Abstract
Alternative splicing is a complex post-transcriptional process that can be regulated by cis-acting elements located within genomic non-coding regions. Recent studies have identified that polymorphic variations in non-coding regions of the α-synuclein gene (SNCA) locus are associated with an increased risk for developing Parkinson’s disease (PD). The underlying mechanism(s) for this susceptibility may involve changes in α-synuclein mRNA expression and alternative splicing. As a first step towards understanding the biology of α-synuclein splice variants in PD, we characterized the levels of the full-length SNCA-140 mRNA transcript and SNCA-126, -112, and -98 alternatively spliced variants in different neuronal regions from PD patients or transgenic mice overexpressing human α-synuclein (ASO). In human post-mortem tissue, α-synuclein spliced transcripts were expressed in a region-specific manner in cortex, substantia nigra, and cerebellum. We observed increased nigral SNCA-140 and SNCA-126 transcript levels in PD patients when compared to neurologically unaffected cases. Human α-synuclein splicing changes were also found to occur in a region-specific manner in ASO mice. Here, SNCA-126, -112, and -98 transcript levels did not increase proportionally with SNCA-140 levels, or parallel the region-specific mouse transcript ratios seen in wild-type (WT) littermates. While most transcripts were elevated in ASO mice when compared to WT mice, the most prominent increase was found in the ventral midbrain of 15-month-old ASO mice. These results demonstrate region-specific human α-synuclein transcript level abnormalities in PD patients and in a transgenic mouse model of α-synucleinopathy. This study is relevant to understanding the normal, adaptive, or pathological role(s) of α-synuclein splice variants.
Keywords: Parkinson’s disease, alpha-synuclein, SNCA, alterative splicing, isoform, substantia nigra
INTRODUCTION
α-Synuclein is a small, natively unfolded protein that is enriched in pre-synaptic nerve termini (Bisaglia et al., 2009), exhibiting broad, steady state neuronal expression with a capacity to interact with phospholipid membranes (Davidson et al., 1998) to promote SNARE complex assembly (Burre et al., 2010). Pathologically, α-synuclein is a component of Lewy body inclusions present within affected neurons of Parkinson’s disease (PD) patients (Spillantini and Goedert, 2000). A genetic link exists between familial PD and α-synuclein gene (SNCA) abnormalities, including missense mutations and genomic multiplication (Nuytemans et al., 2010), and abnormal α-synuclein expression and post-translational modification is found in both familial and sporadic PD (Kim and Lee, 2008; Oueslati et al., 2010). Recent genome-wide association studies have shown that polymorphic variations in non-coding regions of the SNCA locus contribute to the etiology of sporadic PD (Satake et al., 2009; Simon-Sanchez et al., 2009). While the mechanism(s) underlying this association remains unknown, PD-associated polymorphisms located within an SNCA 5′ and 3′ linkage disequilibrium block are associated with differential α-synuclein mRNA expression (Cronin et al., 2009; Fuchs et al., 2008), which may be mediated through changes in α-synuclein alternative splicing (Beyer et al., 2007; McCarthy et al., 2010).
Alternative splicing is a critical regulatory mechanism that augments transcriptome diversity by increasing the coding capacity of a single gene (Keren et al., 2010). Lacking a stable secondary structure, α-synuclein is an intrinsically disordered protein that relies on an ensemble of alternative conformations for functional activity (Uversky, 2003). In addition to a number of post-translational modifications, including phosphorylation, nitration, cleavage, and ubiquitination (Oueslati et al., 2010), at least four α-synuclein spliced mRNA transcripts have been identified in humans: the full-length isoform, SNCA-140, which is encoded by all six exons of SNCA (Ueda et al., 1993), and three alternative variants, SNCA-126, -112, and -98, which are generated by in-frame excision of exons 3, 5, or both, respectively (Beyer et al., 2008b; Campion et al., 1995). While the biological and pathological significance of α-synuclein isoforms remains unknown, alterations in α-synuclein isoform stoichiometry have been associated with intracellular aggregation (Kalivendi et al., 2009; McLean and Hyman, 2002) and mRNA transcripts that give rise to these isoforms have been shown to be differentially expressed in human α-synucleinopathies, including PD and dementia with Lewy bodies (Beyer et al., 2008a; Neystat et al., 1999).
To date, a comparison among the expression levels of all four α-synuclein spliced transcripts in different neuronal regions has not been performed. In the current study, using carefully designed primers, we evaluate α-synuclein spliced transcript expression in different neuronal regions from PD post-mortem tissue and in transgenic mice overexpressing human α-synuclein (ASO).
MATERIALS AND METHODS
Animals and Tissue Preparation
All experimental and surgical procedures were performed in accordance with McLean Hospital’s Institutional Animal Care and UseCommittee guidelines. Mice were housed at the Animal Care Facility at McLean Hospital with ad libitum access to water and food. Colony lighting followed a full spectrum 12/12 hr light/dark cycle with the onset of lights at 0800 hr. Transgenic overexpression of human α-synuclein under the Thy1 promoter in ASO mice has been previously described (Rockenstein et al., 2002). ASO mice were maintained on a mixed C57BL/6-DBA/2 background by breeding female hemizygous mice with male BDF1 hybrids (Charles River, Wilmington, USA). Genotypes were verified by polymerase chain reaction (PCR) using tail DNA. Non-transgenic wild-type (WT) littermates were used as the source of control tissue. Frozen post-mortem human tissue from frontal cortex, substantia nigra, and cerebellum of male neurologically unaffected control (CTRL) and PD cases were provided by the Harvard Brain Tissue Resource Center (McLean Hospital, Belmont, MA) (Table 2). We analyzed six CTRL and PD cases for each tissue section, except for the CTRL substantia nigra, where only five cases were available. Of note, there is an average age difference of ~9.8 years between PD and CTRL cases.
Table 2.
Parkinson’s Disease and Control Case Information
| Case | Diagnosis/Cause of death | Gender | Disease Onset (years) | Age at Death (years) | Family History | PMI (hours) | Pathological Features |
|---|---|---|---|---|---|---|---|
| AN05440 | PD, advanced/seizure | M | 70 | 77 | Yes | 18 | PD w/limbic stage diffuse LBD; Braak Stage III; cortical non-neuritic plaques; dense NFTs in proximal CA1; SN 3–4+ pallor, severe neuronal loss, and LB positive. |
| AN10333 | PD, advanced/unknown | M | 66 | 74 | n/a | 30 | PD w/limbic stage diffuse LBD; Braak Stage II; agyrophilic grains in CA1/CA2; SN 3–4+ pallor, severe neuronal loss, and LB positive. |
| AN10634 | PD, advanced/renal failure | M | 74 | 86 | n/a | 20 | PD w/limbic stage diffuse LBD; Braak Stage III; early limbic stage AD; sparse cortical NFTS & LB; SN 4+ pallor, severe neuronal loss, and LB positive. |
| AN06461 | PD, advanced/unknown | M | 70 | 87 | Yes | 28 | PD w/limbic stage diffuse LBD; Braak Stage I; SN 4+ pallor, severe neuronal loss, and LB positive. |
| AN05419 | PD, advanced/respiratory arrest (pneumonia) | M | 69 | 76 | Yes | 13 | PD w/neocortical stage LBD; Braak Stage III; sparse neocortical amyloid plaques; SN 3–4+ pallor, severe neuronal loss, and LB positive. |
| AN18571 | PD, advanced/unknown | M | n/a | 70 | No* | 20 | PD w/diffuse LBD; Braak Stage I; mild hippocampal neuronal loss w/agyrophilic grains; SN 4+ pallor, severe neuronal loss, and LB positive; non-paralytic polio at age 5. |
Normal control cases: AN02888: myocardial infarction, M, 77, (PMI: 29); AN02690: myocardial infarction, M, 62 (PMI: 26); AN12544: cardiac arrest, M, 55 (PMI: 24); AN04211: pancreatic cancer, M, 63 (PMI: 20); AN10236: pancreatic cancer, M, 76 (PMI: 24); AN06646, cardiac arrest, M, 78 (PMI: 19)
CA1/2, cornu Ammonis 1/2; LB, Lewy body; LBD, Lewy body disease; n/a, not available; NFTs, neurobibrillary tangles; PD, Parkinson’s disease; PMI, post-mortem interval; SN, Substantia nigra;
paternal history of tremor. All tissues were obtained from the Harvard Brain Tissue Resource Center with pathological diagnosis. Cause of death indicated on State Death Certificate or by correspondence w/pathologist.
