j.neuron.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modha D. S., Singh R. (2010). Network architecture of the long-distance pathways in the macaque brain. Proc. Natl. Acad. Sci. U.S.A. 107, 13485–13490 10.1073/pnas.1008054107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara S., Inoue K., Witter M. P., Iijima T. (2009). Untangling neural networks with dual retrograde transsynaptic viral infection. Front. Neurosci. 3:3. 10.3389/neuro.01.032.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropireddy D., Ascoli G. A. (2011). Potential synaptic connectivity of different neurons onto pyramidal cells in a 3D reconstruction of the rat hippocampus. Front. Neuroinform. 5:5. 10.3389/fninf.2011.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ T. A., Ramakrishnan C., Hovy E. H., Bota M., Burns G. A. (2011). Knowledge engineering tools for reasoning with scientific observations and interpretations: a neural connectivity use case. BMC Bioinformatics 12, 351. 10.1186/1471-2105-12-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem K. S., Pauls J. M., Augath M., Trinath T., Prause B. A., Hashikawa T., Logothetis N. K. (2002). Magnetic resonance imaging of neuronal connections in the macaque monkey. Neuron 34, 685–700 10.1016/S0896-6273(02)00718-3 [DOI] [PubMed] [Google Scholar]
- Schmahmann J. D., Pandya D. N., Wang R., Dai G., D’Arceuil H. E., de Crespigny A. J., Wedeen V. J. (2007). Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain 130, 630–653 10.1093/brain/awl359 [DOI] [PubMed] [Google Scholar]
- Segall J. M., Allen E. A., Jung R. E., Erhardt E. B., Arja S. K., Kiehl K., Calhoun V. D. (2012). Correspondence between structure and function in the human brain at rest. Front. Neuroinform. 6:10. 10.3389/fninf.2012.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. (2011). The human connectome: a complex network. Ann. N. Y. Acad. Sci. 1224, 109–125 10.1111/j.1749-6632.2010.05888.x [DOI] [PubMed] [Google Scholar]
- Sporns O., Tononi G., Kotter R. (2005). The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42. 10.1371/journal.pcbi.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger J. F., Zuschratter W., Luhmann H. J., Schubert D. (2009). Local circuits targeting parvalbumin-containing interneurons in layer IV of rat barrel cortex. Brain Struct. Funct. 214, 1–13 10.1007/s00429-009-0225-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K. E., Kamper L., Bozkurt A., Burns G. A., Young M. P., Kotter R. (2001). Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1159–1186 10.1098/rstb.2001.0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K. E., McIntosch A. R., Hilgetag C. C. (2010). In memoriam: Rolf Kötter (1961-2010). PLoS Comput. Biol. 6, e1000965. 10.1371/journal.pcbi.1000965 [DOI] [Google Scholar]
- Sugar J., Witter M. P., van Strien N. M., Cappaert N. L. (2011). The retrosplenial cortex: intrinsic connectivity and connections with the (para)hippocampal region in the rat. An interactive connectome. Front. Neuroinform. 5:7. 10.3389/fninf.2011.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson L. W. (2004). Brain Maps: Structure of the Rat Brain. Amsterdam: Elsevier Science B.V. [Google Scholar]
- Tallis M., Thompson R., Russ T. A., Burns G. A. (2011). Knowledge synthesis with maps of neural connectivity. Front. Neuroinform. 5:24. 10.3389/fninf.2011.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. H., Swanson L. W. (2010). Hypothesis-driven structural connectivity analysis supports network over hierarchical model of brain architecture. Proc. Natl. Acad. Sci. U.S.A. 107, 15235–15239 10.1073/pnas.1001999107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini G. (2011). Rabies virus as a transneuronal tracer of neuronal connections. Adv. Virus Res. 79, 165–202 10.1016/B978-0-12-387040-7.00010-X [DOI] [PubMed] [Google Scholar]
- Van der Linden A., Verhoye M., Van Meir V., Tindemans I., Eens M., Absil P., Balthazart J. (2002). In vivo manganese-enhanced magnetic resonance imaging reveals connections and functional properties of the songbird vocal control system. Neuroscience 112, 467–474 10.1016/S0306-4522(02)00070-2 [DOI] [PubMed] [Google Scholar]
- Van Essen D. C., Ugurbil K. (2012). The future of the human connectome. Neuroimage. [Epub ahead of print]. 10.1016/j.neuroimage.2012.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien N. M., Cappaert N. L., Witter M. P. (2009). The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat. Rev. Neurosci. 10, 272–282 10.1038/nrn2614 [DOI] [PubMed] [Google Scholar]
- Wouterlood F. G., Hartig W., Groenewegen H. J., Voorn P. (2011). Density gradients of vesicular glutamate- and GABA transporter immunoreactive boutons in calbindin- and mu-opiod receptor defined compartments in the rat striatum. J. Comp. Neurol. [Epub ahead of print]. 10.1002/cne.23031 [DOI] [PubMed] [Google Scholar]
- Yendiki A., Panneck P., Srinivasan P., Stevens A., Zöllei L., Augustinack J., Wang R., Salat D., Ehrlich S., Behrens T., Jbabdi S., Gollub R., Fischl1 B. (2011). Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Front. Neuroinform. 5:23. 10.3389/fninf.2011.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakiewicz I. M., van Dongen Y. C., Leergaard T. B., Bjaalie J. G. (2011). Workflow and atlas system for brain-wide mapping of axonal connectivity in rat. PLoS ONE 6, e22669. 10.1371/journal.pone.0022669 [DOI] [PMC free article] [PubMed] [Google Scholar]