RNA Isolation and RT-PCR
Male WT and ASO mice (n = 4–6 per group) were anaesthetized with sodium pentobarbital (i.p., 130 mg/kg) and transcardially perfused with ice cold heparin saline (pH 7.4). Tissues were quickly hand dissected with the assistance of a McIlwain Tissue Chopper (Surrey, UK), set at 750 μM slices, from the cortex, hippocampal formation, striatum, ventral midbrain, cerebellum, and heart on an ice-cold surface, snap frozen, and stored at −80°C. Total RNA from human and mouse tissue was obtained using the RNeasy Lipid Tissue Mini Kit (Qiagen, Valencia, CA) as per the manufacturers instructions and included the optional DNase digestion procedure. cDNA was generated using the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). One-tenth of the reverse transcription (RT) reaction was used as the template for PCR amplification. We performed semi-quantitative PCR throughout the study in order to visually inspect each amplicon and to rule out the possibility of amplifying other, unknown spliced transcripts. PCR conditions were optimized by systematic evaluation of primer annealing temperatures (see Table 1 for primer details). PCR of α-synuclein transcripts was performed using GoTaq PCR Master Mix (Promega), with an initialization step at 95°C for 5 min, then 33 cycles [(30 cycles for glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] at 95°C for 30 sec, 55°C for 30 sec (52°C and 50°C for SNCA-112 and -98, respectively; and 57°C for human GAPDH), and 72°C for 30 sec, followed by a final extension at 72°C for 5 min. Amplicons were separated by 1.5% agarose gel with SYBR Safe DNA gel stain (Invitrogen) by electrophoresis. α-Synuclein primer specificity was assessed by cloning individual PCR fragments into a topoisomerase vector (pCR 4-TOPO, Invitrogen) and sequence verified (Dana-Farber/Harvard Cancer Center DNA Resource Core, Boston, MA). For SNCA-140 sequence chromatograms, a pCMV6-XL5 plasmid containing human SNCA-140 (SC119919; OriGene, Rockville, MD) was used. For amplicon analysis of SNCA-126 in 15-month-old WT and ASO ventral midbrain tissue, One Taq 2X Master Mix (New England Biolabs, Ipswich, MA) was used, as per the manufacturer’s instructions. Human and mouse alpha-synuclein mRNA sequences used for alignment was obtained from the University of California, Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/).
Table 1.
Primer Details
| Fig. 1 | Primer Name | Sequence | Tm | Target | Product |
|---|---|---|---|---|---|
| A | hSNCA ex3 fwd | 5′-AAA ACC AAG GAG GGA GTG GT | 58.4°C | exon 3 | SNCA-140 SNCA-112 |
| B | hSNCA ex51 rev | 5′-TGT CAG GAT CCA CAG GCA TA | 58.4° | exon 5 | SNCA-140 SNCA-126 |
| C | hSNCA ex2/4 fwd | 5′-CTC TAT GTA GTG GCT GAG | 53.8°C | 3′ exon 2 5′ exon 4 |
SNCA-126 SNCA-98 |
| B | hSNCA ex52 rev | 5′-GCC TCA TTG TCA GGA TCC AC | 60.5°C | exon 5 | SNCA-140 SNCA-126 |
| D | hSNCA ex4/6 rev | 5′-ATA CCC TTC CTT GCC CAA CTG | 61.2°C | 3′ exon 4 5′ exon 6 |
SNCA-112 SNCA-98 |
| CSC | ex2/4 fwd scramb | 5′-TTA GTG TCC ATG GCT GAG | 53.8°C | 5′ exon 4 | none |
| CSC2 | ex2/4 fwd scramb 2 | 5′-CTC TAT GTA GGA TGG CGT | 53.8°C | 3′ exon 2 | none |
| DSC | ex4/6 rev scramb | 5′-CAC TTA CCA CTT GCC CAA CTG | 61.2°C | 5′ exon 4 | none |
| DSC2 | ex4/6 rev scramb 2 | 5′-ATA CCC TTC AGC CAT TCC TGC | 61.2°C | 5′ exon 6 | none |
| mGAPDH fwd | 5′-AAC TTT GGC ATT GTG GAA GG | 56.4°C | GAPDH | ||
| mGAPDH rev | 5′-ACA CAT TGG GGG TAG GAA CA | 58.4°C | GAPDH | ||
| hGAPDH fwd | 5′-ACC ACA GTC CAT GCC ATC AC | 60.5°C | GAPDH | ||
| hGAPDH rev | 5′-TCC ACC ACC CTG TTG CTG TA | 60.5°C | GAPDH |
ex, exon; fwd, sense primer; h, human; m, mouse; rev, antisense primer; scramb, scrambled primer; SNCA, α-synuclein; Tm, melting temperature (salt αadjusted, 50 mM Na+)
Statistical Analyses
Gels were imaged at equal exposure on a FOTO/Analyst transilluminator with PC Image software (Fotodyne, Hartland, WI). Exported images were analyzed using Image J software (National Institutes of Health, Bethesda, MD). Using the integrated density function after background subtraction, we measured the sum of the pixels from the electrophoretic bands. These values were divided by the sum of the pixels representing the baseline housekeeping gene, GAPDH, to obtain relative transcript-specific expression levels. A single 100 kb DNA ladder (New England Biolabs) was used as an internal reference. To avoid omitting weakly expressed bands from quantification, longer exposure times were analysed and statistically validated. Where applicable, bands below detectable limits are indicated by “nd”. Gels presented in this study are from equivalent exposures and optimized to be below saturation limits and within the linear range. Statistical tests were performed with Prism 4.0 (GraphPad software, San Diego, CA). For between- and within-group comparisons of the changes in α-synuclein transcript expression among different tissues, we used one-way analysis of variance (ANOVA) with Tukey’s post-hoc and p < 0.05 considered significant. Where indicated in the figures, * = p < 0.05 between WT and ASO tissues; † = p < 0.05 among WT tissues; and # = p < 0.05 among ASO tissues.
RESULTS
Mutually exclusive primer design eliminates false positive amplification of α-synuclein spliced transcripts
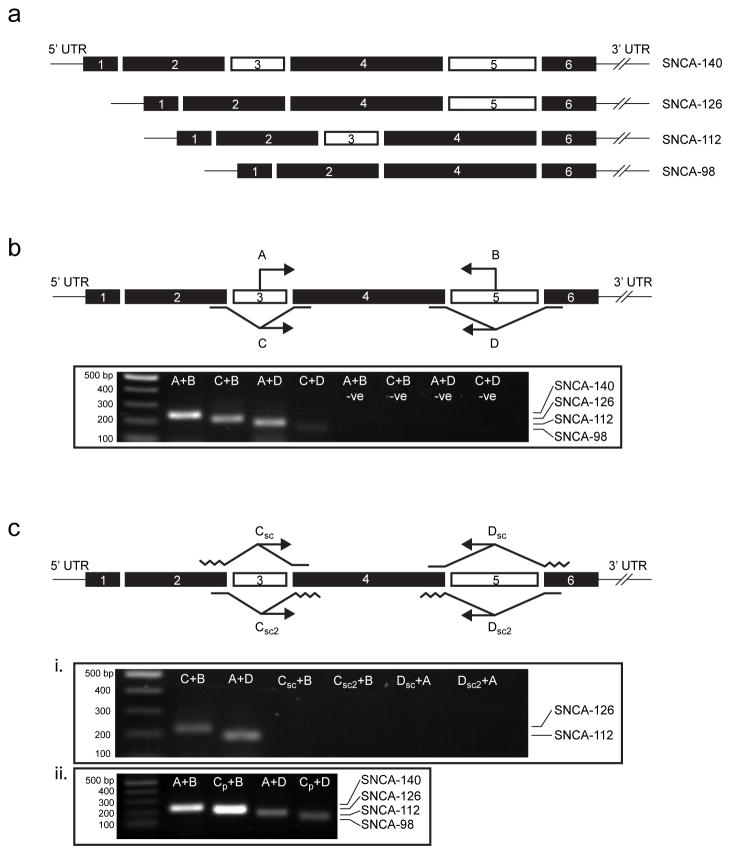
Previous studies focusing on human α-synuclein alternative splicing have used primers or probes located upstream or downstream of the splicing event to amplify individually spliced mRNA transcripts (Beyer et al., 2008a; Beyer et al., 2008b; Beyer et al., 2006; McCarthy et al., 2010; Ueda et al., 1994). Because of the pattern of α-synuclein exon 3 and/or 5 exclusion that generates each alternative transcript (Fig. 1a), the use of primers not specific to these splicing sites may permit the co-amplification of other transcripts. Additionally, the length of exon skipping primers bridging nonconsecutive exons (2/4 or 4/6) should be minimized to prevent non-specific annealing of 5′ or 3′ primer ends; failure to do so may risk false positive amplification of transcripts with exon 3 or 5 inclusion, such as SNCA-140.
Fig. 1.
Derivation of α-synuclein splice-specific primers. (a) Six exons encode the full-length α-synuclein protein, SNCA-140, while three alternatively spliced variants that skip exons 3, 5, or both generate isoforms SNCA-126, -112, and -98, respectively. (b) We designed primers that amplify cDNA encoding specific α-synuclein isoforms: two sense primers, one located in exon 3 (A) and one bridging exons 2 and 4 (C), were used in combination with two antisense primers, one located in exon 5 (B) and one bridging exons 4 and 6 (D), to amplify SNCA-140 (A+B), -126 (C+B), -112 (A+D), and -98 (C+D) from human temporal cortex. Negative controls amplifying each primer set but without cDNA are indicated (-ve). (c) Half-scrambled primers of C and D were used to validate the specificity of our exon skipping primers shown in (b). Compared to SNCA-126 (C+B) and SNCA-112 (A+D), no PCR products were observed when regions corresponding to at least half of the C and D primer sequences flanking exons 3 or 5 were scrambled (Csc, Csc2, Dsc, and Dsc2), indicating that annealing only occurred in the presence of a spliced cDNA template (i). We determined that the forward primer previously used to report α-synuclein exon 3 skipping (Cp) was too long, resulting in non-specific amplification of SNCA-140 and other transcripts with exon 3 inclusion, leading to an overestimation of SNCA-126 and SNCA-98 (compare Fig. 1b with 1c, ii).
To characterize α-synuclein splicing, we designed sense and antisense α-synuclein primers that permit the amplification of specific spliced transcripts while prohibiting the co-amplification of other spliced transcripts in a mutually exclusive fashion. To amplify SNCA-140, primers were targeted to sequences located within exons 3 and 5 (Fig. 1b, ‘A’ and ‘B’). Since in-frame excision of exons 3, 5, or both gives rise to SNCA-126, -112, and -98, respectively, we generated exon skipping primers that recognize the flanking sequences of exons 3 and/or 5: SNCA-126 was detected using an exon skipping sense primer spanning exons 2 and 4 paired with the antisense primer located in exon 5 (Fig. 1b, ‘C’ and ‘B’); SNCA-112 was detected with an exon skipping antisense primer spanning exons 4 and 6 paired with the sense primer located in exon 3 (Fig. 1b, ‘D’ and ‘A’); and SNCA-98 was detected with both exon skipping sense primer and antisense primers described above (Fig. 1b, ‘C’ and ‘D’). Transcript specificity was tested by RT-PCR on tissue from human frontal cortex of a neurologically normal 65-year-old male. We observed single band amplification for each of our primer sets, which were subsequently sequence verified to confirm primer specificity (Supplemental Fig. 1a). We identified SNCA-140 (Fig. 1b, ‘A+B’) as the predominate transcript, followed by SNCA-126 (Fig. 1b, ‘C+B’) and SNCA-112 (Fig. 1b, ‘A+D’), while SNCA-98 (Fig. 1b, ‘C+D’) was present, but minimally visible as saturation limits were approached.
While the design and use of exon skipping primers is frequently described, we used half-scrambled primers to identify and eliminate non-specific annealing that can occur through 5′ or 3′ extrinsic sequences (Fig. 1c). To our knowledge, this is the first description of such a technique. While this technique is not necessary for antibodies developed against exon skipping epitopes, as spliced isoforms can often be distinguished by molecular weight on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), there is growing interest in identifying or targeting exon skipping events at the mRNA level (Baralle and Baralle, 2005; Buratti and Baralle, 2008; Venables, 2004; Wood et al., 2010; Yokota et al., 2009). To validate our primers, the exon skipping sense primer used to generate SNCA-126 and SNCA-98 (Fig. 1b, ‘C’) was half-scrambled either at the 5′ or 3′ end, corresponding to the 3′ end of exon 2 and the 5′ end of exon 4, respectively (Fig. 1c, ‘Csc’ and ‘Csc2’). Similarly, the exon skipping antisense primer used to generate SNCA-112 and SNCA-98 (Fig. 1b, ‘D’) was half-scrambled either at the 3′ or 5′ end, corresponding to the 5′ end of exon 6 and the 3′ end of exon 4, respectively (Fig. 1c, ‘Dsc’ and ‘Dsc2’). Of these primers, Csc needed to be optimized by progressively shortening the 3′ end of the primer sequence (Supplemental Fig. 1b) until we obtained both a negative signal with the scrambled primer (Fig. 1c, ‘Csc+B’, ‘Csc2+B’, ‘Dsc+A’ and ‘Dsc2+A’; Supplemental Fig. 1b) and a positive signal with the shortened unscrambled primer (Fig. 1c, ‘C+B’ and ‘A+D’; Supplemental Fig. 1b), indicating that the primers were only annealing while in the presence of a spliced transcript. Previous reports characterizing α-synuclein exon 3 skipping have used an exon skipping sense primer with long exon 2 and 4 flanking sequences (Beyer et al., 2008a; Beyer et al., 2008b; Beyer et al., 2006). We found that the 5′ and 3′ ends of this primer was particularly susceptible to non-specific binding of SNCA-140, leading to false positive amplification of SNCA-126 and SNCA-98 transcript levels (compare Fig. 1b, C+B and C+D with Fig. 1c, ii, CP+B and CP+D). Details of the primers used in our study are provided in Table 1.
Abnormal elevations in α-synuclein spliced transcript expression in the substantia nigra of PD patients
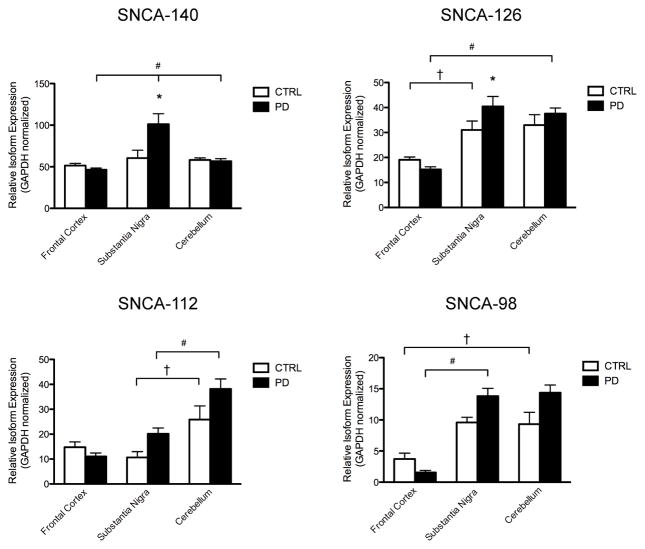
In the current study, we assessed whether region-specific changes in α-synuclein splicing could be observed in the frontal cortex, substantia nigra, and cerebellum of postmortem tissue from CTRL and PD cases (Table 2). No differences in SNCA-140 levels were found among neuronal regions from CTRL cases (Fig. 2, SNCA-140, compare white bars). We found higher SNCA-140 levels in the substantia nigra of PD cases when compared to the substantia nigra of CTRL cases (Fig. 2, SNCA-140, compare black and white bars). This increase in nigral SNCA-140 levels was higher than in other PD tissues (Fig. 2, SNCA-140, compare black bars).
Fig. 2.
RT-PCR of human α-synuclein alternative splicing. Semi-quantitative analysis of SNCA-140, -126, -112, and -98 from CTRL (n = 5–6) and PD (n = 6) post-mortem tissue homogenates of frontal cortex, substantia nigra, and cerebellum. Relative transcript expression is normalized to GAPDH and the results are shown as the mean ± SEM. Significance was determined by one-way ANOVA with post-hoc analysis: * = p < 0.05 between CTRL and PD tissues; † = p < 0.05 among CTRL tissues; and # = p < 0.05 among PD tissues.
We found lower SNCA-126 levels in the cortex of CTRL and PD cases than in the substantia nigra and cerebellum (Fig. 2, SNCA-126, compare white bars and compare black bars, respectively). There was more SNCA-126 in the substantia nigra of PD cases when compared to the substantia nigra of CTRL cases (Fig. 2, SNCA-126, compare black and white bars).
For SNCA-112, the cerebellum of CTRL cases showed higher levels than the substantia nigra of CTRL cases (Fig. 2, SNCA-112, compare white bars). No differences in SNCA-112 levels could be observed between CTRL and PD cases, despite a trend for elevated expression in the substantia nigra and cerebellum of PD cases (Fig. 2, SNCA-112, compare black and white bars). We found higher SNCA-112 levels in the cerebellum of PD cases when compared to the substantia nigra and frontal cortex of PD cases (Fig. 2, SNCA-112, compare black bars).
We found lower SNCA-98 levels in the cortex of CTRL and PD cases than in the substantia nigra and cerebellum (Fig. 2, SNCA-98, compare white bars and compare black bars, respectively). No differences in SNCA-98 levels could be observed between CTRL and PD cases, despite a trend for elevated expression in the frontal cortex of CTRL cases and in the substantia nigra and cerebellum of PD cases (Fig. 2, SNCA-98, compare black and white bars).
Abnormal α-synuclein splicing in 2-month-old transgenic mice overexpressing human α-synuclein
We extended our analysis to identify whether abnormal splicing changes could be observed in an animal model of α-synucleinopathy. ASO mice develop progressive pathological and behavioral deficits reminiscent of PD and dementia with Lewy bodies (Fernagut and Chesselet, 2004). These mice exhibit extensive α-synuclein aggregation throughout the brain (Fernagut et al., 2007; Koob et al., 2010; Rockenstein et al., 2002; Song et al., 2004), as well as enhanced nigral axonal degeneration and mitochondrial pathology in response to MPTP treatment (Song et al., 2004). Deficits in sensorimotor function, motor performance, and coordination are observed as early as 2 months and worsen with age (Fleming et al., 2004).
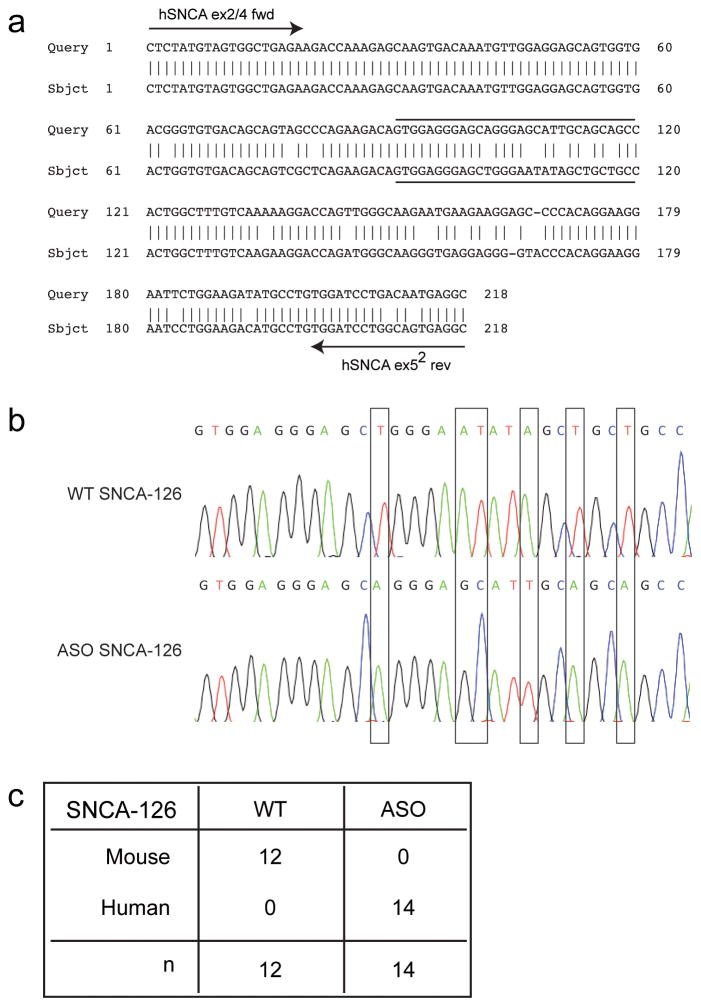
The human α-synuclein transgene in ASO mice is based on the SNCA-140 cDNA construct that shows broad mThy-1-driven human α-synuclein mRNA overexpression in the cortex, hippocampus, susbtantia nigra, and brainstem at 3 months-of-age (Rockenstein et al., 2002). Because the cDNA transgene lacks endogenous promoter and intronic regulatory sequences, yet maintains the capacity to generate the full-length open reading frame for SNCA-126, -112, and -98, we reasoned that the lack of pre-mRNA cis-regulatory elements would result in atypical post-transcriptional mRNA processing. Here, we sought to identify whether these alternative variants were overexpressed and/or misregulated. Splicing of the human cDNA transgene in ASO mice was confirmed by analyzing the SNCA-126 amplicon derived from 15-month-old WT and ASO ventral midbrain tissue (Fig. 3). Human and mouse alpha-synuclein sequence differences (Fig. 3a) were distinguished from individual SNCA-126-positive transformants following TOPO TA cloning (Fig. 3b). Only mouse SNCA-126 was identified in WT mice and only human SNCA-126 was identified in ASO mice (Fig. 3c).
Fig. 3.
Confirmation of human alpha-synuclein splicing in ASO mice. (a) A BLAST alignment of human (top) and mouse (bottom) SNCA-126 amplified by hSNCA 2/4 fwd and hSNCA ex52 rev primers identifies ~90% sequence similarity. (b) Sequence chromatograms show base pair differences (vertical bar comparisons) between mouse and human SNCA-126 amplicons in 15-month-old WT and ASO ventral midbrain tissue after TOPO TA cloning, respectively. Chromatogram sequences are from the overlined and underlined sequences in (a). (c) Several SNCA-126-positive transformants were analyzed to confirm splicing of the cDNA transgene and to determine the abundance of human or mouse SNCA-126 in the ASO amplicon. Only mouse SNCA-126 was identified in WT mice (n = 12) and only human SNCA-126 was identified in ASO mice (n = 14).
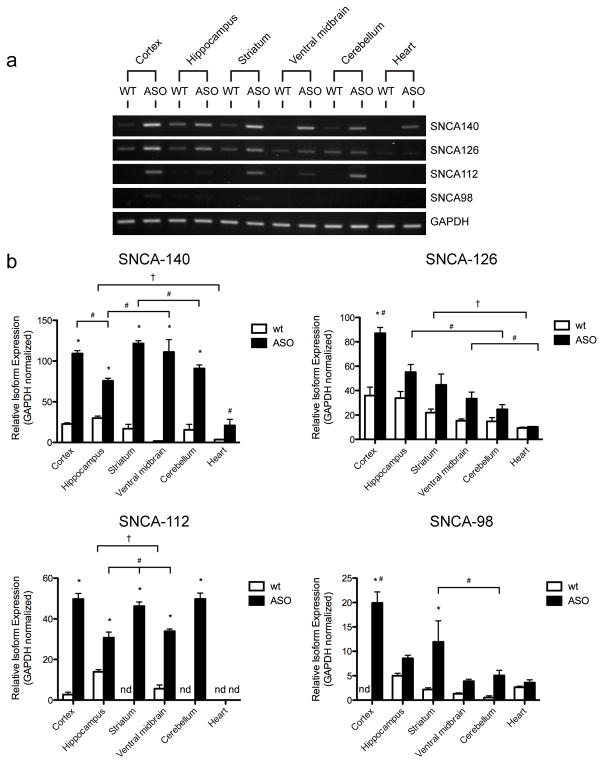
We next evaluated the electrophoretic α-synuclein splicing profile from the cortex, hippocampus, striatum, ventral midbrain, cerebellum, and heart of a representative 2-month-old WT and ASO mouse (Fig. 4a). As previously reported (Rockenstein et al., 2002), we observed region-specific differences in the expression of SNCA-140 transcripts between WT and ASO mice. Alternative transcripts SNCA-126, -112, and -98 also showed region-specific expression, however, in ASO mice, this did not necessarily coincide with the elevated expression of SNCA-140 or the spliced transcript levels found in WT mice
Fig. 4.
RT-PCR of α-synuclein alternative splicing at 2 months-of-age in neuronal regions of WT and ASO mice. (a) Agarose gel-based electrophoretic distribution profile of α-synuclein spliced transcripts in the cortex, hippocampus, striatum, substantia nigra, cerebellum, and heart of a representative 2-month-old WT and ASO mouse. (b) Semi-quantitative analysis of SNCA-140, -126, -112, and -98 from several 2-month-old WT and ASO mice (n = 4–6). Relative transcript expression is normalized to GAPDH and the results are shown as the mean ± SEM. Transcripts not detected are indicated by “nd”. Significance was determined by one-way ANOVA with post-hoc analysis: * = p < 0.05 between WT and ASO tissues; † = p < 0.05 among WT tissues; and # = p < 0.05 among ASO tissues.
We quantified differences in α-synuclein spliced transcript levels from several 2-month-old WT and ASO mice (n = 4–5 per group) (Fig. 4b). Only minor SNCA-140 expression differences were observed among neuronal regions of WT mice (Fig. 4b, SNCA-140, compare white bars). We observed higher SNCA-140 levels in neuronal regions of ASO mice when compared to neuronal regions of WT mice (Fig. 4b, SNCA-140, compare black and white bars). Here, we found that transcript levels ranged from a 2.5 fold increase in the cortex to a 63 fold increase in the ventral midbrain. We found differences in SNCA-140 levels among ASO regions, with the cortex, striatum, and ventral midbrain being higher than in the cerebellum, hippocampus, and heart (Fig. 4b, SNCA-140, compare black bars).
We found differences in SNCA-126 levels among neuronal regions of WT mice (Fig. 4b, SNCA-126, compare white bars). In general, SNCA-126 expression patterns among WT regions paralleled SNCA-126 expression patterns in ASO regions, with higher SNCA-126 levels being found in the cortex, hippocampus, and striatum. Only in the cortex did we find higher SNCA-126 levels in ASO mice when compared to WT mice (Fig. 4b, SNCA-126, compare black and white bars). Differences in SNCA-126 levels were observed among ASO regions, notably in the cortex, which was at least 1.6 fold higher than SNCA-126 levels in other ASO regions (Fig. 4b, SNCA-126, compare black bars).
We observed higher SNCA-112 levels in the hippocampus of WT mice when compared to other WT regions (Fig. 4b, SNCA-112, compare white bars). We did not detect SNCA-112 expression in the striatum, cerebellum, or heart of WT mice, despite SNCA-112 expression in those same regions of ASO mice. Comparisons of SNCA-112 levels between WT and ASO mice revealed 18, 6, and 2 fold increases in the cortex, ventral midbrain, and hippocampus, respectively (Fig. 4b, SNCA-112, compare black and white bars). SNCA-112 fold changes in striatum, cerebellum, and heart could not be determined because SNCA-112 levels were not detected in WT mice. Differences in SNCA-112 levels could be observed among ASO regions, with the cortex, striatum, and cerebellum showing at least a 1.4 fold higher level of expression than in the ventral midbrain or hippocampus (Fig. 4b, SNCA-112, compare black bars).
No differences were found in SNCA-98 levels among neuronal regions of WT mice (Fig. 4b, SNCA-98, compare white bars). We did not detect SNCA-98 expression in the cortex of WT mice, despite SNCA-112 expression in those same regions of ASO mice. Comparisons of SNCA-98 levels between WT and ASO mice revealed higher SNCA-98 levels in the cortex and striatum of ASO mice (Fig. 4b, SNCA-98, compare black and white bars). Here, there was a 5.5 fold increase in the striatum, while fold changes in the cortex could not be determined because SNCA-98 was not detected in the cortex of WT mice. Higher SNCA-98 levels were found in the cortex when compared among other ASO regions (Fig. 4b, SNCA-98, compare black bars).
Significant elevations in α-synuclein spliced transcript expression in the ventral midbrain of ASO mice at 15 months-of-age
Alternative splicing may contribute to tissue-specific changes in α-synuclein mRNA expression with age (Beyer et al., 2007; Ueda et al., 1994). The effects of these changes may have relevance to the development and progression of PD and other α-synucleinopathies. As such, we evaluated α-synuclein splicing in 15-month-old WT and ASO mice to identify whether splicing changes could be observed in parallel with the widespread pathological and behavioral deficits occurring in ASO mice at this age (Hallett et al., 2010).
We evaluated the electrophoretic α-synuclein splicing profile from the cortex, hippocampus, striatum, ventral midbrain, cerebellum, and heart of a representative 15-month-old WT and ASO mouse (Fig. 5a). Whereas SNCA-140, -126, -112, and -98 elevations were observed in all tissues of ASO mice, albeit at varying levels, we were unable to identify SNCA-112 and -98 in the cortex, and SNCA-112 in the striatum, ventral midbrain, and cerebellum from WT mice. Perhaps the most striking example, upon visual inspection, was the increased band intensity of ventral midbrain splice variants in ASO mice when compared to other WT and ASO tissues.
Fig. 5.
RT-PCR of α-synuclein alternative splicing at 15 months-of-age in neuronal regions of WT and ASO mice. (a) Agarose gel-based electrophoretic distribution profile of α-synuclein spliced transcripts in the cortex, hippocampus, striatum, substantia nigra, cerebellum, and heart of a representative 15-month-old WT and ASO mouse. (b) Semi-quantitative analysis of SNCA-140, -126, -112, and -98 from several 15-month-old WT and ASO mice (n=4–6). Relative transcript expression is normalized to GAPDH and the results are shown as the mean ± SEM. Transcripts that were not detected are indicated by “nd”. Significance was determined by one-way ANOVA with post-hoc analysis: * = p < 0.05 between WT and ASO tissues; † = p < 0.05 among WT tissues; and # = p < 0.05 among ASO tissues.
Quantitative analysis of several 15-month-old WT and ASO mice (n = 4–5 per group) revealed significant changes in α-synuclein splicing (Fig. 5b). No differences in SNCA-140 levels were found among WT tissues (Fig. 5b, SNCA-140, compare white bars). We found higher SNCA-140 levels in tissues of ASO mice when compared to tissues of WT mice (Fig. 5b, SNCA-140, compare black and white bars). Here, we found that SNCA-140 levels ranged from a 7 fold increase in the cortex to a 39 fold increase in the heart. Among ASO tissues, the ventral midbrain displayed higher SNCA-140 levels when compared to the striatum, hippocampus, and cerebellum, each of which were, in turn, higher than SNCA-140 levels in the cortex and heart (Fig. 5b, SNCA-140, compare black bars).
We did not identify differences in SNCA-126 levels among WT tissues (Fig. 5b, SNCA-126, compare white bars). Evaluation of SNCA-126 levels between ASO and WT mice revealed higher SNCA-126 levels in the ventral midbrain of ASO mice, which was 4.6 fold higher than in WT mice (Fig. 5b, SNCA-126, compare black and white bars). Among ASO tissues, the ventral midbrain displayed higher SNCA-126 levels than other tissues (Fig. 5b, SNCA-126, compare black bars).
We observed that SNCA-112 expression in WT mice was restricted to the hippocampus and heart (Fig. 5b, SNCA-112, compare white bars). In contrast, we were able to detect SNCA-112 expression in all ASO tissues. As such, we found higher SNCA-112 levels in neuronal tissues from ASO mice (Fig. 5b, SNCA-112, compare black and white bars). Here, the hippocampus of ASO mice was 11.8 fold higher than in WT mice. We could not determine fold changes for the cortex, striatum, ventral midbrain, and cerebellum for lack of SNCA-112 expression in WT mice. Among ASO tissues, the ventral midbrain showed higher SNCA-112 levels than the hippocampus, striatum, cerebellum, and heart (Fig. 5b, SNCA-112, compare black bars).
No differences were found in SNCA-98 levels among neuronal tissues from WT mice (Fig. 5b, SNCA-98, compare white bars). Evaluation of SNCA-98 levels between WT and ASO mice revealed higher SNCA-98 levels in the ventral midbrain of ASO mice (Fig. 5b, SNCA-98, compare white and black bars). Among ASO tissues, SNCA-98 levels in the ventral midbrain were highest, while the cortex showed an intermediate level of expression (Fig. 5b, SNCA-98, compare black bars).
DISCUSSION
In the current study, we used isoform-specific primers to characterize α-synuclein mRNA splicing in PD post-mortem brain tissue and in a transgenic mouse model of α-synucleinopathy. We showed that α-synuclein splicing occurs in a region-specific manner in both normal and diseased human and mouse brains. In our analysis of PD cases, we found a significant increase in the production of SNCA-140 and SNCA-126 transcripts in the substantia nigra when compared to CTRL cases. In ASO mice, transgenic overexpression of human SNCA-140 was accompanied by elevations in SNCA-126, -112, and -98; however, we found that the splicing pattern of the truncated variants significantly differed from that of WT mice and did not necessarily coincide with the increases observed for SNCA-140. In aged ASO mice, we observed marked overexpression of each spliced transcript in the ventral midbrain when compared to WT and other ASO tissues.
Tissue-Specific Variations in α-Synuclein Splicing Suggest Complex Post-transcriptional Requirements
Alternative splicing is a significant determinant of transcriptome diversity and underscores many complex tissue-specific expression patterns during development and aging (Grosso et al., 2008; Keren et al., 2010). In addition to the full-length SNCA-140 transcript, three α-synuclein spliced variants have been identified in humans and are distinguished by their pattern of exon 3 and/or 5 exclusion (Beyer et al., 2008b; Campion et al., 1995; Ueda et al., 1993). In the normal adult brain, however, α-synuclein expression is typically quantified by measurement of the SNCA-140 transcript or by total mRNA levels, which are highest in the hippocampus, olfactory bulb, striatum, and cerebellum relative to other brain and peripheral tissues (Iwai et al., 1995; Maroteaux et al., 1988; Ueda et al., 1993). With age, there is a steady state level of decline in α-synuclein mRNA expression (Adamczyk et al., 2005; Chu and Kordower, 2007; Iwai et al., 1995; Mak et al., 2009; Petersen et al., 1999). A previous study showed differential expression of SNCA-140 and SNCA-112 spliced transcripts in a human fetal brain cDNA library, in whole cerebral brain tissue, and among adult and fetal peripheral tissues (Ueda et al., 1994), indicating that α-synuclein splicing may represent an important regulatory and/or functional role during development. Our evaluation of α-synuclein spliced transcript levels from different human and mouse neuronal regions identifies an adult, region-specific expression pattern for each spliced transcript. Interestingly, in several neuronal regions from 2-month-old WT mice, PCR favoured the amplification of SNCA-126 or SNCA-112 over SNCA-140, which challenges the common belief that the most typically described isoform, SNCA-140, is transcribed at higher levels relative to other transcripts. The relative abundance of SNCA-126 and SNCA-112 in specific neuronal regions suggests there exist physiological requirements for the generation of these spliced transcripts. Moreover, the lack of SNCA-112 and SNCA-98 expression in select neuronal regions from WT mice directly contrasts with their abundant overexpression in ASO mice, supporting the notion that α-synuclein splicing is tightly regulated under normal conditions. The reason for this differential, region-specific expression in the normal adult brain remains unknown and, unfortunately, the current lack of α-synuclein isoform-specific antibodies and an inability to distinguish among their molecular weights during SDS-PAGE separation hampers characterization of α-synuclein isoforms at the protein level (Beyer, 2006). While we recognize the limitation of our study to the mRNA level, it is of note that exons 3 and 5 harbor post-translational phosphorylation, ubiquitination, nitration, and truncation sites of α-synuclein (Beyer, 2006; Oueslati et al., 2010), suggesting that stoichiometric differences in α-synuclein splicing may represent a post-transcriptional “gateway” for further downstream modifications and, thus, physiological function. Differential α-synuclein splicing may be associated with the functional requirements of normal α-synuclein biology or in response to pathological cellular states. For example, in a recent study of cocaine abuse, researchers found that SNCA-112 protein was significantly upregulated in the substantia nigra of cocaine-related excited delirium cases, but not in cocaine abusers or drug-free control subjects, suggesting that α-synuclein isoforms play a distinct functional role in accordance with different cellular adaptive states (Mash et al., 2011).
Abnormal Elevations of α-Synuclein Spliced Transcript Expression in PD Brain Regions
Accumulating evidence suggest that abnormal changes in α-synuclein steady state expression is critical to the pathogenesis of PD (Auluck et al., 2010; McCormack and Di Monte, 2009; Oueslati et al., 2010). Changes in α-synuclein splicing may also have relevance to the pathogenesis of PD (Beyer, 2006). In the current study, we identified select elevations in SNCA-140 and SNCA-126 transcripts in the substantia nigra of PD post-mortem cases. While our results are consistent with previous reports of SNCA-140 elevations in PD affected regions (Chiba-Falek et al., 2006; Grundemann et al., 2008; McCormack and Di Monte, 2009), we found that previous reports may have overestimated cortical SNCA-126 and SNCA-98 transcript levels in PD cases (Beyer et al., 2008a; Beyer et al., 2008b; Beyer et al., 2006). Moreover, a reported decrease in cortical and nigral SNCA-140 and SNCA-112 transcript levels (Neystat et al., 1999) may have arisen as a result of differences in sample preparation, for which region- and cell-specific differences may yield varying changes in α-synuclein mRNA expression (Grundemann et al., 2008). Nevertheless, taken together, these results provide clear evidence that abnormal splicing of α-synuclein occurs in susceptible brain regions of PD patients.
α-Synuclein Splicing Misregulation in a Transgenic Mouse Model of α-Synucleinopathy
We show that in ASO mice, as early as 2 months-of-age, SNCA-126, -112, and -98 are increased, but do not necessarily parallel SNCA-140 overexpression or the splicing ratios found in WT mice. This splicing pattern may arise from the human α-synuclein transgene used in the generation of ASO mice (Rockenstein et al., 2002), which may lack cis-acting splice recognition sites normally located within endogenous intronic and promoter sequences (Barash et al., 2010; Wang and Burge, 2008). This reasoning is supported by two reports identifying positive correlations between SNCA non-coding polymorphisms, including a PD-associated risk allele in the 3′ region and a variable poly-T sequence in the distal end of intron 2, and changes in SNCA-112 and SNCA-126 transcript levels, respectively (Beyer et al., 2007; McCarthy et al., 2010). The importance of these findings lie in the recognition that SNCA allelic variation has been shown to modify tissue-specific α-synuclein mRNA expression with an increased risk of developing PD (Bras and Singleton, 2009; Chiba-Falek and Nussbaum, 2001; Cronin et al., 2009; McCarthy et al., 2010). While we are able to identify splicing of human alpha-synuclein in ASO mice, we can only speculate on the mechanism of transgene splicing, which may occur through the use of exonic enhancer or inhibitor regulatory sequences or through strong splicing definition at the 5′ and 3′ ends of the flanking exons. It is reasonable to suggest that abnormal SNCA-126, -112, and -98 transcript expression may, in part, contribute to the underlying pathology of ASO mice (Beyer, 2006). Surprisingly, spliced transcript expression is seldom addressed in transgenic animal models that use cDNA transgenes lacking endogenous regulatory elements. This raises the possibility that missplicing of alternative transcripts derived from the transgene may play a critical role in the interpretation of the animal model being investigated.
While variability in SNCA cis-acting elements is implicated in PD susceptibility, aging remains the most prominent risk factor for developing PD (Ross and Smith, 2007; Vance et al., 2010). We observed region-specific changes in the pattern of α-synuclein spliced transcript expression between 2- and 15-month-old ASO mice, suggesting that environmental and cellular trans-acting factors modulate the level of α-synuclein splicing in response to neurodegeneration. In 15-month-old ASO mice, we observed a marked increase in the level of α-synuclein splicing in the ventral midbrain, a region including the substantia nigra pars reticulata where we have observed significant pathological changes, including proteinase-K resistant α-synuclein inclusions (Hallett et al., 2010). While defining associations between splicing and pathology are beyond the scope of this study, both human and rat α-synuclein splice variants have been shown to form intracellular, punctate aggregates when overexpressed in vitro (Kalivendi et al., 2009; McLean and Hyman, 2002), indicating that abnormal overexpression of alternative mRNA transcripts may have pathological relevance. In this regard, utilizing ASO mice to understand α-synuclein splicing misregulation may contribute to our understanding of the development of PD, and related α-synucleinopathies. Identifying to what extent α-synuclein spliced transcripts are translated in vivo and an examination of their functional and pathological effects is ongoing (Hallett et al., 2010). Determining the normal physiological or pathological requirements that are associated with differential α-synuclein splicing remains important for our understanding of α-synuclein function.
Supplementary Material
(a) Sequence chromatograms comparing human SNCA-140 with SNCA-126 and SNCA-112 confirm exon 2/4 and 4/6 skipping events, respectively. (b) To identify extrinsic sequences embedded in the half-scrambled exon skipping primer Csc, the unscrambled 3′ end of Csc was progressively truncated until no bands were observed when cDNA from human cortex was subjected to a 35 cycle PCR reaction containing the antisense primer B. (c) Primers less than 18 base pairs (Csc-18 and Csc-17) did not produce any bands, while larger primers (Csc-19–24) were capable of annealing to and amplifying α-synuclein exon 4. (d) When fully unscrambled, only the C-18 primer was capable of amplifying spliced transcripts from cDNA of human cortex after a 35 cycle PCR reaction containing antisense primer B; the C-18 primer was, therefore, used throughout this study and is refered to as “hSNCA ex2/4 fwd” in Table 1.
Acknowledgments
We would like to thank the Harvard Brain Tissue Resource Center (PHS Grant# R24 MH068855; Belmont, MA) for human tissue samples. This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke P50 (Grant NS39793 to O.I.); the U.S. Department of Defense (Grant W81XWH-05-1-0555 to O.I.); the Canadian Institutes of Health Research (Fellowship to J.R.M); the Consolidated Anti-Aging Foundation; the Orchard Foundation; the Hansen family; and the Harold and Ronna Cooper family.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamczyk A, Solecka J, Strosznajder JB. Expression of alpha-synuclein in different brain parts of adult and aged rats. J Physiol Pharmacol. 2005;56:29–37. [PubMed] [Google Scholar]
- Auluck PK, Caraveo G, Lindquist S. alpha-Synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Beyer K. Alpha-synuclein structure, posttranslational modification and alternative splicing as aggregation enhancers. Acta Neuropathol. 2006;112:237–251. doi: 10.1007/s00401-006-0104-6. [DOI] [PubMed] [Google Scholar]
- Beyer K, Domingo-Sabat M, Humbert J, Carrato C, Ferrer I, Ariza A. Differential expression of alpha-synuclein, parkin, and synphilin-1 isoforms in Lewy body disease. Neurogenetics. 2008a;9:163–172. doi: 10.1007/s10048-008-0124-6. [DOI] [PubMed] [Google Scholar]
- Beyer K, Domingo-Sabat M, Lao JI, Carrato C, Ferrer I, Ariza A. Identification and characterization of a new alpha-synuclein isoform and its role in Lewy body diseases. Neurogenetics. 2008b;9:15–23. doi: 10.1007/s10048-007-0106-0. [DOI] [PubMed] [Google Scholar]
- Beyer K, Humbert J, Ferrer A, Lao JI, Carrato C, Lopez D, Ferrer I, Ariza A. Low alpha-synuclein 126 mRNA levels in dementia with Lewy bodies and Alzheimer disease. Neuroreport. 2006;17:1327–1330. doi: 10.1097/01.wnr.0000224773.66904.e7. [DOI] [PubMed] [Google Scholar]
- Beyer K, Humbert J, Ferrer A, Lao JI, Latorre P, Lopez D, Tolosa E, Ferrer I, Ariza A. A variable poly-T sequence modulates alpha-synuclein isoform expression and is associated with aging. J Neurosci Res. 2007;85:1538–1546. doi: 10.1002/jnr.21270. [DOI] [PubMed] [Google Scholar]
- Bisaglia M, Mammi S, Bubacco L. Structural insights on physiological functions and pathological effects of alpha-synuclein. FASEB J. 2009;23:329–340. doi: 10.1096/fj.08-119784. [DOI] [PubMed] [Google Scholar]
- Bras JM, Singleton A. Genetic susceptibility in Parkinson’s disease. Biochim Biophys Acta. 2009;1792:597–603. doi: 10.1016/j.bbadis.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion D, Martin C, Heilig R, Charbonnier F, Moreau V, Flaman JM, Petit JL, Hannequin D, Brice A, Frebourg T. The NACP/synuclein gene: chromosomal assignment and screening for alterations in Alzheimer disease. Genomics. 1995;26:254–257. doi: 10.1016/0888-7543(95)80208-4. [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Lopez GJ, Nussbaum RL. Levels of alpha-synuclein mRNA in sporadic Parkinson disease patients. Mov Disord. 2006;21:1703–1708. doi: 10.1002/mds.21007. [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- Chu Y, Kordower JH. Age-associated increases of alpha-synuclein in monkeys and humans are associated with nigrostriatal dopamine depletion: Is this the target for Parkinson’s disease? Neurobiol Dis. 2007;25:134–149. doi: 10.1016/j.nbd.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Cronin KD, Ge D, Manninger P, Linnertz C, Rossoshek A, Orrison BM, Bernard DJ, El-Agnaf OM, Schlossmacher MG, Nussbaum RL, Chiba-Falek O. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18:3274–3285. doi: 10.1093/hmg/ddp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- Fernagut PO, Chesselet MF. Alpha-synuclein and transgenic mouse models. Neurobiol Dis. 2004;17:123–130. doi: 10.1016/j.nbd.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Fernagut PO, Hutson CB, Fleming SM, Tetreaut NA, Salcedo J, Masliah E, Chesselet MF. Behavioral and histopathological consequences of paraquat intoxication in mice: effects of alpha-synuclein over-expression. Synapse. 2007;61:991–1001. doi: 10.1002/syn.20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, Tichopad A, Golub Y, Munz M, Schweitzer KJ, Wolf B, Berg D, Mueller JC, Gasser T. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundemann J, Schlaudraff F, Haeckel O, Liss B. Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson’s disease. Nucleic Acids Res. 2008;36:e38. doi: 10.1093/nar/gkn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai A, Masliah E, Yoshimoto M, Ge N, Flanagan L, de Silva HA, Kittel A, Saitoh T. The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- Kalivendi SV, Yedlapudi D, Hillard CJ, Kalyanaraman B. Oxidants Induce Alternative Splicing of alpha-Synuclein: Implications for Parkinson’s Disease. Free Radic Biol Med. 2009 doi: 10.1016/j.freeradbiomed.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- Kim C, Lee SJ. Controlling the mass action of alpha-synuclein in Parkinson’s disease. J Neurochem. 2008;107:303–316. doi: 10.1111/j.1471-4159.2008.05612.x. [DOI] [PubMed] [Google Scholar]
- Koob AO, Ubhi K, Paulsson JF, Kelly J, Rockenstein E, Mante M, Adame A, Masliah E. Lovastatin ameliorates alpha-synuclein accumulation and oxidation in transgenic mouse models of alpha-synucleinopathies. Exp Neurol. 2010;221:267–274. doi: 10.1016/j.expneurol.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SK, McCormack AL, Langston JW, Kordower JH, Di Monte DA. Decreased alpha-synuclein expression in the aging mouse substantia nigra. Exp Neurol. 2009;220:359–365. doi: 10.1016/j.expneurol.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mash DC, Xie XB, Qin Y. SNCA isoforms associated with cocaine abuse. Abstract, Society for Neuroscience; Washington, D.C: 2011. Nov 12–16, [Google Scholar]
- McCarthy JJ, Linnertz C, Saucier L, Burke JR, Hulette CM, Welsh-Bohmer KA, Chiba-Falek O. The effect of SNCA 3′ region on the levels of SNCA-112 splicing variant. Neurogenetics. 2010 doi: 10.1007/s10048-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack AL, Di Monte DA. Enhanced alpha-synuclein expression in human neurodegenerative diseases: pathogenetic and therapeutic implications. Curr Protein Pept Sci. 2009;10:476–482. doi: 10.2174/138920309789351912. [DOI] [PubMed] [Google Scholar]
- McLean PJ, Hyman BT. An alternatively spliced form of rodent alpha-synuclein forms intracellular inclusions in vitro: role of the carboxy-terminus in alpha-synuclein aggregation. Neurosci Lett. 2002;323:219–223. doi: 10.1016/s0304-3940(02)00154-4. [DOI] [PubMed] [Google Scholar]
- Neystat M, Lynch T, Przedborski S, Kholodilov N, Rzhetskaya M, Burke RE. Alpha-synuclein expression in substantia nigra and cortex in Parkinson’s disease. Mov Disord. 1999;14:417–422. doi: 10.1002/1531-8257(199905)14:3<417::aid-mds1005>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum Mutat. 2010;31:763–780. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueslati A, Fournier M, Lashuel HA. Role of post-translational modifications in modulating the structure, function and toxicity of alpha-synuclein: implications for Parkinson’s disease pathogenesis and therapies. Prog Brain Res. 2010;183:115–145. doi: 10.1016/S0079-6123(10)83007-9. [DOI] [PubMed] [Google Scholar]
- Petersen K, Olesen OF, Mikkelsen JD. Developmental expression of alpha-synuclein in rat hippocampus and cerebral cortex. Neuroscience. 1999;91:651–659. doi: 10.1016/s0306-4522(98)00596-x. [DOI] [PubMed] [Google Scholar]
- Rockenstein E, Mallory M, Hashimoto M, Song D, Shults CW, Lang I, Masliah E. Differential neuropathological alterations in transgenic mice expressing alpha-synuclein from the platelet-derived growth factor and Thy-1 promoters. J Neurosci Res. 2002;68:568–578. doi: 10.1002/jnr.10231. [DOI] [PubMed] [Google Scholar]
- Ross CA, Smith WW. Gene-environment interactions in Parkinson’s disease. Parkinsonism Relat Disord. 2007;13(Suppl 3):S309–315. doi: 10.1016/S1353-8020(08)70022-1. [DOI] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song DD, Shults CW, Sisk A, Rockenstein E, Masliah E. Enhanced substantia nigra mitochondrial pathology in human alpha-synuclein transgenic mice after treatment with MPTP. Exp Neurol. 2004;186:158–172. doi: 10.1016/S0014-4886(03)00342-X. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Goedert M. The alpha-synucleinopathies: Parkinson’s disease, dementia with Lewy bodies, and multiple system atrophy. Ann N Y Acad Sci. 2000;920:16–27. doi: 10.1111/j.1749-6632.2000.tb06900.x. [DOI] [PubMed] [Google Scholar]
- Ueda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero DA, Kondo J, Ihara Y, Saitoh T. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:11282–11286. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Saitoh T, Mori H. Tissue-dependent alternative splicing of mRNA for NACP, the precursor of non-A beta component of Alzheimer’s disease amyloid. Biochem Biophys Res Commun. 1994;205:1366–1372. doi: 10.1006/bbrc.1994.2816. [DOI] [PubMed] [Google Scholar]
- Uversky VN. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- Vance JM, Ali S, Bradley WG, Singer C, Di Monte DA. Gene-environment interactions in Parkinson’s disease and other forms of parkinsonism. Neurotoxicology. 2010;31:598–602. doi: 10.1016/j.neuro.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MJ, Gait MJ, Yin H. RNA-targeted splice-correction therapy for neuromuscular disease. Brain. 2010;133:957–972. doi: 10.1093/brain/awq002. [DOI] [PubMed] [Google Scholar]
- Yokota T, Takeda S, Lu QL, Partridge TA, Nakamura A, Hoffman EP. A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground. Arch Neurol. 2009;66:32–38. doi: 10.1001/archneurol.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Sequence chromatograms comparing human SNCA-140 with SNCA-126 and SNCA-112 confirm exon 2/4 and 4/6 skipping events, respectively. (b) To identify extrinsic sequences embedded in the half-scrambled exon skipping primer Csc, the unscrambled 3′ end of Csc was progressively truncated until no bands were observed when cDNA from human cortex was subjected to a 35 cycle PCR reaction containing the antisense primer B. (c) Primers less than 18 base pairs (Csc-18 and Csc-17) did not produce any bands, while larger primers (Csc-19–24) were capable of annealing to and amplifying α-synuclein exon 4. (d) When fully unscrambled, only the C-18 primer was capable of amplifying spliced transcripts from cDNA of human cortex after a 35 cycle PCR reaction containing antisense primer B; the C-18 primer was, therefore, used throughout this study and is refered to as “hSNCA ex2/4 fwd” in Table 1.